Abstract
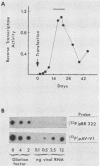
We used a retrovirus shuttle vector to make molecular clones of circular viral DNA from infected cells. One-third of the molecules examined had deletions that started within or near the U5 domain of the long terminal repeat (LTR) region and extended a variable distance toward the gag gene. We present evidence that some of these deletions arose by cleavage of a single LTR unit, in contrast to the cleavage of tandem LTR units associated with the integration reaction. These results suggest that in the formation of defective circular DNA, the U5 domain can be recognized and cleaved in the absence of an adjacent U3 domain. The cleavage of isolated U5 domains may represent an important mechanism responsible for the generation of certain forms of both defective circular DNA and defective integrated DNA.
Full text
PDF
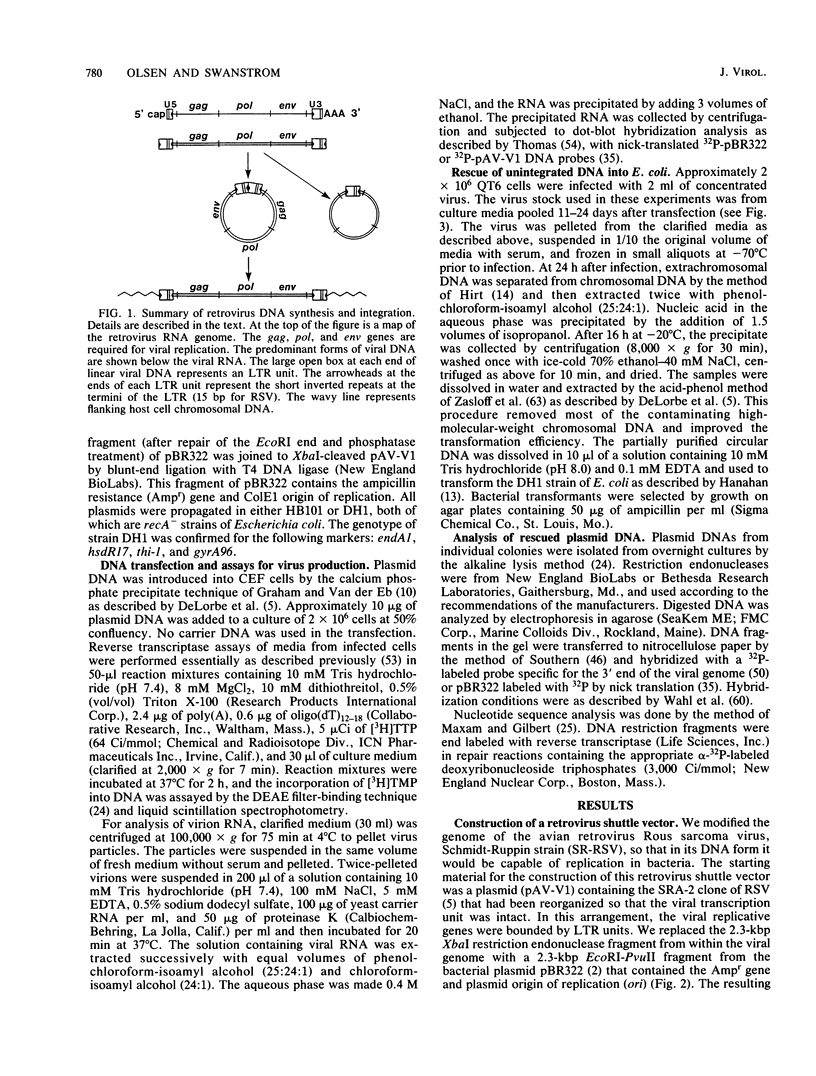
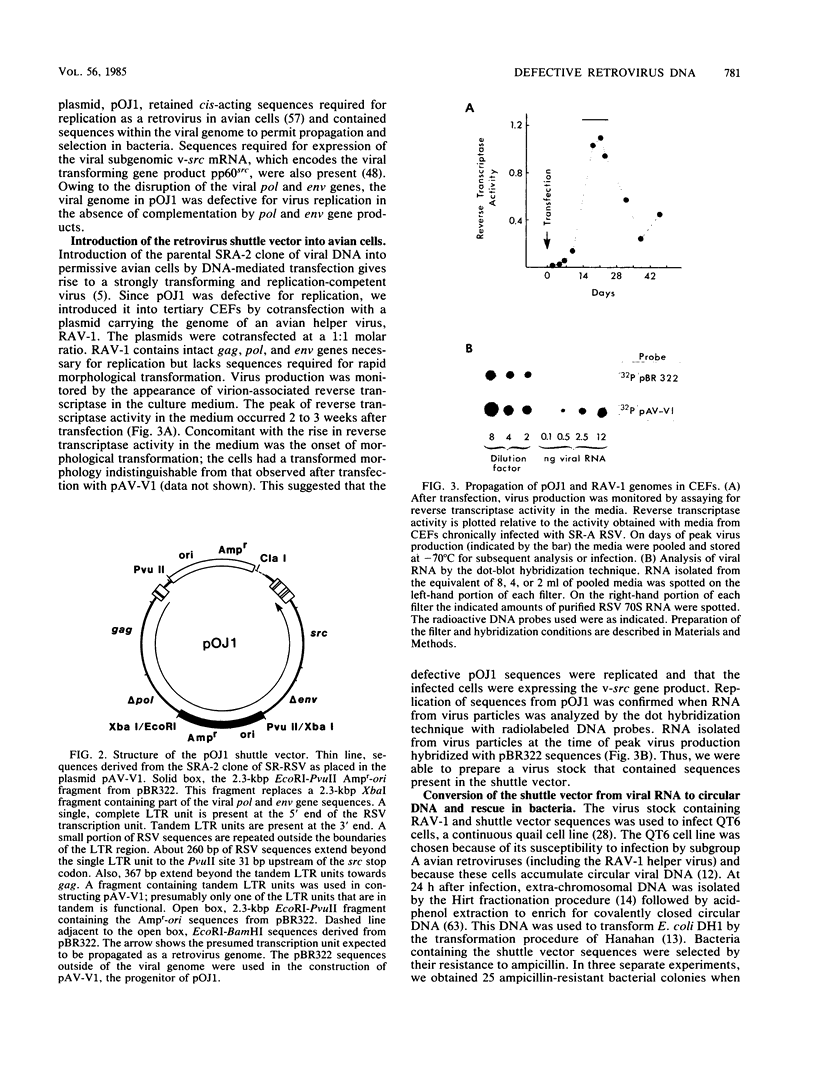
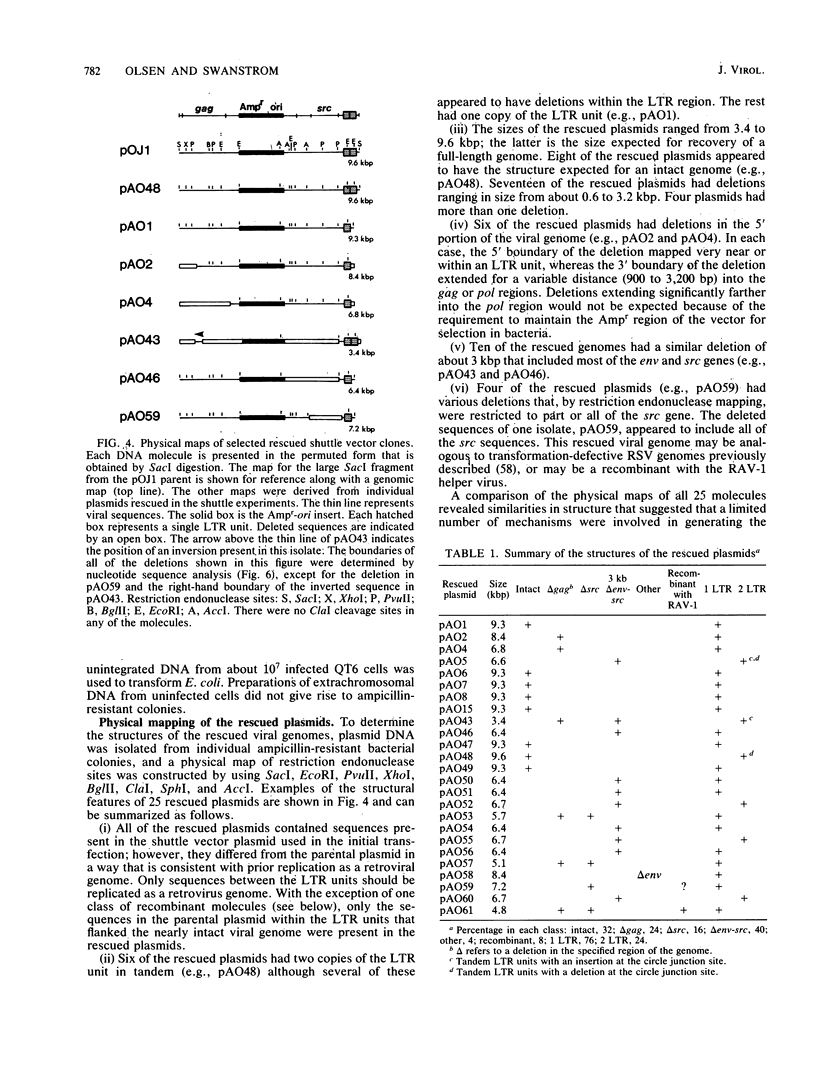




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger S. A., Bernstein A. Characterization of a retrovirus shuttle vector capable of either proviral integration or extrachromosomal replication in mouse cells. Mol Cell Biol. 1985 Feb;5(2):305–312. doi: 10.1128/mcb.5.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cepko C. L., Roberts B. E., Mulligan R. C. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984 Jul;37(3):1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- Chen P. J., Cywinski A., Taylor J. M. Reverse transcription of 7S L RNA by an avian retrovirus. J Virol. 1985 May;54(2):278–284. doi: 10.1128/jvi.54.2.278-284.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorbe W. J., Luciw P. A., Goodman H. M., Varmus H. E., Bishop J. M. Molecular cloning and characterization of avian sarcoma virus circular DNA molecules. J Virol. 1980 Oct;36(1):50–61. doi: 10.1128/jvi.36.1.50-61.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Leis J., Longiaru M., Skalka A. M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the alpha beta form of avian reverse transcriptase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Longiaru M., Cobrinik D., Kowal R., deHaseth P., Skalka A. M., Leis J. Circles with two tandem long terminal repeats are specifically cleaved by pol gene-associated endonuclease from avian sarcoma and leukosis viruses: nucleotide sequences required for site-specific cleavage. J Virol. 1985 Nov;56(2):589–599. doi: 10.1128/jvi.56.2.589-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P., Mason W. Virus-coded DNA endonuclease from avian retrovirus. J Virol. 1981 May;38(2):548–555. doi: 10.1128/jvi.38.2.548-555.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Guntaka R. V., Richards O. C., Shank P. R., Kung H. J., Davidson N. Covalently closed circular DNA of avian sarcoma virus: purification from nuclei of infected quail tumor cells and measurement by electron microscopy and gel electrophoresis. J Mol Biol. 1976 Sep 15;106(2):337–357. doi: 10.1016/0022-2836(76)90090-5. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Omer C. A., Weis J. H., Mitsialis S. A., Faras A. J., Guntaka R. V. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J Virol. 1982 Apr;42(1):346–351. doi: 10.1128/jvi.42.1.346-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Hanafusa H. Genetic recombination with avian tumor virus. Virology. 1972 Jul;49(1):37–44. doi: 10.1016/s0042-6822(72)80005-9. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Harada F., Kawai S. Characterization of a Rous sarcoma virus mutant defective in packaging its own genomic RNA: biochemical properties of mutant TK15 and mutant-induced transformants. J Virol. 1984 Jul;51(1):154–162. doi: 10.1128/jvi.51.1.154-162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Medeiros E., Hayward W. S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell. 1978 Dec;15(4):1371–1381. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Grandgenett D. P., Parsons J. T. Avian retrovirus pp32 DNA-binding protein. I. Recognition of specific sequences on retrovirus DNA terminal repeats. J Virol. 1982 Oct;44(1):330–343. doi: 10.1128/jvi.44.1.330-343.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovici C., Moscovici M. G., Jimenez H., Lai M. M., Hayman M. J., Vogt P. K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977 May;11(1):95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983 Nov 10;306(5939):155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Courtneidge S. A., Crittenden L. B., Fadly A. M., Bishop J. M., Varmus H. E. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981 Feb;23(2):311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Pugatsch T., Stacey D. W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983 Jul 30;128(2):505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- Rassart E., DesGroseillers L., Jolicoeur P. Molecular cloning of B- and N-tropic endogenous BALB/c murine leukemia virus circular DNA intermediates: isolation and characterization of infectious recombinant clones. J Virol. 1981 Jul;39(1):162–171. doi: 10.1128/jvi.39.1.162-171.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Samuel K. P., Papas T. S., Chirikjian J. G. DNA endonucleases associated with the avian myeloblastosis virus DNA polymerase. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2659–2663. doi: 10.1073/pnas.76.6.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Sealy L., Privalsky M. L., Moscovici G., Moscovici C., Bishop J. M. Site-specific mutagenesis of avian erythroblastosis virus: erb-B is required for oncogenicity. Virology. 1983 Oct 15;130(1):155–178. doi: 10.1016/0042-6822(83)90125-3. [DOI] [PubMed] [Google Scholar]
- Shank P. R., Cohen J. C., Varmus H. E., Yamamoto K. R., Ringold G. M. Mapping of linear and circular forms of mouse mammary tumor virus DNA with restriction endonucleases: evidence for a large specific deletion occurring at high frequency during circularization. Proc Natl Acad Sci U S A. 1978 May;75(5):2112–2116. doi: 10.1073/pnas.75.5.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Shank P. R., Linial M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: characterization of a deletion in the provirus. J Virol. 1980 Nov;36(2):450–456. doi: 10.1128/jvi.36.2.450-456.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Varmus H. E. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J Virol. 1978 Jan;25(1):104–104. doi: 10.1128/jvi.25.1.104-104.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Hoffman J., Goff S. P., Baltimore D. Intramolecular integration within Moloney murine leukemia virus DNA. J Virol. 1981 Oct;40(1):164–172. doi: 10.1128/jvi.40.1.164-172.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Parker R. C., Varmus H. E., Bishop J. M. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1983 May;80(9):2519–2523. doi: 10.1073/pnas.80.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Varmus H. E., Bishop J. M. Nucleotide sequence of the 5' noncoding region and part of the gag gene of Rous sarcoma virus. J Virol. 1982 Feb;41(2):535–541. doi: 10.1128/jvi.41.2.535-541.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal J., Kung H. J., Varmus H. E., Bishop J. M. Characterization of DNA complementary to nucleotide sequences adjacent to poly(A) at the 3'-terminus of the avian sarcoma virus genome. Virology. 1977 Jun 1;79(1):183–197. doi: 10.1016/0042-6822(77)90344-0. [DOI] [PubMed] [Google Scholar]
- Tamura T., Takano T. Long terminal repeat (LTR)-derived recombination of retroviral DNA: sequence analyses of an aberrant clone of baboon endogenous virus DNA which carries an inversion from the LTR to the gag region. Nucleic Acids Res. 1982 Sep 11;10(17):5333–5343. doi: 10.1093/nar/10.17.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M. An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes. Biochim Biophys Acta. 1977 Mar 21;473(1):57–71. doi: 10.1016/0304-419x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Tereba A., Murti K. G. A very sensitive biochemical assay for detecting and quantitating avian oncornaviruses. Virology. 1977 Jul 1;80(1):166–176. doi: 10.1016/0042-6822(77)90389-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Vogt P. K. Genetically stable reassortment of markers during mixed infection with avian tumor viruses. Virology. 1971 Dec;46(3):947–952. doi: 10.1016/0042-6822(71)90093-6. [DOI] [PubMed] [Google Scholar]
- Vogt P. K. Spontaneous segregation of nontransforming viruses from cloned sarcoma viruses. Virology. 1971 Dec;46(3):939–946. doi: 10.1016/0042-6822(71)90092-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway D., Payne G., Varmus H. E. Proviral deletions and oncogene base-substitutions in insertionally mutagenized c-myc alleles may contribute to the progression of avian bursal tumors. Proc Natl Acad Sci U S A. 1984 Feb;81(3):843–847. doi: 10.1073/pnas.81.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F. K., Weinberg R. A. Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: discovery of a smaller circular form. Cell. 1979 Feb;16(2):323–332. doi: 10.1016/0092-8674(79)90009-6. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Ginder G. D., Felsenfeld G. A new method for the purification and identification of covalently closed circular DNA molcules. Nucleic Acids Res. 1978 Apr;5(4):1139–1152. doi: 10.1093/nar/5.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]