Abstract
Bloodsucking parasites such as ticks have evolved a wide variety of immunomodulatory proteins that are secreted in their saliva, allowing them to feed for long periods of time without being detected by the host immune system. One possible strategy used by ticks to evade the host immune response is to produce proteins that selectively bind and neutralize the chemokines that normally recruit cells of the innate immune system that protect the host from parasites. We have identified distinct cDNAs encoding novel chemokine binding proteins (CHPBs), which we have termed Evasins, using an expression cloning approach. These CHBPs have unusually stringent chemokine selectivity, differentiating them from broader spectrum viral CHBPs. Evasin-1 binds to CCL3, CCL4, and CCL18; Evasin-3 binds to CXCL8 and CXCL1; and Evasin-4 binds to CCL5 and CCL11. We report the characterization of Evasin-1 and -3, which are unrelated in primary sequence and tertiary structure, and reveal novel folds. Administration of recombinant Evasin-1 and -3 in animal models of disease demonstrates that they have potent antiinflammatory properties. These novel CHBPs designed by nature are even smaller than the recently described single-domain antibodies (Hollinger, P., and P.J. Hudson. 2005. Nat. Biotechnol. 23:1126–1136), and may be therapeutically useful as novel antiinflammatory agents in the future.
Many pathogenic organisms, including viruses, have developed strategies to evade the host immune system based on interference with host cytokine and chemokine proinflammatory mediators (1–3). Inhibitory binding proteins for proinflammatory cytokines, including TBP, IFNAR, and IL-18 BP, have also been described in humans, but no secreted chemokine binding proteins (CHPBs) have so far been identified in vertebrates. However, internal and external parasites have evolved chemokine neutralization strategies that allow them to productively suppress the mammalian immune response to prolong survival on their host. Ticks are bloodsucking parasites whose role in the transmission of pathogens such as Borrelia burgdorferi, which causes Lyme disease, has been the subject of intensive research (4). Ticks feed on their hosts for relatively long periods of time, ranging from several days to weeks, and provided that they do not carry infectious agents, can remain essentially undetected by their hosts. To achieve this, they have evolved an arsenal of antiinflammatory, anticoagulant, and analgesic agents that are used to render the ticks invisible to the immune system (5). Little is known about the molecular nature of such agents, but the cloning and characterization of these factors may shed light on the immunoevasion mechanisms used by these organisms, as well as yield novel proteins of potential therapeutic use in human inflammatory pathologies.
Successful prevention of the activity of families of small chemoattractant proteins called chemokines has been shown for several viral CHBPs (for reviews see [1, 6]), and, more recently, by a CHBP isolated from the parasitic worm Schistosoma mansoni (7). Deletion of the gene encoding the broad spectrum CHPB T1 from the rabbit poxvirus myxoma virus resulted in extensive leukocyte infiltration in the dermis of the infected animal, which was absent upon infection by the wild-type virus (8). This study clearly demonstrated that the virus had evolved a mechanism that prevented the recruitment of leukocytes to the site of infection.
Recently, the presence of antichemokine activities has been reported in tick saliva (9, 10). Tick salivary gland extracts were shown to contain an activity that neutralized CXCL8, and in a later study, neutralization of other chemokines was also demonstrated. Our own analyses of tick saliva harvested during feeding also demonstrated the presence of molecules that bound to CXCL8, CCL3, and CCL5. Using an expression cloning strategy to identify the protein(s) in tick saliva that bind to chemokines, we cloned a CHPB, which we called Evasin-1, that specifically binds to the CC chemokines CCL3, CCL4, and CCL18 (11). We also cloned a second protein of similar size, Evasin-2, for which the ligand remains to be elucidated. Subsequent rescreening of the tick salivary gland cDNA library with a selection of chemokines and cytokines enabled us to identify two further CHBPs. In this study, we report the cloning of a CXC CHPB named Evasin-3 and a second CC CHPB, distinct from Evasin-1, named Evasin-4. Interestingly, Evasin-3 bears no sequence homology to Evasin-1 and -4, which appear to belong to the same structural family. The three proteins are all surprisingly small compared with other chemokine and cytokine binding proteins described to date. Evasin-1 and -3 were crystallized, and their three-dimensional structures have been solved, revealing novel folds with binding modes distinct from that of viral CHBPs. Investigations of their specificity in vitro and their efficacy in vivo in animal models of chronic inflammatory diseases such as pulmonary fibrosis, arthritis, and psoriasis have demonstrated that the Evasins have potent antiinflammatory activities, providing tantalizing new insights into scaffolds evolved by nature to inhibit inflammation.
RESULTS
Evidence for the presence of CHPBs in tick saliva and their expression cloning
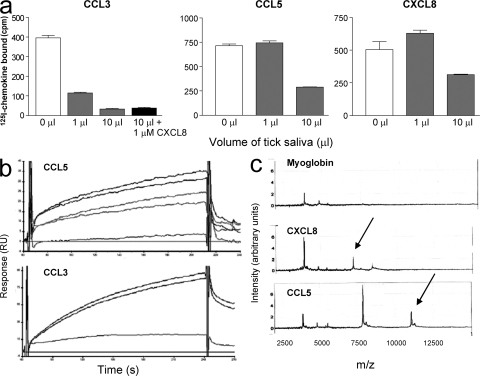
We analyzed the ability of saliva from Rhipicephalus sanguineus (common brown dog tick) to inhibit the binding of the I125-labeled chemokines CCL2, CCL5, and CXCL8, as well as CCL3, to their respective receptors using a scintillation proximity assay. As shown in Fig. 1 a, inhibition of binding was achieved for CCL3, CCL5, and CXCL8, with the most potent effect on CCL3. No inhibition was observed for CCL2 although saliva harvested from another tick species, Amblyomma cajennense, appeared to have anti-CCL2 activity (unpublished data). The possibility that the inhibition was caused by degradation of the radiolabeled chemokines by a proteolytic activity in the saliva was addressed by incubation of the radiolabeled chemokines in saliva for 4 h. Subsequent analysis by SDS-PAGE revealed that no degradation of the chemokine had occurred (unpublished data). We next addressed the question of whether there was more than one binding protein present in the saliva by testing whether an excess of CXCL8 was able to block the activity of the CCL3 binding protein. As shown in Fig. 1 a, a 500-fold excess of CXCL8 had no effect on the ability of the saliva to inhibit CCL3 binding to CCR5, therefore suggesting the presence of at least two binding proteins.
Figure 1.
Chemokine binding activity in tick saliva. (a) Tick saliva inhibits the binding of CCL3, CCL5, and CXCL8 to their cognate receptors. Radiolabeled chemokines were incubated with CHO cell membranes expressing CCR1 for CCL3 and CCL5, and CXCR2 for CXCL8, in the presence of 1 μl, 10 μl, or in the absence of tick saliva. The antichemokine activity is caused by the presence of more than one binding protein because an excess of CXCL8 did not abolish the inhibition of CCL3 binding. Error bars represent the mean ± SEM. (b) Detection of CHPBs in tick saliva by SPR. Sensograms demonstrated binding to CCL5(E66A) and CCL3 immobilized on a CM4 chip. (c) Detection of distinct CHPBs to CCL5 and CXCL8 by Retentate chromatography SELDI of flight mass spectrometry (RC-SELDI-TOF-MS). Tick saliva was incubated on a PS-20 Proteinchip array coated with either CCL5 (bottom), CXCL8 (middle), or the control protein equine myoglobin (top). Specific peaks, indicated by arrows, corresponding to a protein with a mass of 11,052 Daltons binding to CCL5 and of 7,133 Daltons for the binding to CXCL8 were observed.
Incubation of 125I-CCL3 with tick saliva, followed by chemical cross-linking, produces a clear band shift when the reaction products were analyzed on SDS PAGE, and this method was adopted as a screening assay in our attempts to clone the tick CHBPs. We therefore used the band shift assay to screen supernatants from HEK293 cells transfected with a tick salivary gland cDNA library, to identify a cDNA encoding Evasin-1 (11). Surface plasmon resonance (SPR) performed with immobilized CCL3 and CCL5 demonstrated the presence of binding proteins for these two chemokines in tick saliva (Fig. 1 b), and surface enhanced laser desorption ionization (SELDI) identified peaks that bound to immobilized CXCL8 and CCL5 (Fig. 1 c), but surprisingly, no peak was observed for CCL3 (not depicted). The masses identified by SELDI indicated very small proteins with a mass of 7,133 Daltons binding to CXCL8 and a mass of 11,052 Daltons binding to CCL5; these masses are very close to those of the predicted protein cores, as discussed in the following section.
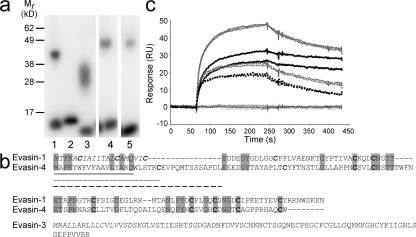
We therefore rescreened the cDNA library using a cocktail of chemokines and cytokines, containing iodinated CXCL8, IL-1β, and IL-2. IL-2 was included because an anti-IL-2 activity has previously been reported in tick salivary gland extracts (9). In the first round of screening, we identified a band shift in one pool of 270 clones. After deconvolution of the positive pool, and cross-linking to the individual iodinated proteins present in the cocktail, we identified a cDNA encoding a CXCL8 binding protein. As we had previously identified a putative CHBP that we named Evasin-2, the function of which remains to be elucidated, we called the CXCL8 binding protein Evasin-3 (Fig. 2 a, lane 3). Competition experiments using a panel of chemokines (CCL3, CXCL1, CXCL4, CXCL7, CXCL8, CXCL9, CXCL10, CXCL11, CXCL12, and CXCL13) indicated that only CXCL1 and CXCL8 were able to compete with radiolabeled CXCL8 for binding to Evasin-3 (unpublished data). We then screened the expression library with a second cocktail consisting of iodinated CCL5, CCL11, and CXCL10. Upon analysis of supernatants from HEK cells transfected with the pools of clones from the cDNA expression library, we observed a band shift migrating at ∼50 kD in several pools. This band was almost masked by a nonspecific band migrating at ∼60 kD detected in all experiments. Deconvolution of the positive pools revealed a cDNA clone that was able to mediate cross-linking to both CCL5 and CCL11 (Fig. 2 a, lanes 4 and 5), but not CXCL10 when tested in HEK293 cells. The CCL5 and CCL11 binding protein was named Evasin-4.
Figure 2.
Identification of Evasin-3 and -4. (a) Identification of Evasin-3 and -4 by cross-linking to radiolabeled chemokine in supernatants from transfected HEK293 cells. Viral CHBP p35 incubated with [125I]-CCL2 as a positive control in the presence (lane 1) or absence (lane 2) of the cross-linker BS3; supernatant from clone 69.19.1 incubated with [125I]-CXCL8 and BS3 (lane 3); supernatant from clone 10.27.1 incubated with [125I]-CCL5 and BS3 (lane 4) or [125I]-CCL11 and BS3 (lane 5). (b) Alignment of the predicted protein sequences of Evasin-1 and -4. Conserved residues are highlighted in gray and cysteines are shown in bold type. The predicted protein sequence of Evasin-3 is shown below the alignment. The signal peptides are shown in italics. Possible signal peptides of Evasin-4 are shown as follows: based on SignalJ prediction, black underline; based on secondary structure prediction, dotted underline; based on similarity to Evasin-1, dashed line. (c) Determination of the selectivity of immobilized recombinant Evasin-3 produced in E. coli by surface plasmon resonance. Sensograms corresponding to CXCL8 (bold black lines) CXCL1 (dotted black lines), MIP-2 (gray lines), and KC (dotted gray lines) showed strong binding of these chemokines to Evasin-3. The sensograms corresponding to CCL1, CCL2, CCL3, CCL4, CCL5, CCL11, CCL18, CCL27, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, and CX3CL1 (black lines) indicated no binding of these chemokines to Evasin-3.
Structural analyses of the Evasins
The cDNA for Evasin-3 encoded an ORF of 92 amino acids that, after signal peptide cleavage, gave rise to a mature protein of 66 amino acids with a predicted molecular mass of 7,005 Daltons. The cDNA for Evasin-4 encoded an ORF of 117 amino acids. Cleavage of the signal peptide sequence predicted that using the Signal P algorithm would generate a mature protein of 110 amino acids with a predicted molecular mass of 12,032 Daltons (Fig. 2 b). Analyses of the amino acid sequences of Evasin-3 and -4 revealed that Evasin-3 was unrelated to Evasin-1, whereas the Cys residues of the mature Evasin-4 sequence demonstrated an almost perfect alignment with those of Evasin-1 (Fig. 2 b). This indicates an identical disulfide topology, suggesting that these two proteins are structurally related, despite their relatively low level of identity at the amino acid level (27% identity over 91 amino acids). Conservation of a homologous fold, despite a low level of identity at the primary sequence level, is well known in protein families such as cytokines and chemokines.
Transient expression of C-terminal 6His tagged Evasin-3 and -4 in HEK293 cells revealed that unlike Evasin-1, these two proteins were poorly secreted. We were able to obtain a small amount of purified Evasin-3 (150 μg), which was extensively glycosylated for in vitro characterization. SPR analysis revealed that Evasin-3, similar to Evasin-1, is a highly selective CHPB in that it was only able to bind to CXCL1 and CXCL8 and their murine counterparts, KC and MIP-2 (Fig. 2 c). The kinetic parameters, as determined by SPR, are shown in Table I. It has proven difficult to produce recombinant 6His-tagged Evasin-4 in HEK293 cells, thus the identification of the N terminus of the mature protein has not yet been experimentally verified. Using the Signal P program to predict the cleavage site of the putative signal peptide sequence indicates that the mature protein sequence would commence with WLSTKC (Fig. 2 b). However, secondary structure predictions show that these amino acids are contained in a helix, suggesting that signal peptide cleavage may in fact be after Cys23, yielding an N terminus of EVPQM, as predicted by the latest version of Signal P. Nevertheless, alignment of the amino acid sequences of Evasin-4 and -1 still shows that Evasin-4 has an extended N terminus compared with Evasin-1, which may be an important determinant of ligand binding specificity. We are currently testing different expression strategies to obtain sufficient Evasin-4 for N-terminal sequence verification and for in vitro and in vivo studies.
Table I.
Binding characteristics of Evasin-1 and -3 as determined by SPR
| CKBP
|
Chemokine
|
ka × 106
|
kd × 10−3
|
Kd × 10−9
|
|---|---|---|---|---|
| M−1s−1 | s−1 | nM | ||
| Evasin-1 | CCL3 | 13.8 ± 3.54 | 2.43 ± 0.61 | 0.12 |
| CCL3L1 | 13.2 ± 2.9 | 0.67 ± 0.13 | 0.05 | |
| mMIP-1α | 15.6 ± 16 | 1.18 ± 1.07 | 0.08 | |
| CCL4 | 4.29 ± 2.58 | 2.2 ± 0.18 | 0.51 | |
| mMIP-1β | 3.84 ± 1.42 | 0.58 ± 0.12 | 0.15 | |
| Evasin-3 (HEK) | CXCL1 | 3.95 ± 4.23 | 1.42 ± 0.36 | 0.34 |
| CXCL8 | 0.41 ± 0.30 | 0.29 ± 0.14 | 0.7 | |
| KC | 0.64 ± 0.58 | 1.44 ± 0.35 | 2.25 | |
| MIP-2 | 0.79 ± 0.9 | 0.33 ± 0.11 | 0.4 | |
| Evasin-3 (E. coli) | CXCL1 | 2.61 ± 0.69 | 2.22 ± 0.49 | 0.85 |
| CXCL8 | 0.84 ± 0.33 | 0.36 ± 0.05 | 0.43 | |
| KC | 0.32 ± 0.17 | 1.85 ± 0.43 | 5.78 | |
| MIP-2 | 1.02 ± 0.56 | 0.65 ± 0.25 | 0.64 |
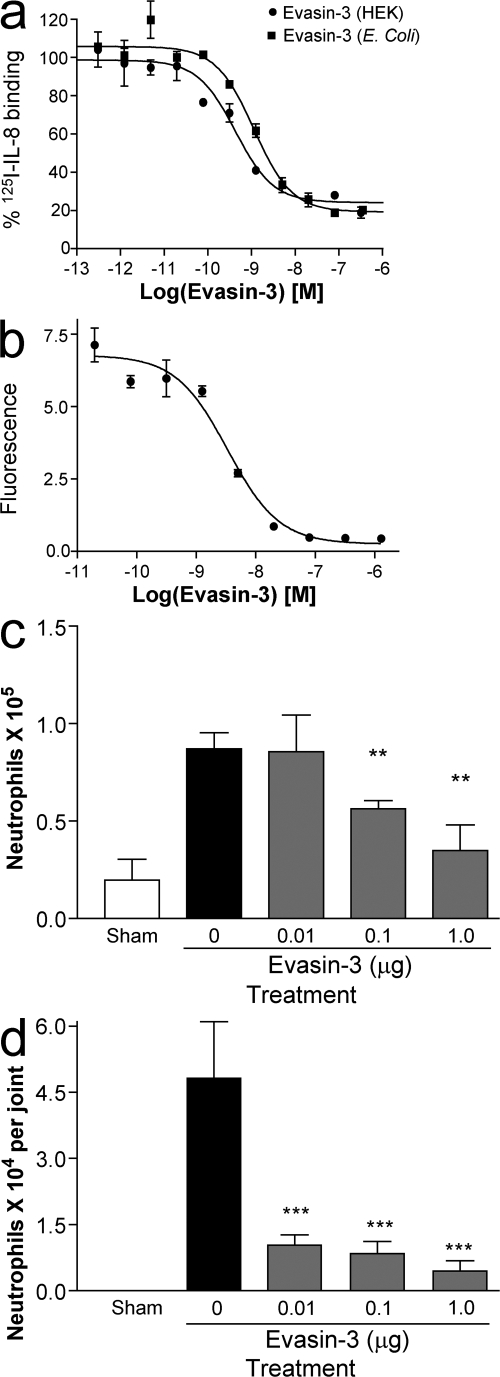
Because the yield of Evasin-3 was poor in the HEK293 expression system, we attempted expression in a prokaryotic host, Escherichia coli. It was very probable that Evasin-3 would be directed into inclusion bodies based on a formula predicting solubility for recombinant expression in E. coli (12). Surprisingly, the protein was expressed at high levels as soluble protein that could be purified to homogeneity in two chromatographic steps (unpublished data). Although the protein produced in E. coli is not glycosylated, it had equivalent activity to Evasin-3 produced in HEK293 cells (Table I; see also Fig. 8 a), suggesting that the glycosylation was not essential for activity.
Figure 8.
Evasin-3 inhibits cell recruitment in vitro and in vivo. (a) Inhibition of 125I-CXCL8 binding to CXCR2 by Evasin-3 produced in HEK293 cells (•) and by Evasin-3 produced in E. coli (▪). (b) Inhibition of CXCL8 induced neutrophil chemotaxis in vitro. (c) Inhibition of KC induced neutrophil recruitment into the peritoneal cavity (c) and into the knee joint (d). There were six mice per group in each experiment. **, P < 0.01; ***, P < 0.001 when compared with PBS-treated group. Error bars represent the mean ± SEM.
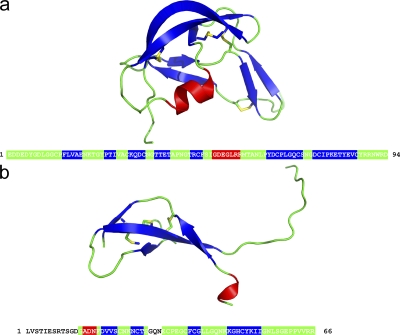
Evasin-3 produced in E. coli crystallized readily, and the structure revealed a novel fold that was totally unrelated to that of Evasin-1 (Fig. 3). The crystallographic structure of Evasin-1, described in detail elsewhere (unpublished data), also revealed a novel protein fold (Fig. 3 a). Neither protein has a structural homologue in the Protein Data Bank database. So although Evasin-1 and -3 are of similar size and function, they are completely unrelated at the amino acid sequence level, as well as in the secondary and tertiary structure (Fig. 3, a and b). Although the mode of binding of Evasin-1 has been elucidated by the determination of the structure of the complex of Evasin-1 with CCL3 (unpublished data), the structure of the Evasin-3 complex remains to be solved. If ticks produce two very different CHPBs, with very different selectivity profiles, it is likely that they bind their substrates in very different way. The elucidation of the structure of the complex of Evasin-3 with its ligand(s) would thus greatly help to understand the selectivity of these different CHPBs.
Figure 3.
Crystal structures of Evasin-1 and -3. Ribbon diagram of Evasin-1 (a) and Evasin-3 (b) with the amino acid sequence depicting residues in loops (green), α-helix (red), or β-sheet (blue).
Evasin-1 inhibits cell recruitment in vitro and in vivo
We have previously determined the binding affinities of Evasin-1 for three chemokines by surface plasmon resonance, and we have extended these analyses to include the second form of CCL3, known as CCL3L1, as well as the murine homologues of CCL3 and CCL4 (Table I). The Kd value for 0.05 nM CCL3L1 is similar to that determined for CCL3, in contrast to their very different potencies on CCR5 and CCR1 (13). Importantly, the affinities for murine MIP-1α and -1β, with Kd values of 0.08 and 0.15 nM, respectively, are similar to those for the human chemokines, allowing subsequent investigation of the antiinflammatory properties of Evasin-1 in murine models.
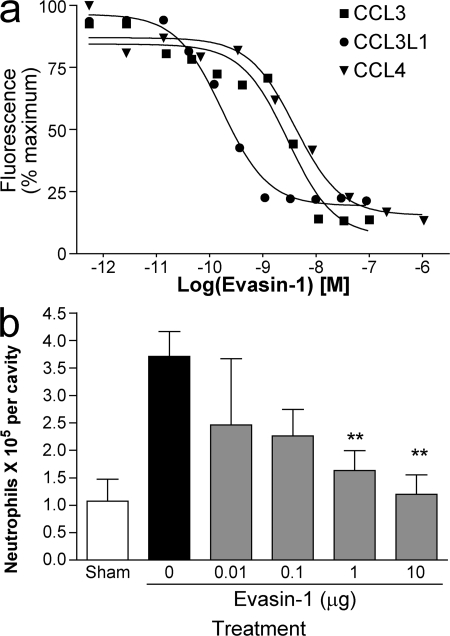
The IC50 values reported for the inhibition of receptor binding, 0.015 nM for CCL3 binding to CCR1 and 3 nM for CCL4 binding to CCR5 (11), are reflected by the ability of Evasin-1 to inhibit the chemotactic activity of these chemokines in vitro and in vivo. We chose the L1.2/CCR5 transfectants for these analyses because we were able to test inhibition of CCL3, CCL3L1, the high-affinity variant for this receptor, and CCL4 in the same assay system. As shown in Fig. 4 a, Evasin-1 showed potent inhibition of ligand-induced chemotaxis of the L1.2/CCR5 transfectants. Using an ED80 ligand concentration of 1 nM for CCL3L1 and CCL4, and 10 nM for CCL3, the IC50 values were 0.17 nM for CCL3L1, 2 nM for CCL3, and 4 nM for CCL4.
Figure 4.
Evasin-1 inhibits cell recruitment in vitro and in vivo. (a) Inhibition of CCL3- (▪), CCL3L1- (•) and CCL4-mediated (▴) chemotaxis of L1.2/CCR5 transfectants. The IC50 values are 2 nM for CCL3, 0.17 nM for CCL3L1, and 4 nM for CCL4. Data are shown as the mean ± the SD as a percentage of maximal recruitment and are representative of three to five individual experiments. (b) Evasin-1 inhibits granulocyte recruitment induced by CCL3 in mice. Evasin-1 was administered s.c. 45 min before the injection of 10 μg of CCL3 i.p. Cells in the peritoneal cavity were counted 18 h after the injection of CCL3. One representative experiment of three is shown, with 8 mice per group. **, P < 0.01 when compared with PBS-treated group. Error bars represent the mean ± SEM.
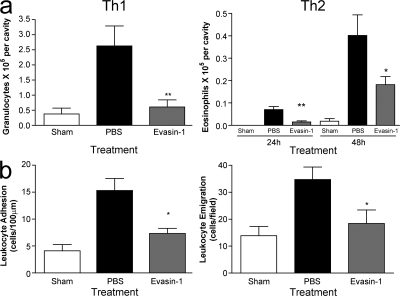
We then tested Evasin-1 in in vivo leukocyte recruitment models in mice. CCL3 and murine MIP-1α were administered into the peritoneal cavity, and the ability of systemically administered Evasin-1 to block leukocyte recruitment was assessed after 18 h. Evasin-1 showed dose-related inhibition of neutrophil recruitment with maximal activity at a dose of 10 μg (Fig. 4 b), with little effect on other cell types (not depicted). We therefore used this dose in subsequent studies. The inhibition of granulocyte recruitment by Evasin-1 was then further investigated in a murine model of a Th1-predominant delayed-type hypersensitivity and a Th2-predominant, late-phase reaction. In the Th1 sensitization model using methylated BSA (mBSA), recruitment of granulocytes into the pleural cavity was significantly impaired (Fig. 5 a). Although the inhibition of neutrophil recruitment was most striking, inhibition of CD3+ lymphocytes was also observed, although not of CD11b+ monocytes as determined by FACS (unpublished data). In the Th2 sensitization model, Evasin-1 inhibited eosinophil recruitment induced by antigen challenge into the lungs of mice immunized with S. mansoni eggs. An analysis of naive mice by intravital microscopy demonstrated that Evasin-1 was able to block adhesion and emigration of leukocytes induced by injection of MIP-1α (Fig. 5 b). Lastly, administration of Evasin-1 alone had no chemoattractant activity (unpublished data).
Figure 5.
Evasin-1 inhibits cellular recruitment in models of Th1 and Th2 inflammation. (a) Evasin-1 inhibits granulocyte recruitment induced by antigen challenge of immunized mice in models of Th1-predominant delayed-type hypersensitivity and Th2-predominant late-phase response. In the Th1 model, 10 μg Evasin-1 was administered s.c. 45 min before intrapleural injection of 10 μg of mBSA in mice immunized with mBSA in Freund's complete adjuvant. Cells in the pleural cavity were counted 48 h after the injection of antigen. In the Th2 model, 10 μg Evasin-1 was administered s.c. 45 min before and 24 h after the intratracheal injection of 10 μg of S. mansoni egg antigen in mice immunized with S. mansoni eggs. Cells in the bronchoalveolar lavage fluid were counted 24 and 48 h after the injection of antigen. There were six mice per group in each experiment. *, P < 0.05; **, P < 0.01 when compared with PBS-treated group. (b) Evasin-1 blocks adhesion and emigration of leukocytes induced by mMIP-1α as assessed by intravital microscopy performed in the cremaster vein of C57BL/6 mice. Evasin-1 was injected s.c. in the right scrotal skin 1 h before exteriorization. Normalized data were analyzed by one-way analysis of variance, and differences between groups were assessed using Student-Newman-Keuls post-test. *, P < 0.05; **, P < 0.01. There were six mice per group in each experiment. Error bars represent the mean ± SEM.
Evasin-1 shows antiinflammatory activity in murine models of disease
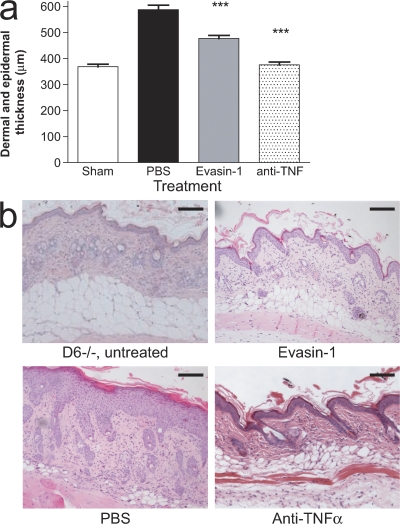
The ability of Evasin-1 to prevent CCL3-/CCL4-dependent cellular recruitment translated into potent antiinflammatory effects in animal models of disease. Skin inflammation in D6−/− mice sensitized with TPA gives rise to a phenotype closely resembling that of the human inflammatory skin disorder, psoriasis, and has been shown to be dependent on the induction of several CC chemokines, including CCL3 and CCL4, which are high-affinity ligands for D6 (14). Evasin-1 administration resulted in a significant reduction in skin inflammation observed in these mice (Fig. 6, a and b), but not complete abrogation as observed with an anti-TNF antibody. This is in accordance with our previous observations and indicates that although several CC chemokines are implicated in this model, targeting CCL3 and CCL4 is sufficient to ameliorate pathology.
Figure 6.
Evasin-1 inhibits cellular recruitment in an animal model of psoriasis. (a) Dermal and epidermal thickness of the skin of WT and D6-deficient (KO) mice after a single cutaneous application of TPA. Evasin-1 (30 μg/mouse) and anti-TNF-α were given daily, and tissues were collected on day 4 for histological assessment. One representative experiment of three is shown, with eight mice per group. ***, P < 0.001 when compared with PBS-treated group. Error bars represent the mean ± SEM. (b) Haematoxylin and eosin staining of sections from untreated skin of sham and TPA-injected D6-deficient mice treated with PBS, Evasin-1, or anti-TNF-α. Skin sections were obtained on day 4 after TPA application and are representative of 10 mice per group per experiment. Bars, 100 μm.
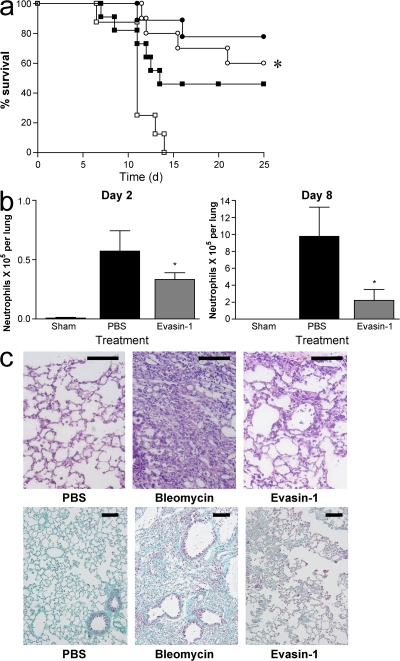
CCL3 has previously been demonstrated to play a role in mediating pulmonary fibrosis (15). We therefore tested the effect of Evasin-1 in a lung injury model induced by intratracheal instillation of bleomycin. Mild or severe disease was induced with 0.0625 or 0.125 U of bleomycin. Evasin-1 reduced lethality in both forms of the disease (Fig. 7 a). Reduction in lethality was associated with a decrease in neutrophil accumulation at 2 and 8 d after bleomycin challenge (Fig. 7 b), and a net decrease in the ensuing fibrosis observed at later stages after the bleomycin challenge. Histopathological analysis of lung tissue confirmed decreased leukocyte infiltration in the lung (Fig. 7 c), which was corroborated by a decrease in the number of CD3+CD4+ and CD3+CD8+ lymphocytes and GR1+ leukocytes in the bronchoalveolar lavage fluid of mice challenged with bleomycin. In bleomycin-instilled mice that did not receive Evasin-1, there was a considerable deterioration of normal pulmonary architecture with loss of alveolar structure and collagen deposition (Fig. 7 c). Treatment with Evasin-1 resulted in the preservation of lung architecture and a decrease in collagen deposition. Morphometric analysis of Gomori's trichrome–stained areas showed that Evasin-1 treatment reduced collagen deposition by 40%.
Figure 7.
Evasin-1 inhibits lethality, cellular recruitment, and fibrosis in a model of bleomycin-induced pulmonary injury. (a) Evasin-1 inhibits lethality associated with pulmonary instillation of bleomycin in C57BL/6 mice. Bleomycin was instilled intratracheally at doses of 0.0625 U (circles) or 0.125 U (squares) and survival rates were evaluated for 25 d. PBS (open symbols) or Evasin-1 (10 μg per animal; closed symbols) were administered s.c. 45 min before and every 12 h after initial bleomycin instillation. There were 12 mice per group in each experiment. *, P < 0.05 when comparing PBS- and Evasin-1–treated animals at 0.125 U. (b) Evasin-1 inhibits neutrophil recruitment induced by bleomycin. Neutrophil recruitment was evaluated at 2 and 8 d after bleomycin (0.125 U) and Evasin-1 (10 μg per animal) was administered s.c. 45 min before and every 12 h after initial bleomycin instillation. There were six mice per group in each experiment. *, P < 0.05 when compared with the PBS-treated group. Error bars represent the mean ± SEM. (c) Evasin-1 inhibits pulmonary inflammation and fibrosis induced by instillation of bleomycin. Hematoxylin and eosin– (top) and Gomori's trichrome–stained (bottom) sections from animals instilled with sodium chloride (0.9% NaCl, 25 μl) or bleomycin (0.125 U). Evasin-1 (10 μg per animal) was administered as previously described and lungs were removed on day 16. Bars: (top) 100 μm; (bottom) 50 μm.
Evasin-3 is a potent inhibitor of neutrophil-mediated inflammation
Evasin-3 has also been shown to have potent antiinflammatory properties. In accordance with the nanomolar affinities observed by SPR analysis (Table I), Evasin-3 produced in HEK293 cells and in E. coli inhibited binding of 125I-CXCL8 to CXCR1 with IC50 values of 0.7 and 1 nM, respectively (Fig. 8 a), demonstrating that glycosylation is not necessary for binding, similar to Evasin-1 (11). CXCL8-induced chemotaxis of neutrophils in vitro was inhibited by Evasin-3 with an IC50 of 3.35 nM (Fig. 8 b). In vivo, Evasin-3 inhibited KC-induced neutrophil recruitment into the peritoneal cavity in a dose-dependent manner, with maximal inhibition at a dose of 1 μg per mouse (Fig. 8 c). We also tested the ability of Evasin-3 to inhibit KC recruitment into the knee joint, and found it to be even more potent in this cavity (Fig. 8 d).
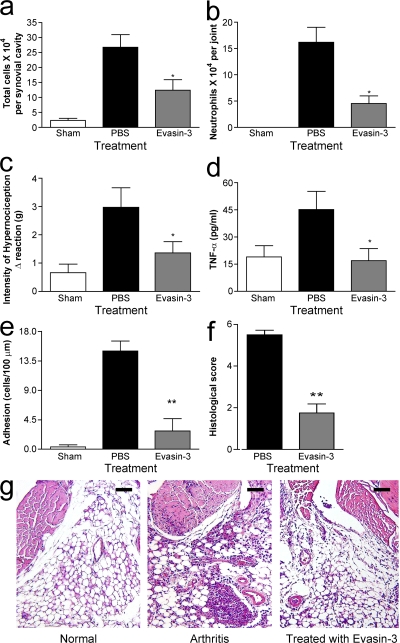
Evasin-3 was then tested in a mouse model of antigen-induced arthritis. In this model, rapid up-regulation of KC and MIP-2 precedes the recruitment of neutrophils into the knee joint. This is followed by CXCR2 up-regulation and TNF-α production, culminating in synovial hyperplasia and inflammatory hypernociception (16). Administration of Evasin-3 resulted in a 50% decrease in the total number of leukocytes in the synovial cavity (Fig. 9 a). The major leukocyte populations affected were neutrophils, which were reduced by 70% in the knee joint (Fig. 9 b). Moreover, Evasin-3 treatment significantly reduced the inflammatory hypernociception associated with this model (Fig. 9 c), and interestingly, the treatment resulted in decreased local production of TNF-α (Fig. 9 d). Intravital microscopy studies of the synovial microvasculature showed that administration of Evasin-3 prevented the adhesion of leukocytes to the synovial endothelium (Fig. 9 e), providing a direct demonstration of the efficacy of Evasin-3 in the prevention of neutrophil influx in an inflammatory setting. Consistent with the aforementioned findings, histopathological analysis of tissue demonstrated overall inhibition of leukocyte influx into the joint and periarticular tissues, decreased edema, and decreased synovial hyperplasia (Fig. 9, f and g).
Figure 9.
Evasin-3 inhibits neutrophil recruitment and ameliorates inflammation and functionality in a mouse model of antigen-induced arthritis. (a–g) Reduction of inflammation in mBSA induced arthritis by Evasin-3 (1 μg per animal). (a) Inhibition of total cell accumulation in the synovial cavity. (b) Inhibition of neutrophil infiltration into the knee joint. (c) Reduction of hypernociception induced by intraarticular antigen challenge of immunized mice. (d) Reduction of TNF-α production in periarticular tissues. (e) Inhibition of adhesion of leukocytes to synovial microvessels as assessed by intravital microscopy. (f and g) Histological evaluation of the tissue sections form the knee joint. All parameters were evaluated 24 h after antigen challenge, and there were 6 mice per group in each experiment. *, P < 0.05; **, P < 0.01. Error bars represent the mean ± SEM. Bars, 100 μm.
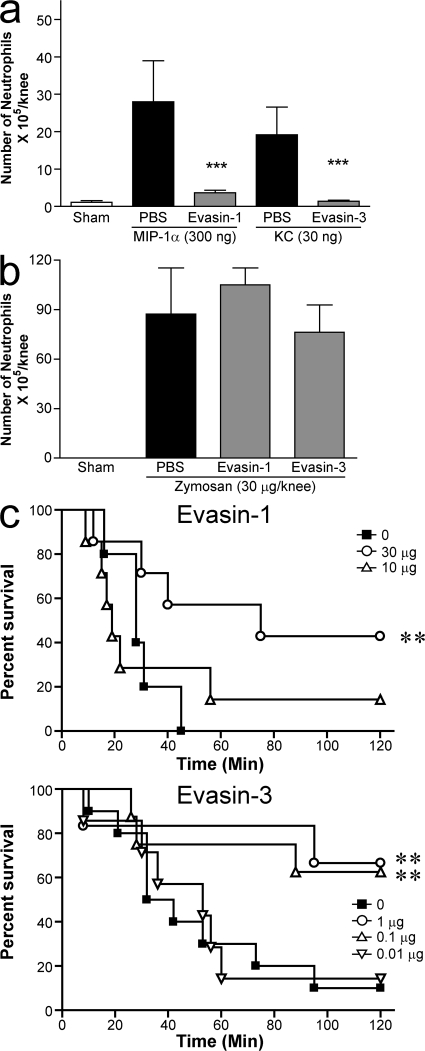
Finally, we tested both proteins in the same recruitment model, chemokine-induced neutrophil in the knee joint, at the doses used in the disease models, and both Evasins were able to completely inhibit the recruitment induced by their respective ligand (Fig. 10 a). The effect of the Evasins was shown to be specific to chemokine inhibition because neither was able to inhibit neutrophil recruitment mediated by zymosan (Fig. 10 b), which has been shown to mediate its effects through chemoattractants such as C5a (17) and LTB4 (18). As both Evasins inhibit neutrophil recruitment mediated by different chemokines, we tested them head to head in the same model, intestinal ischemia. As shown in Fig. 10 c, Evasin-3 was more efficacious, showing significant reduction in lethality at doses of 1 and 0.1 μg, whereas Evasin-1 only showed significant efficacy at the 30 μg dose, with only a slight effect seen at the 10 μg dose.
Figure 10.
Direct comparison of Evasin-3 and -1. (a) Inhibition of chemokine-induced neutrophil recruitment into the knee joint induced by chemokine (a) or by Zymozan (b). Evasin-1 (10 μg) and Evasin-3 (1 μg) were administered 30 min into the knee joint before the administration of 300 ng mMIP-1α or 30 ng KC, respectively, or 30 μg Zymosan. Cells in the joint were counted 6 h after the injection of stimuli. There were five mice per group in each experiment. **, P < 0.01; ***, P < 0.001 when compared with PBS-treated group. (c) Prevention of ischemic reperfusion. Evasin-1 and -3 were administered s.c. at the doses indicated 30 min before the reperfusion of the occluded superior mesenteric artery. There were seven to eight mice in each experimental group. **, P < 0.01 when compared with PBS-treated group. Error bars represent the mean ± SEM.
DISCUSSION
Since the identification of the first members of chemokine family over 20 yr ago, it has become clear that the interaction of these small proteins with their cognate receptors are important targets for therapeutic intervention in several inflammatory, autoimmune, and infectious diseases. Indeed, there are currently many intense drug discovery programs in this area. Yet, although there are numerous reports of the efficacy of small molecule antagonists (19–21), neutralizing antibodies (22, 23), and other antagonists (24) in animal models of disease, it has proven to be more of a challenge to translate these molecules into medicines in the clinic. Although many molecules are currently progressing into late-stage clinical trials, it is possible that alternative approaches may be needed to successfully target the chemokine–chemokine receptor interaction. Ticks have evolved natural antichemokine strategies in the guise of CHPBs, which are able to counter the host defense systems against parasites. Unraveling the molecular nature of these CHBPs may produce novel insights into the design of therapeutically useful chemokine blocking agents.
In this study, we describe the cloning and characterization of a family of CHPBs from tick saliva. In vitro characterization of the family members identified to date indicates that these CHBPs are considerably different from the known viral CHPBs in terms of chemokine selectivity. Many of the viral CHBPs such as myxoma virus M-T7 and murine gammaherpes virus 68 M3 and the recently identified S. mansoni (for review see [3]) tend toward broad selectivity, binding both CC and CXC chemokines; the Evasins described here have restricted selectivity. In the studies presented here, we have shown that Evasins are able to bind and inhibit specific chemokines in vitro, and inhibit leukocyte recruitment induced by their respective ligands in simple models of leukocyte recruitment in mice. Furthermore, the antiinflammatory properties of Evasins were confirmed mechanistically by intravital microscopy where their administration both in naive and inflamed mice abrogated leukocyte adhesion to, and transmigration across, the endothelium. Together, these experiments clearly demonstrated the potential of Evasins to prevent leukocyte recruitment.
We evaluated the Evasins in more complex animal models of human disease. Evasin-1, which is highly selective for CCL3 and CCL4, reduced symptoms in a murine model of skin inflammation that resembles psoriasis in humans, in which several CC chemokines are implicated (14). Because both Evasin-1 and -3 were shown to prevent neutrophil influx, they were subsequently tested in disease models in which neutrophils have previously been shown to play an important role. Thus, in the CCL3-dependent bleomycin-induced lung injury, Evasin-1 reduced leukocyte influx, fibrosis, and lethality, an effect qualitatively and quantitatively similar to experiments in CCL3-deficient mice (15, 25). Blockade of CXCR2 has been reported to prevent leukocyte influx and joint damage in several models of experimental arthritis in rats and mice (26, 27). Evasin-3, which binds to both human and murine CXCR2 ligands, prevented neutrophil influx into the joint in a model of antigen-induced arthritis. Interestingly, this treatment also prevented the local production of TNF-α, a cytokine that has been shown to play an important role in the pathogenesis of arthritis, and which is associated with hypernociception or inflammatory pain in experimental animals. Indeed, blockade of TNF-α with biologicals has been successful in the treatment of rheumatoid arthritis and other rheumatologic conditions (28, 29).
Our initial expectation was that identification of CHBPs in primitive eukaryotic species such as ticks may allow us to identify orthologues in mammalian genomes, based on either primary sequence or on the three-dimensional folds of the CHBPs. However, extensive searching of public databases at both the amino acid and nucleotide sequence level have indicated that there are no human or mammalian orthologues or structural counterparts to the Evasins. On the other hand, by blast analysis of the available tick sequences, we were able to identify an EST in the cattle tick Boophilus microplus that may encode a potential orthologue of Evasin-1 (30% identity over 95 amino acids). Although the level of identity is low, it is comparable to that seen between Evasin-1 and -4. We are now in the process of producing the Boophilus microplus protein to verify if it is indeed a CHPB. It is likely that additional CHPBs from other species of tick will be identified in due course through homology searching, as the number of tick EST sequences appearing in public databases increases from tick salivary gland transcriptome sequencing projects.
Thus, it appears that ticks have specifically developed novel protective CKBPs as they adapted to their chemokine-producing vertebrate hosts. Although mammals do have molecules that can bind and block chemokine function, they are molecularly and structurally distinct from the Evasins. For example, the atypical chemokine receptors such as D6, CCX-CKR, and Duffy (30, 31) are nonsignaling GPCRs that appear to be able to function as mammalian equivalents of the soluble CKBPs expressed by pathogens and parasitic worms. This suggests that, contrary to viral chemokines and chemokine receptors that have been hijacked from the mammalian genome, pathogens have evolved CKBPs through a “convergent” evolution mechanism.
The leukocytes involved in the first line of defense against pathogens and parasites are neutrophils and eosinophils. The CHBPs described here specifically inhibit the chemokines that recruit these cell types; Evasin-1 (in rodents) and Evasin-3 prevent neutrophil accumulation, and preliminary results indicate that Evasin-4 binds the eosinophil-recruiting chemokines CCL5 and CCL11. Moreover, their selectivity profiles highlight the issue of species specificity in the chemokine system. The common brown dog tick feeds on dogs and other domestic animals, as well as rodents, deer, and humans; thus, in spite of chemokine selectivity, one would expect species cross-reactivity. We have performed our studies using human chemokines, and in some cases, their murine counterparts, and have shown that the Evasins are able to bind to chemokines in both species. The cross species reactivity may be an advantage in the development of these molecules as potential therapeutic agents, as one of the pitfalls in the development of small molecule antagonists of human chemokine receptors has been their lack of cross-reactivity with rodent chemokine receptors, which has hampered testing in animal models of disease, PK/PD, and toxicological studies. However, results obtained from in vivo testing of Evasins still needs to be interpreted with caution, as there are known differences in specific chemokine receptor expression on immune cells between the human and murine system, particularly on neutrophils. In humans, the principal mediators of neutrophil recruitment are CXC chemokines 1–8, acting through CXCR2, and CXCL8, which acts on both CXCR1 and CXCR2. On the contrary, recruitment of murine neutrophils is mediated by both CCR1 and CXCR2, and perhaps also the recently identified murine CXCR1 (32), although little is known about the role of the latter in vivo. However, CCR1 is not normally expressed on human neutrophils, although it can be induced by IFN-γ (33). Interestingly, CCL3 has been shown to recruit neutrophils when injected into human skin (34). Thus, Evasin-1 shows activity against CCL3-mediated neutrophil recruitment rather than monocyte recruitment in the mouse, whereas the ability of Evasin-3 to recruit neutrophils is predictable from its binding profile. It should also be noted that although Evasin-1 has a higher affinity for its ligands than Evasin-3, the latter is more efficacious in the murine models we have tested.
Saliva from ticks harvested at different times during their feeding periods has differential chemokine inhibitory activity, suggesting that different CHPBs are produced at different times during feeding, with the aim of selectively inhibiting specific cell populations (35). It is possible that the most potent inhibitor of neutrophil recruitment produced by the tick is Evasin-3, whereas Evasin-1 may be produced to inhibit (later) monocyte recruitment in nonrodent hosts, as well as T lymphocytes, as was observed in the bleomycin lung inflammation model. Unfortunately, the expression profile of chemokine receptors on leukocytes in the dog, which although its name implies is the preferred host, may simply be the most easily available food source, is unknown and hence we cannot substantiate this hypothesis. Lastly, the availability of recombinant Evasin-4 will enable us to address the ability of this CHBP to modulate eosinophil trafficking because both CCL5 and CCL11 have been extensively described to recruit this cell type.
A striking feature of these proteins is their small size. Both viral and mammalian chemokine and cytokine binding proteins have been shown to be much larger, generally ∼30–40 kD. On the other hand, R. sanguineus has developed very small, highly selective CHBPs. Interestingly, these proteins are similar in size, if not smaller, than the naturally occurring single chain antibodies such as shark V-NAR domains (15 kD) or camelid VhH domains (15 kD). Large efforts are currently being undertaken to produce small proteins known as nanobodies, minibodies, or microproteins, created by rational design in silico, which will mimic the binding properties of natural single-domain antibodies (36). The two proteins described here represent highly potent, miniaturized scaffolds that have evolved to selectively bind certain proteins, pivotal to the immune response, and which could be useful in the creation of new scaffolds, paving the way for development of these molecules as novel antiinflammatory agents.
One of the important issues surrounding the use of any protein as a potential therapeutic agent concerns immunogenicity, which could be particularly pertinent for nonhuman proteins. There are already examples of xenoproteins in the clinic (4, 37) and others are entering clinical trials. However, as these proteins are generally administered in a single dose for acute indications, potential immunogenicity is not a major concern. However, treatment of inflammatory and autoimmune diseases with therapeutic proteins such as IFN-β or antibodies, e.g., Raptiva and Etanercept, requires chronic administration, and even fully human proteins can elicit an antibody response that may reduce the efficacy of the treatment. By their very nature, antiinflammatory proteins in tick saliva do not appear to be immunogenic. It is still unclear if this is entirely caused by the inhibition of immune cell recruitment to the attachment site, or indeed whether the protein structure itself or through glycosylation minimizes exposure of antigenic epitopes. Furthermore, bioinformatic analysis of T cell epitopes on Evasins using software developed by Biovation (EMD Serono) revealed that the tick Evasins have relatively few potential immunogenic epitopes compared with other therapeutic proteins currently in the clinic, such as IFN-β. It is also interesting to note that only 1 mouse out of 6 treated for 25 d had an IgG response to Evasin-1 (unpublished data). So although immunogenicity in the murine system will not predict the outcome in humans, further evaluation of Evasins in preclinical models is warranted for their development as potential protein therapeutic agents.
Lastly, the mining of the salivary transcriptomes of the numerous species of ticks will provide a wealth of potential immunomodulatory molecules, and other pharmacologically relevant proteins, which will provide novel molecular entities to enrich the current repertoire of biotherapeutics in the future (5, 38).
MATERIALS AND METHODS
Biochemical analysis of antichemokine activity in saliva.
Collection of R. sanguineus saliva, inhibition of receptor binding and the cross-linking assay were performed as previously described (11). SPR was performed by coating the chemokines on a CM4 chip (BIAcore), with RNaseA as negative protein control, and tick saliva injected after a 10- or 25-fold dilution in HBS-P buffer (BIAcore) for equilibration and 10 mM glycine-HCl, pH 2.5, for regeneration. The sensograms were analyzed using BIAcore 3000 evaluation software after subtraction of nonspecific binding to the control protein. Retentate chromatography SELDI mass spectrometry (RC-SELDI-TOF-MS) was used to detect distinct CHPBs in the tick saliva. CCL5, CCL3, and CXCL8 or the control protein, equine myoglobin, were diluted to 0.1 μg/μl in PBS and immobilized onto separate spots of a PS-20 ProteinChip array according to the manufacturer's instructions (Ciphergen Biosystems, Inc.). After blocking and washing of the array, frozen tick saliva was diluted fivefold in PBS and 5 μl was applied to each spot and incubated for 2 h at room temperature. After washing, the matrix, sinapinic acid, was dried onto each spot on the array and mass analysis was performed by SELDI-TOF-MS, using a calibrated ProteinChip Biology System IIc reader. Spectra were generated using an automated protocol and a positive ion mode with a laser intensity of 220 and deflector sensitivity of 10. The mass optimization range was set between 10 and 50 kD, with a mass focus set to 18 kD. To obtain quantitative data, the mass spectra from each spot were accumulated from 155 laser shots over 31 different and nonoverlapping positions.
Molecular cloning.
Expression cloning was performed by transient transfection of HEK293 cells with a cDNA library constructed from the salivary glands of R. sanguineus, as previously described (11). Supernatants from transfected cells were screened using two cocktails of radiolabeled [I-125] CXCL8, IL-1, and IL-2, or CCL5, CCL11, and CXCL10, respectively, followed by cross-linking and analysis by SDS-PAGE. After deconvolution of the cDNA pools and sequencing of the positive clones, a C-terminal 6His-tagged version of the cDNA encoding the relevant CHBP was subcloned into the mammalian cell expression vector pEAK12d (Edge Biosystems) using the Gateway cloning system (Invitrogen). The protein was purified by Ni-chelate chromatography after transient expression in 500 ml HEK293 cultures. The cDNA encoding Evasin-3 was subcloned into the pET30a vector (Novagen) for expression in E. coli strain BL21(DE3) and produced in a 5-liter fermenter. The protein was purified from the cytosolic fraction by cation exchange chromatography and size exclusion chromatography.
In vitro characterization.
The selectivity of Evasin-3 was determined by SPR, as previously described (11). Kinetic measurements were performed to determine the binding constants. The kinetic values ka and kd, which are shown in Table I, are the mean of three independent experiments performed in triplicate on three different chips. The affinity constant Kd was calculated from theses mean values. Equilibrium competition receptor binding assays were performed using CHO/CXCR1 transfectants using 125I-CXCL8, as previously described (39). Inhibition of chemotaxis in vitro was measured using L1.2/CCR5 transfectants for Evasin-1 or neutrophils purified from buffy coats as previously described (40).
In vivo experiments.
Male BALB/c or C57BL/6 mice (18–22 g) were housed in a temperature-controlled room and had free access to food and water. All experiments received prior approval by the local animal ethics committee. To test the effect of Evasin-1 on cellular recruitment in vivo, mice were administered 0.01–10 μg Evasin-1 in 200 μl PBS s.c. 45 min before the administration of 10 μg CCL3 or 0.3 μg mMIP-1α in 200 μl PBS i.p. Cell recruitment was evaluated 18 h after the stimulus. To test the effect of Evasin-3 on neutrophil recruitment, KC (30 ng/cavity) was injected into the peritoneal cavity or the left knee joint of the mouse and recruitment was evaluated 4 h later. Evasin-3 (0.01–1 μg/mouse) was given s.c. 45 min before KC administration.
Intravital microscopy was performed in unbranched cremasteric venules (25–40 μm in diameter) of C57BL/6 mice, as previously described (41). For each experiment, 0.3 μg of human CCL3 in 100 μl of PBS was administered locally by s.c. injection beneath the right scrotal skin, 1 h before exteriorization. To test inhibition, a solution of Evasin-1 (10 μg per animal) plus CCL3 (0.3 μg) in 100 μl PBS was prepared 15 min before intrascrotal injection.
mBSA-induced DTH reaction in the pleural cavity.
C57BL/6 mice were immunized s.c. with a 1:1 emulsion of mBSA (5 mg/ml in saline) and complete Freund's adjuvant (Sigma-Aldrich), as previously described (42). 14 d after the immunization, antigen (mBSA; 10 μg per pleural cavity) or vehicle was injected into the pleural cavity, and the number of cells per cavity was evaluated 48 h after injection of antigen.
Sensitization and induction of Th2 cellular recruitment into the lungs.
BALB/c mice were immunized i.p. with 2,500 isolated S. mansoni eggs at day 0 and 7 of the protocol, as previously described (43). On day 14, mice were given an intranasal challenge of 10 μg soluble egg antigens (SEA) in 10 μl of PBS to localize the response to the airway. Mice were then rechallenged 6 d later by intratracheal administration of 10 μg SEA in 25 μl of PBS or with PBS alone. To test inhibition, Evasin-1 (10 μg per animal) was administered s.c. 45 min before and 24 h after antigen challenge. The number of cells in the bronchoalveolar lavage fluid was evaluated at 24 and 48 h after stimulation.
Induction of skin inflammation in D6−/− mice.
Skin inflammation was induced as previously described (14). In brief, the phorbol ester TPA was applied three times, at 24-h intervals, to the shaved dorsal skin of 6–8-wk-old D6 (−/−) mice, and the mice were left for 4 d to allow the exaggerated cutaneous inflammation to develop. In addition, mice were administered either PBS or Evasin-1 daily (30 μg/mouse pr day) by i.p. injection or were given daily doses of anti-TNF as previously described (14). Mice were killed at day 4 after TPA treatment, and the skin was harvested for histological assessment. Skin samples were collected into neutral-buffered formalin and processed for hematoxylin and eosin staining as described (14).
Bleomycin-induced lung injury.
0.0625 or 0.125 U blenoxane (Bristol-Meyers Squibb Co.) in 25 μl PBS was instilled under anesthesia into the trachea of C57BL/6j mice. Control animals received PBS alone. To test inhibition, Evasin-1 (3 or 10 μg per animal) was administered s.c. 45 min before, and every 12 h after bleomycin instillation. At 4, 8, or 25 d after instillation, the lungs were washed with 3 × 1 ml of PBS, and the number of infiltrating leukocytes were evaluated. Right lung lobes were collected and inflated with 10% formaldehyde for histological analysis of tissue sections stained with hematoxylin and eosin. For localization of collagen within the lung tissues, Gomori's trichrome-specific staining was used.
Antigen-induced arthritis.
Animals were immunized intradermally (i.d.) at the base of the tail with 500 μg of methylated BSA (mBSA; Sigma-Aldrich) in 100 μl of an emulsion of saline and an equal volume of CFA (Sigma-Aldrich) on day 0, as previously described (44). Challenge of mice with 10 μg of mBSA in 10 μl sterile saline in the left knee joint was performed 14 d later. The knee cavity was washed 24 h later with PBS (2 × 5 μl), and the periarticular tissues were removed for the evaluation of TNF-α and MPO activity. Intravital microscopy was performed in the synovial microcirculation of the left mouse knee, as previously described (45).
Evaluation of mechanical hypernociception.
Inflammatory hypernociception was performed as previously described (46) using an electronic pressure meter. In brief, an increasing perpendicular force is applied to the central area of the plantar surface of the hind paw to induce the dorsal flexion of the femur-tibial joint, followed by paw withdrawal. The flexion-elicited withdrawal threshold (expressed in grams) is recorded before and after the stimulus and is used to infer mechanical hypernociception.
Ischemia and reperfusion.
Ischemia and reperfusion injury were performed as previously described (47). In brief, ischemia was induced by totally occluding the superior mesenteric artery for 60 min. Reperfusion was reestablished, and death was monitored during the indicated time periods.
Animal experimentation.
Skin inflammation experiments in D6−/− mice were approved by a Glasgow ethical review committee and performed under licence number 60/3415 granted by the UK Home Office. Bleomycin-induced lung injury, antigen-induced arthritis, and ischemia and reperfusion injury experiments were evaluated and approved by the Animal Ethics committee of the Universidade Federal de Minas Gerais, Brazil.
Histological assessment.
Sections were graded as previously described (48). In brief, the histological score (0–8) was the sum of the synovial hyperplasia, cellular exudate, and cartilage depletion/bone erosion ranging from 0 (normal) to 3 (severe), and the degree of synovial infiltrate ranging from 0 (normal) to 5 (severe).
Statistical analysis.
All results are presented as the mean ± SEM. Normalized data were analyzed by one-way ANOVA, and differences between groups were assessed using the Student-Newman-Keuls post test. A P value < 0.05 was considered to be significant.
Acknowledgments
We thank M. de Tiani, S. Jaconi, F. Borlat, V. Dechavanne, L. Glez, L. Friedli, F. Bollin, C. Losberger, and L. Chevalet for advice in protein expression and purification.
This work was supported by the European Union FP6 (INNOCHEM, grant number LSHB-CT-2005-518167 to A.E.I. Proudfoot and M.M. Teixeira).
The authors have no conflicting financial interests.
Abbreviations used: CHPB, chemokine binding protein; mBSA, methylated BSA; SELDI, surface enhanced laser desorption ionization; SPR, surface plasmon resonance.
A. Frauenschuh's present address is Novartis Pharma AG, 4056 Basel, Switzerland.
References
- 1.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36–50. [DOI] [PubMed] [Google Scholar]
- 2.McFadden, G., A. Lalani, H. Everett, P. Nash, and X. Xu. 1998. Virus-encoded receptors for cytokines and chemokines. Semin. Cell Dev. Biol. 9:359–368. [DOI] [PubMed] [Google Scholar]
- 3.Fallon, P.G., and A. Alcami. 2006. Pathogen-derived immunomodulatory molecules: future immunotherapeutics? Trends Immunol. 27:470–476. [DOI] [PubMed] [Google Scholar]
- 4.Hovius, J.W., A.P. van Dam, and E. Fikrig. 2007. Tick-host-pathogen interactions in Lyme borreliosis. Trends Parasitol. 23:434–438. [DOI] [PubMed] [Google Scholar]
- 5.Ribeiro, J.M. 1995. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect. Agents Dis. 4:143–152. [PubMed] [Google Scholar]
- 6.Lucas, A., and G. McFadden. 2004. Secreted immunomodulatory viral proteins as novel biotherapeutics. J. Immunol. 173:4765–4774. [DOI] [PubMed] [Google Scholar]
- 7.Smith, P., R.E. Fallon, N.E. Mangan, C.M. Walsh, M. Saraiva, J.R. Sayers, A.N. McKenzie, A. Alcami, and P.G. Fallon. 2005. Schistosoma mansoni secretes a chemokine binding protein with antiinflammatory activity. J. Exp. Med. 202:1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lalani, A.S., J. Masters, K. Graham, L.Y. Liu, A. Lucas, and G. McFadden. 1999. Role of the myxoma virus soluble CC-chemokine inhibitor glycoprotein, M-T1, during myxoma virus pathogenesis. Virology. 256:233–245. [DOI] [PubMed] [Google Scholar]
- 9.Hajnicka, V., P. Kocakova, M. Slavikova, M. Slovak, J. Gasperik, N. Fuchsberger, and P.A. Nuttall. 2001. Anti-interleukin-8 activity of tick salivary gland extracts. Parasite Immunol. 23:483–489. [DOI] [PubMed] [Google Scholar]
- 10.Hajnicka, V., I. Vancova, P. Kocakova, M. Slovak, J. Gasperik, M. Slavikova, R.S. Hails, M. Labuda, and P.A. Nuttall. 2005. Manipulation of host cytokine network by ticks: a potential gateway for pathogen transmission. Parasitology. 130:333–342. [DOI] [PubMed] [Google Scholar]
- 11.Frauenschuh, A., C.A. Power, M. Deruaz, B.R. Ferreira, J.S. Silva, M.M. Teixeira, J.M. Dias, T. Martin, T.N. Wells, and A.E. Proudfoot. 2007. Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus. J. Biol. Chem. 282:27250–27258. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson, D.L., and R.G. Harrison. 1991. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology (N. Y.). 9:443–448. [DOI] [PubMed] [Google Scholar]
- 13.Nibbs, R.J., J. Yang, N.R. Landau, J.H. Mao, and G.J. Graham. 1999. LD78beta, a non-allelic variant of human MIP-1alpha (LD78alpha), has enhanced receptor interactions and potent HIV suppressive activity. J. Biol. Chem. 274:17478–17483. [DOI] [PubMed] [Google Scholar]
- 14.Jamieson, T., D.N. Cook, R.J. Nibbs, A. Rot, C. Nixon, P. McLean, A. Alcami, S.A. Lira, M. Wiekowski, and G.J. Graham. 2005. The chemokine receptor D6 limits the inflammatory response in vivo. Nat. Immunol. 6:403–411. [DOI] [PubMed] [Google Scholar]
- 15.Smith, R.E., R.M. Strieter, S.H. Phan, N.W. Lukacs, G.B. Huffnagle, C.A. Wilke, M.D. Burdick, P. Lincoln, H. Evanoff, and S.L. Kunkel. 1994. Production and function of murine macrophage inflammatory protein-1 alpha in bleomycin-induced lung injury. J. Immunol. 153:4704–4712. [PubMed] [Google Scholar]
- 16.Grespan, R., S.Y. Fukada, H.P. Lemos, S.M. Vieira, M.H. Napimoga, M.M. Teixeira, A.R. Fraser, F.L. Liew, I.B. McInnes, and F.D. Cunha. 2008. Arthritis Rheum. 58:2030–2040. [DOI] [PubMed] [Google Scholar]
- 17.Mullaly, S.C., and P. Kubes. 2007. Mast cell-expressed complement receptor, not TLR2, is the main detector of zymosan in peritonitis. Eur. J. Immunol. 37:224–234. [DOI] [PubMed] [Google Scholar]
- 18.Rao, N.L., P.J. Dunford, X. Xue, X. Jiang, K.A. Lundeen, F. Coles, J.P. Riley, K.N. Williams, C.A. Grice, J.P. Edwards, et al. 2007. Anti-inflammatory activity of a potent, selective leukotriene A4 hydrolase inhibitor in comparison with the 5-lipoxygenase inhibitor zileuton. J. Pharmacol. Exp. Ther. 321:1154–1160. [DOI] [PubMed] [Google Scholar]
- 19.Liang, M., C. Mallari, M. Rosser, H.P. Ng, K. May, S. Monahan, J.G. Bauman, I. Islam, A. Ghannam, B. Buckman, et al. 2000. Identification and characterization of a potent, selective, and orally active antagonist of the CC chemokine receptor-1. J. Biol. Chem. 275:19000–19008. [DOI] [PubMed] [Google Scholar]
- 20.Anders, H.J., V. Vielhauer, M. Frink, Y. Linde, C.D. Cohen, S.M. Blattner, M. Kretzler, F. Strutz, M. Mack, H.J. Grone, et al. 2002. A chemokine receptor CCR-1 antagonist reduces renal fibrosis after unilateral ureter ligation. J. Clin. Invest. 109:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodmerkel, C.M., R. Huber, M. Covington, S. Diamond, L. Hall, R. Collins, L. Leffet, K. Gallagher, P. Feldman, P. Collier, et al. 2005. Discovery and pharmacological characterization of a novel rodent-active CCR2 antagonist, INCB3344. J. Immunol. 175:5370–5378. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy, K.J., R.M. Strieter, S.L. Kunkel, N.W. Lukacs, and W.J. Karpus. 1998. Acute and relapsing experimental autoimmune encephalomyelitis are regulated by differential expression of the CC chemokines macrophage inflammatory protein-1 alpha and monocyte chemotactic protein-1. J. Neuroimmunol. 92:98–108. [DOI] [PubMed] [Google Scholar]
- 23.Lloyd, C.M., M.E. Dorf, A. Proudfoot, D.J. Salant, and J.C. Gutierrez-Ramos. 1997. Role of MCP-1 and RANTES in inflammation and progression to fibrosis during murine crescentic nephritis. J. Leukoc. Biol. 62:676–680. [DOI] [PubMed] [Google Scholar]
- 24.Proudfoot, A.E. 2002. Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishida, Y., A. Kimura, T. Kondo, T. Hayashi, M. Ueno, N. Takakura, K. Matsushima, and N. Mukaida. 2007. Essential roles of the CC chemokine ligand 3-CC chemokine receptor 5 axis in bleomycin-induced pulmonary fibrosis through regulation of macrophage and fibrocyte infiltration. Am. J. Pathol. 170:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barsante, M.M., T.M. Cunha, M. Allegretti, F. Cattani, F. Policani, C. Bizzarri, W.L. Tafuri, S. Poole, F.Q. Cunha, R. Bertini, and M.M. Teixeira. 2008. Blockade of the chemokine receptor CXCR2 ameliorates adjuvant-induced arthritis in rats. Br. J. Pharmacol. 153:992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunha, T.M., M.M. Barsante, A.T. Guerrero, W.A. Verri, Jr., S.H. Ferreira, F.M. Coelho, R. Bertini, C. Di Giacinto, M. Allegretti, F.Q. Cunha, and M.M. Teixeira. 2008. Treatment with DF 2162, a non-competitive allosteric inhibitor of CXCR1/2, diminishes neutrophil influx and inflammatory hypernociception in mice. Br. J. Pharmacol. 154:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilcek, J., and M. Feldmann. 2004. Historical review: cytokines as therapeutics and targets of therapeutics. Trends Pharmacol. Sci. 25:201–209. [DOI] [PubMed] [Google Scholar]
- 29.Tarner, I.H., U. Muller-Ladner, and S. Gay. 2007. Emerging targets of biologic therapies for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 3:336–345. [DOI] [PubMed] [Google Scholar]
- 30.Nibbs, R., G. Graham, and A. Rot. 2003. Chemokines on the move: control by the chemokine “interceptors” Duffy blood group antigen and D6. Semin. Immunol. 15:287–294. [DOI] [PubMed] [Google Scholar]
- 31.Mantovani, A., R. Bonecchi, and M. Locati. 2006. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat. Rev. Immunol. 6:907–918. [DOI] [PubMed] [Google Scholar]
- 32.Fan, X., A.C. Patera, A. Pong-Kennedy, G. Deno, W. Gonsiorek, D.J. Manfra, G. Vassileva, M. Zeng, C. Jackson, L. Sullivan, et al. 2007. Murine CXCR1 is a functional receptor for GCP-2/CXCL6 and interleukin-8/CXCL8. J. Biol. Chem. 282:11658–11666. [DOI] [PubMed] [Google Scholar]
- 33.Bonecchi, R., N. Polentarutti, W. Luini, A. Borsatti, S. Bernasconi, M. Locati, C.A. Power, A.E.I. Proudfoot, T.N.C. Wells, C. Mackay, et al. 1999. Up-regulation of CCR1 and CCR3 and induction of chemotaxis to CC chemokines by IFN-gamma in human neutrophils. J. Immunol. 162:474–479. [PubMed] [Google Scholar]
- 34.Lee, S.C., M.E. Brummet, S. Shahabuddin, T.G. Woodworth, S.N. Georas, K.M. Leiferman, S.C. Gilman, C. Stellato, R.P. Gladue, R.P. Schleimer, and L.A. Beck. 2000. Cutaneous injection of human subjects with macrophage inflammatory protein-1 alpha induces significant recruitment of neutrophils and monocytes. J. Immunol. 164:3392–3401. [DOI] [PubMed] [Google Scholar]
- 35.Vancova, I., M. Slovak, V. Hajnicka, M. Labuda, L. Simo, K. Peterkova, R.S. Hails, and P.A. Nuttall. 2007. Differential anti-chemokine activity of Amblyomma variegatum adult ticks during blood-feeding. Parasite Immunol. 29:169–177. [DOI] [PubMed] [Google Scholar]
- 36.Holliger, P., and P.J. Hudson. 2005. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 23:1126–1136. [DOI] [PubMed] [Google Scholar]
- 37.Barnett, A. 2007. Exenatide. Expert Opin. Pharmacother. 8:2593–2608. [DOI] [PubMed] [Google Scholar]
- 38.Ribeiro, J.M., F. Alarcon-Chaidez, I.M. Francischetti, B.J. Mans, T.N. Mather, J.G. Valenzuela, and S.K. Wikel. 2006. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 36:111–129. [DOI] [PubMed] [Google Scholar]
- 39.van Riper, G., D.W. Nicholson, M.P. Scheid, P.A. Fischer, M.S. Springer, and H. Rosen. 1994. Induction, characterization, and functional coupling of the high affinity chemokine receptor for RANTES and macrophage inflammatory protein-1 alpha upon differentiation of an eosinophilic HL-60 cell line. J. Immunol. 152:4055–4061. [PubMed] [Google Scholar]
- 40.Proudfoot, A.E., T.M. Handel, Z. Johnson, E.K. Lau, P. LiWang, I. Clark-Lewis, F. Borlat, T.N. Wells, and M.H. Kosco-Vilbois. 2003. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. USA. 100:1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cara, D.C., and P. Kubes. 2004. Intravital microscopy as a tool for studying recruitment and chemotaxis. Methods Mol. Biol. 239:123–132. [DOI] [PubMed] [Google Scholar]
- 42.Teixeira, M.M., A. Talvani, W.L. Tafuri, N.W. Lukacs, and P.G. Hellewell. 2001. Eosinophil recruitment into sites of delayed-type hypersensitivity reactions in mice. J. Leukoc. Biol. 69:353–360. [PubMed] [Google Scholar]
- 43.Lukacs, N.W., T.J. Standiford, S.W. Chensue, R.G. Kunkel, R.M. Strieter, and S.L. Kunkel. 1996. C-C chemokine-induced eosinophil chemotaxis during allergic airway inflammation. J. Leukoc. Biol. 60:573–578. [DOI] [PubMed] [Google Scholar]
- 44.Healy, A.M., E. Izmailova, M. Fitzgerald, R. Walker, M. Hattersley, M. Silva, E. Siebert, J. Terkelsen, D. Picarella, M.D. Pickard, et al. 2006. PKC-theta-deficient mice are protected from Th1-dependent antigen-induced arthritis. J. Immunol. 177:1886–1893. [DOI] [PubMed] [Google Scholar]
- 45.Veihelmann, A., G. Szczesny, D. Nolte, F. Krombach, H.J. Refior, and K. Messmer. 1998. A novel model for the study of synovial microcirculation in the mouse knee joint in vivo. Res. Exp. Med. (Berl.). 198:43–54. [DOI] [PubMed] [Google Scholar]
- 46.Cunha, T.M., W.A. Verri Jr., G.G. Vivancos, I.F. Moreira, S. Reis, C.A. Parada, F.Q. Cunha, and S.H. Ferreira. 2004. An electronic pressure-meter nociception paw test for mice. Braz. J. Med. Biol. Res. 37:401–407. [DOI] [PubMed] [Google Scholar]
- 47.Souza, D.G., A.C. Soares, V. Pinho, H. Torloni, L.F. Reis, M.M. Teixeira, and A.A. Dias. 2002. Increased mortality and inflammation in tumor necrosis factor-stimulated gene-14 transgenic mice after ischemia and reperfusion injury. Am. J. Pathol. 160:1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams, A.S., P.J. Richards, E. Thomas, S. Carty, M.A. Nowell, R.M. Goodfellow, C.M. Dent, B.D. Williams, S.A. Jones, and N. Topley. 2007. Interferon-gamma protects against the development of structural damage in experimental arthritis by regulating polymorphonuclear neutrophil influx into diseased joints. Arthritis Rheum. 56:2244–2254. [DOI] [PubMed] [Google Scholar]