Abstract
DNA damage and mutagenesis are suggested to contribute to aging through their ability to mediate cellular dysfunction. The base excision repair (BER) pathway ameliorates a large number of DNA lesions that arise spontaneously. Many of these lesions are reported to increase with age. Oxidized guanine, repaired largely via base excision repair, is particularly well studied and shown to increase with age. Spontaneous mutant frequencies also increase with age which suggests that mutagenesis may contribute to aging. It is widely accepted that genetic instability contributes to age-related occurrences of cancer and potentially other age-related pathologies. BER activity decreases with age in multiple tissues. The specific BER protein that appears to limit activity varies among tissues. DNA polymerase-β is reduced in brain from aged mice and rats while AP endonuclease is reduced in spermatogenic cells obtained from old mice. The differences in proteins that appear to limit BER activity among tissues may represent true tissue-specific differences in activity or may be due to differences in techniques, environmental conditions or other unidentified differences among the experimental approaches. Much remains to be addressed concerning the potential role of BER in aging and age-related health span.
Keywords: base excision repair, aging, DNA damage, mutagenesis, health span
1. Introduction
Long before cellular responses to DNA damage were elucidated, Alexander proposed the DNA damage theory of aging (Alexander, 1967). According to this theory, an accumulation of DNA damage (i.e. an alteration in the normal three dimensional molecular structure of DNA) leads to cellular dysfunction and thereby aging. We now know that DNA damage results in a plethora of responses including inhibition of transcription, inhibition of replication, impairment of cell cycle progression, transcriptional mutagenesis, senescence, cell death and other processes (Andreassen et al., 2006; Campisi and d'Adda di Fagagna, 2007; Essers et al., 2006; Kao et al., 2005). Failla (Failla, 1958) and Szilard (Szilard, 1959) proposed what is known as the Somatic Mutation Theory of Aging. According to this theory a gradual accumulation of mutations (i.e. fixed and heritable changes in DNA sequence) in somatic cells occurs with increased age that in turn leads to cellular dysfunction and is manifest as aging. DNA damage and mutagenesis are distinct but intimately linked processes. Because of this, it has been difficult to determine whether DNA damage or mutagenesis or both play a major role in aging. Transgenic and knockout mice are now being used to more directly test the roles of DNA repair, DNA damage and mutagenesis in aging.
In contrast, genetic instability is now commonly accepted as an important mechanism in the etiology of cancer, an age-related pathology. In addition, genetic instability in gametes is a human health concern with advanced parental age for women and men. Advanced maternal age is associated with chromosomal aneuploidies in offspring (Caron et al., 1999), while several autosomal dominant disorders are associated with increased paternal age and de novo germline mutations (Crow, 2000). Thus, there are clear associations between genetic instability, age and decreased health span.
Because they function to ameliorate DNA damage and minimize mutagenesis, there are multiple DNA repair pathways that may be important in aging including, nucleotide excision repair (see paper by Laura Niedernhofer, this volume), double-strand break repair (see paper by Paul Hasty, this volume), and others. Nuclear and mitochondrial genetic integrity may both play important roles in aging, thus it is important to consider mechanisms that function to maintain genetic integrity in these organelles (see paper by LeDoux, this volume for mitochondrial DNA repair). The DNA base excision repair (BER) pathway is responsible for ameliorating many types of spontaneous DNA damage (i.e. DNA damage that occurs as a result of normal cellular metabolism without known or intentional exposure to agents that damage DNA). Spontaneous DNA damage and mutagenesis are the most likely sources of genetic instability in aging and age-related diseases. Therefore, BER has the potential to have a major effect in sustaining genetic integrity and preventing aging and age-related diseases involving genetic instability. Below we summarize the status of BER in aging and age-associated health span in mammals.
2. BER pathways
2.1. Overview of pathway
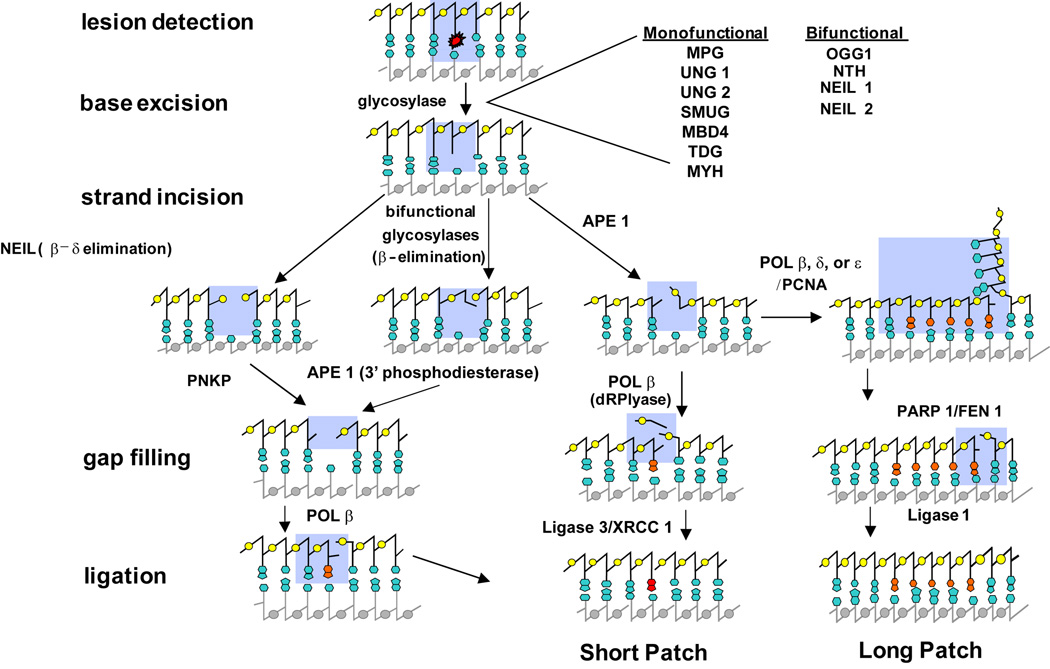
BER is a mechanism whereby cells repair one nucleotide lesions derived from base damage emanating from reactive oxygen species, irradiation, genotoxic chemicals, spontaneous deamination or from naturally occurring abasic (AP) sites (Almeida and Sobol, 2007; Dianov et al., 2001; Hitomi et al., 2007). Because they are formed as repair intermediates in the BER pathway, AP sites and single-strand breaks (SSBs) that require end processing, can be repaired through this pathway. Briefly, BER involves damage recognition by a DNA glycosylase followed by removal of the damaged base (Fig. 1). An AP site is left from the action of the DNA glycosylase, and apurinic/apyrimidinic endonuclease, APE1, incises the strand at the 5’hosphate, leaving a 5’deoxyribosephosphate (5’ dRP) group and 3’ hydroxyl (3’OH) group. If the DNA glycoslyase is bifunctional, it also has an intrinsic AP-lyase activity and can incise the deoxyribosephosphate backbone itself. The ends created by bifunctional DNA glycosylases must be processed to produce a 3’OH and 5’ phosphate before nucleotide insertion by an appropriate DNA polymerase. Repair synthesis replaces the excised DNA and ligation seals the phosphodiester backbone.
Fig 1. Diagram of base excision repair.
Base damage recognition is performed by a monofunctional or bifunctional DNA glycosylase. The damaged base is removed by hydrolyzing the N-glycosidic bond thereby leaving an abasic site. Strand incision occurs at the abasic site primarily through the action of APE1, or alternatively via the lyase activity of a bifunctional DNA glycosylase. The ends of the phosphodiester backbone are processed to render them suitable for DNA repair synthesis. POLβ fills the gap for short-patch repair while a processive polymerase fills the gap in long-patch repair. XRCC1 and DNA ligase 3 seal the phosphodiester backbone in short-ptach BER while DNA ligase 1 completes the repair process in long-patch BER.
2.2 DNA glycosylases and damage recognition
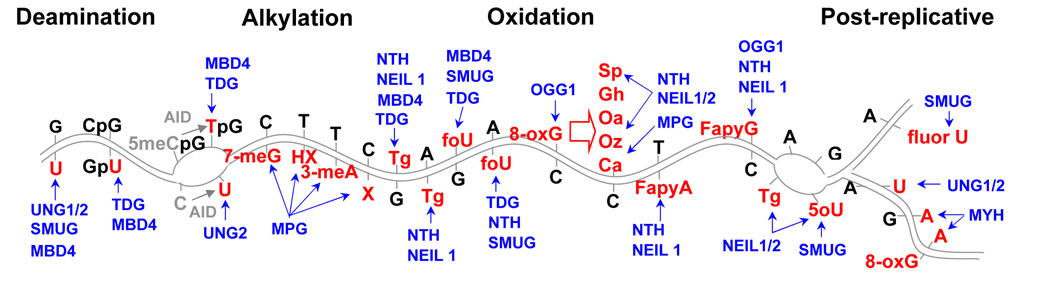
DNA glycosylases recognize and initiate repair for a multitude of lesions in DNA resulting from deamination, spontaneous or enzyme-invoked (Sousa et al., 2007), alkylation and oxidation. In general, aberrant bases distort the DNA helix and are recognized by specific DNA glycosylases (Hitomi et al., 2007). Compared to the damage recognition proteins for other excision repair pathways, DNA glycosylases display limited substrate specificity and tend to show a preference for purine or pyrimidine bases (Hazra et al., 2007). Monofunctional DNA glycosylases hydrolyze the N-glycosidic bond linking the base to the ribose moiety, thereby excising the base and leaving an AP site for further processing by the enzyme APE1. Bifunctional DNA glycosylases have an AP lyase activity in addition to their glycosylase activity and in general excise oxidized bases. A biologically important feature of DNA glycosylases is a redundancy in substrate specificity (Fig 2). Because of this redundancy single DNA glycosylase knockouts in mice typically have very mild consequences.
Fig 2. DNA glycosylases.
Relationship between damaged bases, DNA structure and recognition by specific DNA glycosylases. While each DNA glycosylase has a limited substrate repertoire, there is redundancy in damage recognition among the DNA glycosylases. Damaged bases, deaminated, alkylated or oxidized, are shown in double-stranded DNA, bubble DNA or newly replicated DNA in conjunction with the DNA glycosylases that recognize the damaged base in the particular DNA structure. DNA glycosylases are shown in blue with arrows to the damaged base(s) recognized. Modified bases shown are in red and include U=uracil, X=xanthine, HX=hypoxanthine, 3-meA=3-methyladenine, 7-meG=7 methylguanine, Tg=thymine glycol, foU=5-formyl uracil, 8-oxG=8-oxoguanine; Sp=spiroiminodihydantoin, Gh=guanidinohydantoin, Oa=oxaluric acid, Oz=oxazolone, Ca=cyanuric acid, FapyG=formamidopyrimidine derivative of guanine; FapyA=formamidopyrimidine derivative of adenine, 5oU=5 hydroxy uracil, fluorU= 5 fluorouracil, 5-meCpG= 5-methyl cytosine in the CpG context. The cytosine to uracil and 5-methyl cytosine to thymine reactions catalyzed by activation induced cytosine deaminase, AID, are shown in grey (Bransteitter et al., 2006).
Selected references for the glycosylase repair of mispairs include: G/U, (Akbari et al., 2004; Barrett et al., 1998), CpG/GpU, (Krokan et al., 2002); X/C, HX/T, 3meA/T, 7meG/C, (Chakravarti et al., 1991; Engelward et al., 1997); G/Tg, (Bandaru et al., 2002; Ikeda et al., 1998; Marenstein et al., 2003; Yoon et al., 2003); Tg/A, (Dianov et al., 2001; Hazra et al., 2002b; Marenstein et al., 2003; Rosenquist et al., 2003); foU/G, (Liu et al., 2003); foU/A, (Ikeda et al., 1998; Matsubara et al., 2003; Miyabe et al., 2002); foU/A, (Liu et al., 2003; Miyabe et al., 2002); 8-oxG, (Asagoshi et al., 2000; Laval et al., 1998); oxidized 8-oxG bases Sp, Gh, Oa, Oz, Ca, (Dherin et al., 2004; Hailer et al., 2005); FapyA/T, (Hazra et al., 2002a; Rosenquist et al., 2003); FapyG/C; (Asagoshi et al., 2000; Hazra et al., 2002a; Ide, 2001); TpG/GpC, (Krokan et al., 2002; Sousa et al., 2007); TG in bubbles, (Englander and Ma, 2006; Hazra and Mitra, 2006); 5oU in bubble DNA, (Englander and Ma, 2006; Hazra and Mitra, 2006; Matsubara et al., 2004); fluorU/A, (An et al., 2007); U/A post-replicative, (Nilsen et al., 2000; Parlanti et al., 2007); 8-oxG/A and G/A post replicative, (Ide and Kotera, 2004; McGoldrick et al., 1995).
Exocyclic amine groups of the nucleotide bases are subject to spontaneous, and, as recently reported for immunoglobulin class switching, enzyme-invoked deamination. The resulting purine bases, hypoxanthine and xanthine from guanine and adenine, respectively, are excised by the monofunctional methyl-purine DNA glycosylases, MPG (Miao et al., 1998). Deamination of pyrimidine bases in the DNA creates uracil from cytosine, and more alarmingly, the normal base thymine from 5-methyl-cytosine. At least 4 classes of uracil DNA glycosylases have evolved to counteract this highly mutagenic event. Human UNG1, targeted to the mitochondria, and the nuclear UNG2 act on G/U mispairs. Additionally UNG1/2 can act post-replicatively to remove misincorporated UMP. Thymine DNA glycosylase, TDG, (Cortazar et al., 2007) recognizes U in the context of CpG/GpU pairing, and excises mispaired thymine residues in the context of TpG/GpC resulting from 5-methylcytosine deamination. TDG removes thymine solely on the basis of mispairing and is highly regulated with multiple protein associations reported (Baba et al., 2005; Cortazar et al., 2007; Dianov et al., 2001; Mohan et al., 2007; Parikh et al., 1999; Pearl, 2000; Steinacher and Schar, 2005; Takahashi et al., 2005). Similar to TDG, the MBD4 glycosylase recognizes U or T mispairs in the CpG context (Hendrich et al., 1999). The single-strand selective monfunctional uracil DNA glycosylase (SMUG) excises U from G/U mispairs and is active against a subset of oxidized uracil residues: 5-hydroxyuracil (hoU), 5-hydroxymethyluracil (hmU) and 5-formyluracil (fU) (Matsubara et al., 2004). SMUG also removes 5-fluoro uracil residues (An et al., 2007; Baker et al., 2002). Recent reports suggest the highly efficient UNG2 is adapted for replicating DNA while SMUG might be more important in chromatin interactions (Pettersen et al., 2007).
Alkylating agents attack base nitrogens which lead to 7-methylguanine and 3- methyladenine as the most abundant lesions while 3-methylguanine and 7-methyladenine typically represent minor lesions (Chakravarti et al., 1991; Fortini et al., 2003a). The N-methylpurine DNA glycosylase, MPG, also known as alkylpurine DNA glycosylase, ANPG, is a monofunctional DNA glycosylase that is able to repair these lesions and 1, N6 ethenoadenine, hypoxanthine and xanthine (Chakravarti et al., 1991; Engelward et al., 1997) BER is the major route for removal of most alkylation damage. MPG associates with methylated DNA-binding domain 1, MBD1, a transcriptional repressor. MBD1-MPG binds methlyated promoters. Dissociation of the complex frees MPG to act as a DNA glycosylase (Watanabe et al., 2003). MPG associates with PCNA, APE1 (Xia et al., 2005), and XRCC1 (Campalans et al., 2005), and interacts with RAD23 / hHR23, that function in nucleotide excision repair and ubiquitination pathways (Miao et al., 1998; Miao et al., 2000).
Oxidized bases are generally removed by bifunctional DNA glycosylases. β–elimination is utilized by bifunctional DNA glycosylases OGG1 and NTH, and creates a 3’dRP and the phosphodiesterase domain of APE1 then cleaves the strand leaving a 1 nucleotide (nt) gap. Thymine glycol DNA-glycosylase (NTH) recognizes a wide range of oxidized pyrimidine residues including thymine glycol (Tg) and 5-formyluracil (Ikeda et al., 1998; Marenstein et al., 2003; Miyabe et al., 2002). However, Tg can also be excised by monofunctional MBD4 and TDG (Yoon et al., 2003). NEIL1, 2 and 3 are orthologs of the E.coli Nei DNA glycosylases (Rosenquist et al., 2003; Takao et al., 2002a). While no glycoslyase activity has been found for NEIL3, NEIL1 and 2 recognize oxidized substrates (Hailer et al., 2005; Zhang et al., 2005), and are bifunctional enzymes. A β-δ base elimination and strand incision require polynucleotide kinase/phosphate (PNKP), not APE1, for processing the 3’phosphate ends generated through their lyase activities. These enzymes are also unique in their ability to remove oxidized bases from bubble structures in the DNA, suggesting a critical role for NEIL proteins in base repair during replication and/or transcription (Rosenquist et al., 2003; Takao et al., 2002a). Not surprisingly, NEIL proteins have multiple protein associations. NEIL1 interacts with the checkpoint protein Rad9/Rad1/Hus1 heterotrimer (the 9-1-1 complex) (Guan et al., 2007). Werner syndrome protein, WRN, is a DNA helicase which associates with and stimulates NEIL1 excision activity in bubble DNA (Das et al., 2007a). Y-box binding protein (YB-1) interacts with NEIL2 and improves base excision activity 7-fold (Das et al., 2007b). YB-1 is normally cytoplasmic but translocates to the nucleus under oxidative stress, providing a partner for NEIL2 excision of oxidized substrates.
Human 8-oxoguanine DNA glycosylase, hOGG1, is a bifunctional DNA glycosylase targeting the potentially mutagenic lesion 8-oxoguanine, 8-oxoG, and the formamidopyrimidine derivative of guanine, FapyG. Adenine pairs almost equally well as cytosine with 8-oxoG during replication, creating a scenario for a mutator phenotype in the absence of OGG1 (Nakabeppu, 2001). The human analog of the DNA glycosylase MuY, MYH, recognizes adenine mispaired to 8-oxoG or G and will repair these lesions formed post-replicatively (Gu et al., 2002; Ide and Kotera, 2004; Lee et al., 2004; Russo et al., 2007). The 8-oxoG lesion can itself be oxidized to more complex lesions such as spiroiminodihdantoin and cyanuric acid. These lesions are processed by NTH, NEIL 1/2, and MPG (Dherin et al., 2004; Hailer et al., 2005).
2.3. Strand incision, repair synthesis and ligation
From the initial damage recognition by one of a multitude of DNA glycosylases with overlapping specificities, the next steps in BER require very limited, specific proteins. Strand incision proceeds either from an innate AP-lyase activity in the original bifunctional DNA glycosylase or from one of 2 enzymes, APE1 or PNKP. APE1 performs the majority of strand incision steps in mammalian cells (Almeida and Sobol, 2007; Wilson and Barsky, 2001). XRCC1 acts as a scaffolding protein bringing APE1 into the lesion site as XRCC1 interacts with both DNA glycosylases and APE1 (Vidal et al., 2001). Subsequently POLβ will perform repair synthesis to replace the excised damaged base, and cleave the deoxyribosephosphate moiety with its lyase activity (Dianova et al., 2004; Tomkinson et al., 2001). Lastly an XRCC1/DNA ligase III complex seals the DNA. When a NEIL enzyme is the appropriate damage recognition protein, a 3’ phosphate is produced that requires PNK/P that interacts in the complex with NEIL, POLβ, DNA ligase III and XRCC1 (Campalans et al., 2005; Das et al., 2006; Wiederhold et al., 2004)
2.4. Long-Patch BER
Lesions refractory to short-patch repair, such as a reduced AP site, a C1 oxidized AP site (Fortini and Dogliotti, 2007) or an adenine opposite 8-oxoG (Fortini et al., 2003b; Hashimoto et al., 2004), can undergo long-patch repair. A 5’ blocking lesion appears to result in DNA POLβ release after which PCNA facilitates excision by FEN1 which stimulates strand displacement DNA synthesis (Gary et al., 1999; Liu et al., 2005; Prasad et al., 2000) by POLδ, ε (Dianov et al., 2003). Finally, DNA ligase 1 catalyzes strand ligation. The stoichiometry of BER enzymes may regulate BER. Excess APE1 relative to POLβ stimulates strand displacement by POLβ and allows POLβ to mediate long patch DNA repair (Sukhanova et al., 2004). Under conditions of low energy, poly(ADP-ribose) polymerase 1(PARP1) catalyzes the poly(ADP)ribosylation of proteins (Beneke and Burkle, 2004; Muiras, 2003; Petermann et al., 2003) and stimulates long patch BER (Petermann et al., 2003). An apoptotic fragment of PARP1 can inhibit short-patch BER (Sukhanova et al., 2007). Whether short-patch or long-patch BER is chosen for hypoxanthine repair by MPG is dependent upon sequence context (Xia et al., 2005).
2.5. SSB repair
SSB repair ameliorates nicks in the DNA such as those caused by reactive oxygen species and ionizing radiation (Almeida and Sobol, 2007; Dianov and Parsons, 2007; Huber et al., 2004) or as repair intermediates created by the BER process itself. PARP-1 can bind to an AP-site, if present, at the SSB before any other BER proteins. DNA-PARP1 binding leads to recruitment of XRCC1 and then the typical BER short or long patch pathways prevail. Binding by PARP-1 stimulates automodification with PARP-1 poly(ADP-ribose) polymers and dissociation of PARP-1 from the DNA.
2.6. BER proteins in class switch recombination
In an overview of multifunctional BER and its biological importance, mention must be made of the role the BER machinery plays in class switch recombination in adaptive immunity. Uracil is a normal intermediate of somatic hypermutation and is generated by activation-induced cytosine deaminase (AID) on single stranded DNA. The uracil thus formed is then removed by UNG2 (Bransteitter et al., 2006; Sousa et al., 2007). AID and other members of the apoplipoprotein B editing catalytic polypeptide deamination family (APOBEC) can deaminate 5-methyl cytosine residues to thymine in the non-coding strand of single strand regions with implications in hypermutation, class switch recombination and loss of epigenetic information (Morgan et al., 2004). TDG and MBD4 are thought to initiate repair of the thymine mismatch (Hendrich et al., 1999; Wiebauer and Jiricny, 1989). Spontaneous 5-meCpG deamination can lead to a loss of epigenetic information and may require re-methlyation to complete repair (Walsh and Xu, 2006). A recent report of regarding association of DNA methyltransferase 3a, Dnmt3a, with TDG links BER, re-methylation and epigenetic regulation (Li et al., 2007c).
3. Subcellular localization of BER
BER is found where DNA is located, therefore BER is found in the nucleus and in mitochondria of animal cells (Bogenhagen et al., 2001; Croteau et al., 1999; Mandavilli et al., 2002; Weissman et al., 2007a; Wilson et al., 2003). Indeed, mitochondria are the major source of ROS and are thus in dire need of repair mechanisms such as BER. Differences do exist between nuclear and mitochondrial repair. UNG1 is the mitochondrial targeted variant of UNG. Similarly OGG1 has 4 isoforms produced from differential splicing and 3 isoforms are targeted to mitochondria (Nakabeppu, 2001; Nishioka et al., 1999; Shinmura et al., 2001; Takao et al., 1998). MYH and NTH are also found in mitochondria (Englander et al., 2002; Nakabeppu, 2001; Takao et al., 1998; Wang et al., 2000). APE1 is found in cytoplasm, mitochondria and nuclei (Tell, 2005; Tomkinson et al., 1988; Tsuchimoto et al., 2001) and can vary in location over the life of the animal. Repair synthesis is performed by DNA (POLγ) with ligation by mitochondrial targeted DNA ligase III (Bogenhagen et al., 2001; De and Campbell, 2007). Within mitochondria, the BER enzymes appear localized to the inner membrane, the site of mitochondrial DNA (Stuart et al., 2005).
4. Increased DNA damage and mutagenesis in aging
While a comprehensive review of DNA damage during aging is not the purpose of this review, it is important to ascertain if there is a change in DNA damage relative to age that is repaired through the BER pathway. There is abundant literature regarding DNA damage “accumulation” and age. We would argue that “accumulation” of damage is not the best term to use. Rather, we suggest the terminology of greater steady-state levels of DNA damage. A brief overview of the literature is presented and base damage is considered first (Table 1).
Table 1.
DNA damage and mutagenesis relative to age
| DNA Damage | Model | Ages | DNA Damage | Mutation | Selected References |
|---|---|---|---|---|---|
| 8-oxoguanine | Rat (F344) male | 6, 18, 24 mo* | ↑ in nucl DNA in brain, heart, kidney, liver & muscle; ↑ in mitoDNA in liver | N.D. | Hamilton et al., 2001b |
| Mouse (B62DF1) male | 6, 25 mo | ↑ in nucl DNA in brain, heart, kidney, liver & muscle; ↑ in mitoDNA in liver | N.D. | ||
| Mouse (C57BL/6) male | 6, 26 mo | ↑ in nucl DNA in brain, heart, kidney, & liver. | N.D. | ||
| Mouse (C57BL/6) female | 6, 14, 26 mo | ↑ in nucl DNA in brain, heart, kidney, liver & muscle; ↑ in mitoDNA in liver | N.D. | ||
| AP sites | Rats (Sprague Dawley) | 2–4 mo & 24 mo | ↑ in liver N.C. in brain | N.D. | Atamna et al., 2000 |
| IMR90 cells | PDL*-25–30 (young) PDL 46–54 (senescent) | ↑ in senescent cells | |||
| Human 1° leukocytes | 21–33 yrs* & 58–75 yrs | ↑ in cells from old donors. | |||
| Single Strand Breaks | Mouse (SAMP1) female | 1 d*, 1, 3, 6, 9, 11 mo | ↑ in brain, heart, muscle, liver, lung and intestine; ↑ rate of accumulation in SAMP1, senescence prone mice. | N.D. | Hosokawa et al., 2000 |
| Mouse (SAMR1) (female) | 1 d, 1, 3, 6, 9, 12, 18, 25 mo | N.D. | |||
| SAMP1 SAMR1 | 1, 6, 10 mo | ↑ SSB after NQO in SAMP1, less so in SAMR1 | N.D. | Zahn et al., 2000 | |
| Mutagenesis | Mouse (C57BL/6 LacI) | 1–24 mo | N.D. | ↑ mutation frequency | Lee et al., 1994 |
| Mouse (C57BL/6 LacZ) | 1d, 4–6, 10–24, 25–34 mo. | N.D. | ↑ mutation frequency in liver | Dolle et al., 1997 | |
| ↑ mutation frequency in brain between birth and 3–4 mo | |||||
| Mouse (C57BL/6 LacZ) | 3–4 & 30–33 mo | Genome rearrangements increased in liver at age >27 mo;. | ↑ mutation frequency in small intestine, spleen, liver and heart N.C. in brain | Dolle et al., 2002 | |
| Mouse (C57BL/6 LacI) | 1.5, 6, 12, 18, 25 mo | N.D. | ↑ mutation frequency in liver and bladder | Stuart et al., 2000 | |
| ↑ mutation frequency in brain at early ages | |||||
| Mouse (C57Bl/6 LacZ) | 1–33 mo | N.D. | N.C. in seminiferous tubules | Martin et al., 2001 | |
| Mouse (LacZ MutaMice) | 1 d, 2, 6, 12, 24 mo | N.D. | ↑ mutation frequency in spleen, liver, heart, brain (modest), testis (modest) and skin (modest). | Ono et al., 2000 | |
| Mouse (C6B3F1 LacI) | 2, 15, 28 mo | N.D. | ↑ mutant frequency in spermatogenic cells | Walter et al., 1998 | |
| Mouse (C57Bl/6 LacI) | 4–6, 22–26 mo | N.D. | ↑ mutant frequency in liver | Cabelof et al., 2002 | |
| ↑↑ mutant frequency after DMS | |||||
| Mouse (C57Bl/6N) | 2–22 mo | N.D. | ↑ mutation frequency in mtDNA D-loop in liver | Khaidakov et al., 2003 | |
d=day; mo=month; wk=week; yrs=years; PDL=population doubling level; N.D. = not determined; N.C. = no change; ↑= increase; ↓ = decrease.
Oxidative damage to DNA in the form of 8-oxoG, is the most commonly studied damage relative to age. These studies generally reveal increased levels of oxidative damage (i.e. 8oxodG). These data were questioned when it became commonly known that the method used to isolate the DNA itself caused oxidative damage. Methodology has now been identified for preparing the DNA without detectable induced oxidation (Hamilton et al., 2001a; Helbock et al., 1998). Using such procedures, greater steady-state levels of 8-oxoG were observed in DNA prepared from brain and liver of older rodents (Hamilton et al., 2001b). The vast majority of oxidized bases, including 8-oxoG, would be repaired via the BER pathway.
AP sites can be generated spontaneously via hydrolysis of the N-glycosidic bond, or as a repair intermediate through the action of a DNA glycosylase in the BER pathway. AP sites are repaired largely through the BER pathway. Using human diploid IMR90 cells, formation of AP sites occurs more slowly in young cells than in senescent cells (Atamna et al., 2000). AP sites are also repaired more slowly in senescent IMR90 cells after treatment with H2O2 or methylmethane sulfonate (MMS), a monofunctional alkylating agent. Human leukocytes obtained from old (58- to 75-year-old) individuals display more AP sites than cells from young (22- to 33-year-old) individuals. Analysis of AP sites in old and young rats, yields similar results; a greater prevalence of AP sites are observed in nuclear DNA obtained from old (24-month-old) compared to young (4-month-old) rats.
SSBs are another repair intermediate in the BER pathway and are formed via the action of AP endonuclease (APE1) or from the lyase activity of a bifunctional DNA glycosylase (Almeida and Sobol, 2007). SSBs can also be generated through the attack of reactive oxygen species. Many SSBs can be repaired with BER proteins. PARP1 and PNK are thought to be additionally involved in SSB repair (Dianov and Parsons, 2007; McKinnon and Caldecott, 2007; Wilson and Bohr, 2007). The prevalence of SSBs was examined in senescence-accelerated prone mice, SAMP1, and senescence-accelerated resistant, SAMR1 mice. The prevalence of DNA damage varied among the examined tissues, but was consistently great in SAMP1 tissues (Hosokawa et al., 2000; Zahn et al., 2000). Together, these examples demonstrate that DNA damage that is repaired through the BER pathway increases with age and leads to questions about a possible decline in BER activity with age.
Early mutagenesis studies gave results that supported in some cases and refuted in others, a greater mutant frequency at older ages. These studies were performed using selectable markers (e.g. Hprt) that necessitated maintaining cells in culture. Consequently, the studies were limited essentially to fibroblasts and immortalized cell lines. After the advent of transgenic mouse technology, transgenic lines carrying mutation reporter genes (e.g. lacI, lacZ) were established (Gossen et al., 1989; Kohler et al., 1990). The power of the transgenic models was that virtually any cell or tissue type could be studied for mutagenesis at any age. Analyses of spontaneous mutant frequencies relative to age consistently revealed an increase for liver, and no change in mutant frequency for brain (Dolle et al., 1997; Stuart and Glickman, 2000). The mutant frequency results for spermatogenic cells have varied with some showing no change (Martin et al., 2001) while others observed a significant increase in spermatogenic cells at old age (Walter et al., 1998). The reason for the different results in spermatogenic cells is not clear at present. Although there are tissue specific differences in mutagenesis, there are data generally indicating that mutagenesis increases with age in several tissues. The mutagenesis results combined with the DNA damage results lead to the question of whether BER activity changes with age.
5. BER activity changes with age
The first quantitative assays for the BER pathway, as initiated by uracil-DNA glycosylases (UDGs), revealed differences in activity among mouse tissues (Intano et al., 2001). Spermatogenic cells enriched from the testis displayed the greatest activity followed in order by mitotically active Sertoli cells, mitotically active thymocytes, small intestine, liver and brain. Using a similar assay, significant reductions, i.e. 50–75%, in BER activity were observed using tissues (brain, liver, spleen, testis or spermatogenic cells) collected from old (18- to 20-month-old or 28-months-old) mice compared to young (4- to 6-months-old or 3-months-old) mice (Cabelof et al., 2002; Intano et al., 2003). A reduction of POLβ with increased age was identified in all tested tissues in one study (Cabelof et al., 2002). The reduction of POLβ was observed by RNA abundance, protein abundance and enzyme activity assays. In contrast, POLβ was reduced only in brain in the second study (Intano et al., 2003). Individual BER proteins were added to nuclear extracts prepared from old animals in an effort to identify a protein activity that could restore BER activity to that of young mice. Addition of UNG, APE1, POLβ or DNA ligase 3 had essentially no effect with regard to stimulating BER activity for somatic tissues (Intano et al., 2003). Different genetic backgrounds were used by the groups and may have contributed to the different results. Experimental and/or environmental differences may have contributed to the lack of congruency in the results between the different laboratories.
In a subsequent study, calorie restriction was observed to completely reverse the age-related decline in BER activity and the decline in POLβ expression and activity (Cabelof et al., 2003). Calorie restriction was also found to enhance BER activity in extracts prepared from young mice. A second study examining the effects of calorie restriction on BER similarly found an increase in nuclear BER activity with calorie restriction, i.e. a 26% increase (not significantly different from ad libitum fed mice) in liver, and 42% increase (significantly different from ad libitum fed mice) in kidney (Stuart et al., 2004). The mechanism by which BER activity is modulated through calorie restriction has yet to be definitively determined.
The brain is a complex organ with different regions being involved in age-related neurodegenerative disorders. Some of these disorders are associated with increased oxidative damage, indicating the BER pathway may be important in protecting the brain from such disorders. Initial studies examining the activity of multiple DNA glycosylases in different mouse brain regions has recently been reported (Imam et al., 2006). The incision activities of OGG1, UNG and NTH1 were examined in cerebellum, brain stem, caudate nucleus, frontal cortex and hippocampus. Cerebellum and brain-stem displayed greater levels of OGG1 activity relative to the other studied regions, but also were the only regions showing a significant decline in activity with age. UNG incision activity declined in cerebellum with age. APE1 and DNA ligase III did not change with age in the tested brain regions.
These studies have provided the first insights into the levels of BER activity among mammalian tissues with age. It is clear that the activity varies among tissues and decreases with age. Because tissues consist of multiple cell types, it becomes important to assess BER in defined cell types. Along these lines, primary mouse skeletal muscle satellite cells were prepared, grown and differentiated into myotubes in culture to measure long-patch and short-patch BER activity. Myotubes displayed significantly less BER activity than proliferating cells (Narciso et al., 2007). This suggests that differentiated myotubes may be more likely to sustain greater levels of DNA damage. Indeed, when challenged with H2O2 myotubes displayed greater levels of oxidative base damage than proliferating cells. Sarcopenia is a common phenotype in the elderly and it will be interesting to determine if reduced BER activity contributes to the phenotype. Exercise training in old rats increased OGG and UNG activity but to different extents in red and white muscle fibers, again implying differential regulation of BER proteins to maintain DNA fidelity in oxidatively stressed cells (Radak et al., 2005; Radak et al., 2007).
Primary rat neuronal cultures have been examined for the gap-filling and ligation steps of the BER pathway using adolescent (5-day-old), young adult (6-month-old) and old (≥ 24-month-old) Wistar rats (Krishna et al., 2005). Repair activity declined as age increased. Recombinant POLβ restored activity to the samples from old rats, but repair was more rapid when POLβ and T4 DNA ligase were added. These results suggest that POLβ and a necessary mammalian ligase restrict BER activity in neurons from old rats.
Human peripheral blood lymphocytes collected from individuals ranging in age from newborn to 91-years-old were assayed for OGG1 incision activity. Variation among individuals at each age was observed, but overall OGG1 activity declined significantly with age (Chen et al., 2002a). “Old” IMR90 cells, a human diploid fibroblast line, displayed an approximate 5-fold reduced repair by MPG compared to nonsenescent cells (Atamna et al., 2000). In other studies, late passage IMR90 cells showed reduced OGG1 activity and OGG1 transcription (Shen et al., 2003). The studies with primary human cells and human cell lines indicate that BER activity declines with age in human cells. It will be important to determine the mechanism(s) that mediate the decline in repair activity and to examine additional cell types and tissues.
Subcellular compartmentalization of DNA repair proteins has been a mechanism that potentially contributes to changes in BER activity. Using mouse liver cells, levels of APE1 protein and mRNA were found to be similar for young adult (4-months-old), middle-aged (10-months-old) and old (20-months-old) BALB/c mice. When APE1 distribution in subcellular compartments was examined relative to age, it was found that APE1 activity increased in nuclear and mitochondrial compartments by middle age (Szczesny and Mitra, 2005). APE1 activity was elevated approximately 2-fold by old age in the nuclear compartment. It was suggested that the changes in APE1 activity in subcellular compartments relative to age may reflect levels of oxidative challenge in the involved organelles. The 6-fold increase in APE1 activity seen in mitochondria occurred by 10 months, while the nuclear activity increased more slowly to a final 2-fold increase at 20 months (Szczesny and Mitra, 2005). Once within the nucleus itself, APE1 can be induced to relocate to nuclear speckles by UVA irradiation, co-localizing with OGG1 to facilitate BER (Campalans et al., 2007). In a recent paper (Chaudhry, 2007) whole cell extracts from synchronized HeLa cells were used to examine repair activities of hAPE1, hOGG1 and hNTH1 in different phases of the cell cycle. hAPE1 and hNTH1 were higher in G1 compared to G2 while OGG1 showed no such dependence.
BER activity has been measured by a variety of techniques including assessment of the full pathway and individual BER protein activities (Table 2). With very few exceptions, the results are in agreement that BER activity declines with age. The explanation for decreased BER activity has varied with different BER proteins implicated in different studies and different tissues (Table 3). Such differences may be explained by different techniques used, different strains or species of animals or to responses to different environmental conditions that are not presently understood. However, the overriding consistent feature is that BER activity declines with age.
Table 2.
BER activity with age
| Activity Assayed* | Model | Age | Findings | Selected References |
|---|---|---|---|---|
| UDG-BER | Mouse (CD1) | 5–8 d, 4–6 mo* | Spermatogenic cells > Sertoli cells > thymocytes > liver > brain nuclear extracts | Intano et al., 2001 |
| Mouse (C57Bl/6) | 4–6 mo | Spermatogenic cells > liver > brain nuclear extracts | ||
| Mouse (B6D2F1) | 4–6 mo | Spermatogenic cells > liver > brain nuclear extracts | ||
| UDG-BER | Mouse (CD1) | 6, 8 d, 3 mo | Liver activity > brain nuclear extracts. Activity greatest in neonatal liver and neonatal brain nuclear extracts. | Intano et al., 2003 |
| Mouse (B6D2F1) | 6–8 d, 3, 16, 28 mo | Liver activity ↓ 50% by 16 mo. Brain activity ↓ 60% by 3 mo. | ||
| UDG-BER OGG-BER POLβ | Mouse(C57Bl/6) | 4–6, 22–26 mo | ↓ activity with age in brain, spleen liver and testes nuclear extracts | Cabelof et al., 2002 |
| Testes have highest activity. | ||||
| OGG1 UDG1 EndoG-activity | Rat (Wistar) | 6, 12, 23 mo | Mitochondrial OGG/AP lyase activity ↑ with age in liver and heart; mitochondrial UDG-activity has slight increase in heart. | Souza-Pinto et al., 1999 |
| OGG1 UDG | Mouse (C57Bl/6) | 6, 14, 23 mo | ↑ mitochondrial activity, ↓ nuclear activity with age in liver | de Souza-Pinto et al., 2001b |
| Gap repair | Rat (Wistar) | 5 d, 6 mo, >2 yr | ↓ activity with age in nueronal preparations | Krishna et al., 2005 |
| OGG-BER OGG1 UDG1 APE1 Polγ ligase | Rat (Sprague Dawley) | 17 d embryos, 1, 2, 3 wk 5, 30 mo | Overall BER and individual activities examined in brain mitochondria with sharp decline in OGG1, APE1, POLβ and ligation activities by 5 mo. | Chen et al., 2002a |
| APE1 | Mouse (BALB/c) | 4, 10, 20 mo | ↑ nuclear and mitochondrial activity with age in liver; cytoplasmic activity ↓. | Szczesny and Mitra, 2005 |
| N.C. whole cell activity | ||||
| OGG1 NTH APE1 | IMR90 | PDL *11, 39 | ↓ OGG-activity in nucleus with increased passage number. | Shen et al., 2003 |
| ↓ NTH and APE1 activity in mitochondrial and cytoplasmic fractions. | ||||
| N.C. in nuclear NTH or APE1 activities with passage number | ||||
| OGG1 | Human leukocytes | Newborn-91 yr | ↓ with age | Chen et al., 2003c |
| OGG1 UDG NTH | Mouse (C57Bl/6) | 6 & 18 mo | ↓ in OGG, UDG, NTH activity in mitochondria in all brain regions. Variable patterns seen in nuclei. Cerebelleum with greatest activities and greatest ↓. | Imam et al., 2006 |
| UDG-BER POLβ̃ | Rat (Fisher 344) | 6 wk, 60% calorie restricted; 4–6mo, 22–24 mo | ↓ activity with age in brain, liver and testes nuclear extracts reversed by caloric restriction. | Cabelof et al., 2003 |
BER-refers to complete base excision repair assay; UDG, OGG1, APE1 or NTH alone refers to strand incision activity assay; Pol- refers to gap filling activity; d=day; mo=month; wk=week; yr=years; PDL=population doubling level; ↑= increase; ↓= decreased; N.C.= no change
Table 3.
BER protein abundance with age
| Protein | Models | Ages | Findings | Selected Reference |
|---|---|---|---|---|
| POLβ | Mouse (C57BL/6) | 4–6, 22–26 mo | ↓ levels of POLβ protein in nuclear extracts from brain, kidney, liver, spleen and testes | Cabelof et al., 2002 |
| POLβ | Rats (Fisher 344) rats | at age 6 wk, 60% calorie restricted; 4–6, 22–24 mo | CR completely reverses age related decline in POLβ abundance in nuclear extracts from brain, liver and testes | Cabelof et al., 2003 |
| POLβ OGG1 | Rats (Sprague Dawley) | 17 d embryos, 1, 2, 3 wk 5, 30 mo | Age dependent decrease in protein levels in brain mitochondria. | Chen et al., 2002a |
| APE1 | Mouse (BALB/c) | 4, 10, 20 mo | N.C. with age in total liver extracts. | Szczesny and Mitra, 2005 |
| DNA Lig1 | Mouse (B6D2F1) | 3, 16, 28 mo | N.C. liver or brain or spermatogenic cell nuclear extracts | Intano et al., 2002; Intano et al., 2003 |
| DNA Lig3 | N.C. liver or brain or spermatogenic cell nuclear extracts | |||
| XRCC1 | N.C. liver or brain or spermatogenic cell nuclear extracts | |||
| POLβ | ↓ in brain; N.C. in liver or spermatogenic nuclear extracts | |||
| APE1 | ↓ in spermatogenic cells nuclear extracts; N.C. in liver or brain | |||
| APE1 | Mouse (BALB/c) | 4, 10, 20 mo | No change total abundance; ↑ nuclear and mitochondrial localization with age in liver | Szczesny and Mitra, 2005 |
| APE1 Ligase 3 | Mouse (C57Bl/6) | 6 & 18 mo | N.C. in levels with age in various brain regions | Imam et al., 2006 |
↑ = increase; ↓ = decrease; N.C. = no change.
7. Impact of altered BER on longevity
The field of aging research has been hampered by a paucity of biomarkers that can be used to establish physiological age, therefore longevity has long been used experimentally as a gold standard to functionally assess aging (Masoro, 2003). The natural (i.e. without known or intentional intervention) median and maximal lifespans can be characteristic for a species (e.g. human vs dog) and in some cases, between strains within a species (e.g. different mouse strains). In a classic comparison of lifespan and DNA repair activity Hart and Setlow (Hart and Setlow, 1974) demonstrated a positive correlation between these quantitative traits. The publication of these data reawakened the scientific community to the possible relationship between aging and genetic integrity as first postulated in the 1950’s. Data from multiple laboratories on multiple species was compared and normalized to the laboratory rat in a more recent publication (Cortopassi and Wang, 1996). Animals categorized with a short lifespan (i.e. 1.5–3.3 years; mouse, rat and shrew), medium lifespan (i.e. 3.5–20 years; hamster, rabbit and dog), long lifespan (i.e. 28–50 years; cat, horse, cow and elephant) or extra long lifespan (i.e. 50–120 years; gorilla and human) were positively correlated with DNA repair activity. Notably DNA repair activity measurements cannot be used to accurately predict maximal lifespan suggesting that while DNA repair is an important component to lifespan determination, it is probably not sufficient to determine lifespan.
Assuming that DNA repair is an important component to maximal lifespan, we can ask how alterations in BER activity affect lifespan. The first indication that BER might play an important role in lifespan was revealed in a comparison of longevity with poly(ADP-ribosyl)ation activity among several mammalian species using mononuclear blood cells (Grube and Burkle, 1992). Cells obtained from humans, the longest lived species in the comparison, displayed 5-fold greater activity than rat, the shortest lived species in the comparison. PARP1 protein abundances were similar among the tested species. The full explanation for differences in PARP activity among species is not understood. Two members of the PARP family, PARP1 and PARP2, participate in BER with complementary but not fully overlapping functions (Ame et al., 2004). Mouse models, deficient in PARP1 have been made (de Murcia et al., 1997; Wang et al., 1997). The homozygous null mice are viable, but have a smaller body size than normal littermates. The PARP1 null mice also show notable defects in BER. Surprisingly, no test of longevity for PARP1−/− mice is found in the literature. Therefore, it appears a direct test of PARP1 on longevity is lacking. A PARP2 null mouse was developed which was also sensitive to ionizing radiation and alkylating agents (Menissier de Murcia et al., 2003). The PARP1/PARP2 double mutant was an embryonic lethal (Menissier de Murcia et al., 2003).
UNG is a BER protein that recognizes uracil in DNA and catalyzes the hydrolysis of the N-glycosidic bond to leave an AP site where the uracil was previously located. UNG is the preferred glycosylase to remove uracil formed during somatic hypermutation. Mice deficient in UNG have been made and are viable when homozygous null (UNG−/−) (Nilsen et al., 2000). Longevity was assessed for UNG−/− mice in comparison to UNG+/+ mice. Median lifespan was significantly reduced to 101 weeks of age for UNG −/− mice compared to 116 weeks old for UNG+/+ mice (Nilsen et al., 2003). However, maximum lifespan was not altered in the UNG−/− mice. The old UNG−/− mice displayed more pathology, particularly lymphoproliferation and B-cell lymphoma. The increased prevalence of cancer may have impacted the median lifespan, but did not detectably alter maximal lifespan.
POLβ heterozygous (POLβ+/−) mice are the only other model of impaired BER for which longevity has been published. In contrast to the UNG−/− mice, neither median nor maximal lifespan was impacted (Cabelof et al., 2006). A very modest, but significant difference in mortality rates was detected between POLβ+/− and POLβ+/+ mice. Early and late life differences in mortality between the genotypes likely mediated the modest difference in mortality rates. It is noteworthy that the UNG deficient mice were homozygous null while POLβ deficient mice were heterozygous. It was necessary to use POLβ+/− rather than POLβ−/− because POLβ−/− mice die during the embryonic or early neonatal period (Sugo et al., 2000). POLβ+/− mice developed lymphoid hyperplasia (38% for POLβ+/−, 0% for POLβ+/+), adenocarcinoma (a 2.7-fold increase in POLβ+/− mice) and multiple tumors (20% in POLβ+/− mice and 5% for POLβ+/+ mice). Although POLβ+/− mice had a greater cancer prevalence as detected at old age (i.e. 24- to 26-months-old), median lifespan was unaffected. In this model, unlike the UNG−/− model, increased cancer prevalence did not impact median lifespan.
The very limited information available on lifespan in the background of impaired BER activity has not revealed an effect on maximal longevity. Maximal longevity measurements are typically evaluated at 5–10% survival so that multiple animals can be compared statistically. Regardless, the number of mice remaining is few and may hamper the ability to detect differences. In contrast, at median lifespan sufficient animals are alive in a well planned lifespan study to have meaningful comparisons. UNG−/− mice displayed a reduced median lifespan, while POLβ+/− mice did not. The homozygous state of the UNG mice was likely influential in imparting a detectable difference. Both models clearly demonstrated that reduced BER activity results in a decreased health span, with a greater prevalence of cancer and other pathologies among the UNG−/− and POLβ+/− groups compared to control groups. Health span is unequivocally a critical factor in human longevity. It is possible that lifespan determinants, potentially including BER, exert their effects at different points in the lifespan and BER may impact survival early in the lifespan by improving health span in the first half of the lifespan.
8. Effects of aging and BER on male reproduction and development
8.1. Human studies
The production of mature male reproductive cells starts post-natally at puberty and continues throughout life into old age (Nielsen et al., 1986; Nieschlag et al., 1982; Snyder, 2001). Spermatogenesis, the process that culminates in the production of spermatozoa, is dynamic and complex involving germ cell proliferation and differentiation which is dependent upon extrinsic hormone support and local cellular interactions. The oldest proven paternity reported was for a 94-year-old man (Seymour et al., 1935), thereby indicating that spermatozoa can be biologically functional well into old age. Perhaps it is not surprising that aging effects on spermatogenesis and sperm production are known. Age-related alterations associated with male reproduction include histomorphological changes of the testis (e.g. atrophy, decreased numbers of Leydig cells) (Johnson et al., 1984; Neaves et al., 1984), an increasingly dysfunctional hypothalamic-pituitary-testis axis producing altered hormone concentrations (reviewed by Vermeulen and Kaufman, 1995), decreased semen parameters (e.g. semen volume, sperm count) (Nieschlag et al., 1982; Rolf et al., 1996; Spandorfer et al., 1998), and decreased sperm production (Kidd et al., 2001; Ng et al., 2004), all leading to lower fertility.
An important issue in the assessment of age related changes of male reproduction is the frequency of hereditary diseases in offspring. A positive relationship between paternal age and the incidence of a genetic disorder in offspring was first observed in 1912, when Wilhelm Weinberg noticed that children with achondroplasia, an autosomal dominant skeletal disorder, were more likely to be born later among their siblings (Weinberg, 1912). Since then, many autosomal dominant disorders have been reported to be associated with advanced paternal age (reviewed by Crow, 2000; Risch et al., 1987). Paternally derived base substitutions were found in some age-related disorders, such as achodronplasia (Wilkin et al., 1998), Apert syndrome (Moloney et al., 1996), multiple endocrine neoplasia (MEN) 2A (Mulligan et al., 1994; Zedenius et al., 1994), MEN 2B (Carlson et al., 1994; Kitamura et al., 1995; Mulligan et al., 1994; Zedenius et al., 1994) and progeria (Eriksson et al., 2003). The association between increased age and an increased risk of producing offspring with a genetic disease from a new germline mutation derived from the paternal lineage is known as the paternal age effect. The mechanisms that mediate the paternal age effect are poorly understood and a number of different possibilities have been suggested (Crow, 2000; Glaser et al., 2003; Goriely et al., 2003; Penrose, 1955; Tiemann-Boege et al., 2002). Regardless of the mechanisms by which these mutations are generated, the paternal age associated disorders are a reflection of the increasing mutant frequency in sperm as men age. A few studies have shown an increased mutant frequency in sperm with age (Glaser et al., 2003; Goriely et al., 2003; Tiemann-Boege et al., 2002). However, the slight age-related increase in mutant frequency detected in sperm did not correspond to the exponentially increased incidence of age-related disorders in offspring (Glaser et al., 2003; Goriely et al., 2003; Tiemann-Boege et al., 2002), suggesting that multiple mechanisms exist and that they may work together to increase the number of affected offspring. Because sperm are the fully differentiated cell type generated during spermatogenesis, any adverse effects occurring in precursor cell types could affect sperm quality, including mutant frequency. Thus, monitoring mutation in different spermatogenic cell types during spermatogeneis with age should provide invaluable information on the paternal age effect. Monitoring differentiating cells in the spermatogenic lineage can be challenging for human studies because the procedure to obtain spermatogenic cells from the testis requires an invasive testis biopsy. As a result animal models are used frequently to study spermatogenesis and changes occurring during spermatogenesis.
8.2. Animal studies on germline mutant frequency
Taking advantage of transgenic mice carrying a mutation reporter transgene, i.e. Lac I transgenic mice, mutant frequencies were carefully examined in different spermatogenic cell types and from different aged mice. The spontaneous mutant frequency was elevated in mouse in pachytene spermatocytes, round spermatids and epididymal spermatozoa (Walter, 1998). Further characterization of the mutation spectra has shown that base substitutions accounted for more than 80% of the mutations in germ cells from young (3-to 6-months-old), middle-aged (14- to 16-months-old) and old mice (28-months-old) (Walter et al., 2004). Notably, the mutation spectrum in cells from old mice differed from those in the other two groups (Walter et al., 2004). Five mutation hotspots were identified in spectra for spermatogenic cells from young and middle-aged mice, but no hotspot was found in old mice (Walter et al., 2004). The data on mutation spectra in germ cells from old mice suggest that increased germline mutations in old mice occur as a consequence of different factors than those leading to mutations in young mice. Several mechanisms (e.g. selection, apoptosis, DNA repair) may play roles in regulating germline mutagenesis.
8.3. BER gene knockout animal models and reproductive phenotypes
Knockout mice carrying homozygous inactivation of POLβ APE1, XRCC1 DNA ligase I are embryonic or neonatal lethal (Friedberg and Meira, 2006), indicating that BER is indispensable for early development of mice. The only viable homozygous knockout mouse models in the BER pathway are deficient in one of the DNA glycosylases or PARP1. Table 4 contains a summary of the reproductive phenotypes described for available BER knockout mice including heterozygous and homozygous mouse models. Overall, BER deficiency has little effect on animal fertility, but increases susceptibility to mutagenesis and tumorigenesis. The majority of the BER knockout mice have not been studied with regard to aging so it is unclear whether and/or how the phenotypes change with age.
Table 4.
Selected BER knockout/knockdown mouse models with corresponding phenotypes.
| Gene | BER Activity | Reproductive Phenotypes | Somatic Phenotypes | Reference |
|---|---|---|---|---|
| Mpg | glycosylase | Homozygous mice are fertile. | Mice appear normal. Cell lines derived are more sensitive to alkylating agents. | Engelward et al., 1997; Smith and Engelward, 2000 |
| Ung1/2 | glycosylase | Homozygous mice are fertile | Mice appear normal; accumulate 2000 uracils/cell; display a hyper IgM syndrome with B-cell lymphomas accumulating > 18 mo.; no change in lifespan but decreased median survival. | Hagen et al., 2006; Krokan et al., 2001; Nilsen et al., 2000; Nilsen et al., 2003 |
| Smug | glycosylase | N/A | A mutator phenotype seen with siRNA to Smug. | An et al., 2005 |
| Mbd4 | glycosylase | Homozygous mice are fertile. | Increase in somatic C-->T mutations at CpG sites | Millar et al., 2002; Wong et al., 2002 |
| Nth | glycosylase | Homozygous mice are fertile. | Mice normal to two years of age. | Ocampo et al., 2002; Takao et al., 2002b |
| Neil1 | glycosylase | Homozygous mice are fertile. | Metabolic syndrome at 6- to 10-months-old | Vartanian et al., 2006 |
| Neil3 | glycosylase | Homozygous mice are fertile | No phenotype detected | Torisu et al., 2005 |
| Ogg1 | glycosylase | Homozygous mice are fertile. Mutation frequency was not significantly different in testis compared to wild type. | Increased levels of 8-oxoG seen. Increased prevalence of G to T transversions in the liver | Boiteux and Radicella, 2000; de Souza-Pinto et al., 2001a; Klungland et al., 1999; Le Page et al., 2000; Osterod et al., 2001 |
| Myh | glycosylase | Homozygous mice are fertile. | A mutator phenotype seen in ES cells. No difference survival or tumor incidence to 17 mo. | Hirano et al., 2003; Xie et al., 2004 |
| Myh, Ogg1 | glycosylases | Double knockout mice fertile through at least 2 generations. | Median survival significantly decreased to 10.3 mo., tumor incidence increased; G to T mutation in K-ras in lung tumors | Xie et al., 2004 |
| Ape1 | AP-endonuclease | Ape1+/− animals were fertile, appear normal; but mutant frequency was elevated in spermatogenic cells. | Ape1 −/− is an embryonic lethal. Mutant frequency elevated in liver and spleen of Ape1 +/− mice | Huamani et al., 2004; Meira et al., 2001 |
| Polβ | DNA polymerase | Polβ+/− animals are fertile. | Polβ −/− is an embryonic/neonatal lethal. | Gu et al., 1994; Sugo et al., 2000 |
| Fen1 | Flap endonuclease | Fen +/− animals fertile, appear normal. | Fen −/− is an embryonic lethal. Fen +/− show rapid tumor progression in animals predisposed to cancer by mutation in adenomatous polyposis coli (Apc) gene‥ | Kucherlapati et al., 2002; Larsen et al., 2003 |
| Parp 1 | poly(ADP)-ribosylase | Homozygous mice are fertile. | Mice are viable although hypersensitive to mutagenic agents | de Murcia et al., 1997 |
| Parp2 | poly(ADP)-ribosylase | Null male mice are hypofertile, testis weight is reduced by 70% and the epididymal sperm counts were 0.1% of wild type. | Mice sensitive to ionizing radiation and have an increased post-replicative genomic instability. | Dantzer et al., 2006 Menissier de Murcia et al., 2003 |
| Sir6 | deacetylase | N/A | Progeroid mice with compromised BER, severe metabolic defects and death at 4-weeks-old. | Mostoslavsky et al., 2006 |
N/A = not applicable.
9. BER gene polymorphisms and diseases
9.1. Polymorphisms and cancer
There are currently about 150 known DNA repair genes, and most of them are known to have genetic variation in humans (Wood et al., 2005). Polymorphisms, defined as genetic variants that appear in at least 1% of a population, can be associated with increased risk for a disease or resistance to a specific disease. Over the last few years, a number of studies have probed the role of DNA repair gene polymorphisms with disease susceptibility, particularly with regard to cancer.
BER genes are known to have polymorphisms that are associated with increased risk for certain cancers (Table 5) (Berndt et al., 2007; Goode et al., 2002; Hung et al., 2005; Rybicki et al., 2004). There are several aspects comprising the association between BER gene polymorphisms and the likelihood of developing cancer. First, a multiple of polymorphisms may exist for a gene. For example, at least 20 validated sequence variants have been described to date for the OGG1 gene (Hung et al., 2005). More than 60 validated single nucleotide polymorphisms in XRCC1 are listed in the Ensembl database (Hung et al., 2005). Second, allele frequencies are different in different populations (reviewed by Hung et al., 2005). For example, the frequency of XRCC1 arg399gln polymorphism is 24.1% in the African-American population (Duell et al., 2001), but 36.7% in the Asian population (Chen et al., 2002b) and 46.1% is in the Caucasian population (Han et al., 2003). Third, the strength of the relationship between a polymorphism and predisposition to cancer depends on the position/type of the variant in the gene and the cancer type. For example, homozygous OGG1 cys326cys alleles confer a two-fold increased risk of lung cancer (Goode et al., 2002; Sugimura et al., 1999), and an elevated risk of prostate cancer and nasopharyngeal carcinoma (Chen et al., 2003b; Cho et al., 2003), but not of colon cancer (Hansen et al., 2005). The less frequent OGG1 polymorphism arg154his was found in human gastric cancers (Nohmi et al., 2005). Fourth, effects of polymorphisms may be apparent only in the presence of DNA-damaging agents. For instance, the XRCC1 homozygous gln399gln genotype was related to a significantly reduced risk of squamous cell carcinoma but, the relative risk of squamous cell carcinoma associated with painful sunburn history was significantly higher for homozygous variants (gln399gln and arg399arg) than wild type (Nelson et al., 2002). In addition, arsenic-induced skin lesions (Breton et al., 2007), levels of aflatoxin B1-DNA adducts (Lunn et al., 1999; Matullo et al., 2001) and bleomycin sensitivity (Wang et al., 2003) are associated with polymorphisms of the XRCC1 gene.
Table 5.
BER polymorphisms and cancer associations
| BER gene | Cancer association | Polymorphism | Selected references |
|---|---|---|---|
| MYH | Familial biallelic germline mutations implicated in 1–3% colorectal cancer | Tyr165Cys; Gly382Asp; Tyr90X/1103delC; Gln377X; 1314delA; Pro281Leu | Al-Tassan et al., 2002; Cheadle and Sampson, 2003; Croitoru et al., 2007; Halford et al., 2003; Jones et al., 2002 |
| OGG1 | Variants associated with increased prostate cancer risk. | Ser326Cys | Chen et al., 2003b |
| Variants associated with increased gastric cancer risk. | Arg154His | Nohmi et al., 2005 | |
| No association of variants with risk of adenomas; lowered risk of colorectal cancer. | Ser326Cys | Hansen et al., 2005 | |
| XRCC1 | Polymorphisms implicated in lung cancer | Arg399Gln; Arg194Trp, Arg280His | Kiyohara et al., 2006; Zienolddiny et al., 2006 |
| Homozygous variants with increased risk of squamous cell carcinoma after sun overexposure. | Arg399Gln | Nelson et al., 2002 | |
| Prostate cancer associated if XPD polymorphism also present | Arg399Gln | Rybicki et al., 2004 | |
| OGG1, APE1 | Risk association between single SNP and lung cancer, is minimal | Kiyohara et al., 2006 | |
| OGG1, POLβ, APE1, XRCC1 | Pancreatic cancer risk varies with BER protein polymorphisms. | POLβ A165G and T2133C; OGG1 G2657A; APE1 Asp148Glu; XRCC1 Arg194Trp | Li et al., 2007a |
| OGG1 XRCC1 | OGG1 variant associated with increased risk; XRCC1 variant associated with reduced risk in various cancers. | OGG1 Ser326Cys; XRCC1 Arg194Trp | Goode et al., 2002 |
| OGG1 XRCC1 APE1 | OGG1 variant with increased lung cancer risk; XRCC1 Arg194Trp variant with decreased risk; XRCC1 Arg399Gln with increased risk light smokers, decreased risk heavy smokers; no association of APE1 with lung cancer risk. | OGG1 Ser326Cys; XRCC1 Arg194Trp; XRCC1 Arg399Gln; APE1 Asp148Glu | Hung et al., 2005 |
| OGG1 XRCC1 | Variants with increased risk for nasopharyngeal carcinoma. Presence of both variants results in greater risk. | OGG1 Ser326Cys; XRCC1 Arg280His | Cho et al., 2003 |
| APE1, XRCC1, PARP1 | Polymorphisms in APE1 and PARP1 with decreased risk; polymorphism in XRCC1, and associated with increased risk in advanced colorectal adenoma. | APE1 Gln51His; PARP1 A284A and IVS13+118G>A) XRCC1 Arg399Gln; | Berndt et al., 2007 |
In addition to possible associations with cancer risk, BER gene polymorphisms are found associated with response to the cancer treatment (Stoehlmacher et al., 2001), with azoospermia (Gu et al., 2007) and with rheumatoid arthritis (Koyama et al., 2006). For example, polymorphism of the XRCC1 gene was investigated in patients with metastatic colorectal cancer treated with oxaliplatin and fluorouracil (5-FU). 73% of responders to treatment had an arg399arg genotype, but 66% of non-responders showed a gln399gln or gln399arg genotype (p=0.038). Patients carrying at least one Gln mutant allele were at a 5.2-fold increased risk to fail the 5-FU/oxaliplatin chemotherapy (Stoehlmacher et al., 2001).
BER gene polymorphisms have shown association trends in a variety of human cancers and other diseases, however, conflicting reports weaken any substantial conclusions in epidemiological studies. Therefore a functional study of these polymorphisms will be essential. So far the significance of these polymorphisms is still largely unknown. The most studied BER gene with regard to polymorphisms is perhaps XRCC1, which is required for efficient DNA SSB repair. Takanami et al (2005) has shown that the R280H variant when transfected into a CHO cell line null for the XRCC1 gene product impaired the function of XRCC1. Cell survival after treatment with methyl methanesulfonate (MMS) to induce DNA SSB was not affected. Cells with the R399Q genotype showed no difference in sensitivity to MMS compared to cells transfected with Q399R (Taylor et al., 2002). There was also no difference in the ability to repair MMS-induced SSB in the two isoforms as determined by the comet assay (Taylor et al., 2002). However, Pachkowski et al. (2006) found decreased DNA repair with the R280H variant in cells exposed to chemical stresses of peroxide, MMS or camptothecin in a measure of DNA repair by monitoring NAD(P)H levels. Other studies reported a significant increase in chromosomal deletions in cells with the R399Q genotype treated with UV light (Au et al., 2003) or bleomycin (Qu et al., 2005). Functional evidence of OGG1 gene polymorphism is also equivocal, with some in vitro studies showing the Cys326 variant has identical catalytic activity to wild-type Ser326, and others reporting significant differences (reviewed by (Weiss et al., 2005). These contrasting results on functional studies raise some questions about whether the polymorphisms are completely neutral or harmless.
9.2. Polymorphisms and spermatogenesis
Xrcc1 is expressed in greater abundance in testis in rodents and primates and specifically most abundantly in pachytene spermatocytes and round spermatids of mice (Walter et al., 1994; Walter et al., 1996; Zhou and Walter, 1995). The expression profile of Xrcc1 during mouse spermatogenesis is accompanied with greater BER activity and lower mutant frequencies in these germ cell types (Intano, 2002; Walter et al., 1994). Because Xrcc1 knockout mice die early in embryogenesis (Tebbs et al., 1999), it is unknown how Xrcc1 depletion affects spermatogenesis. Studies of XRCC1 polymorphisms in human studies may provide some clues. The homozygous arg399arg genotype is found to be related to a decreased risk of idiopathic azoospermia in a Chinese population (Gu et al., 2007), indicating that the XRCC1 gene is important for normal spermatogenesis. Patients with an arg399arg genotype may have better BER activity compared to gln399gln and arg399arg genotypes, because the arg399arg genotype experienced more DNA damage after sunlight exposure (Nelson et al., 2002). Direct quantification of XRCC1 functions with different polymorphism genotypes has not been thoroughly examined.
9.3. BER and neurodegenerative disorders
Neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), and ataxia telangiectasia (AT), are characteristically associated with neurological deterioration. Although the etiologies of neurodegenerative diseases are largely unknown, aging is a major risk factor for most, and DNA damage and DNA repair are found to play an important role in these disorders (Table 6). ROS are considered major risk factors for aging and neurodegenerative disorders (Poon et al., 2004). ROS can mediate damage in mitochondrial and nuclear DNA which is largely repaired by BER (Seeberg et al., 1995). Brain uses more oxygen than other organs yet it has less nuclear and mitochondrial BER activity (Intano et al., 2001; Karahalil et al., 2002) than other somatic tissues, rendering the brain more vulnerable to ROS-induced damage. As noted above, BER declines in brain in general and in specific brain regions during aging thereby further increasing the vulnerability of this important tissue to DNA damage.
Table 6.
BER in neurodegeneration
| Disease | Study | Results | Selected References |
|---|---|---|---|
| Alzheimer’s Disease- age related dementia | AD patients and age-matched controls | Increased 8-oxoG in ventricular CSF DNA; decreased free 8-oxoG indicating accumulation of 8-oxoG lesions in patients. | Lovell et al., 1999 |
| Late stage AD patients and early stage with mild cognitive impairment | Elevated 8-oxoG, 8-OHA, and 5,6-diamino-5-formamidopyrimidine in nuclear and mtDNA isolated from vulnerable brain regions in amnestic mild cognitive impairment. | Lovell and Markesbery, 2007 | |
| Brain tissue autopsies | Immunoreactivity to hOGG1-2a is associated with neurofibrillary tangles, dystrophic neurites and reactive astrocytes in AD. | Iida et al., 2002 | |
| Post-mortem autopsies graded for disease progression. | POL-β is increased with increasing severity of disease; decreases with severe neuronal loss. Suggests POL-β involved early in AD pathology. | Copani et al., 2006 | |
| Postmortem AD midfrontal cortex and age-matched controls. | APE1 expression significantly increased in AD subjects. | Davydov et al., 2003 | |
| Post-mortem autopsies sporadic AD, mild cognitive impaired and age-matched controls. | Significant BER deficiencies; decreased OGG-BER, UDG-BER and POLβ activities. No decrease in APE1-BER. Decreases seen in both AD and mild cognitive impairment. | Weissman et al., 2007b | |
| Parkinson’s Disease | Autopsies PD and age-matched controls. | Up-regulation of MYH in the mitochondria of substantia nigra. | Arai et al., 2006 |
| Amyotrophic Lateral Sclerosis | Mutant SOD mice | POLβ and mitochondrial OGG1 is decreased in symptomatic mice. POLγ is decreased in presymptomatic and symptomatic mice. | Murakami et al., 2007 |
| Patients with familial ALS | Identified missense mutations in the APE1 gene. | Olkowski, 1998 | |
| Patients with sporadic ALS and age-matched controls. | APE1 levels and activity are significantly lower in ALS subjects than in controls. | Kisby et al., 1997 | |
| Patients with sporadic ALS and age-matched controls. | Results suggest a possible involvement of the hOGG1 Ser326Cys polymorphism in sporadic ALS pathogenesis. | Coppede et al., 2007 | |
| Ischemia | Mouse and rat models | Ischemia causes transient decrease in BER proteins in the brain. Ischemic preconditioning increases BER capacity. | Chen et al., 2003a; Fujimura et al., 1999; Lan et al., 2003; Li et al., 2007b; Li et al., 2006; Luo et al., 2007 |
| UNG −/− mice | Increased post-ischemic brain injury compared to wild type mice. | Endres et al., 2004 | |
| PARP1 −/− mice | PARP1 critical in ischemia-reperfusion injury. | Shall and de Murcia, 2000 | |
| PARP2 −/− mice | Reduced stroke damage in PARP2 −/− mice. | Kofler et al., 2006 | |
| Huntington’s Disease-trinucleotide instability | HD gene transgenic mice bred with MPG, OGG1 and NTH1 knockouts. | OGG1 excision of 8-oxo-G within CAG repeat DNA can initiate strand displacement and expansion in vitro during BER, creating a toxic oxidation cycle. | Kovtun et al., 2007 |
AD = Alzheimer’s disease; 8-oxoG = 8-oxodeoxyguanine; 8-OHA=8-hydroxyadenine; mtDNA=mitochondrial DNA; hOgg1-2a=mitochondrial splice variant of hOgg1; OGG/UDG/APE-BER=complete base excision repair assay of OGG/UDG/APE substrate; PD=Parkinson’s disease; SOD=superoxide dismutase; ALS= Amyotrophic Lateral Sclerosis; HD=Huntington’s disease
Age-associated decline of BER activity in the central nervous system has been linked to neurodegenerative disorders. Missense mutations in the APE1 gene (Olkowski, 1998), decreased APE1 protein levels and decreased APE1 activity (Kisby et al., 1997) were found in ALS patients. However, contradictory findings were also reported in ALS patients (Shaikh and Martin, 2002). In AD patients, expressions of the mitochondrial β-OGG1 (Iida et al., 2002) and DNA POLβ (Copani et al., 2006) was decreased; whereas expression of APE1 was increased in brain tissues (Davydov et al., 2003). A recent study has demonstrated that BER activity is decreased in both affected and non-affected brain regions in AD patients, resulting from reduced UDG, OGG1 and POLβ activities (Weissman et al., 2007b).
10. Future directions
Lifespan and health span can be extended by a simple procedure of reducing food consumption (Masoro, 2003). However, the molecular mechanisms that mediate the increases in lifespan and health span remain largely undefined. DNA repair, including BER, is clearly important for viability and health span. The extent to which DNA repair contributes to maximal lifespan is less clear. There are now animal models that can be used to directly ask questions about how BER contributes to health span throughout the lifespan of rodents and how BER contributes to lifespan. Although the Ape1, Polβ Xrcc1, Fen1 and Lig I knockouts are lethal, the heterozygotes are viable and can be used to assess health span, repair over the lifespan, mutagenesis over the lifespan and lifespan itself in the background of heterozygosity for a BER gene. The DNA glycosylase knockout mice are viable as nulls and can be used to address the same questions. It is also important to understand the molecular mechanisms that mediate the decline in BER with increased age and the variability among tissues with regard to the apparently different proteins that mediate the decline in activity. Tissue specific knockdowns may also provide insight into the variability among tissues. The full impact of BER on aging and health span may be better displayed when models heterozygous or homozygous null for BER genes are combined with models deficient in apoptosis or cell cycle check point genes. Much remains to be elucidated concerning the role of BER in aging and health span.
Acknowledgements
We wish to acknowledge the valuable contributions from all investigators that have studied the role of BER in aging and regret that only a limited number of publications on this topic could be included in the review. Work cited from the CAW laboratory was supported by AG24364 and AG21163 from the NIA/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Akbari M, Otterlei M, Pena-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, Slupphaug G, Krokan HE. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- Alexander P. The role of DNA lesions in the processes leading to aging in mice. Symp Soc Exp Biol. 1967;21:29–50. [PubMed] [Google Scholar]
- Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- An Q, Robins P, Lindahl T, Barnes DE. C --> T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–945. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Ho GPH, D'Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. doi: 10.1093/carcin/bgi319. [DOI] [PubMed] [Google Scholar]
- Arai T, Fukae J, Hatano T, Kubo S, Ohtsubo T, Nakabeppu Y, Mori H, Mizuno Y, Hattori N. Up-regulation of hMUTYH, a DNA repair enzyme, in the mitochondria of substantia nigra in Parkinson's disease. Acta Neuropathol. 2006;112:139–145. doi: 10.1007/s00401-006-0081-9. [DOI] [PubMed] [Google Scholar]
- Asagoshi K, Yamada T, Terato H, Ohyama Y, Ide H. Enzymatic properties of Escherichia coli and human 7,8-dihydro-8-oxoguanine DNA glycosylases. Nucleic Acids Symp Ser. 2000:11–12. doi: 10.1093/nass/44.1.11. [DOI] [PubMed] [Google Scholar]
- Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc Natl Acad Sci U S A. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ Health Perspect. 2003;111:1843–1850. doi: 10.1289/ehp.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- Baker D, Liu P, Burdzy A, Sowers LC. Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem Res Toxicol. 2002;15:33–39. doi: 10.1021/tx010113b. [DOI] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- Beneke S, Burkle A. Poly(ADP-ribosyl)ation, PARP, and aging. Sci Aging Knowledge Environ 2004. 2004 doi: 10.1126/sageke.2004.49.re9. re9. [DOI] [PubMed] [Google Scholar]
- Berndt SI, Huang WY, Fallin MD, Helzlsouer KJ, Platz EA, Weissfeld JL, Church TR, Welch R, Chanock SJ, Hayes RB. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67:1395–1404. doi: 10.1158/0008-5472.CAN-06-1390. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Pinz KG, Perez-Jannotti RM. Enzymology of mitochondrial base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:257–271. doi: 10.1016/s0079-6603(01)68105-4. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Sneeden JL, Allen S, Pham P, Goodman MF. First AID (activation-induced cytidine deaminase) is needed to produce high affinity isotype-switched antibodies. J Biol Chem. 2006;281:16833–16836. doi: 10.1074/jbc.R600006200. [DOI] [PubMed] [Google Scholar]
- Breton CV, Zhou W, Kile ML, Houseman EA, Quamruzzaman Q, Rahman M, Mahiuddin G, Christiani DC. Susceptibility to arsenic-induced skin lesions from polymorphisms in base excision repair genes. Carcinogenesis. 2007;28:1520–1525. doi: 10.1093/carcin/bgm063. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR. Attenuation of DNA polymerase beta-dependent base excision repair and increased DMS-induced mutagenicity in aged mice. Mutat Res. 2002;500:135–145. doi: 10.1016/s0027-5107(02)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof DC, Yanamadala S, Raffoul JJ, Guo Z, Soofi A, Heydari AR. Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair (Amst) 2003;2:295–307. doi: 10.1016/s1568-7864(02)00219-7. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Ikeno Y, Nyska A, Busuttil RA, Anyangwe N, Vijg J, Matherly LH, Tucker JD, Wilson SH, Richardson A, Heydari AR. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res. 2006;66:7460–7465. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- Campalans A, Marsin S, Nakabeppu Y, O'Connor TR, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst) 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Campalans A, Amouroux R, Bravard A, Epe B, Radicella JP. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci. 2007;120:23–32. doi: 10.1242/jcs.03312. [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Carlson KM, Bracamontes J, Jackson CE, Clark R, Lacroix A, Wells SA, Jr, Goodfellow PJ. Parent-of-origin effects in multiple endocrine neoplasia type 2B. Am J Hum Genet. 1994;55:1076–1082. [PMC free article] [PubMed] [Google Scholar]
- Caron L, Tihy F, Dallaire L. Frequencies of chromosomal abnormalities at amniocentesis: over 20 years of cytogenetic analyses in one laboratory. Am J Med Genet. 1999;82:149–154. doi: 10.1002/(sici)1096-8628(19990115)82:2<149::aid-ajmg10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Ibeanu GC, Tano K, Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- Chaudhry MA. Base excision repair of ionizing radiation-induced DNA damage in G1 and G2 cell cycle phases. Cancer Cell Int. 2007;7:15. doi: 10.1186/1475-2867-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle JP, Sampson JR. Exposing the MYtH about base excision repair and human inherited disease. Hum Mol Genet. 2003;12(Spec No 2):R159–R165. doi: 10.1093/hmg/ddg259. [DOI] [PubMed] [Google Scholar]
- Chen D, Cao G, Hastings T, Feng Y, Pei W, O'Horo C, Chen J. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J Neurochem. 2002a;81:1273–1284. doi: 10.1046/j.1471-4159.2002.00916.x. [DOI] [PubMed] [Google Scholar]
- Chen D, Minami M, Henshall DC, Meller R, Kisby G, Simon RP. Upregulation of mitochondrial base-excision repair capability within rat brain after brief ischemia. J Cereb Blood Flow Metab. 2003a;23:88–98. doi: 10.1097/01.WCB.0000039286.37737.19. [DOI] [PubMed] [Google Scholar]
- Chen L, Elahi A, Pow-Sang J, Lazarus P, Park J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. J Urol. 2003b;170:2471–2474. doi: 10.1097/01.ju.0000087498.23008.bb. [DOI] [PubMed] [Google Scholar]
- Chen S, Tang D, Xue K, Xu L, Ma G, Hsu Y, Cho SS. DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis. 2002b;23:1321–1325. doi: 10.1093/carcin/23.8.1321. [DOI] [PubMed] [Google Scholar]
- Chen SK, Hsieh WA, Tsai MH, Chen CC, Hong AI, Wei YH, Chang WP. Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOgg1), in human aging. J Radiat Res (Tokyo) 2003c;44:31–35. doi: 10.1269/jrr.44.31. [DOI] [PubMed] [Google Scholar]
- Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF, Levine PH, Liu MY, Chen JY, Brinton LA, Cheng YJ, Yang CS. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol Biomarkers Prev. 2003;12:1100–1104. [PubMed] [Google Scholar]
- Copani A, Hoozemans JJ, Caraci F, Calafiore M, Van Haastert ES, Veerhuis R, Rozemuller AJ, Aronica E, Sortino MA, Nicoletti F. DNA polymerase-beta is expressed early in neurons of Alzheimer's disease brain and is loaded into DNA replication forks in neurons challenged with beta-amyloid. J Neurosci. 2006;26:10949–10957. doi: 10.1523/JNEUROSCI.2793-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppede F, Mancuso M, Lo Gerfo A, Carlesi C, Piazza S, Rocchi A, Petrozzi L, Nesti C, Micheli D, Bacci A, Migliore L, Murri L, Siciliano G. Association of the hOGG1 Ser326Cys polymorphism with sporadic amyotrophic lateral sclerosis. Neurosci Lett. 2007;420:163–168. doi: 10.1016/j.neulet.2007.04.067. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Cortopassi GA, Wang E. There is substantial agreement among interspecies estimates of DNA repair activity. Mech Ageing Dev. 1996;91:211–218. doi: 10.1016/s0047-6374(96)01788-5. [DOI] [PubMed] [Google Scholar]
- Croitoru ME, Cleary SP, Berk T, Di Nicola N, Kopolovic I, Bapat B, Gallinger S. Germline MYH mutations in a clinic-based series of Canadian multiple colorectal adenoma patients. J Surg Oncol. 2007;95:499–506. doi: 10.1002/jso.20724. [DOI] [PubMed] [Google Scholar]
- Croteau DL, Stierum RH, Bohr VA. Mitochondrial DNA repair pathways. Mutat Res. 1999;434:137–148. doi: 10.1016/s0921-8777(99)00025-7. [DOI] [PubMed] [Google Scholar]
- Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci U S A. 2006;103:14854–15859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J Biol Chem. 2007a;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, Wilson SH, Hazra TK. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem. 2007b;282:28474–28484. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov V, Hansen LA, Shackelford DA. Is DNA repair compromised in Alzheimer's disease? Neurobiol Aging. 2003;24:953–968. doi: 10.1016/s0197-4580(02)00229-4. [DOI] [PubMed] [Google Scholar]
- De A, Campbell C. A novel interaction between DNA ligase III and DNA polymerase gamma plays an essential role in mitochondrial DNA stability. Biochem J. 2007;402:175–186. doi: 10.1042/BJ20061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001a;61:5378–5381. [PubMed] [Google Scholar]
- de Souza-Pinto NC, Hogue BA, Bohr VA. DNA repair and aging in mouse liver: 8-oxodG glycosylase activity increase in mitochondrial but not in nuclear extracts. Free Radic Biol Med. 2001b;30:916–923. doi: 10.1016/s0891-5849(01)00483-x. [DOI] [PubMed] [Google Scholar]
- Dherin C, Gasparutto D, O'Connor TR, Cadet J, Boiteux S. Excision by the human methylpurine DNA N-glycosylase of cyanuric acid, a stable and mutagenic oxidation product of 8-oxo-7,8-dihydroguanine. Int J Radiat Biol. 2004;80:21–27. doi: 10.1080/09553000310001632976. [DOI] [PubMed] [Google Scholar]
- Di Maso V, Avellini C, Croce LS, Rosso N, Quadrifoglio F, Cesaratto L, Codarin E, Bedogni G, Beltrami CA, Tell G, Tiribelli C. Subcellular localization of APE1/Ref-1 in human hepatocellular carcinoma: possible prognostic significance. Mol Med. 2007;13:89–96. doi: 10.2119/2006-00084.DiMaso. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov GL, Souza-Pinto N, Nyaga SG, Thybo T, Stevnsner T, Bohr VA. Base excision repair in nuclear and mitochondrial DNA. Prog Nucleic Acid Res Mol Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Dianova II, Sleeth KM, Allinson SL, Parsons JL, Breslin C, Caldecott KW, Dianov GL. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res. 2004;32:2550–2555. doi: 10.1093/nar/gkh567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. [Google Scholar]
- Dolle ME, Snyder WK, Dunson DB, Vijg J. Mutational fingerprints of aging. Nucleic Acids Res. 2002;30:545–549. doi: 10.1093/nar/30.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, Eaton A, Mohrenweiser HW, Newman B, Bell DA. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:217–222. [PubMed] [Google Scholar]
- Endres M, Biniszkiewicz D, Sobol RW, Harms C, Ahmadi M, Lipski A, Katchanov J, Mergenthaler P, Dirnagl U, Wilson SH, Meisel A, Jaenisch R. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J Clin Invest. 2004;113:1711–1721. doi: 10.1172/JCI20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander EW, Hu Z, Sharma A, Lee HM, Wu ZH, Greeley GH. Rat MYH, a glycosylase for repair of oxidatively damaged DNA, has brain-specific isoforms that localize to neuronal mitochondria. J Neurochem. 2002;83:1471–1480. doi: 10.1046/j.1471-4159.2002.01259.x. [DOI] [PubMed] [Google Scholar]
- Englander EW, Ma H. Differential modulation of base excision repair activities during brain ontogeny: implications for repair of transcribed DNA. Mech Ageing Dev. 2006;127:64–69. doi: 10.1016/j.mad.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Vermeulen W, Houtsmuller AB. DNA damage repair: anytime, anywhere? Current Opinion in Cell Biology. 2006;18:240–246. doi: 10.1016/j.ceb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Failla G. The aging process and cancerogenesis. Ann N Y Acad Sci. 1958;71:1124–1140. doi: 10.1111/j.1749-6632.1958.tb46828.x. [DOI] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003a;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat Res. 2003b;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair. 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Morita-Fujimura Y, Sugawara T, Chan PH. Early decrease of XRCC1, a DNA base excision repair protein, may contribute to DNA fragmentation after transient focal cerebral ischemia in mice. Stroke. 1999;30:2456–2462. doi: 10.1161/01.str.30.11.2456. discussion 2463. [DOI] [PubMed] [Google Scholar]
- Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J Biol Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Broman KW, Schulman RL, Eskenazi B, Wyrobek AJ, Jabs EW. The paternal-age effect in Apert syndrome is due, in part, to the increased frequency of mutations in sperm. Am J Hum Genet. 2003;73:939–947. doi: 10.1086/378419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- Goriely A, McVean GA, Rojmyr M, Ingemarsson B, Wilkie AO. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science. 2003;301:643–646. doi: 10.1126/science.1085710. [DOI] [PubMed] [Google Scholar]
- Gossen JA, de Leeuw WJ, Tan CH, Zwarthoff EC, Berends F, Lohman PH, Knook DL, Vijg J. Efficient rescue of integrated shuttle vectors from transgenic mice: a model for studying mutations in vivo. Proc Natl Acad Sci U S A. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube K, Burkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci U S A. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Gu AH, Liang J, Lu NX, Wu B, Xia YK, Lu CC, Song L, Wang SL, Wang XR. Association of XRCC1 gene polymorphisms with idiopathic azoospermia in a Chinese population. Asian J Androl. 2007;9:781–786. doi: 10.1111/j.1745-7262.2007.00325.x. [DOI] [PubMed] [Google Scholar]
- Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J Biol Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen L, Pena-Diaz J, Kavli B, Otterlei M, Slupphaug G, Krokan HE. Genomic uracil and human disease. Exp Cell Res. 2006;312:2666–2672. doi: 10.1016/j.yexcr.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Halford SE, Rowan AJ, Lipton L, Sieber OM, Pack K, Thomas HJ, Hodgson SV, Bodmer WF, Tomlinson IP. Germline mutations but not somatic changes at the MYH locus contribute to the pathogenesis of unselected colorectal cancers. Am J Pathol. 2003;162:1545–1548. doi: 10.1016/S0002-9440(10)64288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001a;29:2117–2126. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001b;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Hankinson SE, De Vivo I, Spiegelman D, Tamimi RM, Mohrenweiser HW, Colditz GA, Hunter DJ. A prospective study of XRCC1 haplotypes and their interaction with plasma carotenoids on breast cancer risk. Cancer Res. 2003;63:8536–8541. [PubMed] [Google Scholar]
- Hansen R, Saebo M, Skjelbred CF, Nexo BA, Hagen PC, Bock G, Bowitz Lothe IM, Johnson E, Aase S, Hansteen IL, Vogel U, Kure EH. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005;229:85–91. doi: 10.1016/j.canlet.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Natl Acad Sci U S A. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Tominaga Y, Nakabeppu Y, Moriya M. Futile short-patch DNA base excision repair of adenine:8-oxoguanine mispair. Nucleic Acids Res. 2004;32:5928–5934. doi: 10.1093/nar/gkh909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002a;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002b;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Mitra S. Purification and characterization of NEIL1 and NEIL2, members of a distinct family of mammalian DNA glycosylases for repair of oxidized bases. Methods Enzymol. 2006;408:33–48. doi: 10.1016/S0076-6879(06)08003-7. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxoguanine. Proc Natl Acad Sci U S A. 1998;95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Hirano S, Tominaga Y, Ichinoe A, Ushijima Y, Tsuchimoto D, Honda-Ohnishi Y, Ohtsubo T, Sakumi K, Nakabeppu Y. Mutator phenotype of MUTYH-null mouse embryonic stem cells. J Biol Chem. 2003;278:38121–38124. doi: 10.1074/jbc.C300316200. [DOI] [PubMed] [Google Scholar]
- Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Fujisawa H, Ax S, Zahn-Daimler G, Zahn RK. Age-associated DNA damage is accelerated in the senescence-accelerated mice. Mech Ageing Dev. 2000;118:61–70. doi: 10.1016/s0047-6374(00)00158-5. [DOI] [PubMed] [Google Scholar]
- Huamani J, McMahan CA, Herbert DC, Reddick R, McCarrey JR, MacInnes MI, Chen DJ, Walter CA. Spontaneous Mutagenesis Is Enhanced in Apex Heterozygous Mice. Mol. Cell. Biol. 2004;24:8145–8153. doi: 10.1128/MCB.24.18.8145-8153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- Ide H. DNA substrates containing defined oxidative base lesions and their application to study substrate specificities of base excision repair enzymes. Prog Nucleic Acid Res Mol Biol. 2001;68:207–221. doi: 10.1016/s0079-6603(01)68101-7. [DOI] [PubMed] [Google Scholar]
- Ide H, Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol Pharm Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- Iida T, Furuta A, Nishioka K, Nakabeppu Y, Iwaki T. Expression of 8-oxoguanine DNA glycosylase is reduced and associated with neurofibrillary tangles in Alzheimer's disease brain. Acta Neuropathol. 2002;103:20–25. doi: 10.1007/s004010100418. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J Biol Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol Aging. 2006;27:1129–1136. doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Intano GW, McMahan CA, Walter RB, McCarrey JR, Walter CA. Mixed spermatogenic germ cell nuclear extracts exhibit high base excision repair activity. Nucleic Acids Res. 2001;29:1366–1372. doi: 10.1093/nar/29.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intano GW. Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Molecular & Cellular Biology. 2002;22:2410–2418. doi: 10.1128/MCB.22.7.2410-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intano GW, McMahan CA, McCarrey JR, Walter RB, McKenna AE, Matsumoto Y, MacInnes MA, Chen DJ, Walter CA. Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Mol Cell Biol. 2002;22:2410–2418. doi: 10.1128/MCB.22.7.2410-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intano GW, Cho EJ, McMahan CA, Walter CA. Age-related base excision repair activity in mouse brain and liver nuclear extracts. J Gerontol A Biol Sci Med Sci. 2003;58:205–211. doi: 10.1093/gerona/58.3.b205. [DOI] [PubMed] [Google Scholar]
- Johnson L, Petty CS, Neaves WB. Influence of age on sperm production and testicular weights in men. J Reprod Fertil. 1984;70:211–218. doi: 10.1530/jrf.0.0700211. [DOI] [PubMed] [Google Scholar]
- Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-->T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- Kao J, Rosenstein BS, Peters S, Milano MT, Kron SJ. Cellular Response to DNA Damage. Annals of the New York Academy of Sciences. 2005;1066:243–258. doi: 10.1196/annals.1363.012. [DOI] [PubMed] [Google Scholar]
- Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- Khaidakov M, Heflich RH, Manjanatha MG, Myers MB, Aidoo A. Accumulation of point mutations in mitochondrial DNA of aging mice. Mutat Res. 2003;526:1–7. doi: 10.1016/s0027-5107(03)00010-1. [DOI] [PubMed] [Google Scholar]
- Kidd SA, Eskenazi B, Wyrobek AJ. Effects of male age on semen quality and fertility: a review of the literature. Fertil Steril. 2001;75:237–248. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- Kisby GE, Milne J, Sweatt C. Evidence of reduced DNA repair in amyotrophic lateral sclerosis brain tissue. Neuroreport. 1997;8:1337–1340. doi: 10.1097/00001756-199704140-00004. [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Scavarda N, Wells SA, Jr, Jackson CE, Goodfellow PJ. Two maternally derived missense mutations in the tyrosine kinase domain of the RET protooncogene in a patient with de novo MEN 2B. Hum Mol Genet. 1995;4:1987–1988. doi: 10.1093/hmg/4.10.1987. [DOI] [PubMed] [Google Scholar]
- Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54:267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler J, Otsuka T, Zhang Z, Noppens R, Grafe MR, Koh DW, Dawson VL, de Murcia JM, Hurn PD, Traystman RJ. Differential effect of PARP-2 deletion on brain injury after focal and global cerebral ischemia. J Cereb Blood Flow Metab. 2006;26:135–141. doi: 10.1038/sj.jcbfm.9600173. [DOI] [PubMed] [Google Scholar]
- Kohler SW, Provost GS, Kretz PL, Dycaico MJ, Sorge JA, Short JM. Development of a short-term, in vivo mutagenesis assay: the effects of methylation on the recovery of a lambda phage shuttle vector from transgenic mice. Nucleic Acids Res. 1990;18:3007–3013. doi: 10.1093/nar/18.10.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama A, Kubota Y, Shimamura T, Horiuchi S. Possible association of the X-ray cross complementing gene 1 (XRCC1) Arg280His polymorphism as a risk for rheumatoid arthritis. Rheumatol Int. 2006;26:749–751. doi: 10.1007/s00296-005-0066-3. [DOI] [PubMed] [Google Scholar]
- Krishna TH, Mahipal S, Sudhakar A, Sugimoto H, Kalluri R, Rao KS. Reduced DNA gap repair in aging rat neuronal extracts and its restoration by DNA polymerase beta and DNA-ligase. J Neurochem. 2005;92:818–823. doi: 10.1111/j.1471-4159.2004.02923.x. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, Kane MF, Fan K, Russell R, Brown AM, Kneitz B, Edelmann W, Kolodner RD, Lipkin M, Kucherlapati R. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci U S A. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Li W, Zhang F, Sun FY, Nagayama T, O'Horo C, Chen J. Inducible repair of oxidative DNA lesions in the rat brain after transient focal ischemia and reperfusion. J Cereb Blood Flow Metab. 2003;23:1324–1339. doi: 10.1097/01.WCB.0000091540.60196.F2. [DOI] [PubMed] [Google Scholar]
- Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval J, Jurado J, Saparbaev M, Sidorkina O. Antimutagenic role of base-excision repair enzymes upon free radical-induced DNA damage. Mutat Res. 1998;402:93–102. doi: 10.1016/s0027-5107(97)00286-8. [DOI] [PubMed] [Google Scholar]
- Le Page F, Klungland A, Barnes DE, Sarasin A, Boiteux S. Transcription coupled repair of 8-oxoguanine in murine cells: the ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc Natl Acad Sci U S A. 2000;97:8397–8402. doi: 10.1073/pnas.140137297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AT, DeSimone C, Cerami A, Bucala R. Comparative analysis of DNA mutations in lacI transgenic mice with age. FASEB J. 1994;8:545–550. doi: 10.1096/fasebj.8.8.8181674. [DOI] [PubMed] [Google Scholar]
- Lee HM, Hu Z, Ma H, Greeley GH, Jr, Wang C, Englander EW. Developmental changes in expression and subcellular localization of the DNA repair glycosylase, MYH, in the rat brain. J Neurochem. 2004;88:394–400. doi: 10.1046/j.1471-4159.2003.02164.x. [DOI] [PubMed] [Google Scholar]
- Li D, Li Y, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. Effects of base excision repair gene polymorphisms on pancreatic cancer survival. Int J Cancer. 2007a;120:1748–1754. doi: 10.1002/ijc.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wu H, Yang S, Chen D. Ischemic preconditioning induces XRCC1, DNA polymerase-beta, and DNA ligase III and correlates with enhanced base excision repair. DNA Repair (Amst) 2007b;6:1297–1306. doi: 10.1016/j.dnarep.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Li W, Luo Y, Zhang F, Signore AP, Gobbel GT, Simon RP, Chen J. Ischemic preconditioning in the rat brain enhances the repair of endogenous oxidative DNA damage by activating the base-excision repair pathway. J Cereb Blood Flow Metab. 2006;26:181–198. doi: 10.1038/sj.jcbfm.9600180. [DOI] [PubMed] [Google Scholar]
- Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with baseexcision repair. Nucleic Acids Res. 2007c;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Burdzy A, Sowers LC. Repair of the mutagenic DNA oxidation product, 5-formyluracil. DNA Repair (Amst) 2003;2:199–210. doi: 10.1016/s1568-7864(02)00198-2. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Gabbita SP, Markesbery WR. Increased DNA oxidation and decreased levels of repair products in Alzheimer's disease ventricular CSF. J Neurochem. 1999;72:771–776. doi: 10.1046/j.1471-4159.1999.0720771.x. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Markesbery WR. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer's disease. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59:2557–2561. [PubMed] [Google Scholar]
- Luo Y, Ji X, Ling F, Li W, Zhang F, Cao G, Chen J. Impaired DNA repair via the base-excision repair pathway after focal ischemic brain injury: a protein phosphorylation-dependent mechanism reversed by hypothermic neuroprotection. Front Biosci. 2007;12:1852–1862. doi: 10.2741/2193. [DOI] [PubMed] [Google Scholar]
- Mandavilli BS, Santos JH, Van Houten B. Mitochondrial DNA repair and aging. Mutat Res. 2002;509:127–151. doi: 10.1016/s0027-5107(02)00220-8. [DOI] [PubMed] [Google Scholar]
- Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Substrate specificity of human endonuclease III (hNTH1).Effect of human APE1 on hNTH1 activity. J Biol Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- Martin SL, Hopkins CL, Naumer A, Dolle ME, Vijg J. Mutation frequency and type during ageing in mouse seminiferous tubules. Mech Ageing Dev. 2001;122:1321–1331. doi: 10.1016/s0047-6374(01)00267-6. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Subfield History: Caloric Restriction, Slowing Aging, and Extending Life. Sci. Aging Knowl. Environ. 2003;2003 doi: 10.1126/sageke.2003.8.re2. re2-. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Masaoka A, Tanaka T, Miyano T, Kato N, Terato H, Ohyama Y, Iwai S, Ide H. Mammalian 5-formyluracil-DNA glycosylase. 1. Identification and characterization of a novel activity that releases 5-formyluracil from DNA. Biochemistry. 2003;42:4993–5002. doi: 10.1021/bi027322v. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Tanaka T, Terato H, Ohmae E, Izumi S, Katayanagi K, Ide H. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res. 2004;32:5291–5302. doi: 10.1093/nar/gkh859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S, Piazza A, Vineis P. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)PDNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- McGoldrick JP, Yeh YC, Solomon M, Essigmann JM, Lu AL. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol Cell Biol. 1995;15:989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F, Bouziane M, O'Connor TR. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res. 1998;26:4034–4041. doi: 10.1093/nar/26.17.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F, Bouziane M, Dammann R, Masutani C, Hanaoka F, Pfeifer G, O'Connor TR. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J Biol Chem. 2000;275:28433–28438. doi: 10.1074/jbc.M001064200. [DOI] [PubMed] [Google Scholar]
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- Miyabe I, Zhang QM, Kino K, Sugiyama H, Takao M, Yasui A, Yonei S. Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein. Nucleic Acids Res. 2002;30:3443–3448. doi: 10.1093/nar/gkf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan RD, Rao A, Gagliardi J, Tini M. SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol Cell Biol. 2007;27:229–243. doi: 10.1128/MCB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DM, Slaney SF, Oldridge M, Wall SA, Sahlin P, Stenman G, Wilkie AO. Exclusive paternal origin of new mutations in Apert syndrome. Nat Genet. 1996;13:48–53. doi: 10.1038/ng0596-48. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Muiras ML. Mammalian longevity under the protection of PARP-1's multi-facets. Ageing Res Rev. 2003;2:129–148. doi: 10.1016/s1568-1637(02)00062-4. [DOI] [PubMed] [Google Scholar]
- Mulligan LM, Eng C, Healey CS, Ponder MA, Feldman GL, Li P, Jackson CE, Ponder BA. A de novo mutation of the RET proto-oncogene in a patient with MEN 2A. Hum Mol Genet. 1994;3:1007–1008. doi: 10.1093/hmg/3.6.1007. [DOI] [PubMed] [Google Scholar]
- Murakami T, Nagai M, Miyazaki K, Morimoto N, Ohta Y, Kurata T, Takehisa Y, Kamiya T, Abe K. Early decrease of mitochondrial DNA repair enzymes in spinal motor neurons of presymptomatic transgenic mice carrying a mutant SOD1 gene. Brain Res. 2007;1150:182–189. doi: 10.1016/j.brainres.2007.02.057. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog Nucleic Acid Res Mol Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, Frechet M, Demple B, Crescenzi M, Dogliotti E. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proc Natl Acad Sci U S A. 2007;104:17010–17015. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neaves WB, Johnson L, Porter JC, Parker CR, Jr, Petty CS. Leydig cell numbers, daily sperm production, and serum gonadotropin levels in aging men. J Clin Endocrinol Metab. 1984;59:756–763. doi: 10.1210/jcem-59-4-756. [DOI] [PubMed] [Google Scholar]
- Nelson HH, Kelsey KT, Mott LA, Karagas MR. The XRCC1 Arg399Gln polymorphism, sunburn, and non-melanoma skin cancer: evidence of gene-environment interaction. Cancer Res. 2002;62:152–155. [PubMed] [Google Scholar]
- Ng KK, Donat R, Chan L, Lalak A, Di Pierro I, Handelsman DJ. Sperm output of older men. Hum Reprod. 2004;19:1811–1815. doi: 10.1093/humrep/deh315. [DOI] [PubMed] [Google Scholar]
- Nielsen CT, Skakkebaek NE, Richardson DW, Darling JA, Hunter WM, Jorgensen M, Nielsen A, Ingerslev O, Keiding N, Muller J. Onset of the release of spermatozoa (spermarche) in boys in relation to age, testicular growth, pubic hair, and height. J Clin Endocrinol Metab. 1986;62:532–535. doi: 10.1210/jcem-62-3-532. [DOI] [PubMed] [Google Scholar]
- Nieschlag E, Lammers U, Freischem CW, Langer K, Wickings EJ. Reproductive functions in young fathers and grandfathers. J Clin Endocrinol Metab. 1982;55:676–681. doi: 10.1210/jcem-55-4-676. [DOI] [PubMed] [Google Scholar]
- Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- Nilsen H, Stamp G, Andersen S, Hrivnak G, Krokan HE, Lindahl T, Barnes DE. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Ocampo MT, Chaung W, Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Targeted deletion of mNth1 reveals a novel DNA repair enzyme activity. Mol Cell Biol. 2002;22:6111–6121. doi: 10.1128/MCB.22.17.6111-6121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkowski ZL. Mutant AP endonuclease in patients with amyotrophic lateral sclerosis. Neuroreport. 1998;9:239–242. doi: 10.1097/00001756-199801260-00012. [DOI] [PubMed] [Google Scholar]
- Ono T, Ikehata H, Nakamura S, Saito Y, Hosoi Y, Takai Y, Yamada S, Onodera J, Yamamoto K. Age-associated increase of spontaneous mutant frequency and molecular nature of mutation in newborn and old lacZ-transgenic mouse. Mutat Res. 2000;447:165–177. doi: 10.1016/s0027-5107(99)00200-6. [DOI] [PubMed] [Google Scholar]
- Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis. 2001;22:1459–1463. doi: 10.1093/carcin/22.9.1459. [DOI] [PubMed] [Google Scholar]
- Pachkowski BF, Winkel S, Kubota Y, Swenberg JA, Millikan RC, Nakamura J. XRCC1 genotype and breast cancer: functional studies and epidemiologic data show interactions between XRCC1 codon 280 His and smoking. Cancer Res. 2006;66:2860–2868. doi: 10.1158/0008-5472.CAN-05-3388. [DOI] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- Penrose LS. Parental age and mutation. Lancet. 1955;269:312–313. doi: 10.1016/s0140-6736(55)92305-9. [DOI] [PubMed] [Google Scholar]
- Petermann E, Ziegler M, Oei SL. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair (Amst) 2003;2:1101–1114. doi: 10.1016/s1568-7864(03)00117-4. [DOI] [PubMed] [Google Scholar]
- Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon HF, Calabrese V, Scapagnini G, Butterfield DA. Free radicals: key to brain aging and heme oxygenase as a cellular response to oxidative stress. J Gerontol A Biol Sci Med Sci. 2004;59:478–493. doi: 10.1093/gerona/59.5.m478. [DOI] [PubMed] [Google Scholar]
- Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J Biol Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- Qu T, Morii E, Oboki K, Lu Y, Morimoto K. Micronuclei in EM9 cells expressing polymorphic forms of human XRCC1. Cancer Lett. 2005;221:91–95. doi: 10.1016/j.canlet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Radak Z, Chung HY, Goto S. Exercise and hormesis: oxidative stress-related adaptation for successful aging. Biogerontology. 2005;6:71–75. doi: 10.1007/s10522-004-7386-7. [DOI] [PubMed] [Google Scholar]
- Radak Z, Kumagai S, Nakamoto H, Goto S. 8-Oxoguanosine and uracil repair of nuclear and mitochondrial DNA in red and white skeletal muscle of exercise-trained old rats. J Appl Physiol. 2007;102:1696–1701. doi: 10.1152/japplphysiol.01051.2006. [DOI] [PubMed] [Google Scholar]
- Risch N, Reich EW, Wishnick MM, McCarthy JG. Spontaneous mutation and parental age in humans. Am J Hum Genet. 1987;41:218–248. [PMC free article] [PubMed] [Google Scholar]
- Rolf C, Behre HM, Nieschlag E. Reproductive parameters of older compared to younger men of infertile couples. Int J Androl. 1996;19:135–142. doi: 10.1111/j.1365-2605.1996.tb00451.x. [DOI] [PubMed] [Google Scholar]
- Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- Russo MT, De Luca G, Degan P, Bignami M. Different DNA repair strategies to combat the threat from 8-oxoguanine. Mutat Res. 2007;614:69–76. doi: 10.1016/j.mrfmmm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Conti DV, Moreira A, Cicek M, Casey G, Witte JS. DNA repair gene XRCC1 and XPD polymorphisms and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:23–29. doi: 10.1158/1055-9965.epi-03-0053. [DOI] [PubMed] [Google Scholar]
- Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- Seymour F, Duffy C, Korner A. A case of authentic fertility in a man of 94. JAMA. 1935:1423–1424. [Google Scholar]
- Shaikh AY, Martin LJ. DNA base-excision repair enzyme apurinic/apyrimidinic endonuclease/redox factor-1 is increased and competent in the brain and spinal cord of individuals with amyotrophic lateral sclerosis. Neuromolecular Med. 2002;2:47–60. doi: 10.1007/s12017-002-0038-7. [DOI] [PubMed] [Google Scholar]
- Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat Res. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- Shen GP, Galick H, Inoue M, Wallace SS. Decline of nuclear and mitochondrial oxidative base excision repair activity in late passage human diploid fibroblasts. DNA Repair (Amst) 2003;2:673–693. doi: 10.1016/s1568-7864(03)00006-5. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Yamaguchi S, Saitoh T, Kohno T, Yokota J. Somatic mutations and single nucleotide polymorphisms of base excision repair genes involved in the repair of 8-hydroxyguanine in damaged DNA. Cancer Lett. 2001;166:65–69. doi: 10.1016/s0304-3835(01)00435-9. [DOI] [PubMed] [Google Scholar]
- Smith SA, Engelward BP. In vivo repair of methylation damage in Aag 3-methyladenine DNA glycosylase null mouse cells. Nucleic Acids Res. 2000;28:3294–3300. doi: 10.1093/nar/28.17.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder PJ. Effects of age on testicular function and consequences of testosterone treatment. J Clin Endocrinol Metab. 2001;86:2369–2372. doi: 10.1210/jcem.86.6.7602. [DOI] [PubMed] [Google Scholar]
- Sousa MM, Krokan HE, Slupphaug G. DNA-uracil and human pathology. Mol Aspects Med. 2007;28:276–306. doi: 10.1016/j.mam.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Souza-Pinto NC, Croteau DL, Hudson EK, Hansford RG, Bohr VA. Age-associated increase in 8-oxo-deoxyguanosine glycosylase/AP lyase activity in rat mitochondria. Nucleic Acids Res. 1999;27:1935–1942. doi: 10.1093/nar/27.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandorfer SD, Avrech OM, Colombero LT, Palermo GD, Rosenwaks Z. Effect of parental age on fertilization and pregnancy characteristics in couples treated by intracytoplasmic sperm injection. Hum Reprod. 1998;13:334–338. doi: 10.1093/humrep/13.2.334. [DOI] [PubMed] [Google Scholar]
- Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- Stoehlmacher J, Ghaderi V, Iobal S, Groshen S, Tsao-Wei D, Park D, Lenz HJ. A polymorphism of the XRCC1 gene predicts for response to platinum based treatment in advanced colorectal cancer. Anticancer Res. 2001;21:3075–3079. [PubMed] [Google Scholar]
- Stuart GR, Glickman BW. Through a glass, darkly: reflections of mutation from lacI transgenic mice. Genetics. 2000;155:1359–1367. doi: 10.1093/genetics/155.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GR, Oda Y, de Boer JG, Glickman BW. Mutation frequency and specificity with age in liver, bladder and brain of lacI transgenic mice. Genetics. 2000;154:1291–1300. doi: 10.1093/genetics/154.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. Faseb J. 2004;18:595–597. doi: 10.1096/fj.03-0890fje. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura H, Kohno T, Wakai K, Nagura K, Genka K, Igarashi H, Morris BJ, Baba S, Ohno Y, Gao C, Li Z, Wang J, Takezaki T, Tajima K, Varga T, Sawaguchi T, Lum JK, Martinson JJ, Tsugane S, Iwamasa T, Shinmura K, Yokota J. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1999;8:669–674. [PubMed] [Google Scholar]
- Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova MV, Khodyreva SN, Lavrik OI. Poly(ADP-ribose) polymerase-1 inhibits strand-displacement synthesis of DNA catalyzed by DNA polymerase beta. Biochemistry (Mosc) 2004;69:558–568. doi: 10.1023/b:biry.0000029855.68502.fa. [DOI] [PubMed] [Google Scholar]
- Sukhanova M, Khodyreva S, Lavrik O. Suppression of base excision repair reactions by apoptotic 24kDa-fragment of poly(ADP-ribose) polymerase 1 in bovine testis nuclear extract. DNA Repair (Amst) 2007;6:615–625. doi: 10.1016/j.dnarep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Szczesny B, Mitra S. Effect of aging on intracellular distribution of abasic (AP) endonuclease 1 in the mouse liver. Mech Ageing Dev. 2005;126:1071–1078. doi: 10.1016/j.mad.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Szilard L. On the Nature of the Aging Process. Proc Natl Acad Sci U S A. 1959;45:30–45. doi: 10.1073/pnas.45.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J Biol Chem. 2005;280:5611–5621. doi: 10.1074/jbc.M408130200. [DOI] [PubMed] [Google Scholar]
- Takanami T, Nakamura J, Kubota Y, Horiuchi S. The Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat Res. 2005;582:135–145. doi: 10.1016/j.mrgentox.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002a;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. Embo J. 2002b;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol Cell Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- Tell G, Damante G, Caldwell D, Kelley MR. The Intracellular Localization of APE1/Ref-1.: More than a Passive Phenomenon? 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- Tiemann-Boege I, Navidi W, Grewal R, Cohn D, Eskenazi B, Wyrobek AJ, Arnheim N. The observed human sperm mutation frequency cannot explain the achondroplasia paternal age effect. Proc Natl Acad Sci U S A. 2002;99:14952–14957. doi: 10.1073/pnas.232568699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Bonk RT, Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- Tomkinson AE, Chen L, Dong Z, Leppard JB, Levin DS, Mackey ZB, Motycka TA. Completion of base excision repair by mammalian DNA ligases. Prog Nucleic Acid Res Mol Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic Tissue-Specific Expression of Mouse Neil3 for Endonuclease VIII-Like Protein. J Biochem (Tokyo) 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Kaufman JM. Ageing of the hypothalamo-pituitary-testicular axis in men. Horm Res. 1995;43:25–28. doi: 10.1159/000184233. [DOI] [PubMed] [Google Scholar]
- Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Xu GL. Cytosine methylation and DNA repair. Curr Top Microbiol Immunol. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- Walter CA, Lu J, Bhakta M, Zhou ZQ, Thompson LH, McCarrey JR. Testis and somatic Xrcc-1 DNA repair gene expression. Somat Cell Mol Genet. 1994;20:451–461. doi: 10.1007/BF02255837. [DOI] [PubMed] [Google Scholar]
- Walter CA, Trolian DA, McFarland MB, Street KA, Gurram GR, McCarrey JR. Xrcc-1 expression during male meiosis in the mouse. Biol Reprod. 1996;55:630–635. doi: 10.1095/biolreprod55.3.630. [DOI] [PubMed] [Google Scholar]
- Walter CA. Mutation frequency declines during spermatogenesis in young mice but increases in old mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10015–10019. doi: 10.1073/pnas.95.17.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter CA, Intano GW, McCarrey JR, McMahan CA, Walter RB. Mutation frequency declines during spermatogenesis in young mice but increases in old mice. Proc Natl Acad Sci U S A. 1998;95:10015–10019. doi: 10.1073/pnas.95.17.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter CA, Intano GW, McMahan CA, Kelner K, McCarrey JR, Walter RB. Mutation spectral changes in spermatogenic cells obtained from old mice. DNA Repair (Amst) 2004;3:495–504. doi: 10.1016/j.dnarep.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Wang G, Hazra TK, Mitra S, Lee HM, Englander EW. Mitochondrial DNA damage and a hypoxic response are induced by CoCl(2) in rat neuronal PC12 cells. Nucleic Acids Res. 2000;28:2135–2140. doi: 10.1093/nar/28.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Spitz MR, Zhu Y, Dong Q, Shete S, Wu X. From genotype to phenotype: correlating XRCC1 polymorphisms with mutagen sensitivity. DNA Repair (Amst) 2003;2:901–908. doi: 10.1016/s1568-7864(03)00085-5. [DOI] [PubMed] [Google Scholar]
- Wang Z-Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ichimura T, Fujita N, Tsuruzoe S, Ohki I, Shirakawa M, Kawasuji M, Nakao M. Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin. Proc Natl Acad Sci U S A. 2003;100:12859–12864. doi: 10.1073/pnas.2131819100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg W. Zur Vererbung des Zwergwuchses. Arch. Rassen-u Gesell Biol. 1912:710–718. [Google Scholar]
- Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog. 2005;42:127–141. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007a;145:1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007b;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K, Jiricny J. In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989;339:234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wilkin DJ, Szabo JK, Cameron R, Henderson S, Bellus GA, Mack ML, Kaitila I, Loughlin J, Munnich A, Sykes B, Bonaventure J, Francomano CA. Mutations in fibroblast growth-factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. Am J Hum Genet. 1998;63:711–716. doi: 10.1086/302000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Sofinowski TM, McNeill DR. Repair mechanisms for oxidative DNA damage. Front Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, Edelmann W. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci U S A. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Xia L, Zheng L, Lee HW, Bates SE, Federico L, Shen B, O'Connor TR. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J Mol Biol. 2005;346:1259–1274. doi: 10.1016/j.jmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, Miller JH. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Iwai S, O'Connor TR, Pfeifer GP. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn RK, Zahn-Daimler G, Ax S, Reifferscheid G, Waldmann P, Fujisawa H, Hosokawa M. DNA damage susceptibility and repair in correlation to calendric age and longevity. Mech Ageing Dev. 2000;119:101–112. doi: 10.1016/s0047-6374(00)00170-6. [DOI] [PubMed] [Google Scholar]
- Zedenius J, Wallin G, Hamberger B, Nordenskjold M, Weber G, Larsson C. Somatic and MEN 2A de novo mutations identified in the RET proto-oncogene by screening of sporadic MTC:s. Hum Mol Genet. 1994;3:1259–1262. doi: 10.1093/hmg/3.8.1259. [DOI] [PubMed] [Google Scholar]
- Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst) 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Zhou ZQ, Walter CA. Expression of the DNA repair gene XRCC1 in baboon tissues. Mutat Res. 1995;348:111–116. doi: 10.1016/0165-7992(95)00053-4. [DOI] [PubMed] [Google Scholar]
- Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]