Abstract
Polyunsaturated fatty acids can be converted to lipid hydroperoxides through non-enzymatic and enzymatic pathways. The prototypic ω-6 lipid hydroperoxide 13-hydroperoxy-octadecadienoic acid (13-HPODE) decomposes homolytically to form highly reactiveα,β-unsaturated aldehydes, such as 9,12-dioxo-10(E)-dodecenoic acid (DODE), 4-oxo-2(E)-nonenal (ONE), 4,5-epoxy-2(E)-decenal (EDE), and 4-hydroxy-2(E)-nonenal (HNE), that can form covalent adducts with DNA. Both 4-oxo-2(E)-nonenal and 4-hydroxy-2(E)-nonenal can also modify proteins to form products that can potentially serve as biomarkers of lipid hydroperoxide-mediated macromolecule damage. In this study cytochrome C was used to identify and characterize the modification sites individually for each of these aldehydes and also to determine the most abundant adduct formed following decomposition of 13-HPODE. The adducts were characterized by ESI-TOF/MS analysis of the intact proteins and by a combination of ESI-ion-trap/MSn and quadrupole-TOF/MS/MS analysis of the tryptic and chymotryptic peptides. The major adducts included an HNE-His Michael adduct on H33, EDE-Lys adducts on K7 and K8, ONE-Lys ketoamide adducts on K5, K7, and K8, an apparent ONE-Lys Michael adduct on K5, and DODE-Lys carboxyl ketoamide adducts on K86 and K87. DODE was the most reactive aldehyde toward cytochrome C. The major adduct from this reaction was analogous to the most abundant adduct resulting from the decomposition of 13-HPODE in the presence of cytochrome C.
Introduction
Lipid peroxidation has been implicated in degenerative diseases of aging, specifically cancer, cardiovascular disease, and neurodegeneration. It can occur non-enzymatically through the action of reactive oxygen species (ROS) or enzymatically through the action of cyclooxygenases and lipoxygenases (1,2). The polyunsaturated fatty acid residues of phospholipids are extremely sensitive to oxidation. Thus, phospholipids provide an excellent source for lipid-derived endogenous toxins.
Lipid hydroperoxides that are formed either enzymatically or non-enzymatically can then undergo transition-metal-ion or vitamin C-mediated decomposition to form a number of α,β-unsaturated aldehydes via multiple pathways. One pathway leads to the formation of 4,5-epoxy-2(E)-decenal (EDE) via α-cleavage of an alkoxy radical (3). The second is thought to involve a Hock rearrangement to form 4-hydroperoxy-2(E)-nonenal (HPNE), which is the precursor to 4-oxo-2(E)-nonenal (ONE) and 4-hydroxy-2(E)-nonenal (HNE) (4–8). Recently, it has been shown that 9,12-dioxo-10(E)-dodecenoic acid (DODE) can be formed from 13-hydroperoxy-octadecadienoic acid (13-HPODE), also through a Hock rearrangement, to 9-hydroperoxy-12-oxo-10(E)-dodecenoic acid (HPODD) which then dehydrates (9) (Scheme 1). Similiarly, 9-hydroperoxy-octadecadienoic acid (9-HPODE) was also shown to decompose to DODE (10). Theα,β-unsaturated aldehydes are bifunctional electrophiles that can covalently modify DNA, amino acids, and proteins.
Scheme 1.
Vitamin C-mediated decomposition of linoleic acid- and arachidonic acid-derived lipid hydroperoxides to bifunctional electrophiles.
Protein modifications can potentially serve as biomarkers of lipid hydroperoxide-mediated macromolecule damage in vivo. Since the aldehydes themselves are very reactive, their corresponding protein adducts might provide a more robust dosimeter due to their longer half-lives. HNE-induced protein modifications have been extensively studied. Specifically, HNE-protein adducts were found to occur at the sulfhydryl group of cysteine, the ε-amino group of lysine, and the imidazole of histidine (11,12). More recently, ONE has been shown to be even more reactive than HNE in modifying proteins (13,14).
There has been, however, a lack of studies investigating the modifications induced by the other aldehydes generated from 13-HPODE and none comparing them all with the same protein.
Cytochrome C is an ideal model for these experiments for several reasons, including its small size, i.e., 12 kDa, which allows for easy monitoring of molecular-weight increases of the intact protein. It is also highly conserved between species and is well characterized structurally by X-ray crystallography and NMR (15–18). In addition, its location in the inner membrane of the mitochondria, a highly oxidizing environment, makes it a potential target in vivo.
The goal of this study was to identify and characterize the primary modification site on cytochrome C for each aldehyde and also determine the most abundant adduct formed following decomposition of 13-HPODE.
Materials and Methods
Reagents and supplies
Cytochrome C, ammonium bicarbonate, acetic acid, chymotrypsin, trifluoroacetic acid, and ascorbic acid were purchased from Sigma Chemical Co. (St. Louis, MO). Trypsin was purchased from Promega (Madison, WI). Chelex-100 chelating ion-exchange resin (100–200 mesh size) was purchased from Bio-Rad. Gases were supplied by AirGas (Salem, NH). HNE was purchased from Cayman Chemicals. ONE, EDE, DODE, and 13-HPODE were a generous gift from Ian A. Blair (University of Pennsylvania).
Mass spectrometry
Mass spectrometry experiments were carried out on three instruments: an Agilent ESI-TOF mass spectrometer (Palo Alto, CA), an Applied Biosystems (Framingham, MA) QSTAR XL quadrupole-TOF mass spectrometer equipped with a nanospray source (Proxeon Biosystems, Odense, Denmark), and an Agilent XCT Ultra ion-trap.
The ESI-TOF was typically operated at a needle voltage of 4 kV. Nitrogen was used as the drying (10 l/min), and nebulizer (15 psig) gas with the heated capillary at 325°C. Liquid chromatography was performed using an Agilent 1100 LC system with a Zorbax 300SB-C18 (2.1 × 100mm; 3.5μm; Agilent Technologies Palo Alto, CA) column at a flow rate of 0.2 mL/min. Solvent A was 0.25% acetic acid in water and solvent B was 0.25% acetic acid in 90% acetonitrile with a typical gradient of 5% B at 0 min to 50% B at 45 min, 80% B at 50 min, 5% B at 55 min, and 5% B at 65 min.
MSn analyses were done on theAgilent ion-trap instrument. ESI was conducted using a needle voltage of 3.5 kV. Nitrogen was used as the drying (12 l/min), and nebulizer (10 psig) gas with the heated capillary at 320°C. Targeted MSn was performed using helium as the collision gas at 1.2 V. Liquid chromatography was essentially identical to that described above for the ESI-TOF except for the column dimensions (1 mm × 50 mm, 3.5 μM) and flow-rates (20 μL/minute).
The QStar was used primarily for LC/MS/MS experiments. The electrospray interface used a micro-tee (Upchurch Scientific, Oak Harbor, WA) with a 1-in. piece of platinum rod, inserted into one arm of the micro-tee, to supply the electrical connection. The electrospray voltage was typically 2.3–2.7 kV applied just upstream of the column. Data-dependent MS/MS analysis was performed on the most intense peaks in each full-scan spectrum, using multiply-charged ions (most of the non-peptide background constituents are singly charged). Accumulation time and pulsar frequency were maintained at 3 s and 6.99, respectively; the mass tolerance was 50 mmu and the collision gas pressure setting was 6. The chromatographic system was based on an Agilent capillary 1100 LC binary pump with fused-silica capillary columns (75 μm i.d. × 360 μm o.d.; 14 cm length, tip 8 μm, New Objective, Woburn, MA) that were packed in-house, with 5-μm C18 reverse-phase material (Vydac, Hesperia, CA). The flow rate was 5 μL/minute into a passive splitter that delivered approximately 0.2 μL/minute through the columns. Solvent A was 0.5% acetic acid in water and solvent B was 0.5% acetic acid in 90% acetonitrile. A linear gradient was performed as follows: 2% B at 0 min, 2% B at 3 min, 65% B at 70 min, 80% B at 80 min, 80%B at 90 min, 2%B at 130 min, and 2% B at 170 min.
Reaction of cytochrome C with α,β-unsaturated aldehydes
Cytochrome C (8 nmols, 100 μl in pH 7.0 Chelex-treated 100 mM HEPES buffer) was reacted with each aldehyde at varying concentrations for 24 hrs at 37°C. The samples were filtered (regenerated cellulose 3,000 Da molecular-weight cutoff; Amicon) to remove excess aldehyde. The samples were then brought up in 50 mM ammonium bicarbonate (100 μl, pH 7.0), and an aliquot was removed for intact protein analysis. Trypsin (1:25 at 37°C for 24h) and chymotrypsin (1:10 at 37°C for 3h) were used for proteolytic digestion on the remaining sample. Samples were extracted with Millipore C18 ZipTips (Bedford, MA), washed with 0.1%TFA in water (10 μL × 3) and eluted with 0.1% TFA in 70% acetonitrile (10 μL).
Reaction of cytochrome C or myoglobin with 13-HPODE
Cytochrome C or myoglobin (8 nmols, 100 μl in pH 7.0 Chelex-treated 100 mM HEPES buffer) was reacted with 13-HPODE (224 nmols, 10 μL ethanol) in the presence of vitamin C (2 mM) at 37°C for 24 h (19). The sample was filtered (regenerated cellulose 3,000 Da MWC) to remove unreacted 13-HPODE and vitamin C. The sample was then brought up in 50 mM ammonium bicarbonate (100 μL, pH 7.0). An aliquot for intact protein analysis was removed. Trypsin (1:25 at 37°C for 24h) or chymotrypsin (1:10 at 37°C for 3h) were used for proteolytic digestion on the remaining sample. The samples were extracted with Millipore ZipTipC18 (Medford, MA), washed with 0.1%TFA in water (10 μL × 3) and eluted with 0.1% TFA in 70% acetonitrile (10 μL).
Reaction of peptides with α,β-unsaturated aldehydes
Peptides were reacted with a >5 fold excess of each aldehyde in pH 7.0 chelex-treated 100 mM HEPES buffer for 24 hrs at 37°C. Samples were extracted with Millipore C18 ZipTips (Bedford, MA), washed with 0.1%TFA in water (10 μL × 3) and eluted with 0.1% TFA in 70% acetonitrile (10 μL).
Results
Cytochrome C
Unmodified intact cytochrome C was characterized on the ESI-TOF and quadrupole time-of-flight instruments. The molecular weight of the most abundant molecular form of cytochrome C was found to be 12,358.4168 Da. Using tryptic and chymotryptic digests we were able to obtain 86% coverage of the protein.
DODE Modification of cytochrome C
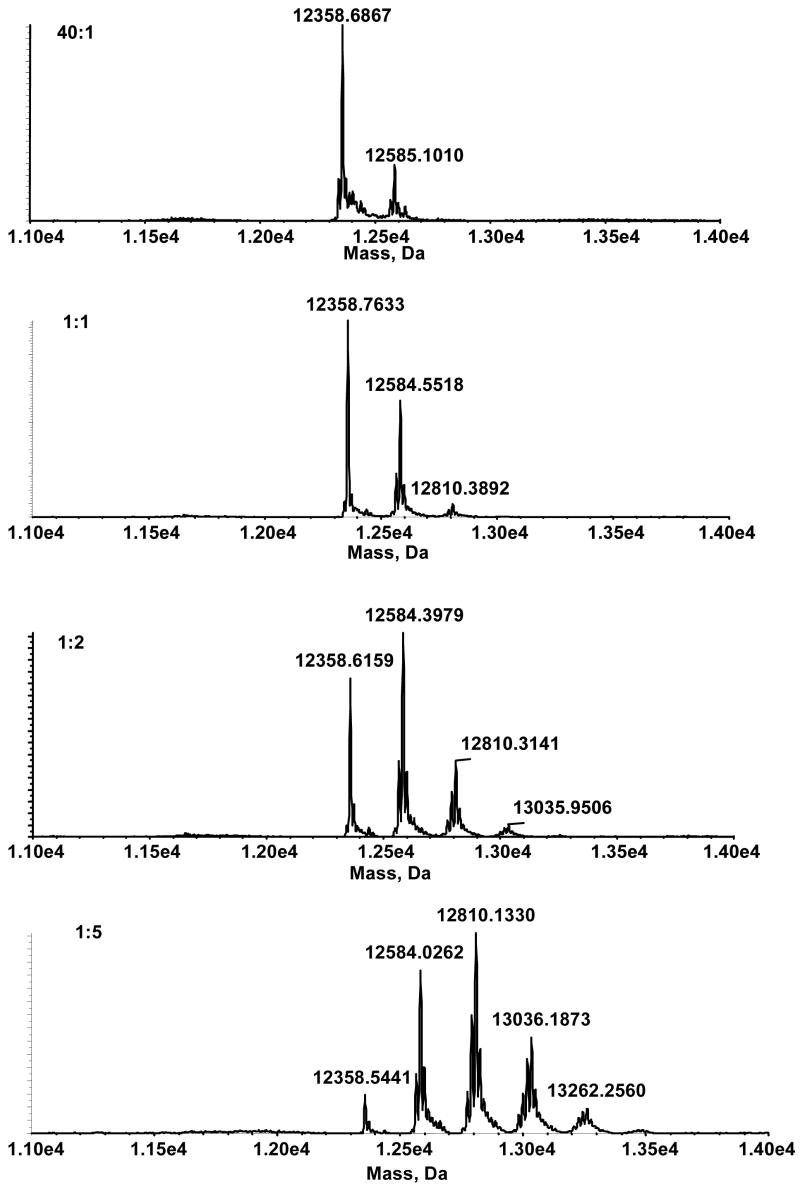
Adduct formation with increasing amounts of DODE in the presence of cytochrome C was followed with the ESI-TOF. Under the conditions used in our experiments, several cytochrome-C-modified species were observed. Beginning at a concentration ratio of 40:1 (cytochrome C:DODE), a peak at m/z 12584.0359 Da, representing a mass shift of 226 Da, was observed (Figure 1a). As the concentration of DODE increased the intensity of m/z 1284.0359 Da increased until a concentration ratio of 1:1 when another peak, m/z 12810.1251 Da, emerged (Figure 1b). At 1:2 a third modified peak, again with a mass shift of 226 Da, emerged at m/z 13036.1685 Da (Figure 1c). Finally, at the highest ratio (1:5,) we observed three substantial peaks and the beginning of a fourth (Figure 1d). As expected, the signal corresponding to unreacted cytochrome C at m/z 12358.4168 Da decreased as the relative concentration of DODE increased; at the highest concentrations of DODE, virtually no unreacted cytochrome C remained.
Figure 1.
Deconvoluted spectra of modified cytochrome C arising from reaction with increasing concentrations of DODE. Ratio of the concentration of cytochrome C:DODE, (a) 40:1 (b) 1:1 (c) 1:2 (d) 1:5.
The adducts were characterized using both tryptic and chymotryptic digests analyzed on the ESI-TOF followed by both data dependent and targeted MS/MS (on the QStar and the ion trap instruments, respectively) of peptides unique to the treated digests. The intact protein data indicated that the [M+H+]+1 ion of a modified peptide should be increased by 226 Da with respect to the unmodified peptide. Three such peptides were detected in the chymotryptic digests.
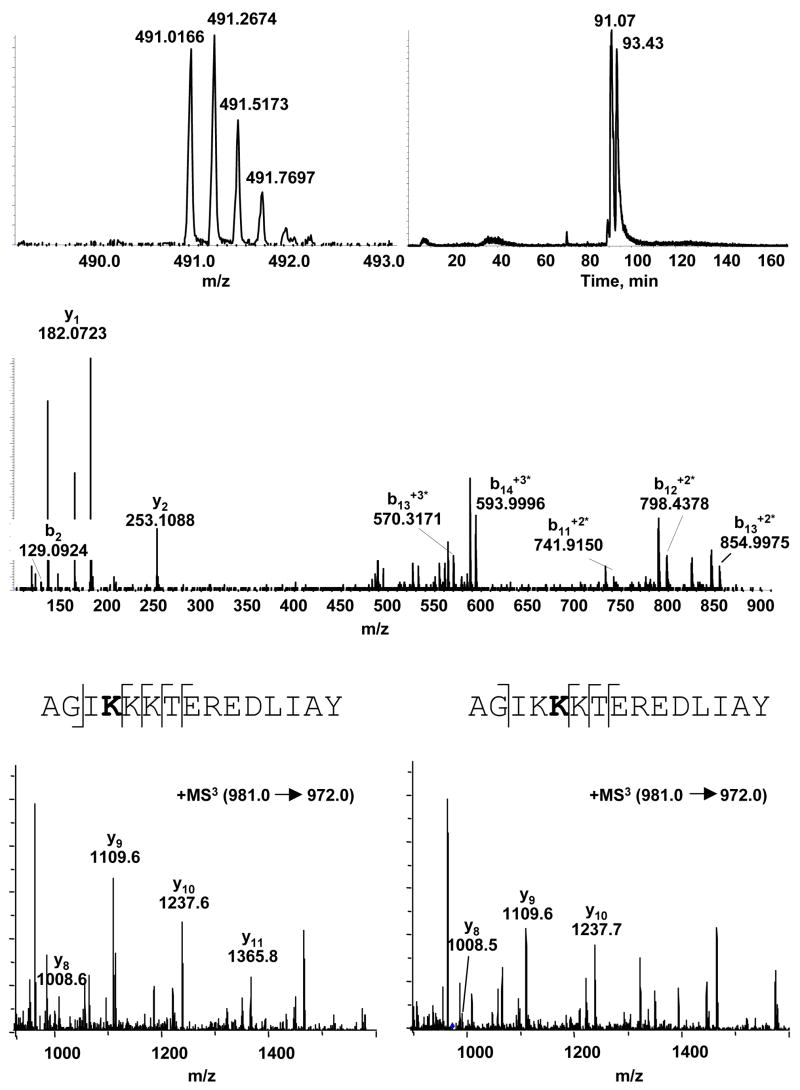
The first modified peptide, observed at ratios of cytochrome C:DODE that yielded only a single modified peak in the analysis of the intact proteins, was 83AGIKKKTEREDLIAY97 ([M+4H+]+4 at m/z = 491.0166; Figure 2a). The extracted ion chromatogram of that peptide revealed the presence of two peaks, at 91.07 minutes and 93.43 minutes, suggesting that more than one amino acid on the peptide was modified (Figure 2b). The MS/MS spectrum (QStar) of AGIKKKTEREDLIAY did not show appropriate fragmentation to allow determination of the site of modification, so targeted MS3 (ion-trap) was conducted on [M+2H+]+2 at m/z = 981 and b15 +2* (m/z = 972) (Figure 2c–e). The combined information from these experiments narrowed the site of adduction to K86 and K87. The presence of y11 in the first peak (91.07 min) and b2 suggested that the adduct was on K86 since isoleucine has no reactive side groups. The second peak (93.43 min) showed a y10 ion but no y11, consistent with an adduct at K87.
Figure 2.
Chymotryptic digest of DODE-modified cytochrome C. (a) LC/MS analysis of modified AGIKKKTEREDLIAY; [M+4H+]+4 at m/z 491.0166 (b) Extracted ion chromatogram of m/z 491 (c) representative LC/MS/MS CID mass spectra of both peaks with m/z 491 (d) LC/MS/MS CID mass spectra of the peak at 91.07 minutes (e) LC/MS/MS CID mass spectra of the 93.43 minute peak.
At higher ratios of DODE-to-protein, two additional modified peptides appeared, i.e., [M+2H+]+2 at m/z 694.8776, corresponding to GDVEKGKKIF (1–10) and [M+2H+]+2 at m/z 515.2842, corresponding to 98LKKATNE104 (Table 1). The extracted ion chromatograms for both peptides revealed the presence of multiple peaks, four peaks for GDVEKGKKIF and two peaks for LKKATNE. The MS/MS spectra of modified GDVEKGKKIF revealed the sites of adduction to be E4 or K5, K5, K7 and K8; those for modified LKKATNE revealed sites of adduction on both K99 and K100.
Table 1.
DODE-modified cytochrome C peptides observed by LC-MS/MS after chymotryptic digestion.
| Sequence (residue numbers) | calc. m/z | obs. m/z | adduct location |
|---|---|---|---|
| AGIKKKTEREDLIAY (83–97) | 490.9998+4 | 491.0166+4 | K86, K87 |
| GDVEKGKKIF (1–10) | 694.8223+2 | 694.8776+2 | K5, K7, K8 |
| LKKATNE (98–104) | 515.2353+2 | 515.2842+2 | K99, K100 |
ONE Modification of cytochrome C
Several cytochrome C modified species were observed in LC/MS experiments on the ESI-TOF mass spectrometer. Beginning at a concentration ratio of 10:1 (cytochrome C:ONE) a peak at 12512.9510 Da, representing a mass shift of 154 Da, was observed. As the concentration of ONE increases the intensity of 12512.9510 Da increases until a concentration of 2:1 when another peak, 12666.8432 Da, emerges. At 1:2 a third modified peak emerges at 12820.5131 Da. Finally, at our highest ratio of 1:5 we observed three substantial peaks, each shifted 154 Da, and the very beginning of a fourth modified peak.
The adducts were characterized using tryptic and chymotryptic digests analyzed by ESI-TOF followed by data-dependent MS/MS (on the QTOF) of peptides unique to the treated protein. The data from the intact modified protein indicated that a modified peptide would be expected to have the [M+H+]+1 ion increased by 154 Da. Three unique peptides, corresponding to the three peaks observed in the intact analysis, were detected in the chymotryptic digests.
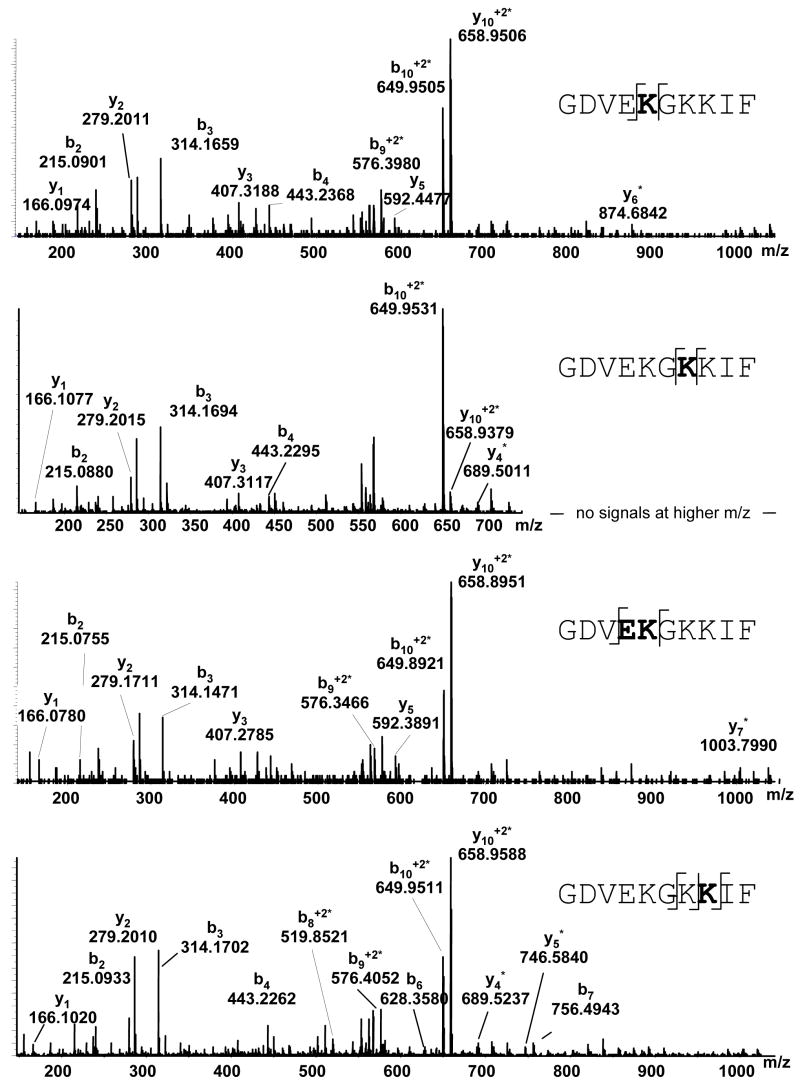
The first modified peptide, observed at ratios of cytochrome C:ONE less than 2:1, was GDVEKGKKIF (1–10) ([M+2H+]+2 at m/z 658.9564). The extracted ion chromatogram on that ion revealed the presence of four peaks, again suggesting that more than one amino acid on the peptide had been modified. The MS/MS spectrum of the other [M+2H+]+2 658.9564 peaks revealed the sites of adduction as K5, K7 and K8. The presence of b4, y5, and y6*in the first peak (103.14 min) confirm that the adduct is on K5. The second peak (104.56 min) has y5 and y7*suggesting that the adduct is on K5 or E4. The third peak (106.72 min) corresponds to an adduct on K7 attributable by the presence of y3 and y4*. The last peak at 107.81 min shows y2, y4*, b7, and b8* verifying that K8 is modified.
At higher concentrations of cytochrome C:ONE two additional modified peptides appeared, i.e., [M+2H+]+2 at m/z 515.2842 corresponding to 98LKKATNE104 and 60KEETLMEY67 with [M+2H+]+2 at m/z 598.8644. The extracted ion chromatogram of LKKATNE revealed the presence of multiple peaks, while KEETLMEY contained only a single peak. The MS/MS spectrum of LKKATNE revealed the sites of adduction as K99 and K100. K60 was found to be the site modified on 60KEETLMEY67.
HNE Modification of cytochrome C
Analyses of adduct formation using increasing amounts of HNE in the presence of cytochrome C were carried out using the ESI-TOF. Under the conditions used in our experiments, a single cytochrome C modified species was observed. Beginning at a concentration of 2:1 (cytochrome C:HNE) a peak at 12515.1502 Da was observed, corresponding to a mass shift of 156 Da. As the concentration of HNE increased the intensity of this signal ncreased, but, even at higher relative concentrations of HNE, a substantial amount of unreacted protein remained.
The adduct was characterized using tryptic and chymotryptic digests analyzed by ESI-TOF followed by data dependent MS/MS (on the QTOF) of peptides unique to the treated spectrum. The modified intact protein data suggested that a modified peptide would have an MH+ increased by 156 Da. A single modified peptide was detected in the tryptic digests: 28TGPNLHGLFGR38 with [M+2H+]+2 at m/z 584.8151. The MS/MS spectrum revealed the site of adduction to be H33 based on the presence of y5 and y6*.
EDE Modification of cytochrome C
As was the case with HNE, a single adducted protein was observed, with a molecular weight of 12409.0593 Da - corresponding to a mass shift of approximately 50 Da. (At the highest HNE:protein ratio of 1:5, another product appeared, but this has not been characterized.)
The major adduct was characterized using tryptic and chymotryptic digests analyzed by ESI-TOF followed by targeted MS/MS (on both the quadrupole-TOF and ESI-Trap) of the peptide unique to the treated spectrum. A single modified peptide with the expected mass shift of 50 Da was detected in the chymotryptic digests, i.e., 1 GDVEKGKKIF10 at [M+2H+]+2 606.8227 (Figure 4a). The extracted-ion chromatogram revealed two doubly-charged peaks at m/z 606.8227 with nearly identical MS/MS spectra (Figure 4b). The first peak (97.7 minutes) has b7 and b8* suggesting the modification is on K8, while the second peak (100.28 minutes) has y3 and y5* revealing the adduct to be on K7 or G6 (Figures 4c–d). Since glycine, however, has no side chain, the site of modification in the second peak appears to be K7.
Figure 4.
Chymotryptic digest of EDE modified cytochrome C. (a) LC/MS analysis of modified GDVEKGKKIF: [M+2H+]+2; m/z 606.8804 (b) Extracted ion chromatogram of m/z 606.9 (ion trap). (c) LC/MS/MS CID mass spectrum of the 97.70 min peak (d) LC/MS/MS CID mass spectrum of the 100.28 minute peak.
13-HPODE modification of cytochrome C
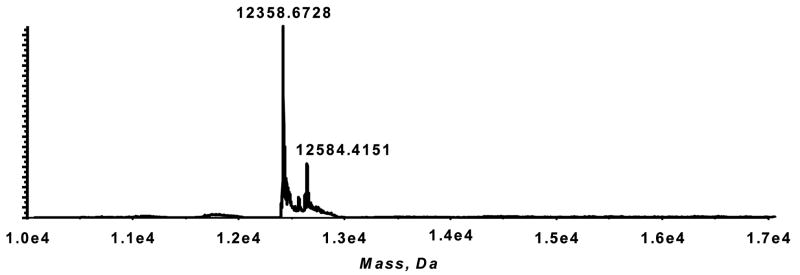
Homolytic decomposition of 13-HPODE was induced by vitamin C in the presence of cytochrome C. A modified protein peak at 12584.4151 Da was observed, representing a mass shift of 226 Da (Figure 5). This is identical to that observed with the reaction of DODE and cytochrome C, as is the modified peptide, i.e., 83AGIKKKTEREDLIAY97 at m/z = 491.0166; [M+4H+]+4 (see supplementary material).
Figure 5.
Decomposition of 13-HPODE in the presence of cytochrome C. Deconvoluted spectrum of cytochrome C modified by 13-HPODE.
The above observations are summarized in Table 3.
Table 3.
Summary matrix of adducts for cytochrome C reactions with all aldehydes in the study.
| 83AGIKKKTEREDLIAY97 | 1GDVEKGKKIF10 | 98LKKATNE104 | 28TGPNLHGLFGRK38 | 60KEETLMEY67 | |
|---|---|---|---|---|---|
| DODE | K86, K87 | K5, K7, K8 | K99, K100 | ||
| ONE | K5, K7, K8 | K99, K100 | K60 | ||
| EDE | K7, K8 | ||||
| HNE | H33 |
Discussion
Many studies have examined the reaction of individual lipid hydroperoxide-derived aldehydic bifunctional electrophiles with proteins. Most of these have focused on HNE, and more recently ONE, as the major products that react with proteins (11–14). However, the most important electrophile involved in protein modification is unknown. We have reacted each electrophile with cytochrome C to determine which is most reactive.
The reaction of cytochrome C with HNE had been examined previously by Isom, et al., who showed that HNE formed several adducts including a Michael adduct on H33 (20). We confirmed their finding, and also showed that this adduct is the initial product formed in the reaction of HNE with cytochrome C. Under our conditions HNE was, along with EDE, the least reactive of the aldehydes; no adduct was observed at molar ratios less than 2:1 HNE:cytochrome C. In addition at a 5:1molar ratio of HNE to cytochrome C, a substantial amount of unreacted protein remained.
As noted above, EDE was also less reactive than the other aldehydes; at a 5-molar excess there was only one substantial adduct and mostly un-reacted cytochrome C. The mass shift of the product was 50 Da, i.e., the reaction did not involve Michael addition. MS/MS analysis identified the adducted peptide as 1GDVEKGKKIF10 with a covalent modification on either K7 or K8. A pathway for the formation of these adducts involves an initial SN2 reaction, followed by cyclization and dehydration to form a Schiff base iminium that can undergo retro-aldol condensation to give a pyrrole adduct (for a net addition of C4H2) and a hexanal (21). To verify these structures the target peptide was synthesized and reacted with EDE. Accurate mass measurements (within 3 ppm) confirmed that the addition was C4H2, in support of the proposed mechanism (data not shown). Since the adduct was formed on a peptide containing multiple lysines in close proximity, we investigated whether this was necessary for the reaction dynamics. Several peptides, with alanines replacing the lysines in different combinations, were synthesized and reacted with EDE. We found, in the peptide model, that multiple lysines in close proximity were not necessary, i.e., EDE reacted with lysines at any position (data not shown).
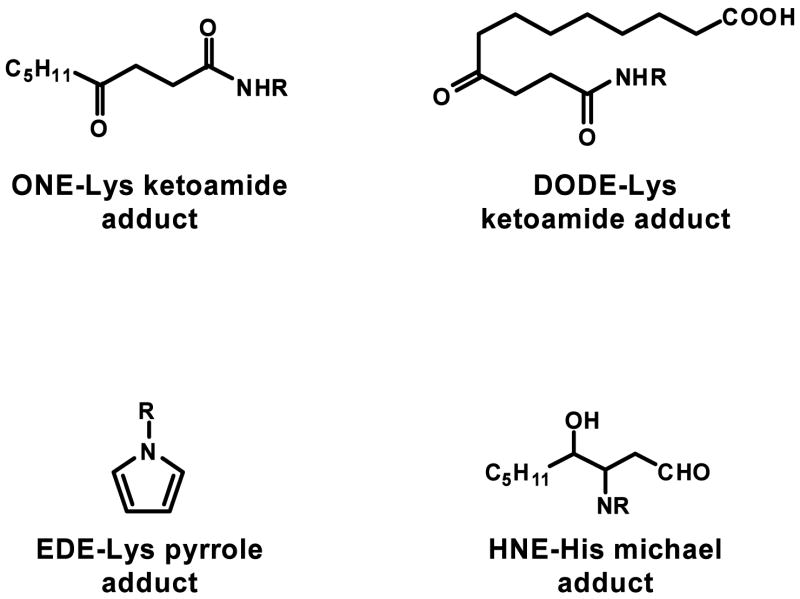
ONE, like EDE, also reacted with the peptide 1GDVEKGKKIF10 although to a much higher degree and with a different mechanism. The adduct formed by cytochrome C and ONE corresponded to an addition of 154 Da, which would be consistent with a Michael adduct. Upon reduction with sodium borohydride, however, there is an addition of 2 Da rather than 4 Da as would be expected for a Michael adduct (data not shown). The addition of only 2 Da suggests that ONE leads to the ketoamide adduct identified and characterized by Sayre et al. (22) (Figure 6). In their proposed mechanism for the ketoamide formation, the trans-α,β-unsaturated aldehyde, in this case ONE, reacts with an amine, R-NH2, to form the corresponding cis-aldehyde or Schiff base, leading ultimately to a 4,5-unsaturated pyrrolidinone intermediate. The addition of water to the double bond of this intermediate, followed by ring opening, results in a ketoamide (22).
Figure 6.
Structures of the α,β-unsaturated aldehyde protein adducts.
At the lowest concentration, ONE formed covalent modifications to 1GDVEKGKKIF10 on E4 or K5, K5, K7, and K8. While it has been shown previously that ONE forms modifications with lysines, a reaction with glutamic acid would be surprising. To test whether the adduct was on E4 or K5, ONE was reacted with the synthesized peptide. Targeted MS/MS analysis was conducted on GDVEKGKKIF (1–10) ([M+2H+]+2 at m/z 658.9564). The second peak (104.56 min) revealed not only y5 and y7* but b4 indicating that the adduct is on K5 not E4 (see supplemental material). Since the first peak and second peak of the extracted ion chromatogram for the peptide correspond to K5 and both peaks correspond to an increase of 154 Da, the peptide appears to form both the ketoamide adduct and a Michael adduct.
DODE reacted with lysine residues in cytochrome C to form stable adducts. These adducts were detectable at concentration ratios as low as 1:40 DODE:cytochrome C, making it the most reactive of the aldehydes in this series. The adducted peptide identified at the lowest DODE concentration was 83AGIKKKTEREDLIAY97 with covalent modifications at K86 or K87. The adducts corresponded to the addition of 226 Da (C12H18O4) from DODE, which, upon sodium borohydride reduction, exhibited an addition of only 2 Da, suggesting a ketoamide adduct (Figure 6; MS data not shown). The DODE ketoamide adduct has also been identified and characterized recently in the Sayre laboratories (L. M. Sayre: personal communication). Person, et al., previously reported adduction by both benzoquinone and glutathione-S-yl-1,4-benzoquinone at sites K86 and K87on cytochrome C, suggesting this is a highly reactive region of the protein (23,24). Pearson, et al., in a study of protein crosslinking agents, also observed reaction at the corresponding locus on bovine cytochrome C, although they declined to speculate on the reasons for activity at these positions (25). Person and co-workers suggest that the enhanced reactivity may reflect conformational flexibility, since lysines 86 and 87 are in loop regions of the protein (23), but this would not explain why only DODE appears to react at these positions. It is possible that the carboxyl group of DODE interacts favorably with the stretch of three basic lysines (K86–K88). As the concentrations of DODE were increased, adducts at other sites were detected - all of which corresponded to the carboxyl ketoamide adduct with lysines (Table 3). DODE was also shown to form both the ketoamide and Michael adduct with K5 on the peptide 1GDVEKGKKIF10 in the same manner as ONE.
We observed that HNE and EDE were essentially equipotent but less reactive than ONE in modifying cytochrome C. DODE, however, appeared to be more reactive than the other three bifunctional electrophiles in forming protein modifications. This correlates with reactions conducted with each of the electrophiles and DNA in vitro. As in our current study, DODE was found to be the most reactive electrophile. The carboxynonanone adduct formed from DODE was found in greater than 2-fold abundance over the next most abundant DNA adduct (4). The DODE-carboxynonanone adduct, however, has never been observed in vivo(26). It has been shown recently that the polar nature of DODE (due to the free carboxyl group) prevents its translocation across the cell membrane, resulting in intracellular accumulation of the reactive aldehyde. This is unlike the other three aldehydes which can easily move across membranes (27). The higher reactivity of DODE compared to ONE, EDE, and HNE, and its ability to accumulate in the cytoplasm, makes its protein modifications important for studying the consequences of lipid hydroperoxide-mediated oxidative stress. In addition we found that the major adduct resulting from the reaction of DODE with cytochrome C was analogous to the most abundant adduct resulting from the decomposition of 13-HPODE in the presence of that protein. The reaction of DODE with myoglobin was also analogous to the most abundant adduct resulting from the decomposition of 13-HPODE (see supplementary materials). This suggests that DODE is the most reactive of this set of aldehydes and that our results are not specific only to cytochrome C.
As noted earlier, DODE may arise from the Hock rearrangement of a bis-hydroperoxide intermediate formed during 13-HPODE decomposition (28). Analogous reactions with the arachidonic acid products, 15-HPETE and 11-HPETE, would not result in the formation of DODE. Therefore, DODE modified proteins could be a specific marker of linoleic acid-derived protein damage. A similar concept has emerged in the studies of leukotriene biosynthesis. A carboxylate-containing DNA adduct, which arises from the reaction between leukotriene A4 and DNA, has been characterized and an LC/MS assay has been developed for its quantitation (29,30).
The preferential sites for lipid hydroperoxide-derived modifications on cytochrome C appear to be primarily lysines. This selectivity could influence the biological function of cytochrome C, since modifications to lysines could have functional effects on the ability of enzyme binding, electron transport efficiency, and apoptosis. Cytochrome C uses various lysine residues including K86 and K87 to bind complexes III and IV in order to shuttle electrons through the electron transport chain (20,31). Disruptions in the binding of these complexes could result in loss of function of mitochondrial respiration. Cytochrome C also uses a number of residues, including K7, in the interaction with Apaf-1. Alteration of this interaction could affect the ability of cytochrome C to initiate apoptosis (32).
Analysis of the decomposition of 13-HPODE in the presence of a single protein provides some new insights concerning potential biomarkers for lipid hydroperoxide-mediated macromolecule damage. Previous studies have focused mainly on HNE as the major lipid hydroperoxide-derived electrophile involved in protein modifications and, more recently, ONE has been shown to be an important electrophile involved in protein modifications. It is now apparent, however, that the major lipid hydroperoxide-derived electrophile involved in these processes may be DODE.
Supplementary Material
1. MS/MS spectrum for the product from the reaction of GDVEKGKIF with ONE. 2. Deconvoluted mass spectra for the products from the reactions of myoglobin with DODE and 13-HPODE. 3. MS data for the 13-HPODE adduct with the synthetic peptide AGIKKKTEREDLIAY for comparison with the data in Figure 2.
Figure 3.
Chymotryptic digest of ONE-modified cytochrome C. (a) LC/MS/MS CID mass spectra of the 103.14 minute peak (b) LC/MS/MS CID mass spectra of the peak at 106.72 minutes (c) LC/MS/MS CID mass spectra of the peak at 104.56 minutes (d) LC/MS/MS CID mass spectra of the peak at 107.81 minutes.
Table 2.
ONE-modified cytochrome C peptides observed by LC-MS/MS after chymotryptic digestion.
| Sequence (residue numbers) | calc. m/z | obs. m/z | adduct location |
|---|---|---|---|
| KEETLMEY (60–67) | 598.7423+2 | 598.8644+2 | K60 |
| GDVEKGKKIF (1–10) | 658.8223+2 | 658.9564+2 | K5, K7, K8 |
| LKKATNE (98–104) | 479.2353+2 | 479.3456+2 | K99, K100 |
Acknowledgments
This work was supported by grants from the Harvard-MIT HST Center for Experimental Pharmacology and Therapeutics, the National Institute of Health (NCI grant CA26731), and the MIT Center for Environmental Health Sciences (NIEHS grant P30 ES002109). We thank Agilent Technologies for access to the XCT Ultra ion trap mass spectrometer. Thanks to Dr. Julie Marr (Agilent) for helpful discussions.
Footnotes
Abbreviations: AA, arachidonic acid; DODE, 9,12-dioxo-10(E)-dodecenoic acid; EDE, 4,5-epoxy-2(E)-decenal; ESI, electrospray ionization; HNE, 4-hydroxy-2(E)-nonenal; HPETE, hydroperoxyeicosatetraenoic acid; HPNE, 4-hydroperoxy-2(E)-nonenal; HPODD, 9-hydroperoxy-12-oxo-10(E)-dodecanoic acid; HPODE, hydroperoxy-octadecadienoic acid; LA, linoleic acid; LC, liquid chromatography; ONE, 4-oxo-2(E)-nonenal; ROS, reactive oxygen species; TOF, time of flight.
References
- 1.Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem. 1999;274(34):23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 2.Laneuville O, Breuer DK, Xu N, Huang ZH, Gage DA, Watson JT, Lagarde M, DeWitt DL, Smith WL. Fatty acid substrate specificities of human prostaglandin-endoperoxide H synthase-1 and -2. Formation of 12-hydroxy-(9Z, 13E/Z, 15Z)- octadecatrienoic acids from alpha-linolenic acid. J Biol Chem. 1995;270(33):19330–19336. doi: 10.1074/jbc.270.33.19330. [DOI] [PubMed] [Google Scholar]
- 3.Lee SH, Oe T, Blair IA. 4,5-Epoxy-2(E)-decenal-induced formation of 1, N(6)-etheno-2′-deoxyadenosine and 1, N(2)-etheno-2′-deoxyguanosine adducts. Chem Res Toxicol. 2002;15(3):300–304. doi: 10.1021/tx010147j. [DOI] [PubMed] [Google Scholar]
- 4.Williams MV, Lee SH, Blair IA. Liquid chromatography/mass spectrometry analysis of bifunctional electrophiles and DNA adducts from vitamin C mediated decomposition of 15-hydroperoxyeicosatetraenoic acid. Rapid Commun Mass Spectrom. 2005;19(6):849–858. doi: 10.1002/rcm.1854. [DOI] [PubMed] [Google Scholar]
- 5.Lee SH, Oe T, Blair IA. Vitamin C-induced decomposition of lipid hydroperoxides to endogenous genotoxins. Science. 2001;292(5524):2083–2086. doi: 10.1126/science.1059501. [DOI] [PubMed] [Google Scholar]
- 6.Lee SH, Blair IA. Characterization of 4-oxo-2-nonenal as a novel product of lipid peroxidation. Chem Res Toxicol. 2000;13(8):698–702. doi: 10.1021/tx000101a. [DOI] [PubMed] [Google Scholar]
- 7.Jian W, Lee SH, Arora JS, Silva Elipe MV, Blair IA. Unexpected formation of etheno-2′-deoxyguanosine adducts from 5(S)-hydroperoxyeicosatetraenoic acid: evidence for a bis-hydroperoxide intermediate. Chem Res Toxicol. 2005;18(3):599–610. doi: 10.1021/tx049693d. [DOI] [PubMed] [Google Scholar]
- 8.Schneider C, Tallman KA, Porter NA, Brash AR. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J Biol Chem. 2001;276(24):20831–20838. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- 9.Lee SH, Silva Elipe MV, Arora JS, Blair IA. Dioxododecenoic acid: a lipid hydroperoxide-derived bifunctional electrophile responsible for etheno DNA adduct formation. Chem Res Toxicol. 2005;18(3):566–578. doi: 10.1021/tx049716o. [DOI] [PubMed] [Google Scholar]
- 10.Gallasch BA, Spiteller G. Synthesis of 9,12-dioxo-10(Z)-dodecenoic acid, a new fatty acid metabolite derived from 9-hydroperoxy-10,12-octadecadienoic acid in lentil seed (Lens culinaris Medik) Lipids. 2000;35(9):953–960. doi: 10.1007/s11745-000-0605-z. [DOI] [PubMed] [Google Scholar]
- 11.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42(4):318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 12.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol Aspects Med. 2003;24(4–5):149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem Res Toxicol. 2003;16(7):901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 14.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15(11):1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 15.Qi PX, Di Stefano DL, Wand AJ. Solution structure of horse heart ferrocytochrome c determined by high-resolution NMR and restrained simulated annealing. Biochemistry. 1994;33(21):6408–6417. doi: 10.1021/bi00187a004. [DOI] [PubMed] [Google Scholar]
- 16.Qi PX, Beckman RA, Wand AJ. Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR. Biochemistry. 1996;35(38):12275–12286. doi: 10.1021/bi961042w. [DOI] [PubMed] [Google Scholar]
- 17.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214(2):585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 18.Banci L, Bertini I, Gray HB, Luchinat C, Reddig T, Rosato A, Turano P. Solution structure of oxidized horse heart cytochrome c. Biochemistry. 1997;36(32):9867–9877. doi: 10.1021/bi970724w. [DOI] [PubMed] [Google Scholar]
- 19.Oe T, Arora JS, Lee SH, Blair IA. A novel lipid hydroperoxide-derived cyclic covalent modification to histone H4. J Biol Chem. 2003;278(43):42098–42105. doi: 10.1074/jbc.M308167200. [DOI] [PubMed] [Google Scholar]
- 20.Isom AL, Barnes S, Wilson L, Kirk M, Coward L, Darley-Usmar V. Modification of Cytochrome c by 4-hydroxy- 2-nonenal: evidence for histidine, lysine, and arginine-aldehyde adducts. J Am Soc Mass Spectrom. 2004;15(8):1136–1147. doi: 10.1016/j.jasms.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Zamora R, Hidalgo FJ. 2-Alkylpyrrole formation from 4,5-epoxy-2-alkenals. Chem Res Toxicol. 2005;18(2):342–348. doi: 10.1021/tx049700y. [DOI] [PubMed] [Google Scholar]
- 22.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and ONE. Drug Metab Rev. 2006;38(4):651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 23.Person MD, Mason DE, Liebler DC, Monks TJ, Lau SS. Alkylation of cytochrome c by (glutathion-S-yl)-1,4-benzoquinone and iodoacetamide demonstrates compound-dependent site specificity. Chem Res Toxicol. 2005;18(1):41–50. doi: 10.1021/tx049873n. [DOI] [PubMed] [Google Scholar]
- 24.Person MD, Monks TJ, Lau SS. An integrated approach to identifying chemically induced posttranslational modifications using comparative MALDI-MS and targeted HPLC-ESI-MS/MS. Chem Res Toxicol. 2003;16(5):598–608. doi: 10.1021/tx020109f. [DOI] [PubMed] [Google Scholar]
- 25.Pearson KM, Pannell LK, Fales HM. Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun Mass Spectrom. 2002;16(3):149–159. doi: 10.1002/rcm.554. [DOI] [PubMed] [Google Scholar]
- 26.Williams MV, Lee SH, Pollack M, Blair IA. Endogenous lipid hydroperoxide-mediated DNA-adduct formation in min mice. J Biol Chem. 2006;281(15):10127–10133. doi: 10.1074/jbc.M600178200. [DOI] [PubMed] [Google Scholar]
- 27.Jian W, Arora JS, Oe T, Shuvaev VV, Blair IA. Induction of endothelial cell apoptosis by lipid hydroperoxide-derived bifunctional electrophiles. Free Radic Biol Med. 2005;39(9):1162–1176. doi: 10.1016/j.freeradbiomed.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Kawai Y, Uchida K, Osawa T. 2′-deoxycytidine in free nucleosides and double-stranded DNA as the major target of lipid peroxidation products. Free Radic Biol Med. 2004;36(5):529–541. doi: 10.1016/j.freeradbiomed.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Hankin JA, Jones DN, Murphy RC. Covalent binding of leukotriene A4 to DNA and RNA. Chem Res Toxicol. 2003;16(4):551–561. doi: 10.1021/tx034018+. [DOI] [PubMed] [Google Scholar]
- 30.Hankin JA, Murphy RC. Mass spectrometric quantitation of deoxyguanosine and leukotriene A4-deoxyguanosine adducts of DNA. Anal Biochem. 2004;333(1):156–164. doi: 10.1016/j.ab.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Poulos TL, Kraut J. A hypothetical model of the cytochrome c peroxidase. cytochrome c electron transfer complex. J Biol Chem. 1980;255(21):10322–10330. [PubMed] [Google Scholar]
- 32.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J Biol Chem. 2001;276(16):13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. MS/MS spectrum for the product from the reaction of GDVEKGKIF with ONE. 2. Deconvoluted mass spectra for the products from the reactions of myoglobin with DODE and 13-HPODE. 3. MS data for the 13-HPODE adduct with the synthetic peptide AGIKKKTEREDLIAY for comparison with the data in Figure 2.