Abstract
Integral membrane proteins are predicted to play key roles in the biogenesis and function of nuclear pore complexes (NPCs). Revealing how the transport apparatus is assembled will be critical for understanding the mechanism of nucleocytoplasmic transport. We observed that expression of the carboxyl-terminal 200 amino acids of the nucleoporin Nup116p had no effect on wild-type yeast cells, but it rendered the nup116 null strain inviable at all temperatures and coincidentally resulted in the formation of nuclear membrane herniations at 23°C. To identify factors related to NPC function, a genetic screen for high-copy suppressors of this lethal nup116-C phenotype was conducted. One gene (designated SNL1 for suppressor of nup116-C lethal) was identified whose expression was necessary and sufficient for rescuing growth. Snl1p has a predicted molecular mass of 18.3 kDa, a putative transmembrane domain, and limited sequence similarity to Pom152p, the only previously identified yeast NPC-associated integral membrane protein. By both indirect immunofluorescence microscopy and subcellular fractionation studies, Snl1p was localized to both the nuclear envelope and the endoplasmic reticulum. Membrane extraction and topology assays suggested that Snl1p was an integral membrane protein, with its carboxyl-terminal region exposed to the cytosol. With regard to genetic specificity, the nup116-C lethality was also suppressed by high-copy GLE2 and NIC96. Moreover, high-copy SNL1 suppressed the temperature sensitivity of gle2–1 and nic96-G3 mutant cells. The nic96-G3 allele was identified in a synthetic lethal genetic screen with a null allele of the closely related nucleoporin nup100. Gle2p physically associated with Nup116p in vitro, and the interaction required the N-terminal region of Nup116p. Therefore, genetic links between the role of Snl1p and at least three NPC-associated proteins were established. We suggest that Snl1p plays a stabilizing role in NPC structure and function.
INTRODUCTION
Nuclear pore complexes (NPCs)1 are large proteinaceous assemblies that provide the only known portals for the nucleocytoplasmic transport of macromolecules. A requisite step in the assembly of an NPC is the formation of a pore through the nuclear envelope, presumably via the regulated fusion of the outer and inner nuclear membranes. The mechanism by which this fusion event occurs and the subsequent factors required for the coordinated assembly of the distinct substructures of an NPC have not been elucidated. High-resolution cryoelectron microscopy and transmission scanning electron microscopy studies have revealed the major structural components of the vertebrate NPC (Ris, 1991; Akey, 1995; Goldberg and Allen, 1995; Pante and Aebi, 1996), with three-dimensional reconstructions resolving the basic framework to ∼10 nm (Hinshaw et al., 1992; Akey and Radermacher, 1993). The overall architecture is based on eight radially symmetrical spokes that are sandwiched between two rings anchoring the filamentous structures on both the cytoplasmic and nuclear faces. The spokes also appear connected to both an inner spoke ring encompassing the cytoplasmic central plug and an outer ring in the nuclear envelope lumen. The latter suggests a requirement for integral membrane proteins to traverse the pore membrane. Moreover, models for NPC biogenesis have suggested that interactions between integral membrane proteins may be required for formation of the pore, and integral membrane proteins are presumed essential for anchoring an NPC in the pore (Macaulay and Forbes, 1996; Goldberg et al., 1997).
On the basis of an estimated molecular mass of more than 108 daltons and the polypeptide complexity of purified yeast NPCs (Reichelt et al., 1990; Rout and Blobel, 1993), an NPC may comprise at least 50 different polypeptides. To date, more than 20 yeast NPC-associated peripheral membrane proteins (nucleoporins) and one yeast pore-associated integral membrane protein have been identified (reviewed in Corbett and Silver, 1997; Doye and Hurt, 1997). A precise understanding of NPC-mediated transport will depend not only on revealing the complete biochemical composition of an NPC, but also on integrating the location and assembly interactions of such proteins into the context of NPC architecture.
Two basic approaches have been used in attempts to identify NPC assembly factors: analysis of yeast nucleoporin mutants for perturbations of NPC structure and characterization of in vitro NPC assembly in Xenopus laevis egg extracts. With regard to the latter, vertebrate cell-free systems have provided excellent models for studying the assembly of nuclear structures (Lohka and Masui, 1983; Burke and Gerace, 1986; Newport, 1987). Mitotic NPC assembly and disassembly can be reconstituted in vitro, and a general framework for the stages of NPC assembly has been revealed. Nuclear pore and NPC formation requires the prior assembly of a double nuclear membrane (Macaulay and Forbes, 1996), and is blocked by the addition of GTPγS, BAPTA, or NEM (Newmeyer and Forbes, 1990; Pfaller et al., 1991; Boman et al., 1992a,b; Newport and Dunphy, 1992; Vigers and Lohka, 1992; Sullivan et al., 1993; Macaulay and Forbes, 1996; Goldberg et al., 1997). Depletion of either vesicular or soluble components from the in vitro extract can also prevent NPC formation (Sheehan et al., 1988; Dabauvalle et al., 1990; Finlay and Forbes, 1990; Finlay et al., 1991; Vigers and Lohka, 1991). These studies suggest that both integral and peripheral membrane proteins are essential mediators of assembly.
A striking array of different NPC structural perturbations has been observed in numerous mutant yeast strain backgrounds (reviewed in Rout and Wente, 1994; Corbett and Silver, 1997; Doye and Hurt, 1997; Wente et al., 1997). Perturbations include clustering of NPCs in localized patches, decreased NPC number per nucleus, the presence of intranuclear annulate lamellae, and extensive lobulation of the nuclear envelope. In previous studies, we characterized an unusual temperature-sensitive structural perturbation of yeast NPCs lacking the nucleoporin Nup116p (Wente and Blobel, 1993). Although the mutant NPCs were still anchored to the inner nuclear membrane, the outer membrane became detached and the inner membrane appeared continuous over the cytoplasmic face of the NPC. Temperature-sensitive alleles of two other genes that encode NPC-associated proteins, npl4–2 and gle2–1, also exhibit similar nuclear membrane/NPC herniations (DeHoratius and Silver, 1996; Murphy and Wente, 1996). The absence or alteration of these proteins may have affected NPC biogenesis or the stability of intact NPCs and the surrounding pore membrane. In our model describing the nup116Δ phenotype, we speculated that a membrane fusion event involving the pore membrane resulted in the herniation structures (Wente and Blobel, 1993). Revealing the structural basis for herniation formation in such mutant cells may provide insight into the pathways for maintaining NPC and nuclear pore structure.
With our long-range goal aimed at determining nucleoporin roles in transport and/or NPC assembly events, recent efforts have focused on revealing the function of each structural region in Nup116p. Nup116p is a member of the GLFG family of nucleoporins, characterized by a region containing 33 repeats of the tetrapeptide glycine-leucine-phenylalanine-glycine (GLFG) (Figure 1A) (Wente et al., 1992; Wimmer et al., 1992). This particular GLFG region is essential for growth at 37°C (Iovine et al., 1995). Several investigations strongly support a role for GLFG regions in mediating nuclear export by interaction with factors containing nuclear export sequences (Stutz et al., 1995; Fritz and Green, 1996; Murphy and Wente, 1996; Stutz et al., 1996; Iovine and Wente, 1997). However, the GLFG region is not sufficient for complete Nup116p function. The flanking amino (N)-terminal and carboxyl (C)-terminal regions are also required, but their functions are unknown (Iovine et al., 1995). The presence of phenylalanine-glycine (FG) repeats similar to those in other nucleoporins suggests a role for the N-terminal region in transport. The C-terminal region of Nup116p displays remarkable homology to regions in two other GLFG nucleoporins: the C-terminal region of Nup100 (Nup100-C), and the middle region of Nup145p (Nup145-M) (Figure 1A) (Wente et al., 1992; Fabre et al., 1994; Wente and Blobel, 1994). These related regions each contain a peptide octamer designated the nucleoporin RNA-binding motif (NRM) that others have suggested is necessary for in vitro binding to homopolymeric RNA of guanine residues [poly(G)] (Fabre et al., 1994). However, the molecular requirement for such poly(G) binding in NPC structure and function remains to be determined.
Figure 1.
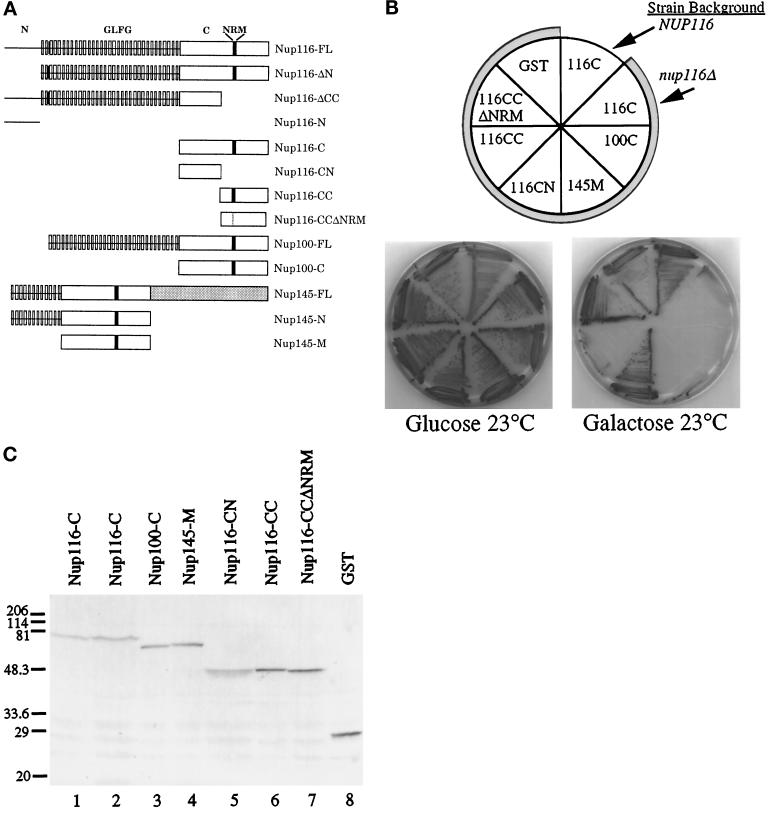
The nup116-C lethal phenotype. (A) Schematic diagrams of the structural regions of Nup116p, Nup100p, and Nup145p and their related deletion/truncation polypeptides as referred to in this study. The N-terminal region of Nup116p spans amino acid residues 1–180, GLFG region residues 181–725, the C-terminal region residues 726-1113, the CN region residues 726–919, and the CC region residues 914-1113. The similarity between the C-terminal regions of Nup116p and Nup100p is 78%, whereas among Nup116-C, Nup100-C, and the middle region of Nup145 (Nup145-M) it is 55% (Wente et al., 1992; Fabre et al., 1994; Wente and Blobel, 1994). The NRM region in Nup116p spans residues 998-1005 (Fabre et al., 1994). FL, full- length protein. (B) The nup116Δ strain is inviable when the homologous Nup116-C, Nup100-C, and Nup145-M regions fused to GST are expressed from a galactose-inducible promoter. The plasmids expressing the indicated protein regions (see Table 1) were transformed into W303α (NUP116) or SWY27 (nup116Δ::HIS3) (shaded sectors). Strains were grown at 23°C for 7 d on SM-trp, 2% glucose, or 2% galactose. Expression of Nup116-C, Nup100-C, and Nup145-M inhibited colony formation of nup116Δ cells. Expression of constructs with a deletion of the NRM (Nup116-CCΔNRM) in the carboxyl-terminal half of Nup116-C or with only the amino-terminal half (Nup116-CN) present resulted in viability. Expression of GST alone did not inhibit growth of the nup116Δ strain. (C) Immunoblot analysis of the strains shown in B. Lane 1, Nup116-C expression in NUP116 cells (W303α). Lanes 2–8, expression in nup116Δ cells (SWY27). Yeast lysates were separated on a 10.5% SDS-polyacrylamide gel and transferred to nitrocellulose for immunoblotting with an affinity-purified rabbit polyclonal antibody recognizing GST. Molecular mass markers are in kDa.
In this article, we have further defined the roles of the N and C-terminal regions of Nup116p. The NPC-associated factor Gle2p directly bound Nup116p in vitro, and the N-terminal region of Nup116p was both necessary and sufficient for the Gle2p interaction. A lethal nup116-C mutant phenotype was characterized that required expression of the C-terminal NRM-containing region. In a genetic selection, a novel high-copy suppressor of the nup116-C phenotype was identified and designated SNL1 (for suppressor of nup116-C lethal). SNL1 encodes an integral membrane protein with a calculated molecular mass of 18.3 kDa. Interestingly, SNL1 is also a high-copy suppressor of both the gle2–1 and nic96-G3 temperature sensitive phenotypes. In addition, the nup116-C phenotype was rescued by overexpression of Gle2p or Nic96p, an essential NPC assembly factor (Zabel et al., 1996). On the basis of these results, we predict that Snl1p plays a stabilizing role in NPC function and biogenesis.
MATERIALS AND METHODS
Strains and Plasmids
The plasmids used in this study are described in Table 1. Bacterial strains were cultured in SOB media and transformed by standard methods (Sambrook et al., 1989). Escherichia coli strain DH5α was used as the bacterial host for all plasmids. The yeast strains were grown in either rich media (YPD; 1% yeast extract, 2% bactopeptone, 2% glucose), or synthetic minimal media (SM) supplemented with appropriate amino acids and 2% of the indicated sugar (glucose, raffinose, or galactose). Yeast transformations were performed using the lithium acetate method (Ito et al., 1983), and general yeast manipulations were conducted as described (Sherman et al., 1986). The haploid yeast strains used in this study include: W303α (MATα ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100), SWY27 [nup116Δ (Wente and Blobel, 1993)], SWY1136 [gle2–1 (Murphy et al., 1996)], SWY1225 [gle2Δ (Murphy et al., 1996)]], SWY1191 [gle1–4 (Murphy and Wente, 1996)], SWY423 (MATα nup133Δ::HIS3 ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100; gift from M. Bucci, Washington University, St. Louis, MO]), PSY826 [npl4–2 (DeHoratius and Silver, 1996)], pom152 null (MATa ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 pom152–2::HIS3, generous gift from R. Wozniak, University of Alberta, Edmunton, Canada), SWY1209 [MATa nup100::HIS3 gle3–1(nic96-G3) ade2–1 ade3::HISG ura3–1 his3–11,15 leu2–3,112 trp1–1 LYS2 + pSW201 (Murphy et al., 1996)], SWY1031 (MATα nup100::HIS3 ade2–1 ade3 ura3–1 his3–11,15 leu2–3, 112 TRP1 lys2 + pSW201), SWY1599 (MATα nup100::HIS3 ade2–1 ade3::HISG ura3–1 his3–11,15 leu2–3,112 TRP1 lys2 NIC96:LEU2), SWY1353 (snl1::HIS3, see below), mtr7–1/acc1–7-1 [MATα mtr7–1 ura3–52 (Schneiter et al., 1996)], MLY1846 [MATa ura3–52 his4–619 sec17–1 (Novick et al., 1980; Latterich and Schekman, 1994)], and MLY1888 [MATa ura3–52 his4–619 sec18–1 (Novick et al., 1980; Latterich and Schekman, 1994)].
Table 1.
Plasmids used in this study
| Plasmid | Construction | Reference |
|---|---|---|
| pBJ382 Backbone | ||
| pSW169 | Fragment encoding amino acids 215–591 of NUP145 locus in BamHI–SacI | This study |
| pSW170 | Fragment encoding amino acids 590–959 of NUP100 locus in BamHI–SacI | This study |
| pSW171 | Fragment encoding amino acids 726–1113 of NUP116 locus in BamHI–SacI | This study |
| pSW556 | Fragment encoding amino acids 726–919 of NUP116 locus in BamHI–SacI | This study |
| pSW557 | Fragment encoding amino acids 914–1113 of NUP116 locus in BamHI–SacI | This study |
| pSW558 | Fragment encoding amino acids 914–1113 of NUP116 locus with 998–1005 replaced by G in BamHI/SacI | This study |
| pCITE Backbone | ||
| pSW691 | Full-length GLE2 with HA fused after C-terminal residue in EcoRI site | This study |
| pSW768 | Full-length NUP116 locus in BamHI/SacI | This study |
| pSW769 | Fragment encoding amino acids 181–1113 of NUP116 locus in BamHI/SacI | This study |
| pSW799 | Fragment encoding amino acids 1–919 of NUP116 locus in BamHI/SacI | This study |
| pSW801 | Fragment encoding amino acids 1–591 of NUP145 locus in BamHI/SacI | This study |
| pSW851 | Fragment encoding amino acids 5–838 of NIC96 locus in BamHI site | This study |
| pSW871 | Fragment encoding amino acids 1–180 of NUP116 locus in BamHI site | This study |
| pRS305 Backbone | ||
| pSW848 | Full-length NIC96 locus | This study |
| pRS315 Backbone | ||
| pSW75 | Full-length NUP116 locus in XbaI/SalI site | Wente et al. (1992) |
| pSW78 | Full-length NUP100 locus in XbaI/SalI site | Wente et al. (1992) |
| pSW278 | Full-length NIC96 locus in BamHI site | This study |
| pSW575 | Full-length SNL1 with NsiI site inserted before stop codon | This study |
| pSW576 | Full-length SNL1 with Protein A inserted at NsiI site of pSW575 | This study |
| pSW939 | Full-length SNL1 with SUC2 inserted at NsiI site of pSW575 | This study |
| pRS316 Backbone | ||
| pSW131 | Full-length NUP116 locus in XbaI/SalI site | Wente and Blobel (1994) |
| pSW201 | Full-length NUP100 locus and ADE3 locus | Murphy et al. (1996) |
| pSW574 | Full-length SNL1 locus in BamHI site | This study |
| pRS425 Backbone | ||
| pSW277 | Full-length NIC96 locus in BamHI site | This study |
| pSW336 | Full-length KAP95 locus in BamHI site | This study |
| pSW544 | Full-length GLE2 locus in SalI/SacI site | This study |
| pSW578 | Full-length SNL1 with protein A insert from pSW576 | |
| pSW586 | Partial p105 locus from library isolate (pSW807) in BamHI site | This study |
| pRS426 Backbone | ||
| pSW573 | Full-length SNL1 locus in BamHI site | This study |
| pLGSD5 Backbone | ||
| pSW418 | Full-length GLE2 locus in BamHI site | This study |
| pSW534 | Full-length SNL1 locus in BamHI site | This study |
| YEp13 Backbone | ||
| pSW847 | Library plasmid containing entire NIC96 locus in BamHI site | This study |
| YEp24 Backbone | ||
| pSW807 | Library plasmid containing entire SNL1 locus in BamHI site | This study |
| pSW853 | Library plasmid containing entire GLE1 locus in BamHI site | This study |
| pSW854 | Library plasmid pSW807 lacking 1885-bp BglII–BglII fragment | This study |
| pGEX-3X Backbone | ||
| pSW552 | Fragment encoding amino acids 36–159 of SNL1 locus in BamHI site | This study |
| pMAL-cRI Backbone | ||
| pSW640 | Fragment encoding amino acids 36–159 of SNL1 locus in BamHI site | This study |
Vector backbone references for pBJ382 for GAL-inducible GST fusion proteins (generous gift from C. Hug, Washington University, St. Louis, MO); pCITE (Novagen); pRS305, pRS315, pRS316, pRS425, and pRS426 (Sikorski and Hieter, 1989); pLGSD5 (Guarente, 1982); YEp13 (Nasmyth and Tatchell, 1980); YEp24 (Carlson and Botstein, 1982); pGEX-3X (Pharmacia); pMAL-cRI (Maina et al., 1988).
Electron Microscopy
Samples were prepared using the protocols described in Wente and Blobel (1993) for preservation of both protein and membrane structures. Briefly, SWY27 cells harboring pSW171 were grown to early logarithmic phase in SM-trp 2% raffinose before shifting to SM-trp 2% glucose, or SM-trp 2% galactose overnight. Samples were fixed by resuspension of the cell pellet in 40 mM potassium phosphate buffer (pH 6.5), 0.5 mM MgCl2, 2% glutaraldehyde, and 2% formaldehyde and were incubated on ice for 30 min. After cell wall digestion and osmium postfixation (Byers and Goetsch, 1991), the samples were embedded in Epon. Thin sections (collected on nickel grids coated with formvar and stabilized with carbon) were contrasted by staining with uranyl acetate and Reynold’s lead. Specimens were visualized with a Zeiss-902 electron microscope, and photographs were taken on Kodak electron microscopy film.
Cloning and Disruption of SNL1
SNL1 was identified in a high-copy suppressor screen of the nup116-C lethal strain (SWY27 harboring pSW171) from a yeast genomic library in vector YEp24 (Carlson and Botstein, 1982). Approximately 61,000 transformants were screened for the ability to grow on SM-trp 2% galactose after 7 d at 23°C. A total of 178 suppressors were identified and the 19 best growers were further analyzed. Two did not grow well after losing pSW171 and were not further analyzed. Five of the strains were identified as harboring an NUP116 plasmid by colony polymerase chain reaction (PCR) with appropriate oligonucleotides complementary to NUP116. The library plasmids from the remaining 12 best growers were recovered from yeast cells and transformed into DH5α cells. After reisolation from bacteria, plasmids were retransformed into the nup116-C lethal strain and only two isolates were able to reconfer high-copy suppression. The ends of the library fragments were sequenced using oligonucleotide primers hybridizing to the tet gene sequence immediately flanking the insertion point of the genome fragments (Sequenase kit version 2.0, United States Biochemical, Cleveland, OH). The resulting DNA sequences were compared with sequences in the yeast genomic sequencing database. One harbored the TRP1 gene and the other contained the fragment shown in Figure 3A.
Figure 3.
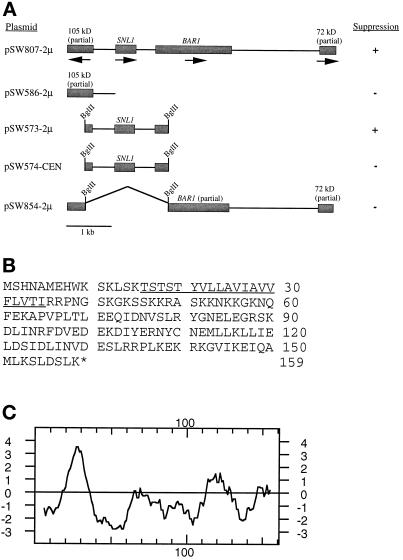
The S. cerevisiae SNL1 gene. (A) A fragment sufficient in high copy for rescuing the nup116-C phenotype was isolated from a yeast genomic library plasmid (pSW807). The library insert contained two complete and two partial ORFs from a region of chromosome IX. The ORF in pSW573 was designated SNL1 (in the yeast genome database as accession number YIL016W). + indicates the plasmid suppressed the nup116-C lethal phenotype, whereas − represents no suppression. The 2μ represents high-copy vector; CEN, low-copy vector. (B) The 159-residue amino acid sequence of the protein encoded by SNL1. A putative transmembrane segment is underlined. (C) Kyte-Doolittle (1982) hydropathy analysis of Snl1p revealed a span of sequence from amino acids 16 to 35 with significant hydropathic character.
The snl1::HIS3 null strain was made according to the method of Baudin et al. (1993) with pBM2815 and two 64-mer oligonucleotides (P18-D5 = GTTGGTGAAAAAATAGCACCAGAAGGGCAATTGTACGTTTCCGTAGGCCTCCTCTAGTACACTC, P18-D3 = TATGAATTCGGCAAGAGCCGTTATCTATAAACTAAAAATACAAACGCGCGCCTCGTTCAGAATG). PCR amplification generated an ∼1100-bp HIS3 fragment flanked on the 5′ end with 45 bp of sequence from −45 to −1 of SNL1 and on the 3′ end with 45 bp of sequence from bp 481 to bp 526. In a similar manner, a kanr fragment flanked by the same SNL1 sequence was generated. The fragments were transformed into either haploid W303α or diploid W303 cells. Isolation of viable haploid null strains [snl1::HIS3 (SWY1353); snl1::kan (SWY1678)] was confirmed by colony PCR and immunoblotting.
Antibodies and Immunoblotting
The C-terminal region of Snl1p was fused in-frame behind glutathione S-transferase (GST) (pSW552), expressed, and purified from DH5α bacteria as follows. Fusion protein synthesis was induced after growth in SOB/amp media (50 μg/ml ampicillin) to logarithmic phase by the addition of isopropyl β-d-thiogalactopyranoside (IPTG) to 0.3 mM, and growth continued at 37°C for 4 h. Bacteria were harvested and lysed by treatment with 1 mg/ml lysozyme in 300 mM NaCl and 50 mM sodium phosphate (pH 8.0) for 30 min on ice and were sonicated for a total of 15 min (cycles of 1-min bursts and 1-min rests). The lysate was centrifuged at 4°C for 30 min at 10,000 × g. The protein concentration was diluted to 50 mg/ml, and the fusion protein was purified using glutathione agarose resin (Sigma Chemical, St. Louis, MO). The antigen was sent to Cocalico Biologicals (Reamstown, PA) for production of rabbit anti-serum WU975. C-terminal Snl1p was also fused in-frame behind the maltose-binding protein. Maltose-binding protein–Snl1p was purified from DH5α cells transformed with pSW640 via induction with 0.3 M IPTG and cell lysis as described above, followed by purification over amylose resin (New England Biolabs, Beverly, MA). Antiserum to Snl1p was subsequently purified by affinity chromatography over a maltose-binding protein–Snl1p Affi-Gel 10 (Bio-Rad, Hercules, CA) column.
Protein samples were separated by electrophoresis in SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Blots were probed with affinity-purified rabbit polyclonal anti-Nup116p C-terminal antibody (Iovine et al., 1995) at a 1:2500 dilution (1 h at room temperature), affinity-purified rabbit polyclonal anti-Snl1p C-terminal antibody at 1:250 (16 h at 4°C), rabbit polyclonal anti-Kar2p antibody (Rose et al., 1989) at 1:20,000 (1 h room temperature), rabbit polyclonal anti-Kex2p C-tail serum KXR-B6 (gift from Robert S. Fuller, University of Michigan, Ann Arbor, MI) at 1:2000 (16 h, 4°C), or affinity-purified rabbit anti-GST antibody at 1:2000 dilution (16 h at 4°C; provided by J. Watkins). All dilutions were made in 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.05% Tween 20 (TBST)/2% nonfat dry milk. After washing in TBST, blots were processed by the ECL system (Amersham, Arlington Heights, IL) according to the manufacturer’s directions or by incubating with affinity-purified alkaline phosphatase conjugated anti-rabbit IgG (Promega, Madison, WI; diluted 1:7500) for 1 h and developing with nitro blue tetrazolium and 5-bromo-4-chloro-3-indoyl-1-phosphate (Promega).
Immunofluorescence Microscopy
Immunofluorescence experiments were performed using a modified method of Kilmartin and Adams (1984; Wente et al., 1992). Wild-type or SWY1354 (snl1::HIS pSNL1-ProtA) yeast cells in early log phase were fixed for 1 min in 3.7% formaldehyde and 10% methanol and were incubated with affinity-purified anti-Snl1p rabbit antibodies at 1:1 or with rabbit anti-mouse IgG (Cappel Laboratories, Organon Teknika Corp., Durham, NC) at 1:250 for 16 h at 4°C. They were then washed with M buffer (40 mM K2HPO4, 10 mM KH2PO4, 150 mM NaCl, 0.1% NaN3. 0.1% Tween 20, 2% nonfat dry milk). Bound antibody was detected by incubation with affinity-purified fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Cappel Laboratories, Organon Teknika Corp.) at a 1:200 dilution for 60 min at room temperature. After additional washes in M buffer and 1% bovine serum albumin/phosphate-buffered saline, cells were mounted in 90% glycerol and 1 mg/ml p-phenylenediamine (pH 8.0), with or without 0.05 μg/ml 4′,6-diamidino-2-phenylindole. Double immunofluorescence labeling experiments were performed as above using a 1:1 mixture of the affinity-purified anti-Snl1p rabbit antibodies and tissue culture supernatant of either mAb414 (Davis and Blobel, 1986) or mAb118C3 (Strambio-de-Castillia et al., 1995). Bound antibodies were detected by incubation with affinity-purified FITC-conjugated goat anti-mouse and Texas red-conjugated goat anti-rabbit antibodies (1:200) (Cappel Laboratories). Photographs were taken for equal exposures using the 100× objective on an Olympus microscope with Kodak T-MAX 400 film.
Subcellular Fractionation: Nuclear Envelope Isolation, Extraction, and Digestion
Total yeast cell extracts were made as described (Yaffe and Schatz, 1984). Yeast spheroplasts were prepared from 12 l of early log phase W303 diploid cells and were lysed by mechanical shearing in PVP solution (8% polyvinylpyrrolidone PVP, 20 mM potassium phosphate, pH 6.5, 0.75 mM MgCl2). The crude nuclei/membrane and cytosol fractions were separated by centrifugation, and the crude nuclei/membrane fraction was further subfractionated on a three-step sucrose/PVP solution gradient (2.0 M, 2.1 M, and 2.3 M sucrose) (centrifuged at 28,000 rpm/Beckman SW28 rotor, 4 h, 4°C, as described in Rout and Kilmartin, 1990). Nuclear envelopes were isolated from the enriched nuclei fraction as described in Strambio-de-Castillia et al. (1995). To extract peripheral proteins from the membranes, 25 mg of purified nuclear envelopes were diluted in 500 μl of 10 mM bisTris (pH 6.5), 0.1 mM MgCl2, 1 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride. One milliliter of 0.2 M Na2CO3 (pH 11) was added to produce a final concentration of 0.1 M Na2CO3 (pH 11) and it was incubated on ice for 20 min. Membranes were pelleted at 436,000 × g in a TLA 100.3 rotor at 4°C. The supernatant fraction was trichloroacetic acid (TCA) precipitated and the membrane pellet was resuspended in 20 μl of SDS sample buffer.
Topology Analysis with Snl1p-Suc2p Fusion
The sequence encoding Suc2p (starting at the third residue of the mature protein) was inserted in-frame in an NsiI site at the C terminus of Snl1p (pSW575, NsiI site inserted immediately before the stop codon). The Snl1p-Suc2p fusion protein was expressed in an snl1Δ strain (SWY1353). The N-linked glycosylation status of the fusion protein was evaluated with a modified protocol of Wilkinson et al. (1996). Cells were grown in SM-leu/glucose to an OD600 of 0.4. Ten A600 units were collected and resuspended in 250 μl of lysis buffer (20 mM Tris, pH 7.5, 5 mM MgCl2, 2% Triton X-100, 150 mM NaCl), and glass beads were added to the meniscus and vortexed for 10 min (70 s on, 30 s rest). The lysate was centrifuged at 1600 rpm for 10 min, and 50 μl of the supernatant were added to 1.2 ml of 50 mM sodium phosphate (pH 6), 0.5 mM phenylmethylsulfonyl fluoride, and the indicated amount of Endoglycosidase H (Boehringer Mannheim, Indianapolis, IN). The mixture was incubated at 37°C for 4 h, TCA precipitated, and boiled in SDS sample buffer for immunoblot analysis.
Cloning of GLE3
The SWY1209 (gle3–1) strain identified in a nup100 synthetic lethal screen was transformed with a yeast genomic library in vector YEp13 (Nasmyth and Tatchell, 1980). Approximately 11,000 transformants were screened for the ability to sector after 7 d of growth at 30°C on SM-leu/glucose plates. Sectoring isolates were screened for the presence of NUP100 by genomic colony PCR with appropriate oligonucleotides. The novel rescuing library plasmids were recovered from the yeast cells and transformed into DH5α cells. After reisolation from bacteria, the ends of the library fragment in rescuing plasmid pSW847 were sequenced. The resulting DNA sequence was compared with the yeast genome database, revealing a ∼6.5-kb pair insert from chromosome VI that harbored NIC96. A subclone expressing only the NIC96 gene was sufficient for complementation of the nonsectoring phenotype (pSW278). To prove that NIC96 was allelic to gle3–1, the NIC96 locus was marked by integration of a NIC96-LEU2 plasmid (pSW848) in strain SWY1031. The resulting NIC96-LEU2 strain (SWY1599) was mated with SWY1209, the diploids were sporulated and dissected, and the products of 13 tetrads were examined. In all cases, the nonsectoring phenotype segregated with leucine auxotrophy, indicating linkage of the synthetic lethal mutation to NIC96. The mutant allele isolated in the nup100 synthetic lethal screen will be subsequently referred to as nic96-G3 (for gle3). Further analysis of the nic96-G3 strain at a variety of growth temperatures revealed a temperature-sensitive defect at 38°C.
In Vitro Translation and Immunoprecipitation
Cell-free translation was performed using purified DNA in the TNT Reticulocyte Lysate System as described below. pSW691 harboring a C-terminally hemagglutinin (HA)-epitope tagged GLE2 -HA (isolated from strain SWY1013, Murphy et al., 1996) or pSW851 harboring NIC96 was cotranslated with the indicated plasmids bearing sequences of NUP116 or NUP145. The resulting mixture of labeled peptides (12.5 μl) was diluted in 100 μl of lysis buffer (20 mM Tris, pH 7.5, 5 mM MgCl2, 2% Triton X-100, 150 mM NaCl). Subsequently, 1 μl of affinity-purified rabbit polyclonal anti-GLFG antibody (provided by J. Watkins and H. Kaplan, Washington University, St. Louis, MO) or 3 μl of mAb12CA5 (anti-HA) tissue culture supernatant and 20 μl of packed protein A-Sepharose beads (Pharmacia Biotech, Uppsala, Sweden) were added. The mixture was incubated for 90 min at 4°C on a rotator. The beads were centrifuged and washed six times with 0.5 ml of ice-cold wash buffer (0.05% Tween, 150 mM NaCl, 50 mM Tris, pH 7.5). Immunoprecipitates were resuspended in 20 μl of SDS loading buffer, boiled, and electrophoresed on 7% SDS-polyacrylamide gels.
RESULTS
Characterization of an nup116-C Lethal Phenotype that Requires the NRM Motif
Complete deletion of NUP116 results in a temperature-sensitive growth phenotype at 37°C (Wente et al., 1992). In a previous study, we examined the requirements for the different Nup116p structural regions with a plasmid shuffle assay in the nup116 null (Δ) strain (Iovine et al., 1995). Expression of only the C-terminal region of Nup116p (nup116-C) under control of the NUP116 promoter did not rescue the temperature sensitivity of the nup116Δ strain, and surprisingly, the nup116Δ strain expressing nup116-C was inviable at all growth temperatures. Expression of this C-terminal region by either the NUP116 or GAL10 promoters in wild-type NUP116 cells did not inhibit cell growth (Iovine et al., 1995). These results suggested that the expression of the C-terminal region alone had a gain-of-function, lethal perturbation in the absence of full-length (FL) Nup116p. To test this possibility, the sequence encoding the C-terminal region (amino acids 726-1113) was fused in-frame to GST and placed under the control of the inducible GAL10 promoter. The plasmid was transformed into both NUP116 and nup116Δ haploid strains, and the relative expression level of the fusion protein was detected by immunoblotting (Figure 1C). The effect on cell growth was monitored on plates containing either galactose (inducing) or glucose (repressing) as a carbon source. As shown in Figure 1B, expression of GST-Nup116-C rendered the nup116Δ strain inviable at 23°C, whereas at apparently similar expression levels the wild-type NUP116 strain was viable and formed colonies.
To test whether expression of the homologous regions from Nup100p and Nup145p had similar lethal perturbations in nup116Δ cells, sequences encoding the Nup100-C and Nup145-M regions were fused to GAL10-GST. The plasmids were transformed into the nup116Δ strain, and the cells were analyzed for growth perturbations and protein expression levels (Figure 1, B and C). The presence of either Nup100-C or Nup145-M inhibited nup116Δ cell growth at 23°C. This similar capacity for growth inhibition of nup116Δ cells supported the hypothesis that the Nup116-C, Nup100-C, and Nup145-M regions perform analogous roles in NPC function (Wente et al., 1992; Fabre et al., 1994; Wente and Blobel, 1994).
The nup100Δ and nup145ΔN strains are viable at all growth temperatures, and these mutant cells do not exhibit the membrane herniations found in nup116Δ cells (Wente and Blobel, 1993, 1994). The NPCs in nup100Δ cells appear identical to wild type, whereas the NPCs in nup145ΔN cells are clustered. To determine whether the lethal effect of Nup116-C expression was specific to the nup116Δ genetic background, we tested for growth perturbations in nup100Δ or nup145ΔN strains harboring the GAL10-GST-nup116-C plasmid. At 23°C, the expression of Nup116-C was not toxic to nup100Δ or nup145ΔN cells as reflected by colony growth on galactose plates (our unpublished results). Thus, the nup116-C lethal phenotype required the absence of FL Nup116p.
To further define the region in the Nup116-C polypeptide responsible for the nup116-C lethal phenotype, each half of the C-terminal region was fused to GST and expressed from the GAL10 promoter in nup116Δ cells (Figure 1, B and C). The amino-terminal half of Nup116-C (Nup116-CN: amino acids 726–919) did not inhibit cell growth at 23°C. In contrast, expression of the carboxyl-terminal half of the Nup116-C region (Nup116-CC; amino acids 914-1113) resulted in lethality. Since the NRM resides in the Nup116-CC region, we tested directly for its role by replacing the sequence encoding the NRM octamer with that for a single glycine residue (designated Nup116-CCΔNRM). Expression of Nup116-CCΔNRM in nup116Δ cells did not confer lethality (Figure 1, B and C). Therefore, the NRM was required for the lethal nup116-C phenotype.
Nuclear Membrane Herniations Are Present in the nup116-C Cells
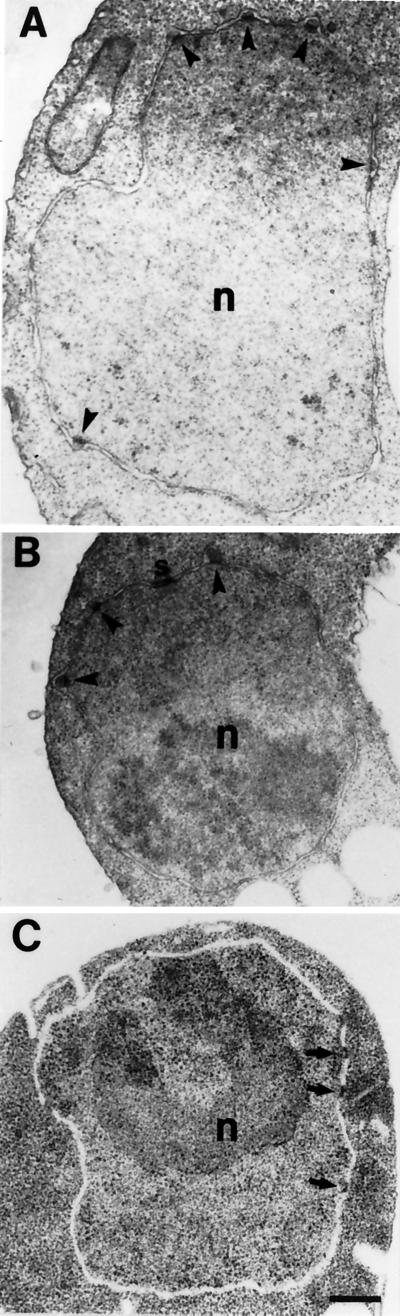
The nup116Δ temperature-arrested cells have herniations of the nuclear envelope associated with NPCs (Wente and Blobel, 1993). To test whether the lethal nup116-C phenotype was related to the null phenotype, the nuclear envelope of nup116Δ cells expressing Nup116-C was examined by thin section electron microscopy. After induction of Nup116-C expression in galactose containing media for 5 h at 23°C, the cells were fixed and processed. As shown in Figure 2, A and B, the nuclear envelope of nup116-C lethal cells exhibited nuclear membrane herniations remarkably similar in structure to those observed in nup116Δ cells at 37°C. At the nucleoplasmic base of each herniation, an electron-dense structure presumably representing the NPC was present. The inner nuclear membrane appeared to be anchored to this NPC structure, but the membrane was continuous over the cytoplasmic face of the NPC. The outer nuclear membrane was not attached to these NPCs and resided over the herniated inner membrane. Such herniations were not observed when the nup116-C cells were grown in repressing glucose media (Figure 2C). Therefore, the presence of the C-terminal region appeared to further destabilize NPC and pore membrane structure in nup116Δ cells.
Figure 2.
The nup116-C lethal strain exhibits NPC-associated herniations of the nuclear envelope. SWY27 cells harboring the galactose-inducible Nup116-C plasmid (pSW171) were grown in SM-trp 2% raffinose at 23°C and then shifted to SM-trp 2% galactose (A and B) or SM-trp 2% glucose (C) for 15 h. Cells were subsequently processed for thin section electron microscopy. The arrowheads in A and B point to representative nuclear membrane herniations that are attached to NPC-like structures at the nuclear base. The arrows in C indicate wild-type NPCs. Bar, 250 nm.
Identification of SNL1 in a Screen for High-Copy Extragenic Suppressors of the nup116-C Lethal Phenotype
We predicted that the nup116 lethal phenotype might be rescued by overexpression of factors that stabilize the NPC and pore membrane. The nup116-C mutant appeared superior to nup116Δ for genetic analysis because the nup116-C phenotype was dependent on the presence of a defined protein region (and not the absence of a protein). In addition, the nup116-C lethal phenotype may be more easily suppressed at 23°C than suppression of temperature sensitivity at 37°C. Thus, a genetic screen was conducted for high-copy suppressors of the nup116-C lethal phenotype at 23°C. A yeast 2 μ genomic library was transformed into nup116Δ cells harboring a plasmid expressing nup116-C under control of the GAL10 promoter. Among approximately 61,000 possible transformants, 178 isolates that grew on galactose containing media at 23°C were classified according to their doubling rates at 23°C. A subset of seven strains whose growth on galactose was both dependent on the presence of the library plasmid and similar to that of wild-type cells was further analyzed.
Five of the suppressing plasmids were identified as harboring NUP116 by genomic colony PCR with appropriate oligonucleotides. The presence of a high-copy NUP116 plasmid would effectively rescue the nup116Δ phenotype to wild type, in which background the expression of Nup116-C is not lethal. The sixth suppressor plasmid was identified as harboring TRP1 and it most likely mediated growth by allowing loss of the GAL10-nup116-C/TRP1 plasmid. Immunoblot analysis confirmed that the strain with the TRP1 library plasmid was not expressing Nup116-C (our unpublished results). The remaining strong suppressor plasmid appeared to be novel and specific. By DNA sequence analysis and comparison to the yeast genome database, the library plasmid insert contained the region of chromosome IX shown in Figure 3A. Four open reading frames (ORFs) were present: the FL BAR1 gene, which encodes a protease for α factor mating pheromone; an FL ORF for a hypothetical 18.3-kDa protein (YIL016W in the yeast genomic database); and partial genes for hypothetical proteins of 105 and 79 kDa.
High-copy plasmids harboring a DNA fragment encoding only the 105-kDa protein did not suppress the nup116-C phenotype. In addition, a plasmid with a 1885-kb BglII fragment removed (between the BAR1 and 105-kDa genes) was not capable of suppression. However, when the BglII fragment alone was tested in high-copy plasmids, growth of the nup116-C cells was rescued. The BglII fragment in a low-copy CEN vector did not support growth of the nup116-C strain. This suggested that the ORF encoding a putative protein with a predicted molecular mass of 18.3 kDa was both necessary and sufficient for mediating high-copy suppression of the nup116-C lethal phenotype. Therefore, this gene was designated SNL1 (for suppressor of nup116-C lethal).
The 159-amino acid residue sequence for Snl1p is shown in Figure 3B. Using BLAST programs (Altschul et al., 1990), no significant homology to any other proteins in the yeast genome database was revealed. Interestingly, analysis of the Snl1p sequence identified a span of 20-amino acid residues with sufficient hydropathy to function as a transmembrane segment (Figure 3C). This region from residues 16 to 35 was immediately flanked on either side by charged, basic residues. Pom152p is the only reported integral membrane protein associated with NPC function (Wozniak et al., 1994). Therefore, the ALIGN program (Dayhoff et al., 1983) was used to directly examine Snl1p and Pom152p for structural similarities. As shown in Figure 4, FL Snl1p overlapped with a region of the Pom152p sequence from residues 170 through 326. The C-terminal 41 residues of Snl1p in particular were 70% similar to Pom152p (13/41 identical and 16/41 with similarity). We have detected no significant homology between Snl1p and gp210 or Pom121p, the two vertebrate NPC-associated integral membrane proteins (Gerace et al., 1982; Wozniak et al., 1989; Greber et al., 1990; Hallberg et al., 1993). The previously reported 19-amino acid segment of similarity shared between yeast Pom152p and rat Pom121p is separate from the Pom152p homology with Snl1p (Wozniak et al., 1994).
Figure 4.
Alignment of the Snl1p and Pom152p sequences. An ALIGN analysis between the FL proteins revealed significant homology (Dayhoff et al., 1983). The center line designates the identical (capital letter) and conserved (:, .) residues. The boxed residues in the Snl1p sequence (16–35) and Pom152p (177–195) note the position of the respective predicted transmembrane segments. Pom152p is predicted to be oriented as a type II integral membrane protein (Wozniak et al., 1994); however, Pom152p topology has not been experimentally determined. Snl1p is positioned with its C-terminal region cytoplasmically exposed (Figure 8B). If the sequence similarity is significant, this suggests a similar, potential type I orientation for Pom152p.
SNL1 and POM152 Null Alleles Are Not Synthetically Lethal
To test whether SNL1 encoded an essential gene product, the chromosomal allele of SNL1 was replaced by homologous recombination with HIS3, or kanr resulting in a null (Δ) mutant allele. Sporulation and dissection of a heterozygous diploid null strain resulted in the recovery of four viable spores (our unpublished results). The absence of Snl1p in snl1Δ::HIS3 haploids was confirmed with anti-Snl1p antibodies (see below). The disruption conferred no obvious growth defects as compared with wild-type cells at 14, 23, 30, or 37°C in rich media. Therefore, SNL1 is not required for cell growth under these conditions. The observation that pom152 null mutants are viable suggested that an unidentified yeast integral membrane protein(s) functionally compensates for its absence (Wozniak et al., 1994). To test for functional links, the phenotype of a haploid strain harboring both the snl1 and pom152 null alleles was examined. A doubly disrupted, heterozygous diploid strain (snl1Δ::HIS3/SNL1 pom152Δ:: HIS3/POM152) was induced to sporulate and subjected to tetrad analysis. In all cases, all four spores from each asci were viable (our unpublished results). Furthermore, for tetrads where the HIS3 markers cosegregated, the His+ colonies were viable at all tested growth temperatures (14, 23, 30, and 37°C). This indicated that the snl1 and pom152 null mutations were not lethal in combination. The lack of synthetic lethality between snl1 and pom152 null mutants possibly reflects their redundant function in cells with yet unidentified integral membrane proteins.
Snl1p Is a Type I Integral Membrane Protein Localized to the Nuclear Envelope and Endoplasmic Reticulum
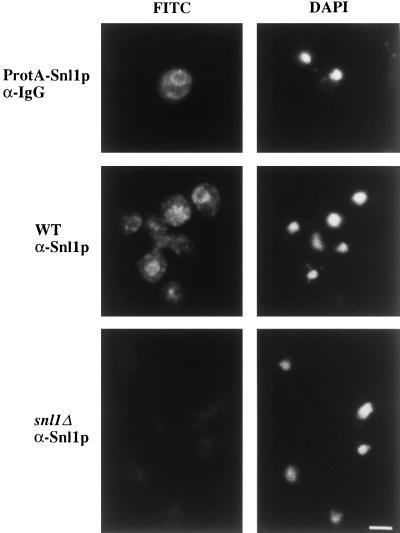
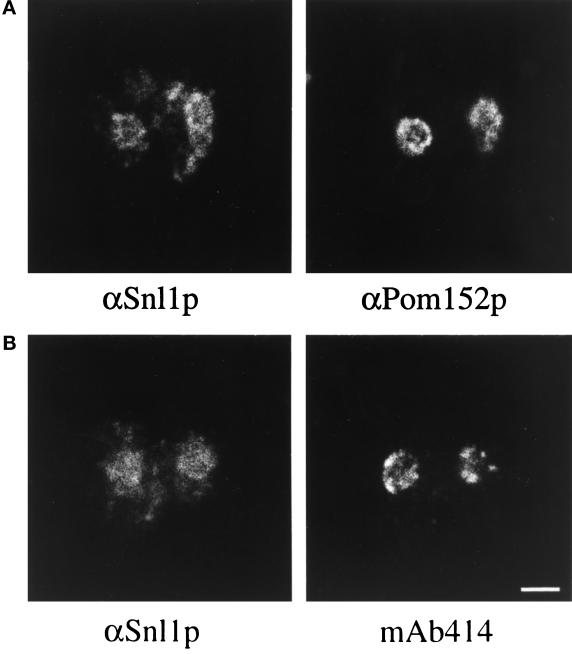
To further characterize Snl1p, the localization of Snl1p was determined by both indirect immunofluorescence microscopy and subcellular fractionation. Sequences encoding five tandem IgG binding domains from Staphylococcus aureus protein A were fused in-frame before the stop codon for SNL1. The epitope-tagged gene, under control of the endogenous promoter, was expressed in the snl1Δ strain from both high-copy and low-copy vectors. Localization of the C-terminally tagged protein A–Snl1p was visualized in fixed cells by the binding of FITC-conjugated antibody. As shown in Figure 5, the staining was predominantly on the nuclear envelope as well as on the endoplasmic reticulum, which is continuous with the outer nuclear membrane. The variable protein A–Snl1p staining level between different cells in Figure 5, upper panel, was probably due to plasmid-copy number differences. A similar pattern but extremely faint staining was observed with expression from a low-copy vector. To confirm that the localization was not an artifact of overexpression, rabbit polyclonal antibodies were raised against a bacterially expressed GST fusion protein with residues 36–159 of Snl1p. The antibodies were affinity purified and were tested on both wild-type SNL1 cells and snl1Δ cells. The staining pattern in wild-type SNL1 cells for endogenous protein was identical to that observed with protein A–Snl1p (Figure 5, middle panel). In snl1Δ cells, only diffuse background staining was present, demonstrating the specificity of the polyclonal anti-Snl1p antibodies (Figure 5, lower panel).
Figure 5.
Snl1p is localized to the nuclear envelope/endoplasmic reticulum. snl1Δ cells alone (bottom panels) or harboring a high-copy plasmid expressing a protein A–Snl1p fusion (top panels) and wild-type W303α cells (middle panels) were grown to early log phase in YPD at 30°C before they were fixed in methanol/formaldehyde as described (see MATERIALS AND METHODS). Fixed cells were incubated with rabbit anti-mouse IgG (αIgG) or affinity- purified rabbit polyclonal antibodies raised against Snl1p (αSnl1p) as designated. Binding was detected with an FITC-conjugated goat anti-rabbit IgG. Perinuclear and peripheral staining was observed in wild-type cells, but not in cells lacking Snl1p. The DAPI staining delineating the location of the nucleus is shown on the right. Bar, 2.5 μm.
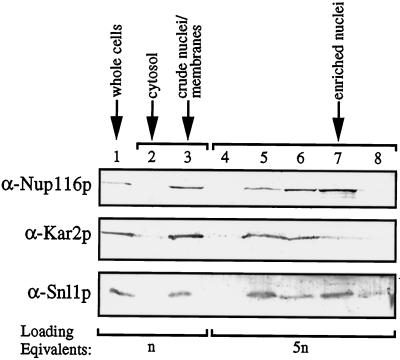
To confirm that the apparent nuclear envelope and endoplasmic reticulum staining for Snl1p was distinct from the punctate nuclear rim staining reported for nucleoporins, double immunofluorescence experiments were conducted. Wild-type yeast cells were double labeled with affinity- purified rabbit anti-Snl1p antibodies and mouse monoclonal antibodies recognizing either nucleoporins (mAb414) (Davis and Blobel, 1986; Rout and Blobel, 1993) or Pom152p (mAb118C3) (Strambio-de-Castillia et al., 1995). Both the nucleoporin (Figure 6B, right) and the anti-Pom152p (Figure 6A, right) staining were predominantly confined to the circumference of the nuclear envelope. In both cases, the anti-Snl1p staining overlapped with the nuclear envelope staining (corresponding left panels of Figure 6). However, the anti-Snl1p nuclear localization was not strictly punctate on the nucleus, and it also extended throughout the presumptive endoplasmic reticulum. To further analyze the subcellular distribution of Snl1p, yeast cells were fractionated and tested by immunoblotting with antibodies specific for the lumenal endoplasmic reticulum protein Kar2p, the nucleoporin Nup116p, and Snl1p (Figure 7). All three proteins coenriched with the crude nuclei/membrane fraction from lysed yeast spheroplasts (Figure 7, lane 3). As previously reported (Strambio-de-Castillia et al., 1995), when this crude fraction was further subfractionated on a three-step sucrose gradient, the nuclear envelope and endoplasmic reticulum markers became separated. The majority of Kar2p was observed in lane 5, whereas the majority of Nup116p was observed in lane 7. Interestingly, Snl1p distribution was unique, with approximately equivalent levels peaking in both lanes 5 and 7. These results suggested that Snl1p was equally distributed between the nuclear envelope and endoplasmic reticulum membranes, and was therefore not exclusively localized at the NPC.
Figure 6.
Double immunofluorescence comparison of Snl1p localization to Pom152p and peripheral nucleoporins. Diploid W303 cells were processed for indirect immunofluorescence microscopy as described in MATERIALS AND METHODS. Fixed cells were incubated with affinity-purified rabbit polyclonal anti-Snl1p antibodies (αSnl1p) and with mouse monoclonal anti-Pom152p antibodies (αPom152p, mAb118C3) or mouse monoclonal anti-nucleoporin antibodies (mAb414). The antibodies were detected with Texas red goat anti-rabbit IgG and FITC-labeled goat anti-mouse IgG. Identical fields of cells are shown in A and B. Bar, 3 μm.
Figure 7.
Immunoblot analysis of yeast subcellular fractionation. Samples of total yeast cells (lane 1), crude cytosol (lane 2), crude nuclei/membranes (lane 3), and sucrose gradient fractions resolving the crude nuclei/membrane fraction (lanes 4–8) were prepared as described in MATERIALS AND METHODS, separated electrophoretically on a 13% SDS-polyacrylamide gel, and transferred to nitrocellulose. The blot was divided in thirds, and the top portion was probed with an affinity-purified antibody recognizing Nup116p, the middle portion with antibody for Kar2p, and the bottom portion with antibody for Snl1p. All three proteins enriched in the crude nuclei/membrane fraction. Loading equivalents represent the number of cell equivalents that were used as starting material to prepare each of the samples.
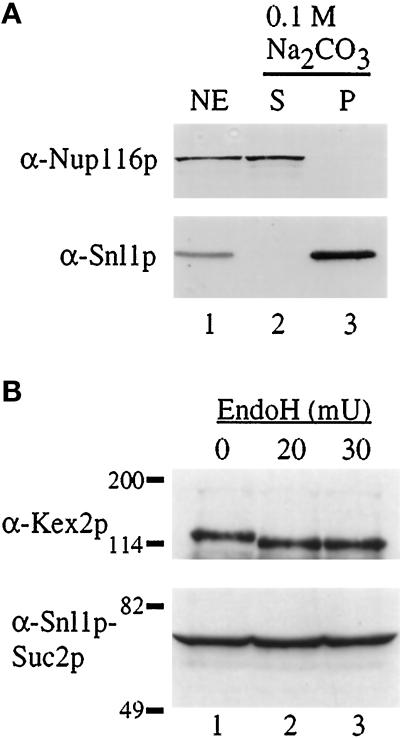
Because Snl1p was localized to both the nuclear and endoplasmic reticulum membranes and the hydropathy plot predicted a putative transmembrane span, we tested by subcellular fractionation whether Snl1p behaved as an integral membrane protein. Nuclei were purified from wild-type cells, and a nuclear envelope fraction was isolated using a sucrose flotation gradient (Strambio-de-Castillia et al., 1995). The nuclear membranes were extracted with 0.1 M sodium carbonate (pH 11) and peripheral versus integral membrane proteins were separated by centrifugation into supernatant (s) and pellet (p) fractions. Samples of the fractions were analyzed by immunoblotting with the anti-Snl1p and anti-Nup116p antibodies (Figure 8A). Snl1p resisted high pH extraction as reflected by its exclusive association with the pellet fraction. In contrast, Nup116p (a peripheral nucleoporin) was fully extracted and in the supernatant fraction.
Figure 8.
Snl1p is an integral membrane protein, oriented with the C-terminal region accessible to the cytoplasm. (A) Purified nuclear envelopes (NE, lane 1) from W303 diploid nuclei were treated with 0.1 M Na2CO3 (pH 11) and separated into supernatant (S, lane 2) and pellet (P, lane 3) fractions by centrifugation. Samples were separated on a 13% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunoblotted. Strips corresponding to the molecular mass of Nup116p (top) and Snl1p (bottom) were cut from the same blot and probed with the respective antibodies. (B) Cell extracts from a strain expressing an Snl1p–Suc2p fusion protein were either mock treated (0, lane 1) or treated with EndoH (lane 2, 3) as described in MATERIALS AND METHODS (mU = milliunits). The samples were TCA precipitated and resuspended in SDS sample buffer for separation on a 7% SDS-polyacrylamide gel. Immunoblot analysis was performed for Kex2p (top) and Snl1p–Suc2p (bottom). Molecular mass markers are indicated in kDa.
The single transmembrane span near the N terminus of Snl1p predicts that the majority of the polypeptide will be either lumenally or cytoplasmically exposed. To define the Snl1p membrane topology, we based our studies on the previous analysis of yeast endoplasmic reticulum membrane proteins (Sengstag et al., 1990; Feldheim et al., 1992; Wilkinson et al., 1996). The fusion of Suc2p (invertase) to protein regions that are lumenally exposed results in extensive N-linked glycosylation of the Suc2p region with a coincident 20- to 26-kDa increase in apparent molecular mass. In contrast, cytoplasmic localization of Suc2p results in a nonglycosylated polypeptide that migrates at its predicted molecular mass. We therefore fused Suc2p to the C terminus of Snl1p. If the region C-terminal to the Snl1p membrane span is exposed to the cytosol, the Snl1p-Suc2p hybrid would not be glycosylated and would migrate with a predicted mass of ∼76 kDa. Alternatively, if the C-terminal region is lumenal, a larger glycosylated Snl1p-Suc2p polypeptide should be observed. The Snl1p-Suc2p fusion protein was expressed in a snl1Δ strain, and cell extracts were prepared, treated with EndoH, and analyzed by immunoblotting. As a control, the glycosylation state of endogenous Kex2p was monitored. As previously reported (Fuller et al., 1989; Wilcox and Fuller, 1991), Kex2p was sensitive to treatment with EndoH as reflected by the shift in apparent molecular mass between lanes 1 and 2 (Figure 8B, top). In contrast, migration of the Snl1p-Suc2p polypeptide was not affected by EndoH treatment (Figure 8B, bottom). Moreover, the apparent molecular mass of the Snl1p-Suc2p hybrid was exactly the size predicted without glycosylation (∼76 kDa). The lack of glycosylation suggested that the C-terminal region of Snl1p was exposed to the cytoplasm. Immunofluorescence analysis showed that the Snl1p-Suc2p protein was correctly targeted to membranes (our unpublished results). Correct targeting was also observed with biochemical fractionation and membrane extraction analysis of an Snl1p–protein A fusion, wherein the protein A domain was inserted in the same position as the Suc2p (our unpublished results). Interestingly, the type I topology for Snl1p is the opposite orientation to that predicted for Pom152p. Therefore, either the limited sequence similarities between Pom152p and Snl1p are not significant, or Pom152p is also oriented as a type I membrane protein. Pom152p topology has not been experimentally determined (Wozniak et al., 1994). Overall, these data strongly suggest that Sn1lp is a novel integral membrane protein that functionally interacts with NPCs.
Specific Genetic Linkage between SNL1, GLE2, NIC96, and NUP116 Function
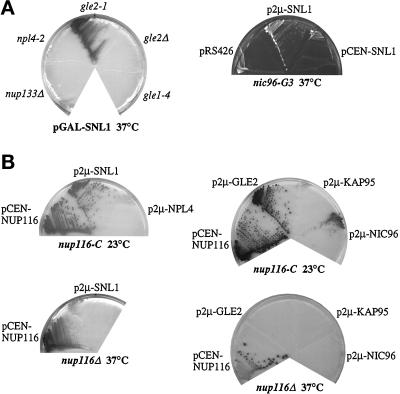
To examine whether high-copy suppression by SNL1 was allele specific, other conditional mutant strains were transformed with a high-copy GAL10-SNL1 plasmid and tested for colony growth at the restrictive temperature on galactose-containing media. High-copy SNL1 expression did not suppress the temperature-sensitive lethal phenotypes of nup133Δ, gle1–4, and npl4–2 mutants (Figure 9A) or mtr7–1/acc1–7-1, sec17–1, and sec18–1 mutants (our unpublished results). However, overexpression of Snl1p did rescue the temperature sensitivity of gle2–1 and gle3–1 cells. We previously identified gle2–1 and gle3–1 in a synthetic lethal genetic screen with a nup100 null mutant (Murphy et al., 1996). GLE2 encodes an NPC-associated protein that coimmunoprecipitates in a complex with Nup116p, the nuclear import factor Kap95p, and other unidentified nuclear proteins (Murphy et al., 1996; Iovine and Wente, 1997). At 37°C, the gle2–1 mutant is lethal, inhibits RNA export, and results in clusters of herniated NPCs similar in structure to the herniations in nup116Δ and nup116-C cells (Murphy et al., 1996). The wild-type GLE3 gene is allelic to NIC96 (see MATERIALS AND METHODS), a nucleoporin with an essential role in NPC assembly (Grandi et al., 1993; Aitchison et al., 1995; Grandi et al., 1995a,b; Nehrbass et al., 1996; Zabel et al., 1996). The mutant nic96 allele from the nup100 synthetic lethal screen will be referred to henceforth as nic96-G3. In a further test, overexpression of Snl1p did not suppress the gle2Δ or nup116Δ temperature- sensitive phenotypes (Figure 9), suggesting that Snl1p required the presence of the mutant Gle2p protein or the Nup116-C polypeptide to exert its suppression activity. Overall, the high-copy SNL1 suppression was specific to a subset of conditional NPC mutant phenotypes and did not appear to be related to the general endoplasmic reticulum mutants tested.
Figure 9.
Genetic interactions among SNL1, GLE2, NUP116, and NIC96. (A) High-copy SNL1 suppresses the temperature sensitivity of the gle2–1 mutant and the nic96-G3 mutant. The indicated mutant strains were transformed with either pGAL-SNL1 (pSW534), p2μ-SNL1 (pSW807), pCEN-SNL1 (pSW574), or pRS426 (empty 2μ URA3). The strains were streaked to SM-ura 2% galactose to induce overexpression of Snl1p (left) or on SM-ura-leu 2% glucose (right) and grown for 5 d at 37°C. The gle1–4 cells expressing high-copy SNL1 were also inviable when grown at 30°C (lowest nonpermissive growth temperature). The nic96-G3 colonies are white (right), whereas the other strains are red/pink (left and in B). Therefore, different photographic contrasts were used to document the results. (B) Overexpression of SNL1, GLE2, and NIC96 suppresses the nup116-C lethal phenotype. nup116Δ cells alone or harboring pGAL10-GST-nup116-C (pSW171) (nup116-C) were transformed with a CEN pNUP116 plasmid as a control or with the designated 2μ plasmids harboring SNL1, GLE2, NIC96, KAP95, or NPL4. To select for the respective plasmids, the nup116-C cells were grown on SM-trp-ura 2% galactose (left) or SM-trp-leu 2% galactose (right) for 7 d at 23°C. The nup116Δ cells were grown on SM-ura 2% glucose (left) or SM-leu 2% glucose (right) for 7 d at 37°C. Strains harboring a 2μ GLE1 plasmid behaved the same as those with pKAP95 (no colony formation; our unpublished results).
High-copy vectors harboring GLE1, GLE2, NIC96, NPL4, and KAP95 were tested for their ability to suppress the nup116-C lethal phenotype (data for GLE1, our unpublished results). Others have reported that high-copy NPL4, which encodes an NPC-associated protein, can partially suppress the growth defect of nup116Δ cells at 37°C (DeHoratius and Silver, 1996); however, overexpressed NPL4 did not suppress the nup116-C lethal phenotype (Figure 9B). The import factor Kap95p physically interacts with Nup116p, whereas the nuclear export factor Gle1p genetically interacts with Nup116p (Iovine et al., 1995; Murphy et al., 1996; Murphy and Wente, 1996; Iovine and Wente, 1997), but neither high-copy KAP95 nor GLE1 suppressed the nup116-C growth defect. Interestingly, high-copy GLE2 and NIC96 rescued the nup116-C lethal phenotype at 23°C. Like SNL1, GLE2 and NIC96 did not allow growth of nup116Δ cells at 37°C. Therefore, an additional genetic link between the functions of NUP116, GLE2, NIC96, and SNL1 was demonstrated. Moreover, only a subset of factors with reported genetic and/or physical associations with Nup116p were either rescued by SNL1 or prevented nup116-C lethality. These specific genetic interactions imply a possible functional or physical interaction among these proteins.
Gle2p Interacts In Vitro with the N-Terminal Region of Nup116p
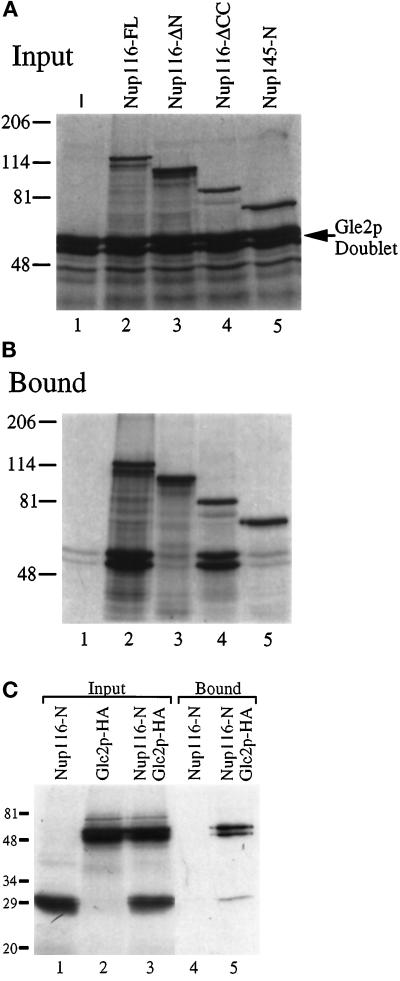
To test whether there was a physical basis for the genetic interactions between NUP116, GLE2, and NIC96, an in vitro analysis for direct protein–protein interactions was conducted. Various combinations of 35S-labeled FL Gle2p [tagged at the C terminus with the HA epitope (Murphy et al., 1996)], FL Nic96p, and FL or deleted/truncated Nup116p proteins were cotranslated in reticulocyte lysates. The proteins were translated with approximately equal efficiency. Gle2p was observed as a doublet (Figure 10A, lane 1, and C, lane 2), possibly reflecting either differential in vitro translational initiation at the multiple methionines present in the N-terminal region of Gle2p or posttranslational modification. We have also observed endogenous Gle2p as a doublet when isolated from enriched, detergent-solubilized NPCs (Murphy et al., 1996). To detect interactions, mixtures of in vitro expressed proteins were incubated with either affinity-purified rabbit polyclonal antibodies generated against the GLFG region of Nup116p (Figure 10, A and B) or mouse mAb12CA5 recognizing the Gle2p-HA epitope (Figure 10C). Bound proteins were isolated with protein A–Sepharose resin and analyzed using SDS-PAGE and autoradiography. The doublet from Gle2p was quantitatively isolated only in the presence of Nup116p. Gle2p was not immunoprecipitated in the absence of Nup116p (Figure 10B, lane 1) or with the first 591 residues of Nup145 (Figure 10B, lane 5). Similar experiments with Nic96p did not detect any specific coimmunoprecipitation with either Nup116p alone or with the Nup116p-Gle2p complex (our unpublished results). In a recent study, Nup116p and Gle2p copurified from total yeast nuclei lysates in a complex along with Kap95p and several other unidentified nuclear proteins (Iovine and Wente, 1997). Therefore, these results suggested a probable direct interaction between FL Nup116p and Gle2p.
Figure 10.
The amino-terminal region of Nup116p is required for in vitro interaction with Gle2p. Gle2p-HA was cotranslated in vitro with the indicated Nup116p and Nup145p polypeptides (see Figure 1A). (A) Autoradiograph of a 7% SDS-polyacrylamide gel with aliquots of the respective cotranslation input fractions used for immunoprecipitation in B. Lane 1 is a translation of Gle2p alone. The position of the Gle2p doublet is noted by the arrow. The upper bands correspond to the respective Nup116p or Nup145p products. (B and C) The 35S-labeled polypeptide mixtures were immunoprecipitated with affinity-purified rabbit polyclonal anti-GLFG antibody (B) or mAb12CA5 (anti-HA/Gle2p, C) and protein A– Sepharose beads. The bound fraction was eluted by boiling in SDS sample buffer and separated by electrophoresis on either a 7% (B) or 13% (C) SDS-polyacrylamide gel. 35S-labeled proteins were detected by autoradiography. Molecular mass markers are noted in kDa.
Because high-copy GLE2 rescued the nup116-C phenotype, Gle2p could mediate this activity by directly interacting with the C-terminal region of Nup116p. To determine the region of Nup116p that mediated in vitro binding to Gle2p, plasmids encoding N-terminal and C-terminal Nup116p deletions were tested (Figure 10). Nup116p lacking the N-terminal 180-amino acid residues did not immunoprecipitate Gle2p (Figure 10B, lane 3), whereas Nup116p, lacking the CC-region (last 194 amino acids), was capable of binding Gle2p (Figure 10B, lane 4). These results correlate with our previous tests in the two-hybrid assay, wherein FL Gle2p did not interact with the GLFG or C-terminal regions of Nup116p (Murphy et al., 1996). Interestingly, a fragment of the N-terminal region alone (first 180 residues) specifically bound Gle2p (Figure 10C, lane 5). Therefore, the N-terminal region of Nup116p was both necessary and sufficient for the in vitro Gle2p–Nup116p interaction.
DISCUSSION
To identify mediators of NPC structure and function, we characterized a lethal nup116-C mutant phenotype in the yeast Saccharomyces cerevisiae. We report here the genetic isolation of SNL1 as a high-copy suppressor of the nup116-C lethality. SNL1 encodes a novel integral membrane protein localized to both the nuclear and endoplasmic reticulum membranes. Further analysis has suggested specific links between the role of Snl1p and at least three soluble NPC-associated proteins: Nup116p, Gle2p, and Nic96p. Since relatively little is known about the factors required for the biogenesis and maintenance of NPC structure, these results may have important implications for NPC function.
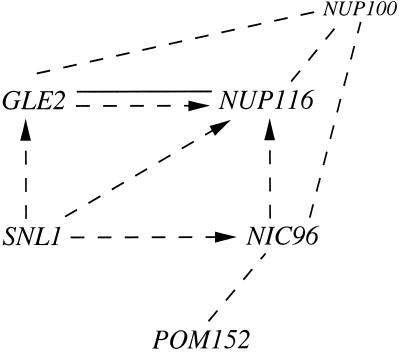
The hypothesis that Snl1p has a role in NPC structure is based on several observations. First, high-copy SNL1 specifically rescues the growth of mutants with perturbed NPC/nuclear envelope structure. In Figure 11, the genetic and physical interactions highlighted by the results in this report are diagrammed. The actions of SNL1, NUP116, GLE2, NIC96, and POM152 have been linked for the first time in several different ways. High-copy SNL1, GLE2, and NIC96 all suppress nup116-C lethality, and high-copy SNL1 suppresses the temperature sensitivity of gle2–1 and nic96-G3 mutants. The nic96-G3 (previously reported as gle3) and gle2–1 mutant alleles are synthetically lethal with a null allele of the closely related nup100 (Murphy et al., 1996). POM152 is included in this scenario based on a report by others that a nic96 mutant allele is synthetically lethal with the pom152 null mutant (Aitchison et al., 1995). The specificity of these genetic interactions and the fact that the nup116 and gle2 null alleles are not high-copy suppressed strongly suggests that the function of the encoded polypeptides is closely connected. Second, Snl1p is only the second reported yeast integral membrane protein with a functional connection to NPCs, the other is Pom152p (Wozniak et al., 1994), and critical roles for integral membrane proteins are implicit in the proposed models for NPC function and biogenesis. The entire length of Snl1p is homologous to a portion of Pom152p that flanks the single membrane-spanning segment (Figure 4). Interestingly, our results have shown that Snl1p is oriented as a type I membrane protein, whereas Pom152p topology is predicted to be type II (Wozniak et al., 1994). If the sequence homology is significant, Pom152p may in fact be a type I membrane protein with the bulk of its mass cytoplasmically exposed. Experimental determination of Pom152p topology will be required to test this model.
Figure 11.
Schematic diagram of molecular links between the functions of SNL1, GLE2, NUP116, NUP100, NIC96, and POM152. The dashed lines indicate a synthetic lethal phenotype between mutant alleles, the dashed lines with arrowheads indicate high-copy suppression of a mutant allele by a wild-type gene (with the orientation pointing toward the conditional phenotype of the gene being suppressed), and the solid line indicates direct in vitro protein–protein interactions.
It is intriguing that nup116-C and gle2–1 mutants exhibit similar herniation phenotypes under lethal growth conditions, and that they are both suppressed by overexpression of a gene encoding an integral membrane protein, SNL1. In addition, although nic96 mutants do not exhibit any NPC/nuclear envelope herniated structures, nup188 mutants that are synthetically lethal with nic96 have extensive nuclear envelope perturbations and herniation-like structures (Nehrbass et al., 1996; Zabel et al., 1996). At this point, we predict that the increased levels of the integral membrane protein Snl1p have a stabilizing influence on these particular NPC mutants. If overexpressed Snl1p stabilizes mutant NPC structures that are predisposed to forming herniation-like structures, endogenous Snl1p may have a similar stabilizing role during NPC biogenesis. We predict that Snl1p will be both spatially and temporally poised for stabilization of either the pore membrane fusion event, the newly formed pore, or the assembling NPC substructures.
Alternatively, Snl1p may have a general endoplasmic reticulum function that is indirectly required for normal nuclear envelope and NPC function. The localization of Snl1p in both endoplasmic reticulum and nuclear membranes is consistent with this hypothesis. Moreover, three other proteins with primary roles in endoplasmic reticulum function have reported connections to NPC structure and function. A mutant allele of ACC1 (mtr7–1/acc1–7-1), encoding an enzyme required for fatty acid biosynthesis, inhibits RNA export and perturbs nuclear envelope structure (Mishina et al., 1980; Schneiter et al., 1996). Nuclear transport defects are also observed with SEC63 mutants (npl1), a protein required during polypeptide translocation across the endoplasmic reticulum (Rothblatt et al., 1989; Sadler et al., 1989). A fraction of Sec13p, a component of the COPII coatomer complex, is coprecipitated with a subcomplex of NPC proteins (Barlowe et al., 1994; Siniossoglou et al., 1996). However, in terms of SNL1 function, the lack of mtr7–1/acc1–7-1, sec17–1, and sec18–1 suppression by high-copy SNL1 highlights a potential functional specificity for Snl1p at the NPC.
We previously predicted that the different structural regions of Nup116p would have distinct roles in NPC function (Wente et al., 1992). Moreover, we proposed that the respective FG and GLFG regions are functionally unique from one another (Rout and Wente, 1994; Iovine et al., 1995). The GLFG region may be involved in nuclear export processes by interacting with exporting factors (Stutz et al., 1995; Fritz and Green, 1996; Murphy and Wente, 1996; Stutz et al., 1996; Iovine and Wente, 1997; Powers et al., 1997). In particular, a nuclear export sequence-dependent interaction of the nuclear import factor Kap95p with the GLFG region of Nup116p has been characterized (Iovine and Wente, 1997), and supports models for the GLFG region serving as a docking site for Kap95p recycling and nuclear exit. In contrast, the N-terminal region of Nup116p was both necessary and sufficient for binding Gle2p in vitro (Figure 10). This suggests that the N-terminal region is required for docking Gle2p at the NPC and may mediate the role of Gle2p in RNA export (Murphy et al., 1996). Overall, our results suggest that both the N-terminal FG and the GLFG regions of Nup116p mediate nuclear export steps via interaction with distinct factors. These interactions likely have significance in vivo because Nup116p, Gle2p, and Kap95p coimmunoprecipitate in a complex from total yeast nuclei lysates (Iovine and Wente, 1997).
There are at least two possible mechanisms by which Snl1p could be connected to the function of Nup116p, Gle2p, and Nic96p: 1) Snl1p could directly interact with one or more of these peripheral membrane proteins or 2) Snl1p could associate with NPCs through an unidentified, intervening factor(s). Our preliminary membrane topology results suggest that the vast majority of the Snl1p is cytoplasmically exposed, and therefore direct protein–protein interactions between Snl1p and peripheral nucleoporins are possible. Because the SNL1 genetic suppression is specific to the nup116-C and gle2–1 mutant alleles (it does not suppress the corresponding null mutants), Snl1p is a good candidate for interaction with either the C-terminal region of Nup116p or with Gle2p, and direct tests are in progress. Determining which proteins directly interact with Snl1p will be critical for understanding its function.
If Snl1p does not interact with the C-terminal region of Nup116p, the factor that directly interacts with this region may be one of the remaining high-copy suppressors of the nup116-C lethal phenotype. GLE2 and NIC96 were not identified in the original analysis, reflecting the fact that the high-copy genetic screen for nup116-C suppressors was not saturating. Indeed SNL1 was identified from characterization of only 19 of the original 178 isolates. Revealing the factors that interact with the C-terminal region of Nup116p may provide insight into the maintenance of NPC structure and NPC–pore membrane interactions.
ACKNOWLEDGMENTS
We are indebted to our colleagues K. Iovine and J. Watkins for noting the lethal nup116-C phenotype and to R. Murphy for isolation of gle3. We thank J. Watkins for excellent technical assistance; Lori LaRose and Marilyn Levi for assistance with the electron microscopy; and numerous colleagues for generously sharing reagents: A. Tartakoff for the mtr7–1/acc1–7-1 strain; M. Latterich and R. Schekman for the sec17–1 and sec18–1 strains; P. Silver for the npl4 reagents; R. Wozniak for the pom152 strain; M. Bucci for the nup133 strain; M. Rose for the Kar2p antibody; R. Fuller for the Kex2p antibody; and C. Strambio-de-Castillia, G. Blobel, and M. Rout for the mAb118C3 antibody. K. Iovine, R. Murphy, M. Bucci, C. Nicchitta, D. Schnell, and K. Blumer provided valuable comments on the article and critical discussions. This work was supported by a grant from the National Institutes of Health, GM-51219, to S.R.W.
Footnotes
Abbreviations used: BAPTA, 1,2-bis-(O-Aminophenoxy)-ethane-N,N,N′,N′-tetra acetic acid; FG, phenylalanine-glycine; GLFG, glycine-leucine-phenylalanine-glycine; GST, glutathione S-transferase; NEM, N-ethylmaleimide; NPC, nuclear pore complex; NRM, nucleoporin RNA-binding motif; Δ, null.
REFERENCES
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama NR, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by the Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier KO, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Delannoy MR, Wilson KL. GTP hydrolysis is required for vesicle fusion during nuclear envelope assembly in vitro. J Cell Biol. 1992a;116:281–294. doi: 10.1083/jcb.116.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Taylor TC, Melancon P, Wilson KL. A role for ADP-ribosylation factor in nuclear vesicle dynamics. Nature. 1992b;358:512–514. doi: 10.1038/358512a0. [DOI] [PubMed] [Google Scholar]
- Burke B, Gerace L. A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle MC, Loos K, Scheer U. Identification of a soluble precursor complex essential for nuclear pore assembly in vitro. Chromosoma. 1990;100:56–66. doi: 10.1007/BF00337603. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- DeHoratius C, Silver PA. Nuclear transport defects and nuclear envelope alterations are associated with mutations of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz CC, Green MR. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- Fuller RS, Brake AJ, Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989;246:482–486. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- Gerace L, Ottaviano Y, Kondor-Koch C. Identification of a major polypeptide of the nuclear pore complex. J Cell Biol. 1982;95:826–837. doi: 10.1083/jcb.95.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. Structural and functional organization of the nuclear envelope. Curr Opin Cell Biol. 1995;7:301–309. doi: 10.1016/0955-0674(95)80083-2. [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Wiese C, Allen TD, Wilson KL. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci. 1997;110:409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with ’GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995a;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995b;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 1990;9:1495–502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Yocum RR, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA. 1982;79:7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Latterich M, Schekman R. The karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP-γ-S, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina CV, Riggs PD, Grandea A, III, Slatko BE, Moran LS, Tagliamonte JA, McReynolds LA, Guan CD. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Mishina M, Roggenkamp R, Schweizer E. Yeast mutants defective in acetyl-coenzyme A carboxylase and biotin:apocarboxylase ligase. Eur J Biochem. 1980;111:79–87. doi: 10.1111/j.1432-1033.1980.tb06077.x. [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a S. cerevisiae homologue of the S. pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Wente S. An RNA export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth KA, Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer DD, Forbes DJ. A N-ethylmaleimide-sensitive cytosolic factor necessary for nuclear protein import: requirement in signal-mediated binding to the nuclear pore. J Cell Biol. 1990;110:547–557. doi: 10.1083/jcb.110.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Newport JW, Dunphy W. Characterization of the membrane binding and fusion events during nuclear envelope assembly using purified components. J Cell Biol. 1992;116:295–306. doi: 10.1083/jcb.116.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- Pfaller R, Smythe C, Newport JW. Assembly/disassembly of the nuclear envelope membrane: cell cycle-dependent binding of nuclear membrane vesicles to chromatin in vitro. Cell. 1991;65:209–217. doi: 10.1016/0092-8674(91)90155-r. [DOI] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EJ, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris H. The three dimensional structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull. 1991;21:54–56. [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2642–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Sadler I, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch E-V, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C, Stirling C, Schekman R, Rine J. Genetic and biochemical evaluation of eukaryotic membrane topology: multiple transmembrane domains of Saccharomyces cerevisiae 3-hydroxyl-3-methylglutaryl coenzyme A reductase. Mol Cell Biol. 1990;10:672–680. doi: 10.1128/mcb.10.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:665–675. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E, Mattaj IW, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Sullivan KM, Busa WB, Wilson KL. Calcium mobilization is required for nuclear vesicle fusion in vitro: implications for membrane traffic and IP3 receptor function. Cell. 1993;73:1411–1422. doi: 10.1016/0092-8674(93)90366-x. [DOI] [PubMed] [Google Scholar]
- Vigers GP, Lohka MJ. A distinct vesicle population targets membranes and pore complexes to the nuclear envelope in Xenopus eggs. J Cell Biol. 1991;112:545–556. doi: 10.1083/jcb.112.4.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigers GP, Lohka MJ. Regulation of nuclear envelope precursor functions during cell division. J Cell Sci. 1992;102:273–284. doi: 10.1242/jcs.102.2.273. [DOI] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Gasser SM, Caplan AJ. The nucleus and nucleocytoplasmic transport in Saccharomyces cerevisiae. In: Broach JR, Jones E, Pringle J, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 471–546. [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Fuller RS. Posttranslational processing of the prohormone-cleaving Kex2 protease in the Saccharomyces cerevisiae secretory pathway. J Cell Biol. 1991;115:297–307. doi: 10.1083/jcb.115.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson BM, Critchley AJ, Stirling CJ. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J Biol Chem. 1996;271:25590–25597. doi: 10.1074/jbc.271.41.25590. [DOI] [PubMed] [Google Scholar]
- Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt EC. A new subclass of nucleoporins that functionally interact with nuclear pore protein NSP1. EMBO J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Bartnik E, Blobel G. Primary structure analysis of an integral membrane glycoprotein of the nuclear pore. J Cell Biol. 1989;108:2083–2092. doi: 10.1083/jcb.108.6.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G, Rout MP. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]