Abstract
Myomesin is a 185-kDa protein located in the M-band of striated muscle where it interacts with myosin and titin, possibly connecting thick filaments with the third filament system. By using expression of epitope-tagged myomesin fragments in cultured cardiomyocytes and biochemical binding assays, we could demonstrate that the M-band targeting activity and the myosin-binding site are located in different domains of the molecule. An N-terminal immunoglobulin-like domain is sufficient for targeting to the M-band, but solid-phase overlay assays between individual N-terminal domains and the thick filament protein myosin revealed that the unique head domain contains the myosin-binding site. When expressed in cardiomyocytes, the head domains of rat and chicken myomesin showed species-specific differences in their incorporation pattern. The head domain of rat myomesin localized to a central area within the A-band, whereas the head domain of chicken myomesin was diffusely distributed in the cytoplasm. We therefore conclude that the head domain of myomesin binds to myosin but that this affinity is not sufficient for the restriction of the domain to the M-band in vivo. Instead, the neighboring immunoglobulin-like domain is essential for the precise incorporation of myomesin into the M-band, possibly because of interaction with a yet unknown protein of the sarcomere.
INTRODUCTION
During differentiation each cell type expresses a specific set of proteins, which then have to be targeted to their proper location within the cell to be assembled. Targeting and assembly can be investigated especially well during the formation of myofibrils in striated muscle. In myofibrils, two filament systems, the thin and thick filaments, are responsible for generating active shortening of the sarcomere, whereas a third filament system, composed of the giant protein titin (Trinick, 1991; Labeit and Kolmerer, 1995), mediates passive elasticity and also acts as a sarcomeric ruler during myofibrillogenesis (Horowits et al., 1986; Trinick, 1994; Gautel and Goulding, 1996). The three filament systems are connected by a host of additional proteins into highly regular, paracrystalline structures, the so-called sarcomeres (for review, see Small et al., 1992). It is important to understand how the complex sarcomeric structure is assembled from its building blocks and how it is maintained. Very likely, the information for the assembly is contained within the sarcomeric proteins that are expressed and continuously assembled into sarcomeres as the cells grow (Fürst et al., 1989). If this assembly is to occur in an ordered manner, every protein has to be sorted to its proper place within the growing myofibril. Indeed, domains that are responsible for sorting have been identified in several sarcomeric proteins (Soldati and Perriard, 1991; Gilbert et al., 1996; Komiyama et al., 1996; Obermann et al., 1998; Stolz and Wallimann, 1998).
The Z-disk and the M-band are structures situated in the centers of the thin and thick filaments, respectively, and are thought to play an important role in the assembly of the actomyosin filament systems during myofibrillogenesis. Four proteins located in the M-band have been identified: myomesin (Grove et al., 1984, 1985), M-protein (Masaki and Takaiti, 1974), skelemin (Price, 1987), and the muscle isoform of creatine kinase (Turner et al., 1973; Wallimann et al., 1983). Myomesin and M-protein are both implicated in anchoring thick filaments to the elastic third filament system, because both proteins have an affinity for the thick filament component myosin (Mani and Kay, 1978; Obermann et al., 1995) as well as for titin (Nave et al., 1989; Obermann et al., 1997). Myomesin might be the primary link between thick and elastic filaments, because in contrast to M-protein, it is found in all types of adult striated muscle (Grove et al., 1989). Myomesin is a member of the immunoglobulin superfamily (Bantle et al., 1996) and is composed mainly of immunoglobulin-like and fibronectin type III domains (see Figure 1A). Several other thick filament–associated proteins, also called myosin-binding proteins (MyBPs), belong to this family, such as M-protein (Noguchi et al., 1992; Vinkemeier et al., 1993), myosin-binding protein C (MyBP-C)1 (Offer et al., 1973; Einheber and Fischman, 1990), 86K or H-protein (Bähler et al., 1985; Starr et al., 1985), and titin (Labeit and Kolmerer, 1995). The observation that many thick filament–associated proteins contain immunoglobulin-like and fibronectin type III domains points toward a role of these domains in mediating interactions with myosin (Fürst and Gautel, 1995; Bantle et al., 1996).
Figure 1.
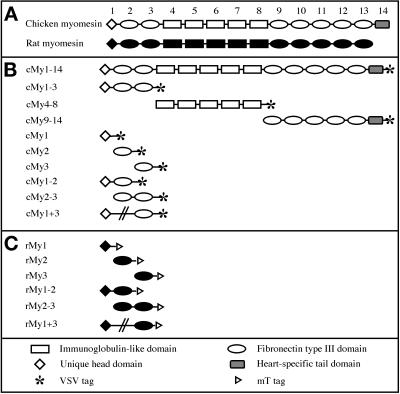
Chicken and rat myomesin truncation and deletion mutants. (A) Domain pattern derived from the cDNAs of chicken and rat myomesin. A head domain with no homology to other known proteins is followed by a conserved arrangement of immunoglobulin-like and fibronectin type III domains. The heart isoform of chicken myomesin contains an additional unique domain at the C terminus. (B) Chicken myomesin truncation and deletion mutants. The VSV epitope was fused C-terminally to all constructs. (C) Rat myomesin truncation and deletion mutants. The mT epitope was fused C-terminally to all constructs. The deletion mutants are depicted by an interrupted line connecting domains one and three in B and C. Abbreviations used in this article to indicate the constructs are shown on the left-hand side.
Despite several studies demonstrating interactions of myomesin with myosin and titin using biochemical assays, nothing is known about how these interactions direct myomesin to its assembly site in the M-band in vivo. We have constructed a panel of epitope-tagged fragments of chicken and rat myomesin to identify domains of myomesin that are involved in M-band targeting. We show that an immunoglobulin-like domain in the N-terminus of myomesin is crucial for the proper targeting of the protein to the M-band. Furthermore, we have identified a species-specific difference in M-band sorting of the N-terminal head domain. On the basis of targeting properties of the two head domains of chicken and rat myomesin, we show that although it is sufficient to bind myosin in solid-phase assays, the head domain has only a limited targeting specificity for the thick filament when expressed in cultured cardiomyocytes. The information that is necessary for the protein to find its precise location in the M-band is located in the adjacent immunoglobulin-like domain. Thus, although the affinity for myosin is a characteristic property of myomesin, it is not sufficient to direct the protein to its correct assembly place in the sarcomere.
MATERIALS AND METHODS
Chicken and Rat Myomesin Expression Constructs
Several cDNAs obtained by screening a chicken heart expression library (Bantle et al., 1996) were combined using standard procedures (Sambrook et al., 1989) to yield a cDNA covering the complete ORF of chicken myomesin. An epitope tag encoding 11 amino acids of the G-protein of the vesicular stomatitis virus (VSV) epitope (Gallione and Rose, 1985; Soldati and Perriard, 1991) followed by a stop codon was joined to the 3′ end of the coding region using PCR. The fragment was subcloned into the eukaryotic expression vector pSCT (Soldati and Perriard, 1991), resulting in the complete coding sequence of chicken heart myomesin with the VSV epitope fused to the C terminus (see Figure 1B, cMy1–14). Chicken constructs were generated by PCR using cMy1–14 as a template. In constructs containing the head domain, the original start codon was used; for internal constructs, a start codon was inserted according to the Kozak consensus (Kozak, 1989). Antisense primers incorporated the sequence for the VSV epitope followed by a stop codon. The chicken myomesin constructs are shown in Figure 1B. Rat myomesin constructs were generated by PCR, using a cDNA clone that contains the 5′ part of the rat myomesin sequence (Bantle et al., 1996) as template. Sense and antisense primers were as described for chicken constructs, except that the sequence encoding a 7mer peptide from the mediumT (mT) antigen of polyoma virus (mT epitope) (Grussenmeyer et al., 1985) was used instead of the VSV epitope. The rat myomesin fragments are shown in Figure 1C. Because the stability of immunoglobulin-like domains depends primarily on the size of the N-terminal extension (Politou et al., 1994), linker sequences upstream and downstream of the domains were included to decrease the chances of incorrect folding. PCR fragments were subcloned into pSCT either by using the pDirect system (Clontech, Palo Alto, CA; chicken constructs) or by incorporating BamHI and SalI sites into sense and antisense primers (rat constructs). The green fluorescent protein (GFP)-cMy2 fusion was generated by inserting the second domain of chicken myomesin into the EcoRI and SalI sites of pEGFP-N1 and pEGFP-C1 vectors (Clontech), resulting in a continuous reading frame with the GFP fused either N- or C-terminally to the second domain.
Neonatal Rat Cardiomyocyte Cultures
Primary cultures of neonatal rat cardiomyocytes (NRCs) were prepared as described previously (Sen et al., 1988; Komiyama et al., 1996). Cells were seeded at a density of 0.4 × 106 cells per 35-mm dish in plating medium. The plating medium consisted of 68% DMEM (Amimed AG, Basel, Switzerland), 17% Medium M199 (Amimed), 10% horse serum (Life Technologies, Grand Island, NY), 5% FCS (Life Technologies), 4 mM glutamine (Amimed), and 1% penicillin–streptomycin (Amimed). Before transfection, cells were grown for 24 h in 10% CO2. Plasmid DNA was prepared with the Nucleobond Plasmidprep kit (Macherey Nagel AG, Dueren, Germany). Transfections were performed using a modified CaPO4 transfection protocol (Komiyama et al., 1996). After transfection, cells were kept in maintenance medium for 72 h, followed by fixation and staining. Maintenance medium consisted of 78% DMEM (Amimed), 20% Medium M199 (Amimed), 1% horse serum (Life Technologies), 1% penicillin–streptomycin (Amimed), 4 mM glutamine (Amimed), and 10−4 M phenylephrine (Sigma Chemical Co., St. Louis, MO). For live observations, cells were plated on laminin-coated glass-bottom culture dishes (MatTek Corp., Ashland, MA) and treated as described above.
Immunofluorescence and Microscopy
Cells were washed once with PBS and fixed with 3% paraformaldehyde in PBS for 15 min. After they were washed with PBS, the cells were treated with 0.1 M glycine in PBS for 3 min and permeabilized with 0.2% Triton X-100 in PBS for 15 min. Blocking of unspecific interactions was performed with 5% normal goat serum (Sigma), 1% BSA (Fluka, Buchs, Switzerland) in PBS for 20 min. Antibodies were diluted in 1% BSA in PBS. Primary antibodies were polyclonal rabbit anti-VSV (Soldati and Perriard, 1991), monoclonal mouse anti-mT (Grussenmeyer et al., 1985) (a kind gift of Dr. G. Walter, University of California, San Diego, CA), polyclonal rabbit anti-heart MyBP-C (Bähler et al., 1985), monoclonal mouse antisarcomeric α-actinin EA53 (Sigma), and monoclonal mouse antimyomesin B4 (Grove et al., 1984); secondary antibodies were FITC-coupled goat anti-rabbit IgG (Cappel, West Chester, PA) and Cy3-coupled goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). Incubations with primary and secondary antibodies were for 1 h each, followed by several washes in PBS. Cells were embedded in Tris-buffered glycerol containing 50 mg/ml N-propyl gallate (Sigma) as antifading reagent, as described previously (Messerli et al., 1993a). Analysis of the stained cells was performed using a Leica confocal unit equipped with a DM/IRB E inverse microscope, an argon/krypton mixed gas laser, and a Leica PL APO 63×/1.4 NA objective lens (Leica, Heidelberg, Germany). Image processing was performed on a Silicon Graphics Workstation using the program Imaris (Messerli et al., 1993b) (distributed by Bitplane AG, Zürich, Switzerland). Video imaging of living cells was performed 48 h after transfection. Videos were recorded using a Zeiss (Oberkochen, Germany) Axiovert microscope and a Plan-Neofluar 100×/1.3 NA objective lens connected to a Kappa CF8/1 FMMC CCD camera and a standard video recorder (Gloor Instruments, Uster, Switzerland). Video images were digitized using Adobe Premiere Software (Adobe Systems, San Jose, CA) and a Miro Motion DC30 digitizing board.
Solid-Phase Binding Assay
[35S]-labeled protein was generated from cDNA constructs using a T7-coupled transcription/translation system (Promega, Madison, WI) following the manufacturer’s instructions. For some constructs, RNA was first synthesized in a standard transcription reaction as described previously (Schäfer and Perriard, 1988) and then added directly to the transcription/translation system. [35S]-labeled protein was analyzed on 8–22% SDS-PAGE gradient gels (Matsudaira and Burgess, 1978). The amount of labeled protein was normalized by comparing the signal intensity on autoradiographies of serial dilutions, and the labeled proteins were diluted so that all proteins gave signals of equal strength after overnight exposure to autoradiography film. Therefore, the actual amount of cMy1–3 or rMy1–3 protein is smaller than for single-domain constructs, because the three-domain constructs incorporate more [35S]methionine.
Rabbit muscle light meromyosin (LMM) (1 μg and 100 ng; Sigma) was spotted onto nitrocellulose strips prewetted with PBS and left for 10 min to dry. Unspecific binding was blocked for 1 h with 3% BSA, 0.2% Tween 20 (Fluka) in overlay buffer (100 mM KCl, 1 mM DTT, 20 mM imidazole/HCl, pH 7.0), and the nitrocellulose strips were incubated for 1 h with the labeled myomesin fragments in binding buffer (overlay buffer supplemented with 1% BSA, 0.2% Tween 20). After three washes in binding buffer, nitrocellulose strips were dried for 10 min and exposed to autoradiography film for 12 h. Firefly luciferase cDNA was expressed in the coupled transcription/translation system as described above and was used in the solid-phase binding experiment to assay unspecific binding to myosin.
RESULTS
Generation of Chicken and Rat Myomesin Deletion Mutants
In a first step, we assembled a clone covering the complete coding sequence of the heart isoform of chicken myomesin as described in MATERIALS AND METHODS (Figure 1A). A VSV epitope was fused to the C terminus of the coding region to distinguish the exogenously expressed myomesin construct from endogenous myomesin present in cardiomyocytes. The cDNA fragment was subcloned into the eukaryotic expression vector pSCT (Soldati and Perriard, 1991), which contains the cytomegalovirus (CMV) promoter that yields strong, constitutive expression in most eukaryotic cell types. Domain constructs of chicken and rat myomesin were generated as described in MATERIALS AND METHODS. A VSV epitope was fused C-terminally to all chicken constructs; an mT epitope was used in the case of rat constructs. All domain constructs were cloned into pSCT. Chicken and rat constructs are shown in Figure 1, B and C, respectively.
Recombinant Full-Length Chicken Myomesin
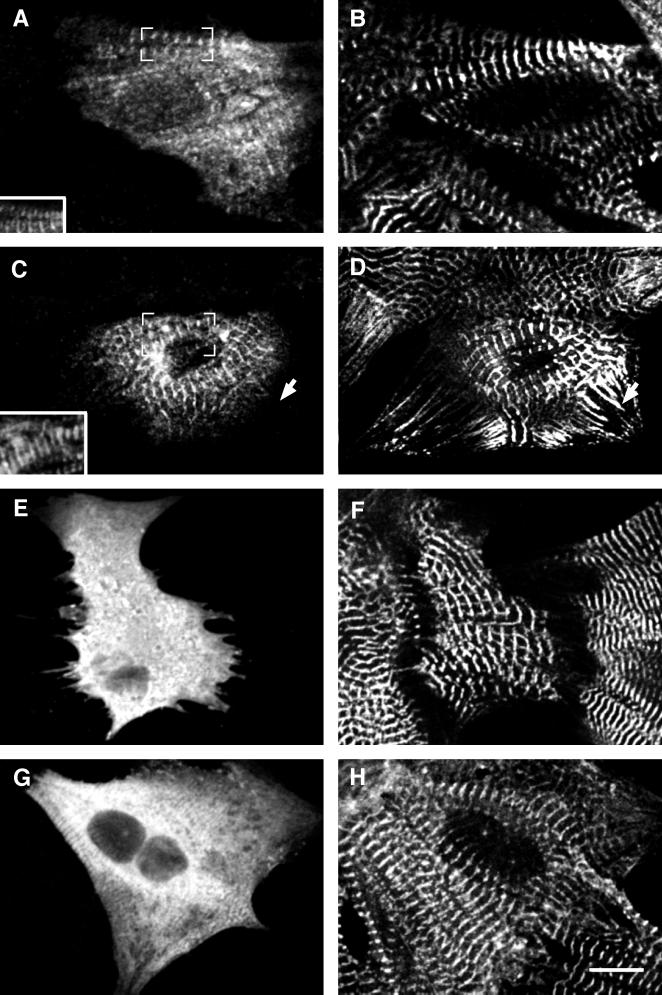
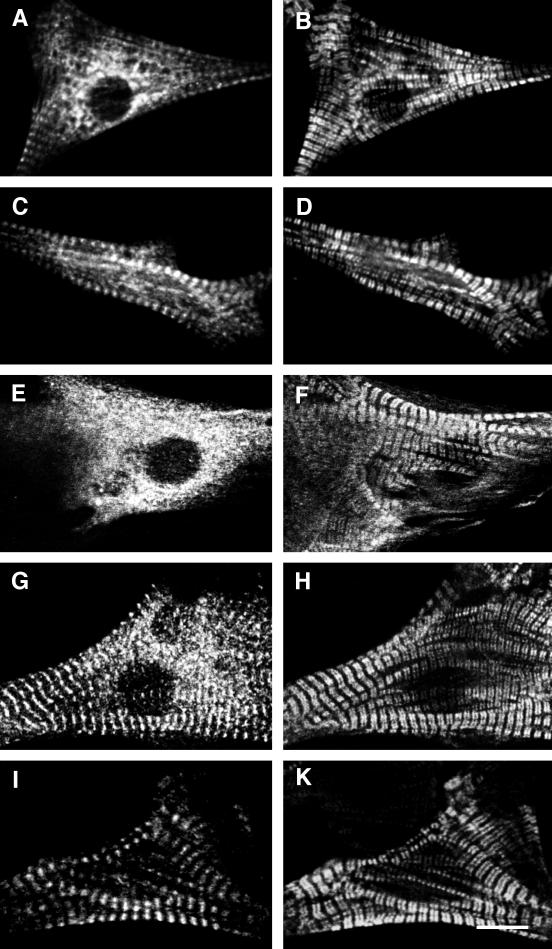
The myomesin constructs were expressed in primary cultures of neonatal rat cardiomyocytes. These cells are well suited for studying the incorporation of exogenously expressed proteins into myofibrils because they spread well in culture and assemble large arrays of myofibrils. Cardiomyocytes transfected with cMy1–14 incorporated the exogenously expressed protein into their myofibrils in narrow stripes ∼2 μm apart (Figure 2A). We used an antibody against sarcomeric α-actinin, which is located in the Z-disk of the sarcomere, to check the localization of the exogenously expressed proteins. Figure 2B shows the pattern of α-actinin, which is visible as a series of narrow stripes representing the Z-discs of the myofibrils. By comparing the signals for the VSV epitope and α-actinin in the superimposition (Figure 2A, inset), it is evident that the VSV epitope lies in the center between two neighboring Z-discs, where the M-band is located. Thus, exogenously expressed full-length chicken myomesin was incorporated at its proper location in the M-band of the sarcomere.
Figure 2.
Full-length chicken myomesin and truncation mutants expressed in NRC. Confocal images of neonatal rat cardiomyocytes expressing full-length chicken heart myomesin cMy1–14 (A and B) and truncation mutants cMy1–3 (C and D), cMy4–8 (E and F), and cMy9–13 (G and H). Cells were stained with an antibody against the VSV epitope (A, C, E, G) and with either an antibody against sarcomeric α-actinin (B, D, H) or an antibody against myomesin (F). Only cMy1–14 and cMy1–3 are incorporated into the M-band region of the myofibrils, as shown by the superimposition of VSV epitope and α-actinin signals in insets A and C. The constructs cMy4–8 and cMy9–14 are diffusely distributed in the cytoplasm. In G, some unspecific interaction of the recombinant protein with the myofibrils can be seen, which probably is an effect of high expression levels. The arrow in D marks NSMFs that incorporate sarcomeric α-actinin but not myomesin. The antibody against myomesin used in F is directed against an epitope in domain 11 and therefore recognizes only endogenous myomesin but not the truncation cMy4–8. Bar, 10 μm.
Chicken Myomesin Truncation Mutants
To investigate which parts of myomesin direct the incorporation of the protein into the M-band, we generated three constructs: cMy1–3 encoded the N-terminal head domain and two immunoglobulin-like domains, cMy4–8 encoded the five fibronectin type III domains in the central part of myomesin, and cMy9–14 encoded the five C-terminal immunoglobulin-like domains followed by the heart-specific tail domain (Figure 1B). In Figure 2, cardiomyocytes transfected with the three constructs are shown. In cells expressing cMy1–3, the recombinant protein was incorporated into the myofibrils in a striated pattern (Figure 2C). Comparison with the α-actinin signal confirmed the M-band localization of cMy1–3 (Figure 2C, inset). In the α-actinin staining of the transfected cardiomyocyte, continuously labeled nonstriated myofibrils (NSMFs) can be seen running toward the border of the cell (Figure 2D, arrow). In contrast to sarcomeric α-actinin, myomesin is never found in NSMFs (Schultheiss et al., 1990), further supporting the specificity of interaction of cMy1–3 with the M-band. The truncations cMy4–8 and cMy9–13 did not incorporate into the myofibrils; instead they were distributed in a diffuse manner in the cytoplasm (Figure 2, E and G). Because the α-actinin and myomesin stainings showed that myofibrils were still present in transfected cells, it results that cMy4–8 and cMy9–14 have no major antimorphic effects on the myofibrils. Therefore, we ascribe the diffuse localization of the two constructs to a lack of interaction with the myofibrils.
Because only the truncation cMy1–3 was correctly targeted, a potential binding site directing myomesin to the M-band must be present within the three N-terminal domains. We therefore generated expression constructs encoding each N-terminal domain to determine whether a single domain was responsible for targeting. We found that cMy2, encoding the N-terminal immunoglobulin-like domain of chicken myomesin, was incorporated in a striated pattern (Figure 3C). Comparison with α-actinin (Figure 3D) showed that the single domain was correctly targeted to the M-band (Figure 3C, inset). In contrast, cMy1 and cMy3, encoding the head domain and the third domain, respectively, were not incorporated into the myofibrils of the cardiomyocytes but showed a diffuse localization throughout the cytoplasm (Figure 3, A and E). Again, the presence of myofibrils as shown with the α-actinin staining (Figure 3, B and F) excluded any antimorphic effects of the constructs; however, in many cells with high expression levels of cMy1, nuclear staining was observed (Figure 3A). Two other constructs that included the second domain, cMy1–2 and cMy2–3, were also targeted to the M-band (our unpublished results). Thus, the second domain is sufficient for M-band targeting of chicken myomesin, because the single domain and all fragments containing this domain were found to be targeted to the M-band in rat cardiomyocytes.
Figure 3.
N-terminal chicken myomesin truncation mutants expressed in NRC. Confocal images of neonatal rat cardiomyocytes expressing constructs cMy1 (A and B), cMy2 (C and D), and cMy3 (E and F). Cells were stained with an antibody against the VSV epitope (A, C, E) and with an antibody against sarcomeric α-actinin (B, D, F). Only the construct cMy2 is targeted to the M-band, whereas cMy1 and cMy3 are diffusely distributed in the cytoplasm of the cells. The inset in C shows a superimposition of the VSV epitope (C) and the α-actinin (D) signals. Bar, 10 μm.
To exclude any species-specific effects, the truncation cMy2 was also tested in embryonic chicken cardiomyocytes. The single immunoglobulin-like domain was targeted to the M-band, and constructs containing the second domain were also targeted to the M-band in these cells (our unpublished results). The identical targeting behavior of the second domain in rat and chicken cardiomyocytes is further proof of its importance for the incorporation of myomesin into the M-band.
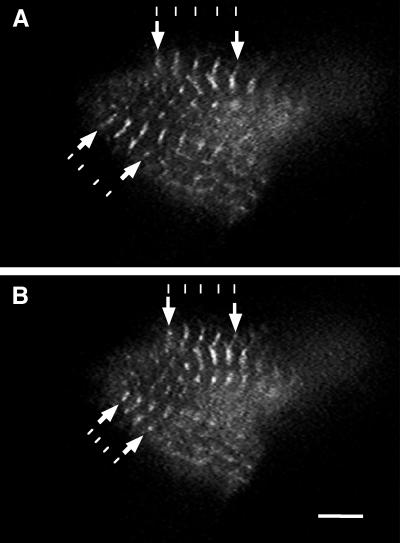
In an attempt to investigate antimorphic effects of cMy2 expression such as inhibition of contractility, we fused cMy2 to GFP (Chalfie et al., 1994). The fusion protein (cMy2 fused N-terminally to GFP [cMy2–GFP]) was expressed in NRC and was targeted to the M-band (Figure 4). Transfected cardiomyocytes continued to beat while expressing cMy2–GFP, which enabled us to observe contraction of the sarcomeres in the living cells. The decrease in distance between neighboring M-bands during contraction of the sarcomeres was readily visible (Figure 4 and accompanying video recording). In relaxed sarcomeres (Figure 4A), the distance between M-bands was ∼1.3 times longer than in contracted ones (Figure 4B). Cardiomyocytes expressing the fusion protein did not show any difference in beating activity as compared with neighboring untransfected cardiomyocytes, and staining of fixed cells with antibodies against various sarcomeric proteins revealed no defects in myofibril structure (our unpublished results). The fusion of cMy2 to the C terminus of GFP (GFP–cMy2) showed no differences in incorporation pattern as compared with cMy2–GFP. Thus, the GFP has no negative effect on the targeting of the second domain. We conclude that expression of cMy2 has no visibly antimorphic effects on the myofibrils and does not inhibit the contractile activity of cardiomyocytes.
Figure 4.
Living cardiomyocyte expressing cMy2–GFP. Single images taken from a video recording of a cardiomyocyte expressing cMy2–GFP showing relaxed (A) and contracted (B) states. cMy2–GFP localizes to the M-bands of the sarcomere and does not inhibit the contractile activity of the cardiomyocyte. The contraction of the sarcomeres can be seen by comparing the decrease in distance between neighboring M-bands in the contracted and uncontracted state (arrows). The lines are drawn to visualize the shortening of the distance between neighboring M-bands. Bar, 5 μm.
Rat Myomesin Truncation Mutants
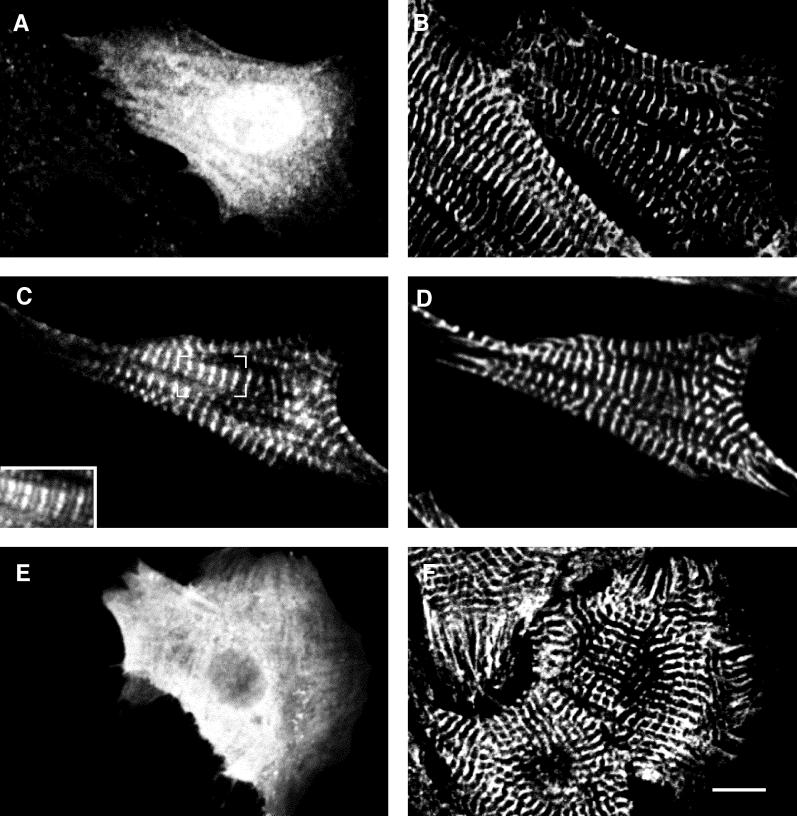
It has previously been shown that only N-terminal fragments of human myomesin that include the head domain interact with myosin in biochemical binding assays (Obermann et al., 1997). Assuming that the interaction between myomesin and the thick filament protein myosin would be sufficient to target myomesin to the M-band in cardiomyocytes, an M-band targeting of the head domain in transfected cardiomyocytes should have been observed. As shown in Figure 3A, cMy1 was not incorporated into myofibrils in rat cardiomyocytes, and no incorporation was observed in chicken cardiomyocytes either (our unpublished results). We speculated that species-specific differences in the head domains of chicken and human myomesin might be responsible for the observed discrepancy because their amino acid sequences are strongly divergent (Bantle et al., 1996). Therefore, we used part of the rat myomesin cDNA, which has a high identity with human myomesin on the amino acid level (Bantle et al., 1996), to generate constructs encoding different combinations of the three N-terminal domains (Figure 1C). The boundaries and length of the constructs were identical to the corresponding constructs of chicken myomesin. Expression of the constructs in neonatal rat cardiomyocytes showed that there is indeed a difference in the incorporation behavior between N-terminal chicken and rat myomesin domains. The truncation mutant rMy1, encoding the head domain of rat myomesin, and the truncation rMy2, encoding the second domain, were both incorporated into the myofibrils in a striated pattern (Figure 5, A and C). Comparison with an antibody against MyBP-C, a protein localized in the region of the sarcomeric A-band containing myosin heads, showed that rMy1 localized to a broad area around the M-band. This limited targeting to an area within the A-band (compare the width of the signals in Figure 5, A and B) was accompanied by the presence of significant amounts of unincorporated protein in the cytoplasm of transfected cardiomyocytes. In cardiomyocytes expressing the construct rMy2, the recombinant protein was localized in the M-band, but transfected cells also had comparatively high levels of unincorporated protein (Figure 5C). The construct rMy3 did not incorporate into the M-band and was diffusely distributed in the cytoplasm (Figure 5E). Consequently, we expressed a construct encoding only the two N-terminal immunoglobulin-like domains of rat myomesin (Figure 1C, rMy2–3). Exogenously expressed protein was localized in sharp striations in the M-band of the sarcomeres, and almost no unincorporated protein was present (Figure 5G). Because the constructs cMy3 and rMy3 were never observed to target to the M-band (Figures 3E and 5E), we attributed the reduction of unincorporated material in the cytoplasm to the increased stability of the two-domain construct. Therefore, we tested whether a similar enhancement in the targeting of the head domain could be achieved by linking it directly to the third domain. Cardiomyocytes expressing the deletion mutant rMy1 + 3 had considerably lower levels of unincorporated protein in the cytoplasm, but the incorporated protein was still localized in broad stripes along a subset of the A-band (Figure 5I). It is evident that the presence of another domain has a beneficial effect on either the domain stability or the folding of both rMy1 and rMy2, because the “stabilized” constructs rMy2–3 and rMy1 + 3 showed decreased levels of unincorporated protein. Yet, targeting to an area within the A-band was observed for rMy1 as well as for rMy1 + 3. We therefore conclude that the single head domain of rat myomesin is not targeted to the M-band but localizes to a broad area within the A-band. It is important to note that rMy1 and rMy1 + 3 did not localize to the entire A-band when compared with a staining against the A-band protein MyBP-C. In contrast, the second domain of rat myomesin shows proper M-band sorting and thus confirms the results found with the second domain of chicken myomesin.
Figure 5.
N-terminal rat myomesin deletion mutants expressed in NRC. Confocal images of neonatal rat cardiomyocytes expressing constructs rMy1 (A and B), rMy2 (C and D), rMy3 (E and F), rMy2–3 (G and H), and the deletion mutant rMy1 + 3 (I and K). Cells were stained with an antibody against the mT epitope (A, C, E, G, I) and an antibody against MyBP-C (B, D, F, H, K). The constructs rMy1 and rMy2 are targeted to an area within the A-band and to the M-band, respectively, but significant levels of unincorporated protein are present in the cytoplasm. The amount of unincorporated protein is reduced in cells expressing the two-domain constructs rMy2–3 and rMy1 + 3. Bar, 10 μm.
Interaction of the N-terminal Domains with Myosin
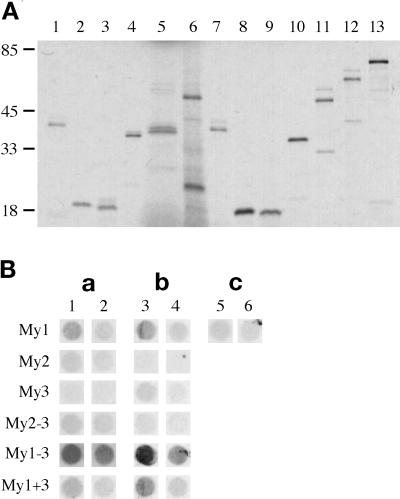
The association of rMy1 with thick filaments in cardiomyocytes raises the question of whether only the N-terminal head domain of myomesin interacts with myosin or whether the immunoglobulin-like domain My2 is involved in myosin binding as well. We therefore tested the N-terminal fragments of rat and chicken myomesin for interaction with the LMM portion of myosin using a solid-phase binding assay. N-terminal constructs were expressed in a coupled transcription/translation system in the presence of [35S]methionine. SDS-PAGE analysis of the radioactively labeled proteins resulted in bands of the expected size for all constructs (Figure 6A). With some of the larger proteins, lower bands, probably representing either products of alternative initiation events or degradation products, could be observed. The radioactively labeled proteins were incubated with two different concentrations of LMM fragment bound to nitrocellulose. The results of the solid-phase binding assay are shown in Figure 6B. The constructs rMy1–3 and cMy1–3 had a strong affinity for myosin, which confirms previous reports about a myosin-binding site in the N-terminal part of myomesin (Obermann et al., 1995, 1997). cMy1 and rMy1 bound to myosin as well, albeit very weakly; however, cMy2 and rMy2 did not bind to myosin, nor did cMy3 and rMy3. To exclude the possibility that the second domain does not bind because of improper folding, the constructs cMy2–3 and rMy2–3 were also tested for myosin interaction. Neither cMy2–3 nor rMy2–3 showed significant binding to myosin. The deletion mutant rMy1 + 3 showed stronger binding to myosin than the single head domain rMy1, but a similar enhancement in binding to myosin was not observed for cMy1 + 3. It has to be noted, however, that all chicken myomesin fragments tested in the assay showed weaker binding to myosin than their rat myomesin counterparts. The signal obtained with cMy1–3 and rMy1–3 was stronger than that obtained with cMy1 and rMy1, which is an indication that the three N-terminal domains bound more strongly to myosin than the head domains alone.
Figure 6.
Myosin binding of N-terminal truncation and deletion mutants. (A) SDS-PAGE (8–22%) of chicken and rat constructs expressed in the coupled transcription/translation system. Lanes 1–6, chicken mutants: lane 1, cMy1; lane 2, cMy2; lane 3, cMy3; lane 4, cMy2–3; lane 5, cMy1 + 3; lane 6, cMy1–3. Lanes 7–12, rat mutants: lane 7, rMy1; lane 8, rMy2; lane 9, rMy3; lane 10, rMy2–3; lane 11, rMy1 + 3; lane 12, rMy1–3. Lane 13, luciferase. Molecular weight standards are in kilodaltons. All constructs express proteins of the expected size, although in the case of longer constructs, additional lower bands representing either alternative initiation events or proteolytic degradation products can be observed. (B) Solid-phase binding assay of chicken and rat constructs. The rows are labeled after the domain nomenclature used in Figure 1. a, Chicken constructs; b, rat constructs; c, luciferase control. Different amounts of LMM fragment were used: 1 μg LMM fragment (columns 1, 3, 5) and 100 ng LMM fragment (columns 2, 4, 6).
The results of the solid-phase binding assay strongly support our findings on the localization of the rat myomesin head domain in cardiomyocytes. The single domain rMy1 localizes to an area within the A-band in vivo and shows myosin binding in vitro. In contrast, the second domain does not interact with myosin in our solid-phase binding assays but nevertheless is the only N-terminal domain that is specifically targeted to the M-band in both chicken and rat myomesin. We therefore conclude that the N-terminal head domain has a basic affinity for the thick filament protein myosin but that this affinity is not sufficient to target myomesin to its assembly site in the sarcomere. For a proper interaction of myomesin with the M-band, the second domain is absolutely necessary.
DISCUSSION
Myomesin belongs to the family of so-called myosin-binding proteins whose distinguishing property is their interaction with the thick filament protein myosin (Weber et al., 1993). Although myosin filaments stretch across the entire A-band in a staggered manner, the myosin-binding proteins have restricted localization patterns. Analysis by electron microscopy has shown that myomesin is located exclusively in the central area of the M-band (Obermann et al., 1996), whereas MyBP-C, for example, is localized in several consecutive stripes in the A-band (Craig and Offer, 1976). Therefore, it is likely that apart from the basic myosin-binding activity, interactions with other sarcomeric components may be needed for the precise targeting of these proteins.
Our results show that in cultured cardiomyocytes, two domains in the N-terminal part of rat myomesin are targeted to specific sites in the sarcomere. The unique head domain of rat myomesin localized to a central area within the A-band, and the adjacent immunoglobulin-like domain localized to the M-band. The other parts of myomesin did not interact with the sarcomere and were diffusely distributed in the cytoplasm. So far, no binding partner of the C-terminal immunoglobulin-like domains of myomesin has been identified, but the fibronectin type III domains 4–6 were shown to bind titin in a solid-phase overlay assay (Obermann et al., 1997). Therefore, it is surprising that the construct cMy4–8 did not localize to the M-band in cardiomyocytes. A suggested function of titin is that of a sarcomeric ruler with which other proteins can interact to find their proper place in the sarcomere (Trinick, 1994). The interaction of myomesin with titin would be well suited to localize the protein to the M-band, but our epitope-tagging experiments show that this is not the case. It is possible that in the mature sarcomere not all binding sites are accessible for exogenously expressed fragments or that the interaction is regulated by additional factors such as phosphorylation. For example, phosphorylation of the exogenous fragment cMy4–8 may prevent its interaction with titin and therefore its incorporation into the M-band, because it has been shown that the in vitro interaction of myomesin with titin is abolished by phosphorylation of a linker connecting two of the fibronectin type III domains (Obermann et al., 1997).
In both species investigated, the second domain of myomesin was targeted to the M-band. Furthermore, it was absolutely essential for M-band targeting because only fragments containing this domain localized to the M-band; however, it was observed that the fragment rMy2 localized in slightly broader stripes than the fragment cMy2, whereas the fragment rMy2–3 clearly localized to the M-band. The somewhat enhanced targeting of the two-domain fragment may simply be due to a stabilization of the interacting domain rMy2 by its neighboring domain, especially in view of the fact that the corresponding chicken myomesin fragment cMy2 was clearly targeted to the M-band. On the other hand, the enhanced targeting may result from a cooperative interaction of both immunoglobulin-like domains. The need for cooperative interactions to ensure targeting to sarcomeric sites has been observed before. In the case of MyBP-C, the C-terminal immunoglobulin-like domain bound to myosin in biochemical assays (Okagaki et al., 1993), but only a larger fragment encoding the three C-terminal domains was correctly incorporated into the A-bands of primary chicken myotubes (Gilbert et al., 1996). Interestingly, this three-domain fragment also bound to titin (Freiburg and Gautel, 1996), which suggests that MyBP-C needs both interactions for its incorporation into the A-band. In M-protein, the cooperative action of two immunoglobulin-like domains is needed for binding to myosin (Obermann et al., 1998).
In addition, the observation that in the solid-phase overlay assay the complete N-terminal fragment had a stronger affinity for myosin than the unique head domain alone suggests that the second domain may bind cooperatively with the head domain to myosin. Thus, it may be needed to restrict myomesin to the M-band. The binding site of myomesin was mapped to the C-terminal portion of the myosin rod (Obermann et al., 1997). The rod portions of myosin filaments from two adjacent A-bands overlap in an antiparallel manner in the M-band region (Huxley, 1963). No myosin heads are present in this region of overlap, which is also called the “bare zone.” The unique configuration of antiparallel, overlapping myosin rods may lead to binding sites that are present only in the bare zone and not in the cross-bridge zone of the thick filament. The binding of the unique head domain to this region could explain the wider localization pattern of the constructs rMy1 and rMy1 + 3. Studies using electron microscopy will be needed to prove this hypothesis.
The unique head domain of chicken myomesin did not localize to the bare zone but instead was diffusely distributed in the cytoplasm. Although this may be a result of misfolding or degradation, the observation that cMy1 retained its ability to bind myosin when expressed in vitro suggests that the domain is capable of folding. Therefore, the difference in targeting behavior of the unique head domains in cultured cardiomyocytes may be due to a species-specific difference that cannot be explained by the biochemical binding assays. A comparison of the head domain sequences of different species reveals strong differences, such as a repeat motif composed of the residues KQSTAS that is present in varying copy numbers in the head domains of human, mouse, and rat myomesin but is absent from the head domain of chicken myomesin (Bantle et al., 1996). The repeat motif may be involved in myosin binding, and its absence in chicken myomesin may be responsible for the lack of targeting observed with cMy1. Possibly, such species-specific differences in sarcomeric proteins may lead to different ultrastructures of the M-band (Carlsson and Thornell, 1987; Pask et al., 1994).
None of the constructs containing the second domain were incorporated into the NSMFs present in cardiomyocytes. NSMFs are filamentous extensions of myofibrils that contain some nonsarcomeric and sarcomeric proteins, such as titin and sarcomeric α-actinin, but no myomesin or MyBP-C (Schultheiss et al., 1990). Therefore, NSMFs probably do not yet have all the binding sites that are present in mature sarcomeres. The fact that the second domain did not localize to NSMFs may indicate either that they do not contain the binding partner or alternatively that the binding partner is not yet in the configuration required to bind myomesin. For example, in mature myofibrils, titin filaments overlap in an antiparallel manner in the M-band (Obermann et al., 1996). This overlap may lead to unique binding sites that are needed for the interaction with M-band proteins and may not be present in precursor structures such as the NSMFs. Several domains of human myomesin have been mapped for binding to titin using solid-phase overlay assays, and no affinity for titin was found in the case of the second domain (Obermann et al., 1997); however, because the second domain does not bind to titin present in NSMFs either, a complex of overlapping titin molecules and/or other M-band proteins may be needed for an interaction with myomesin. This hypothesis is supported by studies performed in differentiating human skeletal muscle cells in vitro where evidence for a rearrangement of the C-terminal portion of titin on integration into the M-band of mature sarcomeres has been found (van der Ven and Fürst, 1997).
Clearly, the interactions between myomesin and other sarcomeric proteins that take place during the assembly of myofibrils cannot be explained by simply studying their binding behavior in biochemical assays; therefore, assays that investigate the interactions in the cellular environment are indispensable. These studies may lead to the identification of a yet unknown protein that is necessary for restriction of myomesin to the M-band. The question of whether myomesin is essential as a linker between the filament systems and whether it has additional properties can be answered only by combining both approaches. It is hoped that further characterization of the second domain and its binding partners will lead to a better understanding of the role of myomesin in the assembly and maintenance of myofibrils.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ms. E. Perriard for outstanding technical assistance, as well as Drs. H.M. Eppenberger, D. Helfman, J.P. Magyar, and V. Taylor for helpful discussions, and Dr. G. Walter for donation of antibodies. This work was supported by a grant from the Swiss National Science Foundation (grants 31.37537/93 and 31.52417/97) and by a predoctoral training grant from the Federal Institute of Technology.
Abbreviations used:
- GFP
green fluorescent protein
- LMM
light meromyosin
- mT
mediumT antigen
- MyBP-C
myosin-binding protein C
- NRC
neonatal rat cardiomyocytes
- NSMFs
non-striated myofibrils
- VSV
vesicular stomatitis virus
Footnotes
Online version of this article contains video material for Figure 4. Online version available at www.molbiolcell.org.
REFERENCES
- Bähler M, Eppenberger HM, Wallimann T. Novel thick filament protein of chicken pectoralis muscle: the 86 kd protein. II. Distribution and localization. J Mol Biol. 1985;186:393–401. doi: 10.1016/0022-2836(85)90113-5. [DOI] [PubMed] [Google Scholar]
- Bantle S, Keller S, Haussmann I, Auerbach D, Perriard E, Mühlebach S, Perriard J-C. Tissue-specific isoforms of chicken myomesin are generated by alternative splicing. J Biol Chem. 1996;271:19042–19052. doi: 10.1074/jbc.271.32.19042. [DOI] [PubMed] [Google Scholar]
- Carlsson E, Thornell LE. Diversification of the myofibrillar M band in rat skeletal muscle during postnatal development. Cell Tissue Res. 1987;248:169–180. doi: 10.1007/BF01239978. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc R Soc Lond B Biol Sci. 1976;192:325–332. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- Einheber S, Fischman DA. Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proc Natl Acad Sci USA. 1990;87:2157–2161. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C: implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- Fürst DO, Gautel M. The anatomy of molecular giant: how the sarcomere cytoskeleton is assembled from immunoglobulin superfamily molecules. J Mol Cell Cardiol. 1995;27:951–959. doi: 10.1016/0022-2828(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Weber K. Myogenesis in the mouse embryo: differential onset of expression of myogenic proteins and the involvement of titin in myofibril assembly. J Cell Biol. 1989;109:517–527. doi: 10.1083/jcb.109.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallione CJ, Rose JK. A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J Virol. 1985;54:374–382. doi: 10.1128/jvi.54.2.374-382.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M, Goulding D. A molecular map of titin/connectin elasticity reveals two different mechanisms acting in series. FEBS Lett. 1996;385:11–14. doi: 10.1016/0014-5793(96)00338-9. [DOI] [PubMed] [Google Scholar]
- Gilbert R, Kelly MG, Mikawa T, Fischman DA. The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J Cell Sci. 1996;109:101–111. doi: 10.1242/jcs.109.1.101. [DOI] [PubMed] [Google Scholar]
- Grove BK, Cerny L, Perriard J-C, Eppenberger HM. Myomesin and M-protein: expression of two M-band proteins in pectoral muscle and heart during development. J Cell Biol. 1985;101:1413–1421. doi: 10.1083/jcb.101.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove BK, Cerny L, Perriard J-C, Eppenberger HM, Thornell L-E. Fiber type-specific distribution of M-band proteins in chicken muscle. J Histochem Cytochem. 1989;37:447–454. doi: 10.1177/37.4.2926123. [DOI] [PubMed] [Google Scholar]
- Grove BK, Kurer V, Lehner C, Doetschman TC, Perriard J-C, Eppenberger HM. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grussenmeyer T, Scheidtmann KH, Hutchinson MA, Eckhart W, Walter G. Complexes of polyoma virus medium T antigen and cellular proteins. Proc Natl Acad Sci USA. 1985;82:7952–7954. doi: 10.1073/pnas.82.23.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowits R, Kempner ES, Bisher ME, Podolsky RJ. A physiological role for titin and nebulin in skeletal muscle. Nature. 1986;23:160–164. doi: 10.1038/323160a0. [DOI] [PubMed] [Google Scholar]
- Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- Komiyama M, Soldati T, von Arx P, Perriard J-C. The intracompartmental sorting of myosin alkali light chain isoproteins reflects the sequence of developmental expression as determined by double epitope-tagging competition. J Cell Sci. 1996;109:2089–2099. doi: 10.1242/jcs.109.8.2089. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Mani RS, Kay CM. Interaction studies of the 165,000 dalton protein component of the M-line with the S2 subfragment of myosin. Biochim Biophys Acta. 1978;536:134–141. doi: 10.1016/0005-2795(78)90059-4. [DOI] [PubMed] [Google Scholar]
- Masaki T, Takaiti O. M-Protein. J Biochem. 1974;75:367–380. doi: 10.1093/oxfordjournals.jbchem.a130403. [DOI] [PubMed] [Google Scholar]
- Matsudaira P, Burgess DR. SDS microslab linear gradient PAGE. Anal Biochem. 1978;87:386–396. doi: 10.1016/0003-2697(78)90688-7. [DOI] [PubMed] [Google Scholar]
- Messerli JM, Eppenberger-Eberhardt ME, Rutishauser BM, Schwarb P, von Arx P, Koch-Schneidemann S, Eppenberger HM, Perriard J-C. Remodelling of cardiomyocyte cytoarchitecture visualized by three-dimensional (3D) confocal microscopy. Histochemistry. 1993a;100:193–202. doi: 10.1007/BF00269092. [DOI] [PubMed] [Google Scholar]
- Messerli JM, van der Voort HTM, Rungger-Brändle E, Perriard J-C. Three-dimensional visualization of multi-channel volume data: the amSFP algorithm. Cytometry. 1993b;14:725–735. doi: 10.1002/cyto.990140705. [DOI] [PubMed] [Google Scholar]
- Nave R, Fürst DO, Weber K. Visualization of the polarity of isolated titin molecules: a single globular head on a long thin rod as the M band anchoring domain? J Cell Biol. 1989;109:2177–2187. doi: 10.1083/jcb.109.5.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi J, Yanagisawa M, Imamura M, Kasuiya Y, Sakurai T, Tanaka T, Masaki T. Complete primary structure and tissue specific expression of chicken pectoralis M-protein. J Biol Chem. 1992;267:20302–20310. [PubMed] [Google Scholar]
- Obermann WM, Gautel M, Weber K, Fürst DO. Molecular structure of the M band: mapping of titin- and myosin-binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16:211–220. doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Plessmann U, Weber K, Fürst DO. Purification and biochemical characterization of myomesin, a myosin- and titin-binding protein, from bovine skeletal muscle. Eur J Biochem. 1995;233:110–115. doi: 10.1111/j.1432-1033.1995.110_1.x. [DOI] [PubMed] [Google Scholar]
- Obermann WMJ, Gautel M, Steiner F, van der Veen PFM, Weber K, Fürst DO. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein and the 250-kDa carboxy-terminal region of titin by immunoelectron microscopy. J Cell Biol. 1996;134:1441–1453. doi: 10.1083/jcb.134.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WMJ, van der Ven PFM, Steiner F, Weber K, Fürst DO. Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band. Mol Biol Cell. 1998;9:829–840. doi: 10.1091/mbc.9.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extraction, purification and characterization. J Mol Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Weber FE, Fischman DA, Vaughan KT, Mikawa T, Reinach FC. The major myosin-binding domain of skeletal muscle MyPB-C (C-protein) resides in the COOH-terminal, immunoglobulin C2 motif. J Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask HT, Jones KL, Luther PK, Squire JM. M-band structure, M-bridge interactions and contraction speed in vertebrate cardiac muscles. J Muscle Res Cell Motil. 1994;15:633–645. doi: 10.1007/BF00121071. [DOI] [PubMed] [Google Scholar]
- Politou AS, Gautel M, Joseph C, Pastore A. Immunoglobulin-type domains of titin are stabilized by amino-terminal extension. FEBS Lett. 1994;352:27–31. doi: 10.1016/0014-5793(94)00911-2. [DOI] [PubMed] [Google Scholar]
- Price MG. Skelemins: cytoskeletal proteins located at the periphery of M-discs in mammalian striated muscle. J Cell Biol. 1987;104:1325–1336. doi: 10.1083/jcb.104.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schäfer BW, Perriard J-C. Intracellular targeting of isoproteins in muscle cytoarchitecture. J Cell Biol. 1988;106:1161–1170. doi: 10.1083/jcb.106.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss T, Lin ZX, Lu MH, Murray J, Fischman DA, Weber K, Masaki T, Imamura M, Holtzer H. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J Cell Biol. 1990;110:1159–1172. doi: 10.1083/jcb.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Dunnmon P, Henderson SA, Gerard RD, Chien KR. Terminally differentiated neonatal rat myocardial cells proliferate and maintain specific differentiated functions following expression of SV40 large T antigen. J Biol Chem. 1988;263:19132–19136. [PubMed] [Google Scholar]
- Small V, Fürst DO, Thornell L-E. The cytoskeletal lattice of muscle cells. Eur J Biochem. 1992;208:559–572. doi: 10.1111/j.1432-1033.1992.tb17220.x. [DOI] [PubMed] [Google Scholar]
- Soldati T, Perriard JC. Intracompartmental sorting of essential myosin light chains: molecular dissection and in vivo monitoring by epitope tagging. Cell. 1991;66:277–289. doi: 10.1016/0092-8674(91)90618-9. [DOI] [PubMed] [Google Scholar]
- Starr R, Almond R, Offer G. Location of C-protein, H-protein and X-protein in rabbit skeletal muscle fiber types. J Muscle Res Cell Motil. 1985;6:227–256. doi: 10.1007/BF00713063. [DOI] [PubMed] [Google Scholar]
- Stolz M, Wallimann T. Myofibrillar interaction of cytosolic creatine kinase (CK) isoenzymes: allocation of N-terminal binding epitope in MM-CK and BB-CK. J Cell Sci. 1998;111:1207–1216. doi: 10.1242/jcs.111.9.1207. [DOI] [PubMed] [Google Scholar]
- Trinick J. Elastic filaments and giant proteins in muscle. Curr Opin Cell Biol. 1991;3:112–119. doi: 10.1016/0955-0674(91)90173-v. [DOI] [PubMed] [Google Scholar]
- Trinick J. Titin and nebulin: protein rulers in muscle? Trends Biochem Sci. 1994;19:405–409. doi: 10.1016/0968-0004(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Turner DC, Wallimann T, Eppenberger HM. A protein that binds specifically to the M-line of skeletal muscle is identified as the muscle form of creatine kinase. Proc Natl Acad Sci USA. 1973;70:702–705. doi: 10.1073/pnas.70.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ven PF, Fürst DO. Assembly of titin, myomesin and M-protein into the sarcomeric M band in differentiating human skeletal muscle cells in vitro. Cell Struct Funct. 1997;22:163–171. doi: 10.1247/csf.22.163. [DOI] [PubMed] [Google Scholar]
- Vinkemeier U, Obermann W, Weber K, Fürst DO. The globular head domain of titin extends into the center of the sarcomeric M band. J Cell Sci. 1993;106:319–330. doi: 10.1242/jcs.106.1.319. [DOI] [PubMed] [Google Scholar]
- Wallimann T, Moser H, Eppenberger HM. Isoenzyme specific localization of M-line-bound creatine kinase in myogenic cells. J Muscle Res Cell Motil. 1983;4:429–441. doi: 10.1007/BF00711948. [DOI] [PubMed] [Google Scholar]
- Weber FE, Vaughan KT, Reinach FC, Fischman DA. Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. Eur J Biochem. 1993;216:661–669. doi: 10.1111/j.1432-1033.1993.tb18186.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.