Abstract
The Bcr oligomerization domain, from the Bcr-Abl oncoprotein, is an attractive therapeutic target for treating leukemias because it is required for cellular transformation. The domain homodimerizes via an antiparallel coiled coil with an adjacent short, helical swap domain. Inspection of the coiled-coil sequence does not reveal obvious determinants of helix-orientation specificity, raising the possibility that the antiparallel orientation preference and/or the dimeric oligomerization state are due to interactions of the swap domains. To better understand how structural specificity is encoded in Bcr, coiled-coil constructs containing either an N- or C-terminal cysteine were synthesized without the swap domain. When crosslinked to adopt exclusively parallel or antiparallel orientations, these showed similar circular dichroism spectra. Both constructs formed coiled-coil dimers, but the antiparallel construct was ~16 °C more stable than the parallel to thermal denaturation. Equilibrium disulfide-exchange studies confirmed that the isolated coiled-coil homodimer shows a very strong preference for the antiparallel orientation. We conclude that the orientation and oligomerization preferences of Bcr are not caused by the presence of the swap domains, but rather are directly encoded in the coiled-coil sequence. We further explored possible determinants of structural specificity by mutating residues in the d position of the coiled-coil core. Some of the mutations caused a change in orientation specificity, and all of the mutations led to the formation of higher-order oligomers. In the absence of the swap domain, these residues play an important role in disfavoring alternate states and are especially important for encoding dimeric oligomerization specificity.
The Bcr-Abl oncoprotein has been extensively studied due to its important role in several leukemias. The protein is formed by a reciprocal chromosomal translocation that fuses the oligomerization domain of Bcr to the tyrosine kinase domain of Abl. In mechanisms that are just beginning to be understood, this leads to dysregulation of Abl through relief of autoinhibition and consequently to aberrant signaling that is responsible for ~95% of chronic myeloid leukemias and 17–30% of acute lymphoblastic leukemias (1–3). The small-molecule kinase inhibitor Gleevec is an effective therapy for many patients, but resistance to the drug can develop quickly, and it is only moderately effective in later stages of disease (4). Alternative treatments that could be used in conjunction with Gleevec would represent a significant advance.
The Bcr oligomerization domain is an attractive therapeutic candidate. It is essential for transformation (5), and exogenous peptides corresponding to the oligomerization domain can suppress the transformed phenotype and increase sensitivity to Gleevec (6). A high-resolution crystal structure of the oligomerization domain reveals a homoteteramer assembled via helix-helix interactions (Figure 1A). Two 36-residue two-stranded antiparallel coiled coils are at the core of the structure. In each monomer, the coiled-coil region is preceded by a shorter helix that associates as a swap domain with the coiled-coil helix of its dimeric partner (Figure 1B). Two such dimers assemble face-to-face to form the tetramer.
FIGURE 1.
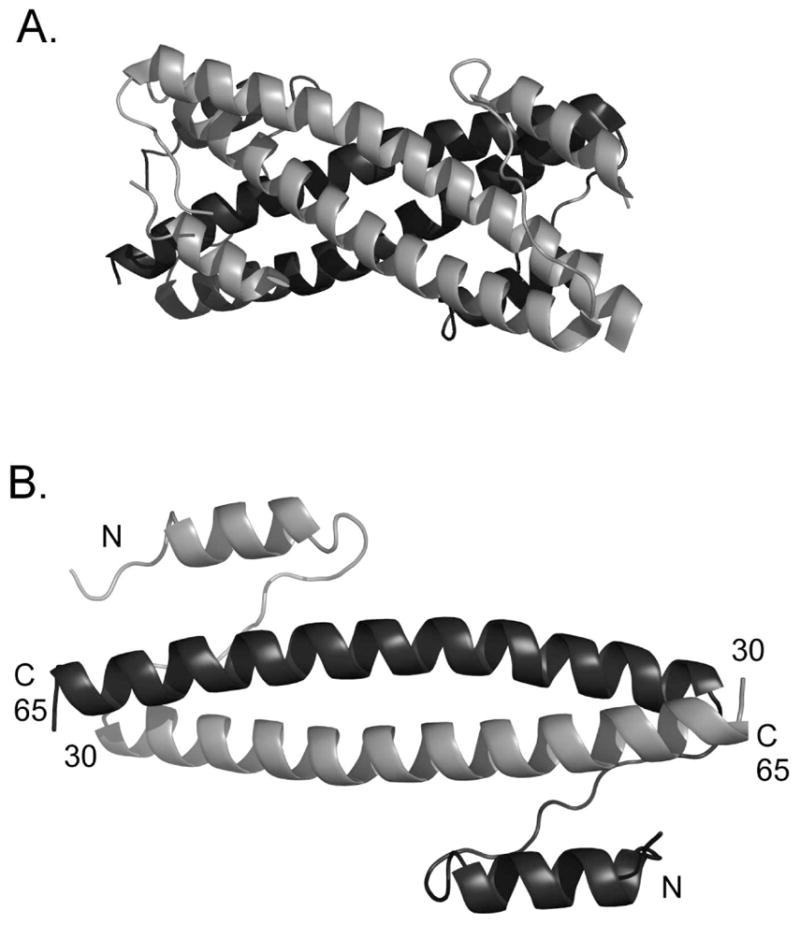
(A) The Bcr oligomerization domain, PDB ID 1K1F (51). (B) View of one of the homodimers that make up the tetramer.
The α-helical coiled-coil motif, which forms the core of the Bcr oligomerization interface, is among the best-studied protein-protein interaction domains (7, 8). It is reasonable to expect that our rich knowledge about coiled coils may aid the search for small-molecule or peptide inhibitors of Bcr-Abl oligomerization. Nevertheless, the coiled-coil sequence of Bcr is unusual, and it is difficult to rationalize how the sequence encodes the structure. In this work we explore a puzzling aspect of Bcr, that is, how the protein specifies formation of a dimeric, antiparallel coiled coil. Because the α-helical coiled coil is among the most common protein motifs found in nature, and because it has proven valuable for understanding interaction specificity in other, non-coiled coil proteins, our studies of basic sequence-structure relationships in Bcr promise to have broad applicability.
The coiled-coil motif consists of two to five α-helices packed together with a left-handed superhelical twist; the helices can associate in either a parallel or antiparallel orientation. Coiled coils play a fundamental functional role in many different proteins, including transcription factors, SNARE complexes, and proteins that mediate viral membrane fusion (7, 8). For many of these proteins, knowledge of whether the helices are arranged in a parallel or antiparallel orientation can be critical in determining how they function. For example, the bZIP transcription factors contain a parallel coiled-coil oligomerization domain (8, 9), but there are other DNA binding proteins that dimerize in an antiparallel orientation (10). Determining the orientation of coiled coils in DNA binding proteins can help in elucidating function and can establish what surfaces are available for other proteins to bind. Structural maintenance of chromosomes (SMC) proteins, which are involved in chromosome condensation, sister chromatid cohesion, gene dosage, and DNA recombination, were originally expected to contain a parallel coiled-coil motif (11–13). After electron micrographs and crystallographic analyses revealed that the helices are actually antiparallel, however, proposed mechanisms had to be re-evaluated (14, 15). In another example, models of coiled coil-mediated membrane fusion are strongly dependent on the orientation of the proteins involved. Mitochondrial fusion proteins contain antiparallel coiled coils (16), whereas SNARE coiled coils are parallel (17, 18). Viral membrane fusion proteins contain a parallel coiled-coil trimer that forms antiparallel interactions with additional helices (19). Understanding the orientation of the coiled coils in each of these cases has precipitated models of membrane fusion (16, 17, 19).
The coiled-coil motif contains a characteristic repeating heptad pattern of amino acids, (abcdefg)n. Hydrophobic a and d residues create a stripe down one side of the α-helix and form the core of a coiled-coil oligomer. Frequently, charged residues at e and g form inter- and/or intrahelical salt bridges (8). In parallel coiled coils, charge complementarity at the e and g positions can impart heterooligomeric specificity (20, 21), and this general principle has been expanded in the design of non-coiled coil proteins (22–24). Similarly, polar residues at a positions in parallel coiled-coil cores, although destabilizing, can provide orientation and oligomerization specificity (25–28). Following this finding, polar residues have been found to impart specificity in globular folds (29–32).
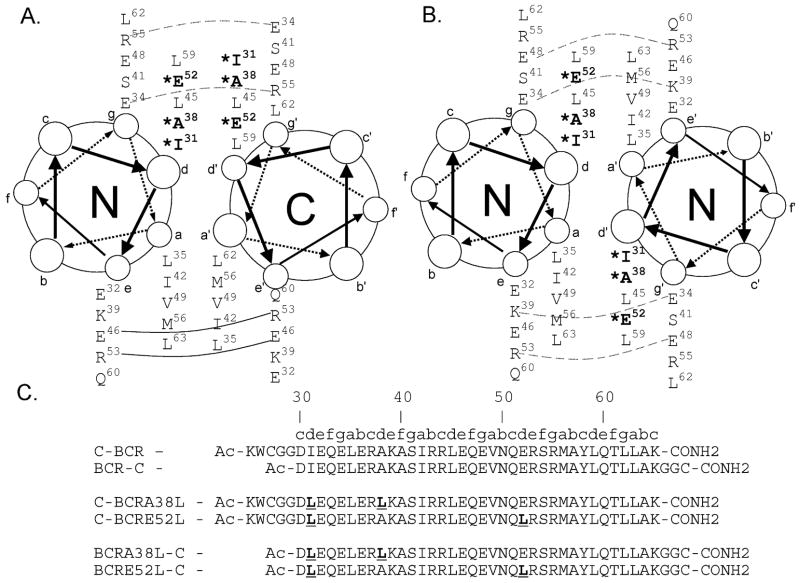
Sequence elements that influence coiled-coil helix-orientation preference are not well understood (33). Parallel and antiparallel coiled coils have very different interactions in the core, with a to a’ and d to d’ for parallel, versus a to d’ for antiparallel coiled coils (where the prime indicates a residue on an opposing helix). Parallel and antiparallel coiled coils also have different interactions on the surface, with g to e’ interactions in parallel versus e to e’ and g to g’ in antiparallel structures (Figure 2A and B) (33). Despite an abundance of both parallel and antiparallel x-ray crystal structures, antiparallel examples are frequently short and intramolecular (34), making it unclear whether the coiled-coil sequence actually encodes helix orientation in these cases.
FIGURE 2.
Helical-wheel diagram of Bcr in an antiparallel (A) and parallel (B) orientation. Mutated positions are shown in bold with an asterisk. (A) Solid lines represent interhelical salt bridges that always form in the crystal structure. Dashed lines represent potential interhelical salt bridges that form in half of the copies in the asymmetric unit. (B) Dashed lines represent potential g to e’ salt bridges that could form in a parallel dimer. (C) Peptides used in this study. C-BCR and BCR-C have the same sequence for residues 30–65 as was used in the crystal structure. Mutated residues are shown in bold and underlined.
A handful of studies have addressed the determinants of helix-orientation specificity and established that the a, d, e, and g residues can each play a role (33). One theory suggests antiparallel coiled coils favor small hydrophobics in the a and d positions because they allow tighter van der Waals packing between the α-helices (35), and another suggests a “steric matching” argument in which helix orientation can be established by the juxtaposition of small and large residues (36–39). Polar residues in the core may participate in specific hydrogen bonding interactions in one state preferentially to another, leading to an orientation preference (40). Charge complementarity of the surface e and g positions can also play a role in determining helix orientation (33, 41–43). Antiparallel coiled coils have been successfully designed by exploiting general principles, such as steric matching and charge complementarity (41, 42, 44, 45). Nevertheless, orientation cannot be accurately predicted from amino-acid sequence for most coiled coils.
Oligomerization specificity is understood somewhat better than helix-orientation specificity, and a few computer programs have been developed that can distinguish two- from three-stranded coiled coils with good success, particularly for parallel examples (46, 47). Some structural features that contribute to oligomerization specificity are well understood. Harbury et al. have demonstrated how hydrophobic packing of β-branched residues can impart strong preferences (28), and the roles of asparagine (25, 26, 28, 48) and lysine (49) residues at the a position and arginine at the d position (50) have also been characterized. Nevertheless, predictions of the oligomerization state of the Bcr domain using the program MULTICOIL incorrectly assign the coiled-coil region as trimeric rather than dimeric (46).
The Bcr oligomerization domain is an example of an important and medically relevant protein for which orientation and oligomerization state cannot be predicted from the amino-acid sequence. The determinants of structural specificity in the Bcr coiled-coil dimers are not obvious in the structure (51) and cannot easily be explained using known general principles. It is possible that the N-terminal swap domain establishes the geometry of the oligomerization interface. It is also possible that the coiled-coil domain adopts a different structure in solution than is observed in the crystal structure. Indeed, solution characterization and crystallographic studies of coiled coils can sometimes yield different results, as for spectrin (52), Coil-Ser (53, 54), and an alanine-zipper peptide (55). Domain swapped proteins have also been reported to change oligomerization state and/or swap region depending upon crystallization conditions (56).
In the first part of this study, we investigated the intrinsic orientation preference of the Bcr coiled coil in solution and found that it is dimeric and maintains a strong preference for the antiparallel orientation, even without the swap domain. To explore the origins of this preference, computational methods, statistical analysis, and structural evaluation were employed. In the second part, we mutated several residues in d positions that are unusual and non-optimal for parallel coiled coils and found that these play an important role in establishing the interaction specificity of Bcr.
MATERIALS AND METHODS
Modeling of Parallel and Antiparallel Bcr and Mutants
The Bcr crystal structure (1K1F, shown in Figure 1) was used for general structural examination. Additionally, the Bcr sequence and numerous mutants were modeled as dimers in both parallel and antiparallel orientations. Several different parallel and antiparallel backbones were created using Crick’s parameterization of coiled coils (57, 58). Native side chains were placed onto these backbones in their optimal conformations with a DEE/A* algorithm, using energies calculated with a molecular mechanics energy function, as in a previous study (23). Coordinates for a model of the native sequence in the parallel orientation are included in the supporting information, Table S1. Structures for the mutants A38L and E52L were based on model Bcr parallel and antiparallel structures, but side chains near the mutation site were reoptimized.
Peptide Design, Synthesis, and Cleavage
Native and mutant Bcr peptides were synthesized using standard Fmoc synthesis and consisted only of the Bcr coiled-coil region (residues 30–65 1K1F). As in the crystal structure, the “native” sequence had several mutations from the wildtype: Cys 38 was mutated to alanine to prevent undesired disulfide bonds; Ile 57 was changed to alanine to disfavor the formation of Bcr tetramers; Phe 54 was changed to serine to eliminate hydrophobic surface exposed as a result of removing the swap domains. All mutant peptides were purchased from Bio-Synthesis, Inc, Lewisville, TX. The peptides were acetylated and amidated to eliminate charges on the termini, and N-terminal KWCGG or C-terminal GGC were added (as shown in Figure 2C). Reduced peptides are indicated with “C” in their name. For example, C-BCRA38L is a peptide with a reduced N-terminal cysteine and position 38 mutated from Ala to Leu. All mutant peptides have Ile 31 changed to Leu. Disulfide-bonded peptides are indicated with a superscript P or AP to indicate parallel or antiparallel helix orientation, respectively. C-BCRA38LP is the oxidized form of C-BCRA38L, whereas BCRA38LAP indicates a construct in which C-BCRA38L and BCRA38L-C are disulfide bonded. Cap-C-BCRA38L indicates C-BCRA38L with the N-terminal cysteine alkylated.
Alkylation of Cysteine Thiol groups
For some experiments, thiol groups were alkylated with iodoacetamide. Peptides were dissolved in 50 mM sodium phosphate buffer, pH 7.2, 2 mM TCEP for 30 minutes. At least a 10-fold molar excess of iodoacetamide was added to the reduced peptide. The solution was stirred in the dark at room temperature for 2 hours. Peptides were HPLC purified following the reaction, and the presence of the acetamide group was confirmed with electrospray mass spectrometry to 2 Da.
Purification and Handling
Prior to purification, peptides were either reduced in 0.2 M Tris/HCl and 100 mM DTT, pH 8.8, for 30 minutes or oxidized in 0.2 M Tris/HCl overnight to form disulfide-linked dimers. Both the reduction and oxidation reactions were quenched by adding acetic acid to a final concentration of 5%, yielding a final pH around 2. Reduced or oxidized peptides were purified on a C18 reverse phase HPLC column using a 0.1%/min acetylnitrile/water gradient with 0.1% trifluoroacetic acid. Samples were immediately frozen with liquid nitrogen after eluting from the HPLC, then lyophilyzed. All peptide solutions were made in an anaerobic chamber with degassed solvents to minimize oxygen exposure. The solutions were kept in the anaerobic chamber until immediately before use. This careful handling procedure eliminated most methionine oxidation problems. The purity of each peptide was greater than 95 percent by analytical HPLC. Masses for the native Bcr peptides were correct to within 1 Da using electrospray mass spectrometry, and the masses for the mutant Bcr peptides were verified by the supplier. Concentrations were determined using the method of Edelhoch (59).
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra from 300 to 200 nm were collected at 25 °C in triplicate on an Aviv circular dichroism spectrometer Model 202 using strain-free quartz cells with a path length of 0.1 cm and an averaging time of 5 seconds. Disulfide-linked peptides were dissolved in degassed buffer (50 mM sodium phosphate, 150 mM NaCl, pH 7.2) in an anaerobic chamber. Thermal unfolding experiments involved monitoring θ222 using a 30 second averaging time, 90 second equilibration time, and temperature increments of 2 °C from 5 to 85 °C. Melts were done in the presence of 2 M GdnHCl. Several consecutive melts were done on the same sample and compared. The Tm, the midpoint of the thermal unfolding curve, was estimated by extrapolating the pre- and post-transition baselines and then determining the temperature for which the CD signal was half of the difference. The Tm for each peptide was reproducible within ~ 1 to 2 °C.
Analytical ultracentrifugation
Alkylated and disulfide-linked peptides, dialyzed against reference buffer (50 mM sodium phosphate, 150 mM NaCl, pH 7.2) in an anaerobic chamber, were spun at 25 °C in a Beckman XL-I analytical ultracentrifuge at 17,000, 20,000, and 23,000 rpm for BCRA38LAP, BCRA38L-C-cap, and cap-C-BCRE52L and 28,000, 31,000, and 34,000 rpm for BCRAP and BCR-CP, for approximately 24 hours at each speed. The following concentrations were used: BCRAP and BCR-CP (5 μM, 25 μM, and 50 μM), BCRA38LAP (25 μM and 50 μM), BCRA38L-C-cap (30 μM, 50 μM, and 100 μM), cap-C-BCRE52L (30 μM, 50 μM, and 100 μM). The contents of each cell were confirmed to be at equilibrium prior to increasing the speed. Data were analyzed using the programs NONLIN (60) and SEDPHAT (61, 62). Various association models were fit, including a single, ideal species, and monomer-oligomer equilibria. The results reported are from single-species fits. Partial specific volumes were calculated from the amino acid sequence (63). Solvent density was calculated by SEDNTERP from its composition (63).
Disulfide-Exchange Experiment
Helix orientation was determined using an equilibrium disulfide-exchange assay (28, 40, 64). Different starting reactants were used to ensure that the products were not kinetically trapped. Peptides were allowed to equilibrate in an anaerobic chamber at room temperature in 50 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA, pH 7.2. The total monomer peptide concentration was 50 μM. Samples were quenched by adding acetic acid to a final concentration of 5% after the reaction reached equilibrium. Samples were considered to be at equilibrium when consecutive HPLC traces showed no change. C-BCRA38LP was sparingly soluble in phosphate buffer, so the assay was performed in the presence of 0.25 M urea and 5% acetonitrile. For the BCRA38L disulfide-exchange experiment, acetonitrile was added to a final concentration of 20% immediately before acetic acid was used to quench the reaction. The products were run on an analytical HPLC, monitored at 229 nm, and the identities of the peaks were confirmed by liquid chromatography-mass spectrometry. Adjustments to the absorbance were made for the difference in the number of amide bonds in the peptides, and an equilibrium constant was calculated.
RESULTS
Biophysical Characterization of the Bcr coiled coil
To probe determinants of oligomerization state and helix-orientation specificity in the Bcr oligomerization domain, peptides corresponding to Bcr residues 30–65 were constructed with either GGC on the C-terminus (BCR-C) or KWCGG on the N-terminus (C-BCR) (64), as shown in Figure 2C. The convention for our abbreviations is defined in the Materials and Methods section. The glycine residues on the peptides provide flexibility in disulfide bond formation, while the tryptophan and lysine residues provide a means to separate and identify the various peptides by HPLC. These constructs contain only the coiled-coil portion of the oligomerization domain, lacking the linker and short “swap domain” helix. The cysteine residues on the termini allow the peptides to be constrained in a parallel or antiparallel orientation via a disulfide bond.
Both the parallel and antiparallel disulfide-constrained peptides are highly helical in phosphate buffer, as shown in Figure 3A. The ratio of the minima at 208 and 222 nm indicates that the helices are associating and is typical of spectra observed for coiled coils (65). The antiparallel orientation is slightly more helical than the parallel orientation and is much more stable to thermal denaturation (Figure 3B). The Tm of the antiparallel construct is ~53 °C, whereas that of the parallel construct is ~37 °C. Equilibrium sedimentation experiments run at and around concentrations used for other characterization studies indicated that both the disulfide-constrained parallel and antiparallel peptides had the molecular weight expected for a two-stranded coiled coil (Figure 4A, 4B, and Table 1).
FIGURE 3.
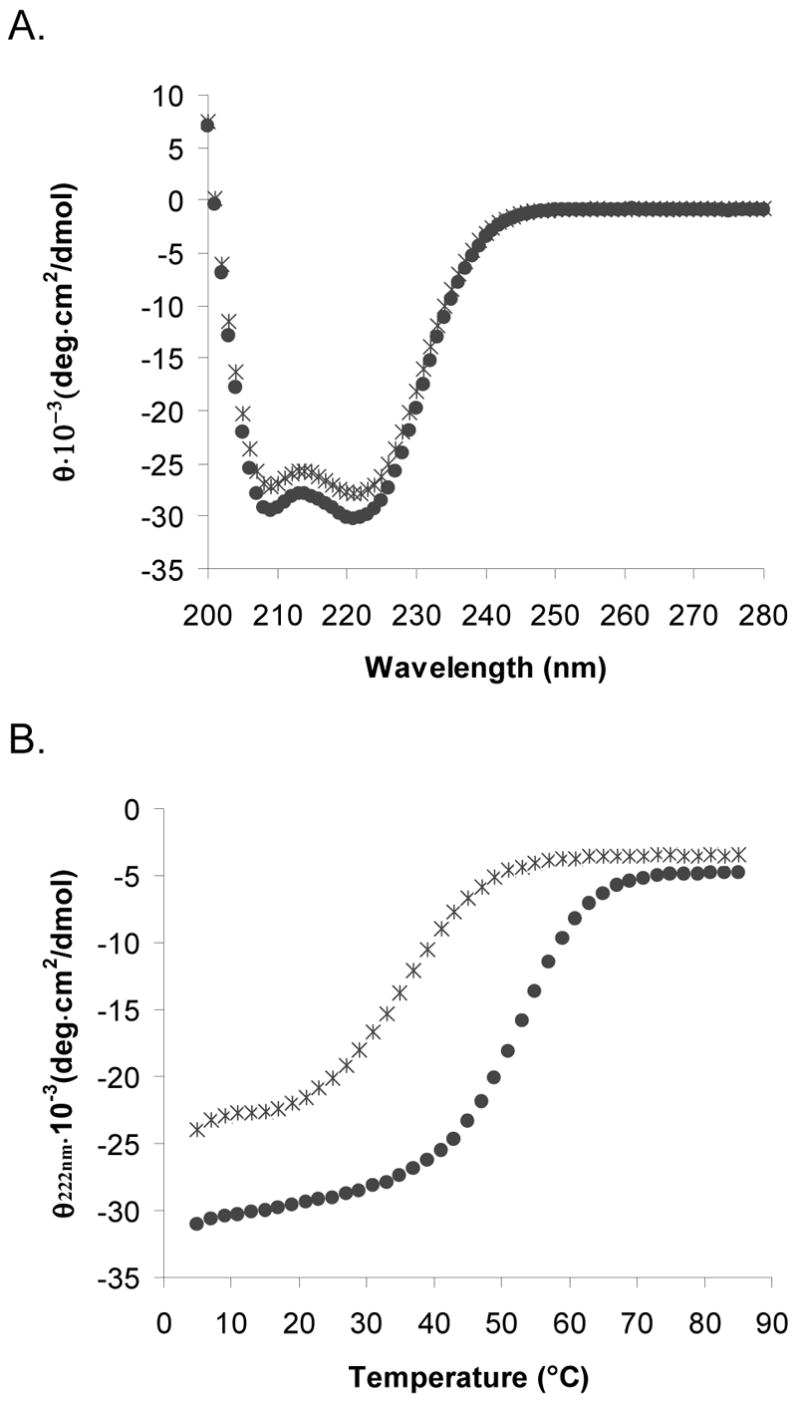
(A) Circular dichroism spectra of BCR-CP (parallel, *) and BCRAP (antiparallel, ●) (25 μM peptide concentration, 50 mM sodium phosphate, 150 mM NaCl, pH 7.2) at 25 °C. (B) Thermal denaturation of BCRAP (antiparallel, ●) and BCRP (parallel, *) monitored at 222 nm in 33 mM sodium phosphate, 100 mM NaCl, 2 M GdnHCl, pH 7.2. Melting temperatures indicate that the antiparallel conformation is more stable than the parallel by ~16 °C.
FIGURE 4.
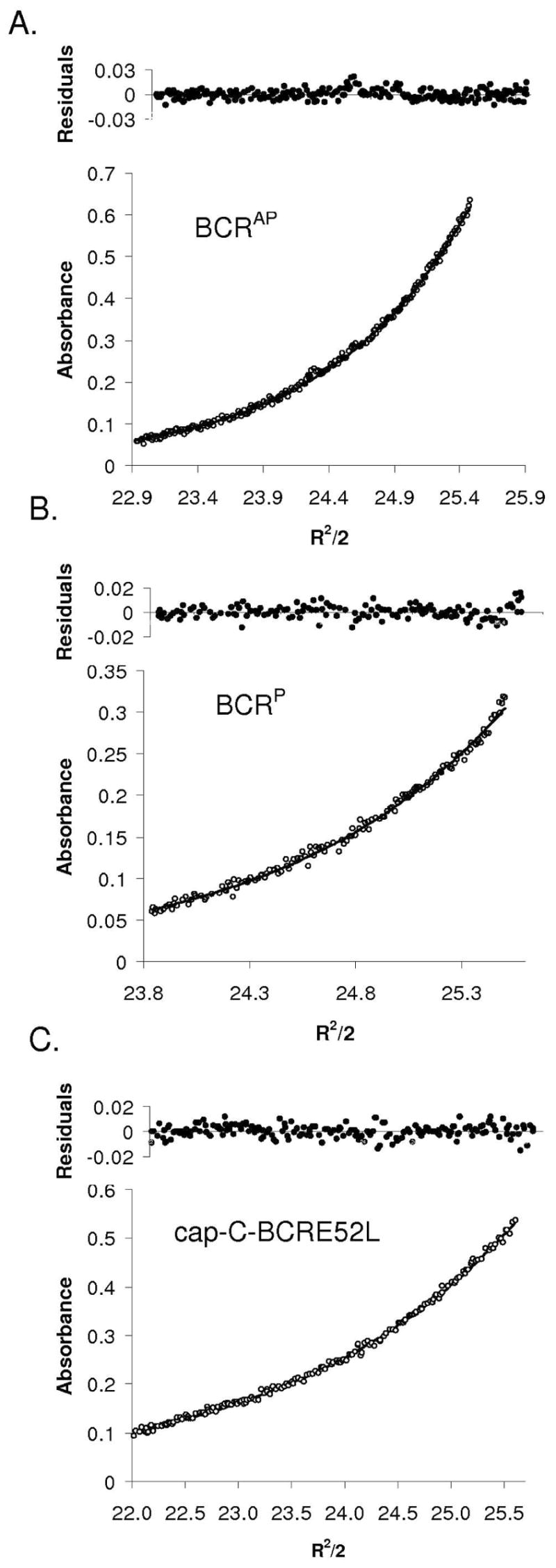
Analytical ultracentrifugation data for Bcr coiled coils. Global fits to data collected at three speeds and three concentrations are shown with representative experimental traces and residuals to the fit. Data shown were collected at 17,000 rpm with 50 μM total monomer concentration. (A) BCRAP and (B) BCRP are both two-stranded coiled coils at 25 μM, in 50 mM sodium phosphate, 150 mM NaCl, pH 7.2; (C) cap-C-BCRE52L is a single-species three-stranded coiled coil at 50 μM in 50 mM sodium phosphate, 150 mM NaCl, pH 7.2. All data were fit with WinNonLin.
Table 1.
Analytical Ultracentrifugation Data Fit to a Single Species Model for Bcr peptides
| BCRAP | BCR-CP | BCRA38LAP | BCRA38L-C-cap | cap-C-BCRE52L | |
|---|---|---|---|---|---|
| v(cm3/g)a | 0.73055 | 0.72908 | 0.734 | 0.73055 | 0.73780 |
| MWcalc | 4731.38 | 4573.19 | 4773.46 | 4733.27 | 4930.61 |
| WinNONLIN | |||||
| MWobs | 10042 | 10395 | 25608c | 26031c | 14093.97 |
| Stoichiometryb | 2.12 | 2.27 | 5.36 | 5.50 | 2.86 |
| SEDPHAT | |||||
| MWobs | 9995 | 10245 | 28029 | 25698.2 | 14111 |
| Stoichiometryc | 2.11 | 2.24 | 5.87 | 5.43 | 2.86 |
Partial specific volumes were calculated as previously described (77). Partial specific volumes of disulfide bonded mixtures are averages of the components.
Stoichiometry is calculated as (MWobs/[MWcalc for a single helix]).
Fit with non-random residuals
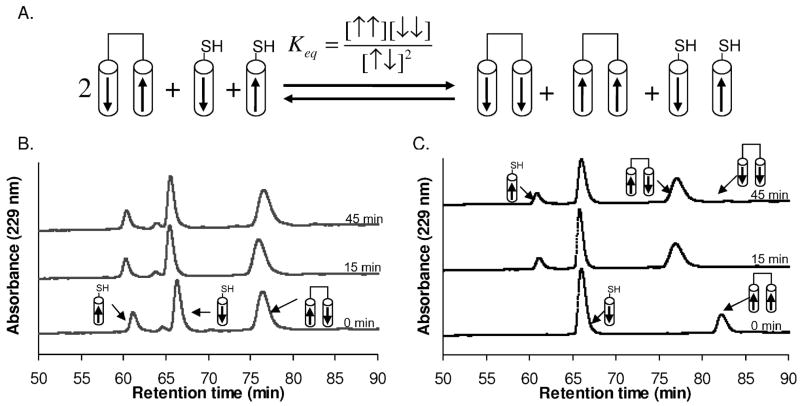
To confirm that the Bcr coiled-coil peptide prefers an antiparallel orientation, oxidized and reduced peptides were mixed together and allowed to equilibrate in an anaerobic atmosphere. A clear antiparallel preference was evident from two experiments with different starting conditions. The first experiment was performed by mixing BCRAP (12.5 μM), C-BCR (12.5 μM), and BCR-C (12.5 μM) at pH 7.2 (Figure 5B). Exchange took place within a few minutes and showed a preference for an antiparallel orientation with Keq = 1.3x10−3. To ensure that the reactants were not kinetically trapped, the disulfide-exchange experiment was repeated by mixing BCR-CP (12.5 μM) with C-BCR (25 μM) (Figure 5C). This experiment gave Keq = 1.5x10−3. These experiments establish that although the Bcr peptides can, in fact, fold as both parallel and antiparallel two-stranded coiled coils, the antiparallel orientation is preferred and both oligomerization state and helix-orientation preference in full-length Bcr are not determined by the swap domain.
FIGURE 5.
Equilibrium disulfide-exchange experiment used to determine the helix orientation of the Bcr coiled coil. (A) N-terminal (C-BCR) and C-terminal (BCR-C) cysteine peptides are represented by cylinders containing arrows that run from N to C terminus. Combinations of disulfide-bonded and reduced peptides were mixed together and allowed to equilibrate at 25 °C, then the reaction was quenched with acetic acid and run on reverse phase HPLC. (B) & (C) Disulfide-exchange reactions were initiated from two conditions. The concentration of different species was monitored as a function of time by HPLC. Both experiments gave similar Keq values, 1.3x10−3 in (B) and 1.5x10−3 in (C), confirming that the reaction had reached an equilibrium strongly favoring the antiparallel species. Integrated peak areas are provided as supporting information in Table S2.
Sequence-based and structural analysis of Bcr
The role of charged residues at e and g positions has been extensively studied as a source of interaction specificity for coiled coils. Surface electrostatics alone have been used successfully to impart orientation specificity (42, 43), and charge complementarity is now a reliable technique for designing an antiparallel coiled coil (37, 38, 44, 45). In Bcr, however, inspection of the e- and g-position residues does not suggest any strongly preferred helix orientation. A helical-wheel diagram (Figure 2A and B) can be used to assess potential interactions in each state. In the parallel orientation, salt bridges typically occur between the g position of one helix and the next e position of a partner helix (e’+). Four such interhelical salt bridges are possible in a parallel Bcr structure, and there are no putative g to e’+ electrostatic repulsions in the parallel state (Figure 2B). The antiparallel x-ray structure of Bcr also contains four interhelical salt bridges and no potentially repulsive g to g’+ or e to e’+ interactions (Figure 2A). Thus, a simple sequence-based analysis of charged surface residues does not strongly favor either orientation.
Coiled-coil orientation specificity is determined by the relative stability of the parallel and antiparallel states. Several d-position residues in the Bcr crystal structure, Ile 31, Ala 38, and Glu 52, are unusual for parallel, dimeric coiled coils, suggesting that these residues may specify an antiparallel state by destabilizing the parallel one. We analyzed the unusual d-position residues by constructing computational models of the native and mutant Bcr sequences on both parallel and antiparallel backbones and by examining the SOCKET database of coiled coils with greater than 15 amino acids. SOCKET identifies coiled coils in the PDB automatically by detecting packing interactions and can be used to derive the frequency with which different amino acids occur in certain heptad positions (34).
The Bcr crystal structure contains an isoleucine (Ile 31) at a d position near both ends of the coiled coil that packs between two hydrophobic leucine residues on the opposing chain. Harbury et al. have demonstrated that β-branched residues, such as isoleucine, do not pack well at the d position in a dimeric parallel orientation and typically lead to the formation of higher-order oligomers (28). Thus, Ile 31 could be an element of negative design, favoring an antiparallel orientation of Bcr by destabilizing the parallel state. On the other hand, the effect of Ile 31 may be mitigated by its location at the end of a helix, where fraying of the ends of the coiled coil may cause it not to be subject to stringent packing requirements. To remove one possible factor disfavoring a parallel state, position 31 was mutated to a leucine in all mutants made. Leucine was chosen because it is found more frequently than any other residue in the d positions of both dimeric parallel and antiparallel coiled coils (34, 66). Modeling on parallel and antiparallel backbones showed that leucine is easily accommodated in the cores of both parallel and antiparallel Bcr dimers.
Ala 38 is another unusual residue at a d position in the Bcr crystal structure that may provide a natural negative design element disfavoring the parallel orientation. In the structure, it interacts weakly with a methionine residue on the opposite chain. In the parallel orientation, two alanines would be directly across from one another, leaving a cavity in the core. The residues above and below the pair of alanines, Ile 31 and Leu 45, are not large enough to reach into the cavity and fill the void space. Analysis of coiled coils in the PDB supports the idea that Ala is highly unfavorable at the d position of parallel coiled coils and may be better accommodated in antiparallel structures. In dimeric coiled coils with greater than 15 amino acids, there are five times more alanines at d positions in antiparallel coiled coils compared to parallel in the SOCKET database (34). Further, our molecular mechanics calculations suggest that although an Ala to Leu mutation at position 38 stabilizes both orientations, it stabilizes the parallel orientation more. There are also more alanine residues at the d position in antiparallel dimers and trimers than in parallel trimers, suggesting an additional possible role in establishing specificity. Thus, we chose to mutate Ala 38 to Leu to test the effects of this change on structure. Leu was again chosen because it is so common in the d position of all coiled coils (34).
Glu 52 is a third d-position residue that may provide an element of negative design disfavoring the parallel orientation. In the crystal structure of Bcr, the glutamates at position 52 reach into solvent and form an intrahelical salt bridge with Arg 55, with the hydrophobic parts of the side chains making some interhelical contacts in the core. In a model parallel structure, the glutamates behave similarly, but pack less well because they are positioned directly across from each other. Interestingly, there are no glutamates in d positions in parallel coiled coils within the SOCKET database, but glutamates are found nearly as often as hydrophobic residues such as Ile, Ala, Met, and Tyr at d positions in antiparallel coiled coils. In our models, changing Glu 52 to Leu improved core packing in the parallel orientation, but was not as easily accommodated in an antiparallel orientation. Using idealized backbones, a leucine at position 52 can be modeled in parallel structures with rotamer conformations that are common in the PDB (67). However, the most energetically favorable packing of leucine in antiparallel structures forces at least one side chain to adopt a statistically uncommon rotamer conformation, indicating that leucine cannot be accommodated without some strain. Therefore, Glu 52 may favor the antiparallel orientation of Bcr via greater destabilization, relative to Leu, of the parallel vs. the antiparallel state. A role for Glu in establishing oligomerization specificity is suggested by the total absence of this residue at the d position of trimers and tetramers in the SOCKET database.
Helix-orientation specificity and characterization of the mutants
To investigate the role of d-position residues Ile 31, Ala 38, and Glu 52, two variants of C-BCR and BCR-C were synthesized. All mutants replaced Ile 31 with Leu. C-BCRA38L and BCRA38L-C additionally contained Leu at position 38, and C-BCRE52L and BCRE52L-C contained Leu at position 52. Sequences are given in Figure 2C.
BCRA38L remained antiparallel, with an equilibrium constant of ~10−4 (Figure 6A) in a disulfide-exchange assay. Because BCRA38L exhibited such a strong antiparallel preference, we characterized the disulfide-linked peptide BCRA38LAP and compared it with BCRAP. Figure 7A shows that BCRA38LAP is slightly more helical than BCRAP at 25 °C, and the ratio of mean residue ellipticities at 208 and 222 nm is typical of coiled coils (65). BCRA38LAP is more stable than BCRAP to thermal denaturation (Figure 7B). The Tm for BCRA38LAP is about 17 °C higher than that for BCRAP.
FIGURE 6.
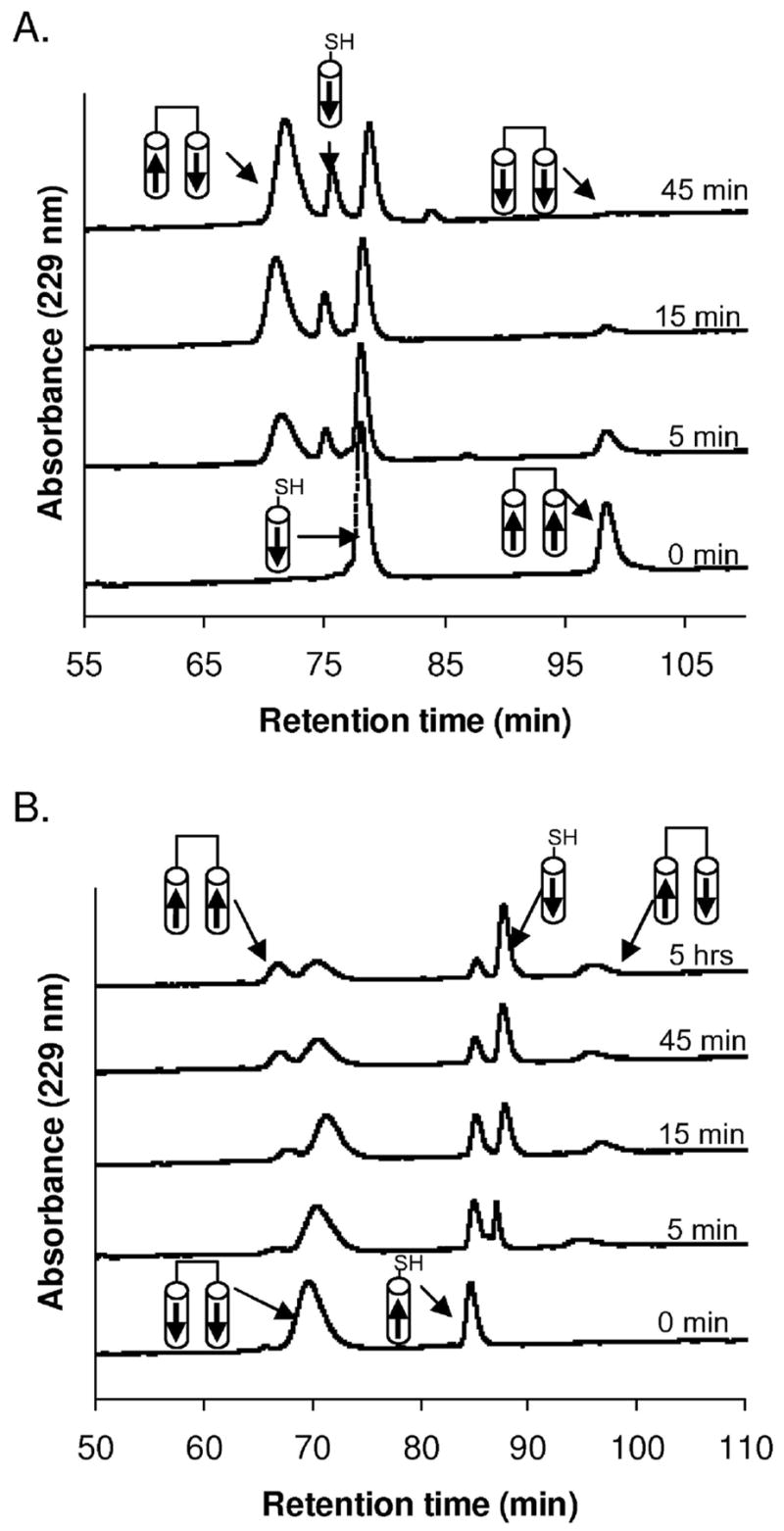
Helix orientation of Bcr mutants determined as in Figure 5. Representative disulfide-exchange data in 50 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA pH 7.2, 25 °C. (A) BCRA38L-CP (12.5 μM) and C-BCRA38L (25 μM); Keq ≈ 10−4. (B) C-BCRE52LP (12.5 μM) and BCRE52L-C (25 μM); Keq ≈ 2.2. Integrated peak areas are provided as supporting information in Table S2.
FIGURE 7.
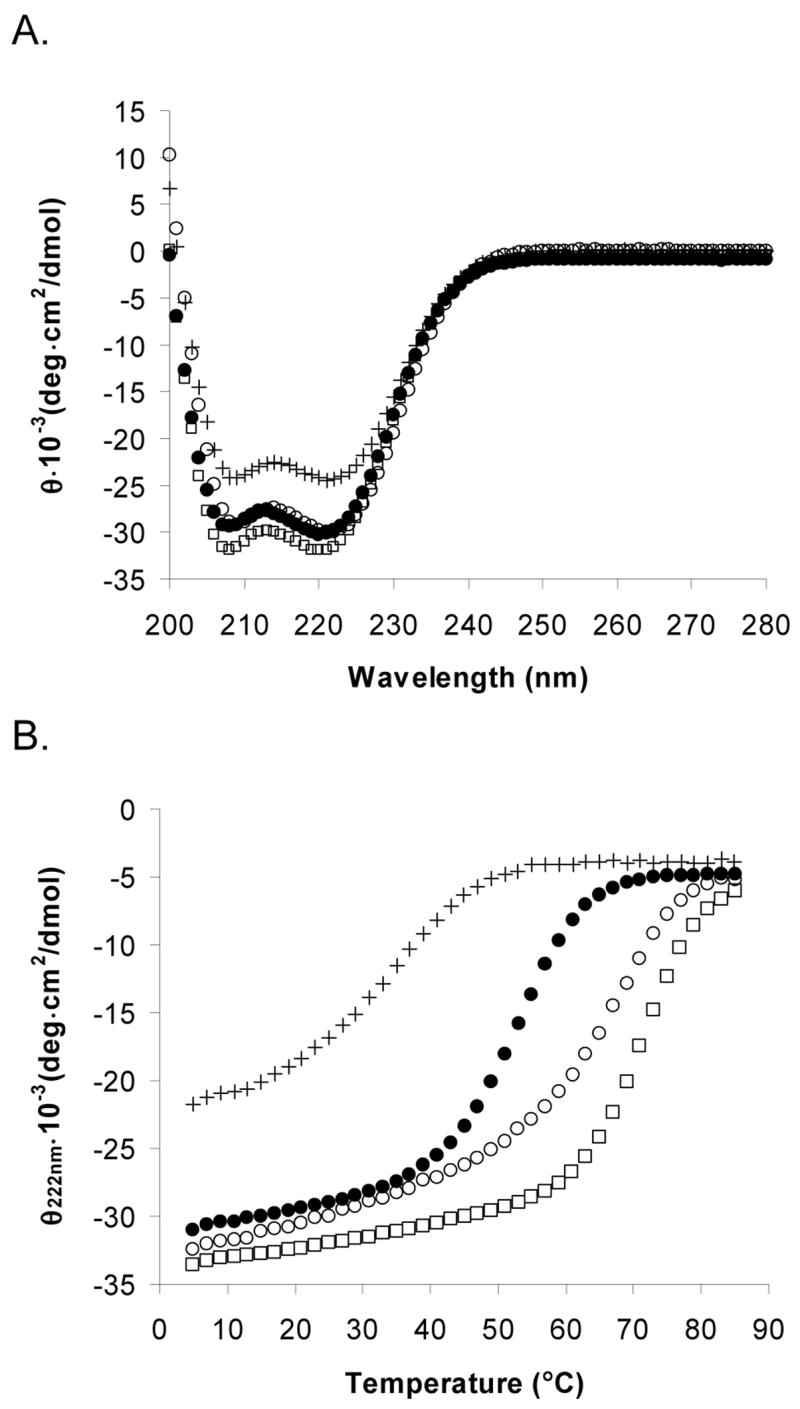
(A) Circular dichroism spectra of BCRA38LAP (□), BCRA38L-C-cap (+), cap-C-BCRE52L (○), compared to BCRAP (●). The peptide concentration was 50 μM for alkylated peptides and 25 μM for disulfide bonded peptides in 50 mM sodium phosphate, 150 mM NaCl, pH 7.2 at 25 °C. (B) Thermal melt of BCRA38LAP (□), BCRA38L-C-cap (+), cap-C-BCRE52L (○), compared to BCRAP (●) monitored at 222 nm. BCRAP was in 33 mM sodium phosphate, 100 mM NaCl, 2 M GdnHCl, pH 7.2. All other peptides were in 50 mM sodium phosphate, 150 mM NaCl, pH 7.2 at 25 °C.
The A38L mutation caused oligomerization specificity to be lost. AUC data for BCRA38LAP did not fit a single-species or two-state model well and gave a weight average close to that of a six-helix species (Table 1). BCRA38L-C-cap still did not fit a single-species model or two-state equilibrium model and had a single-species weight close to BCRA38LAP (Table 1). BCRA38L-C-cap was less helical than BCRA38LAP, as shown in Figure 7A, and was much less stable than either disulfide-bonded BCRA38LAP or BCRAP to thermal denaturation.
In disulfide-exchange experiments, BCRE52L showed a loss of orientation specificity (Figure 6B). The equilibrium constant was determined to be ~2.2, favoring the parallel orientation slightly. Because there was no clear orientation specificity, the cysteine group was capped for biophysical analysis to preclude the formation of a disulfide bond. Cap-C-BCRE52L was as helical as BCRAP, shown in Figure 7A, and the Tm was ~64 °C in the presence of 2 M GdnHCl, indicating this complex was much more stable than BCRAP (Figure 7B). The Glu to Leu mutation also caused a change in oligomerization state; AUC data for cap-C-BCRE52L fit well to a single-species trimer with random residuals (Figure 4C).
DISCUSSION
In this study, we established that even in the absence of the N-terminal swap domain, Bcr constructs remained dimeric and showed a strong preference for an antiparallel orientation. This indicated that structural specificity is encoded directly in the coiled-coil part of the sequence. We went on to examine the effects of several d-position mutations that could potentially be providing a negative-design element disfavoring the competing parallel dimer state or favoring dimers relative to other oligomers (23, 45, 68–71). Elements of negative design have been used to both manually and computationally design coiled coils and helical bundles that adopt different orientations and oligomerization states (23, 37, 44, 45, 68, 71, 72). Further, such features are also important for establishing specificity in native coiled coils (26, 73).
Based on previous studies and our current modeling, isoleucine (or any β-branched amino acid) at d cannot be easily accommodated in parallel dimers due to geometric packing restraints (28). This makes Ile 31 a possible negative design element disfavoring a parallel Bcr dimmer, and we mutated it to leucine in all mutants tested. We did not isolate the effect of this point mutation because we believe that its effect at the end of the coiled coil is probably minimal. It remains possible, therefore, that a single change of Ile to Leu at 31 could give rise to parallel dimers. It is unlikely that the Ile 31 to Leu mutation alone was responsible for the significant changes in oligomerization state that we observed in this study, however. Leucine is very common in both parallel and antiparallel dimers, and it was easily accommodated in models of both states for Bcr.
Poor packing in the core, arising from the opposition of small residues, could provide an element of negative design against parallel helix orientations or higher-order oligomers. There is precedent for both types of effects. The patterning of large and small core residues can change coiled-coil orientation in a tetramer (39), and an antiparallel coiled-coil trimer has been designed using steric matching (pairing alanine with a large non-natural residue) to give the desired orientation (36). Subtle changes in core packing can also cause a change in oligomerization state (28, 74). In a study by Monera et al., the relative placement of Ala and Leu residues was important for oligomerization specificity. Two alanine residues on the same layer in an antiparallel coiled coil formed a dimer, but when the positions of the alanines were staggered such that one layer contained Ala-Leu and the other Leu-Ala, a tetramer formed (74). This suggests it can be more favorable to form two smaller cavities than one large one.
To test the role of a d-position Ala in the context of Bcr, we mutated position 38 to leucine. Position 38 is alanine in the crystal structure of Bcr and in the constructs that we characterized in solution. Note, however, that this residue is cysteine in the native protein. Mutation of Ala 38 to Leu did not change the preference for an antiparallel helix orientation in disulfide-exchange reactions, but it did lead to a change in oligomerization specificity and stability. Ala at position 38 of Bcr may preclude higher-order oligomerization due to an energetic penalty for large cavities in the core, consistent with the formation of higher-order oligomers upon mutation to Leu. BCRA38LAP was also more helical and stable than BCRAP. Increasing hydrophobic content in proteins is frequently stabilizing (75), and indeed, changing an alanine to a leucine at a d position in a model coiled-coil homodimer designed by Moitra et al. stabilized the complex by 9.2 kcal/mol and increased the Tm by ~ 30 °C (76). In Bcr, an Ala to Leu substitution apparently stabilized antiparallel helix arrangements more than parallel ones, as reflected by the somewhat increased antiparallel preference measured in the disulfide-exchange reactions.
Along with constraints on hydrophobic packing, polar residues in the core can be critical for disfavoring an undesired state. For example, Oakley and Kim showed that if the location of an a-position Asn is moved such that a buried Asn-Asn hydrogen bond can form only in the antiparallel orientation, this state is strongly preferred (40). However, asparagines are not found in the cores of antiparallel dimeric coiled coils very often (34), and Asn-Asn interactions at the a position have not been observed in any antiparallel coiled-coil crystal structures thus far (33). Asn-Asn hydrogen bonds are seen very frequently in parallel coiled coils and probably play a large role in giving some coiled coils a parallel orientation.
Polar or charged residues other than Asn could also provide a mechanism for negative design against a particular orientation and, therefore, be important for establishing helix orientation. For example, this idea has been explored by Campbell and Lumb in a model dimeric coiled coil. In their study, a Lys in an a position interacting with a charged residue at g’ gave oligomerization specificity. However, this combination of residues did not impart a specific helix orientation, due to the favorable interactions lysine could make in both the parallel and antiparallel states (49). In the context of a different model heterodimer, a buried Arg at a d position, with the potential to interact with a Glu at g’, was not sufficient to specify helix orientation, but did give a specific dimer (50). Another study took an antiparallel coiled-coil mitochondrial fusion protein with two glutamates in d positions and mutated one at a time to leucine. Membrane fusion was adversely affected by these mutations. The cause of the loss of function was not apparent, however, as no structural analysis of the mutants was done (16).
Glutamate is common at the d position in antiparallel coiled-coil dimers, but is rarely found in parallel dimers or in any trimers; thus, it may have a special structural role. In C-BCRE52L, Glu 52 was replaced by Leu, and both dimerization specificity and the propensity to adopt a unique helix orientation were lost. Although Ile 31 was also changed to Leu in this mutant, the I31L mutation did not alter orientation preference in the context of BCRA38L. Therefore, the change in orientation specificity seen in BCRE52L is likely due to removal of the core glutamate. Cap-C-BCRE52L was a single-species trimer by analytical ultracentrifugation. An up-up-down trimer, in which one α-helix is oriented antiparallel relative to two adjacent parallel helices, is consistent with all of the biophysical data. With a helical bundle in this topology, it would be possible to get approximately equal amounts of parallel and antiparallel helix pairs in the disulfide-exchange experiments. Alternatively, cap-C-BCRE52L may form a mixture of trimeric species with different helix orientations.
Glu 52 is clearly important for establishing structural specificity in the Bcr oligomerization domain. It is easy to understand why glutamate residues at d positions might prevent the formation of higher-order complexes. In a dimer, the glutamate carboxyl groups can reach at least partially out of the core and interact with water. However, these charged atoms would be almost completely buried in a coiled-coil trimer or tetramer, at a significant cost in solvation energy. Whether or how Glu 52 plays a negative design role in disfavoring a parallel dimer state relative to an antiparallel one is less clear, despite the fact that there is a statistical preference for d-position glutamate residues to be antiparallel (34). In the x-ray structure of Bcr, Glu 52 is partially solvent exposed and interacts with Arg 55 on the same helix; the hydrophobic part of the side chain packs between Ile 42 and Ser 41 on the opposite chain. In models of parallel dimeric versions of Bcr, this residue can interact with Arg 55 on the same helix or with Arg 53 on the opposite helix, and the hydrophobic parts of the two Glu 52 residues interact weakly in the core. It is likely a tradeoff between exposing the charged carboxlate vs. forming good hydrophobic contacts in the core that makes glutamate unfavorable at parallel d positions. By visual inspection, interdigitation of a- and d-position residues in antiparallel structures appears to improve packing while maintaining solvent exposure.
Numerous studies have used protein design as a way to explore determinants of coiled-coil structural specificity, and this has been a powerful and effective approach (21, 28, 33, 39, 40, 49, 50, 68, 74). However, general principles uncovered in model systems do not always explain the specificity observed in many native coiled-coil proteins. Some interactions that are effective in designed proteins are rare or even unprecedented in naturally occurring ones (40, 68). In this work, we examined determinants of structural specificity in the Bcr oligomerization domain and found them to be subtle, with some residues playing multiple roles. The influence of charged residues, such as Glu 52, is likely to be context dependent, varying according to the precise interactions that can be formed in multiple competing states. Improved methods for predicting helix-orientation specificity and coiled-coil oligomerization state may need to address this complexity explicitly.
Supplementary Material
Supporting Information Available Coordinates for a model of the native sequence in the parallel orientation and integrated peak areas for the disulfide-exchange experiments are provided as supplemental information. This information is available free of charge via the Internet at: http://pubs.acs.org.
Acknowledgments
We acknowledge the use of the MIT Computational and Systems Biology Initiative Proteomics/Structural Biology Core and High-Performance Computing Facility, and the MIT Center for Environmental Health Sciences Bioanalytical Facility. We thank P. Kim and M. Burgess for BCR-C and C-BCR. K. Taghizadeh, D. Pheasant, M. Ali, J. Glover, D. Pamuk, D. Lee, E. Oakes, G. Grigoryan and D. Mujumdar for experimental and computational assistance and/or helpful discussions. We thank members of the Keating Lab, G. Hersch, T. Keating, and T. Schwartz for helpful comments on the manuscript.
Footnotes
This work was supported by a grant from the NIH (GM67681). C.M.T is an Anna Fuller predoctoral fellow.
Abbreviations: TCEP, (Tris(2-carboxyethyl)phosphine hydrochloride); DTT, (dithiothreitol); BCR, (breakpoint cluster region); SMC, (structural maintenance of chromosomes); Tm, (melting temperature); CD, (circular dichroism); GdnHCl, (guanidinium hydrochloride); AUC, (analytical ultracentrifugation); DEE, (dead-end elimination algorithm); CML, (chronic myeloid leukemia); ALL, (acute lymphoblastic leukemia); HPLC, (high performance liquid chromatography)
References
- 1.Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–40. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 2.Catovsky D. Ph1-positive acute leukaemia and chronic granulocytic leukaemia: one or two diseases? Br J Haematol. 1979;42:493–8. doi: 10.1111/j.1365-2141.1979.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 3.Smith KM, Yacobi R, Van Etten RA. Autoinhibition of Bcr-Abl through its SH3 domain. Mol Cell. 2003;12:27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nardi V, Azam M, Daley GQ. Mechanisms and implications of imatinib resistance mutations in BCR-ABL. Curr Opin Hematol. 2004;11:35–43. doi: 10.1097/00062752-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 5.McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol. 1993;13:7587–95. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beissert T, Puccetti E, Bianchini A, Guller S, Boehrer S, Hoelzer D, Ottmann OG, Nervi C, Ruthardt M. Targeting of the N-terminal coiled coil oligomerization interface of BCR interferes with the transformation potential of BCR-ABL and increases sensitivity to STI571. Blood. 2003;102:2985–93. doi: 10.1182/blood-2003-03-0811. [DOI] [PubMed] [Google Scholar]
- 7.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–8. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 8.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–6. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 9.O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray Structure of the GCN4 Leucine Zipper, a Two-Stranded, Parallel Coiled Coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 10.Bussiere DE, Bastia D, White SW. Crystal structure of the replication terminator protein from B. subtilis at 2.6 A. Cell. 1995;80:651–60. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- 11.Peterson CL. The SMC family: novel motor proteins for chromosome condensation? Cell. 1994;79:389–92. doi: 10.1016/0092-8674(94)90247-x. [DOI] [PubMed] [Google Scholar]
- 12.Saitoh N, Goldberg I, Earnshaw WC. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. Bioessays. 1995;17:759–66. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- 13.Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–58. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 14.Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe J, Cordell SC, van den Ent F. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J Mol Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- 16.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–62. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 17.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 18.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–53. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 19.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 20.O'Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- 21.O'Shea EK, Lumb KJ, Kim PS. Peptide 'Velcro*': design of a heterodimeric coiled coil. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 22.Hendsch ZS, Nohaile MJ, Sauer RT, Tidor B. Preferential heterodimer formation via undercompensated electrostatic interactions. J Am Chem Soc. 2001;123:1264–5. doi: 10.1021/ja0032273. [DOI] [PubMed] [Google Scholar]
- 23.Ali MH, Taylor CM, Grigoryan G, Allen KN, Imperiali B, Keating AE. Design of a heterospecific, tetrameric, 21-residue miniprotein with mixed alpha/beta structure. Structure. 2005;13:225–34. doi: 10.1016/j.str.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Nohaile MJ, Hendsch ZS, Tidor B, Sauer RT. Altering dimerization specificity by changes in surface electrostatics. Proc Natl Acad Sci U S A. 2001;98:3109–14. doi: 10.1073/pnas.051624498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lumb KJ, Kim PS. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez L, Jr, Woolfson DN, Alber T. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 27.Akey DL, Malashkevich VN, Kim P. Buried polar residues in coiled-coil interfaces. Biochemistry. 2001;40:6352–6360. doi: 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- 28.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 29.Efimov AV, Kondratova MS. [A comparative analysis of interhelical polar interactions of various alpha-helix packings in proteins] Mol Biol (Mosk) 2003;37:515–21. [PubMed] [Google Scholar]
- 30.Bolon DN, Mayo SL. Polar residues in the protein core of Escherichia coli thioredoxin are important for fold specificity. Biochemistry. 2001;40:10047–53. doi: 10.1021/bi010427y. [DOI] [PubMed] [Google Scholar]
- 31.Hendsch ZS, Tidor B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994;3:211–26. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozwarski DA, Gronenborn AM, Clore GM, Bazan JF, Bohm A, Wlodawer A, Hatada M, Karplus PA. Structural comparisons among the short-chain helical cytokines. Structure. 1994;2:159–73. doi: 10.1016/s0969-2126(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 33.Oakley MG, Hollenbeck JJ. The design of antiparallel coiled coils. Curr Opin Struct Biol. 2001;11:450–7. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 34.Walshaw J, Woolfson DN. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001;307:1427–50. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 35.Gernert KM, Surles MC, Labean TH, Richardson JS, Richardson DC. The Alacoil: a very tight, antiparallel coiled-coil of helices. Protein Sci. 1995;4:2252–60. doi: 10.1002/pro.5560041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnarr NA, Kennan AJ. Strand orientation by steric matching: a designed antiparallel coiled-coil trimer. J Am Chem Soc. 2004;126:14447–51. doi: 10.1021/ja047496v. [DOI] [PubMed] [Google Scholar]
- 37.Betz SF, DeGrado WF. Controlling topology and native-like behavior of de novo-designed peptides: design and characterization of antiparallel four-stranded coiled coils. Biochemistry. 1996;35:6955–62. doi: 10.1021/bi960095a. [DOI] [PubMed] [Google Scholar]
- 38.Gurnon DG, Whitaker JA, Oakley MG. Design and characterization of a homodimeric antiparallel coiled coil. J Am Chem Soc. 2003;125:7518–9. doi: 10.1021/ja0357590. [DOI] [PubMed] [Google Scholar]
- 39.Monera O, Zhou N, Lavigne P, Kay C, Hodges R. Formation of parallel and antiparallel coiled-coils controlled by the relative positions of alanine residues in the hydrophobic core. J Biol Chem. 1996;271:p3995–4001. doi: 10.1074/jbc.271.8.3995. [DOI] [PubMed] [Google Scholar]
- 40.Oakley MG, Kim PS. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry. 1998;37:12603–10. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- 41.Monera OD, Zhou NE, Kay CM, Hodges RS. Comparison of antiparallel and parallel two-stranded alpha-helical coiled-coils. Design, synthesis, and characterization. J Biol Chem. 1993;268:19218–27. [PubMed] [Google Scholar]
- 42.Monera O, Kay C, Hodges R. Electrostatic interactions control the parallel and antiparallel orientation of alpha-helical chains in two-stranded alpha-helical coiled-coils. Biochemistry. 1994;33:p3862–71. doi: 10.1021/bi00179a010. [DOI] [PubMed] [Google Scholar]
- 43.McClain DL, Binfet JP, Oakley MG. Evaluation of the energetic contribution of interhelical Coulombic interactions for coiled coil helix orientation specificity. J Mol Biol. 2001;313:371–83. doi: 10.1006/jmbi.2001.5044. [DOI] [PubMed] [Google Scholar]
- 44.McClain DL, Woods HL, Oakley MG. Design and characterization of a heterodimeric coiled coil that forms exclusively with an antiparallel relative helix orientation. J Am Chem Soc. 2001;123:3151–2. doi: 10.1021/ja004099l. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh I, Hamilton A, Regan L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 46.Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woolfson DN, Alber T. Predicting oligomerization states of coiled coils. Protein Sci. 1995;4:1596–607. doi: 10.1002/pro.5560040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akey DL, Malashkevich VN, Kim PS. Buried polar residues in coiled-coil interfaces. Biochemistry. 2001;40:6352–6360. doi: 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- 49.Campbell KM, Lumb KJ. Complementation of buried lysine and surface polar residues in a designed heterodimeric coiled coil. Biochemistry. 2002;41:7169–75. doi: 10.1021/bi025559l. [DOI] [PubMed] [Google Scholar]
- 50.McClain DL, Gurnon DG, Oakley MG. Importance of potential interhelical salt-bridges involving interior residues for coiled-coil stability and quaternary structure. J Mol Biol. 2002;324:257–70. doi: 10.1016/s0022-2836(02)01072-0. [DOI] [PubMed] [Google Scholar]
- 51.Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat Struct Biol. 2002 doi: 10.1038/nsb747. [DOI] [PubMed] [Google Scholar]
- 52.Yan Y, Winograd E, Viel A, Cronin T, Harrison SC, Branton D. Crystal structure of the repetitive segments of spectrin. Science. 1993;262:2027–30. doi: 10.1126/science.8266097. [DOI] [PubMed] [Google Scholar]
- 53.Lovejoy B, Choe S, Cascio D, McRorie DK, DeGrado WF, Eisenberg D. Crystal structure of a synthetic triple-stranded alpha-helical bundle. Science. 1993;259:1288–93. doi: 10.1126/science.8446897. [DOI] [PubMed] [Google Scholar]
- 54.O'Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–51. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Lu M. An alanine-zipper structure determined by long range intermolecular interactions. J Biol Chem. 2002;277:48708–13. doi: 10.1074/jbc.M208773200. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–99. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harbury PB, Tidor B, Kim PS. Repacking protein cores with backbone freedom: structure prediction for coiled coils. Proc Natl Acad Sci U S A. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crick FHC. The packing of a-helices: simple coiled coils. Acta Cryst. 1953;6:689–697. [Google Scholar]
- 59.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 60.Johnson ML, Correia JC, Yphantis DA, Halvorson HR. Analysis of Data from the Analytical Ultracentrifuge by Nonlinear Least-Squares Techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuck P. On the Analysis of Protein Self-Association by Sedimentation Velocity Analytical Ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 62.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Roualt TA, Schuck P. Sedimentation Equilibrium Analysis of Protein Interactions with Global Implicit Mass Conservation Constraints and Systematic Noise Decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 63.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, U. K: 1992. Computer aided interpretation of analytical sedimentation data for proteins; pp. 90–125. [Google Scholar]
- 64.O'Shea EK, Rutkowski R, Stafford WFd, Kim PS. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245:646–8. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- 65.Zhou NE, Zhu BY, Kay CM, Hodges RS. The two-stranded alpha-helical coiled-coil is an ideal model for studying protein stability and subunit interactions. Biopolymers. 1992;32:419–26. doi: 10.1002/bip.360320419. [DOI] [PubMed] [Google Scholar]
- 66.Hu JC, O'Shea EK, Kim PS, Sauer RT. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–3. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 67.Dunbrack RL, Jr, Karplus M. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J Mol Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 68.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 69.Pokala N, Handel TM. Energy functions for protein design: adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J Mol Biol. 2005;347:203–27. doi: 10.1016/j.jmb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 70.Park S, Yang X, Saven JG. Advances in computational protein design. Curr Opin Struct Biol. 2004;14:487–94. doi: 10.1016/j.sbi.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 71.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. Computational de novo design, and characterization of an A(2)B(2) diiron protein. J Mol Biol. 2002;321:923–38. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 72.Nautiyal S, Woolfson DN, King DS, Alber T. A designed heterotrimeric coiled coil. Biochemistry. 1995;34:11645–51. doi: 10.1021/bi00037a001. [DOI] [PubMed] [Google Scholar]
- 73.Campbell KM, Sholders AJ, Lumb KJ. Contribution of buried lysine residues to the oligomerization specificity and stability of the fos coiled coil. Biochemistry. 2002;41:4866–71. doi: 10.1021/bi0159276. [DOI] [PubMed] [Google Scholar]
- 74.Monera O, Sonnichsen F, Hicks L, Kay C, Hodges R. The relative positions of alanine residues in the hydrophobic core control the formation of two-stranded or four-stranded alpha-helical coiled-coils. Protein Eng. 1996;9:p353–63. doi: 10.1093/protein/9.4.353. [DOI] [PubMed] [Google Scholar]
- 75.Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C. A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry. 2002;41:14122–31. doi: 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- 76.Moitra J, Szilak L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36:12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 77.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-Aided Interpretation of Analytical Sedimentation Data for Proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available Coordinates for a model of the native sequence in the parallel orientation and integrated peak areas for the disulfide-exchange experiments are provided as supplemental information. This information is available free of charge via the Internet at: http://pubs.acs.org.