Abstract
Squalene epoxidase, encoded by the ERG1 gene in yeast, is a key enzyme of sterol biosynthesis. Analysis of subcellular fractions revealed that squalene epoxidase was present in the microsomal fraction (30,000 × g) and also cofractionated with lipid particles. A dual localization of Erg1p was confirmed by immunofluorescence microscopy. On the basis of the distribution of marker proteins, 62% of cellular Erg1p could be assigned to the endoplasmic reticulum and 38% to lipid particles in late logarithmic-phase cells. In contrast, sterol Δ24-methyltransferase (Erg6p), an enzyme catalyzing a late step in sterol biosynthesis, was found mainly in lipid particles cofractionating with triacylglycerols and steryl esters. The relative distribution of Erg1p between the endoplasmic reticulum and lipid particles changes during growth. Squalene epoxidase (Erg1p) was absent in an erg1 disruptant strain and was induced fivefold in lipid particles and in the endoplasmic reticulum when the ERG1 gene was overexpressed from a multicopy plasmid. The amount of squalene epoxidase in both compartments was also induced approximately fivefold by treatment of yeast cells with terbinafine, an inhibitor of the fungal squalene epoxidase. In contrast to the distribution of the protein, enzymatic activity of squalene epoxidase was only detectable in the endoplasmic reticulum but was absent from isolated lipid particles. When lipid particles of the wild-type strain and microsomes of an erg1 disruptant were mixed, squalene epoxidase activity was partially restored. These findings suggest that factor(s) present in the endoplasmic reticulum are required for squalene epoxidase activity. Close contact between lipid particles and endoplasmic reticulum may be necessary for a concerted action of these two compartments in sterol biosynthesis.
INTRODUCTION
In eukaryotic cells, sterols are important determinants of membrane properties such as fluidity and permeability, which are critical parameters for transmembrane transport and activity of membrane-bound enzymes. Biosynthesis of sterols is a complex oxygen-dependent process and involves similar enzymatic reactions in unicellular and multicellular eukaryotic organisms. In multicellular eukaryotes, most enzymes involved in cholesterol biosynthesis are localized in the endoplasmic reticulum (Reinhart et al., 1987). The predominant sterol of the yeast Saccharomyces cerevisiae is ergosterol, which is structurally and functionally related to cholesterol of mammalian cells. Yeast enzymes catalyzing the transformation of squalene, the polyisoprene precursor, to ergosterol are also believed to be microsomal proteins (Mercer, 1984; Paltauf et al., 1992), but the precise subcellular localization has not yet been described.
In yeast cells (Zinser et al., 1993), as in higher eukaryotes (Lange et al., 1989), the highest concentration of free sterols is present in the plasma membrane. Fatty acyl esters of ergosterol and its precursors, however, are exclusively stored in lipid particles (Zinser et al., 1993; Leber et al., 1994). Esterification of sterols with long-chain fatty acids occurs in microsomes, and steryl ester hydrolase activity is mainly detectable in the plasma membrane (Zinser et al., 1993). Spatial separation of the sites of biosynthesis and cleavage of steryl esters requires efficient and well-regulated transport mechanisms to ensure proper supply of free sterols to cellular membranes. Steryl esters stored in lipid particles were shown to serve as the source of free sterols in the absence of endogenous synthesis caused by specific metabolic inhibitors of sterol-synthesizing enzymes (Leber et al., 1995).
Thus far, lipid particles in yeast have been considered solely as a depot for ergosterol and fatty acids (Clausen et al., 1974). Recent studies in our laboratory (Zinser et al., 1993; Leber et al., 1994, 1995) and other laboratories (Lum and Wright, 1995), however, ascribe these structures a new and more complex role in lipid biosynthesis, metabolism, degradation, and trafficking. High specific activities of glycerophosphate acyltransferase and sterol Δ24-methyltransferase were detected in lipid particles, and sterol Δ24-methyltransferase, encoded by the ERG6 gene, was identified as one of the major lipid particle proteins (Leber et al., 1994).
To obtain more insight into the physiological role(s) of yeast lipid particles, we started characterizing the proteins of this organelle. We focused our interest on a 55-kDa protein that was overproduced in cells treated with terbinafine, an inhibitor of fungal squalene epoxidase. Herein we present evidence for the identification of the 55-kDa protein of lipid particles as squalene epoxidase, the product of the ERG1 gene (Jandrositz et al., 1991). Squalene epoxidase is an FAD-containing mixed-function oxygenase (monooxygenase) and introduces molecular oxygen into the 2,3-position of squalene, which requires a yet unidentified reductase activity (Ryder, 1990). Squalene synthase and squalene epoxidase act together as regulatory enzymes in ergosterol biosynthesis (Bonaventure et al., 1989; Ryder, 1991). Because of its key role in the synthesis of essential sterol compounds, elimination of ERG1 function by gene disruption is lethal, unless ergosterol is supplied to cells growing under anaerobic conditions (Landl et al., 1996). Squalene epoxidase of fungi is the target of a class of compounds, termed allylamines (e.g., terbinafine), that have significant pharmaceutical impact as antifungal drugs (Paltauf et al., 1982; Ryder, 1991).
In this article we demonstrate that squalene epoxidase, Erg1p, is present in both lipid particles and the endoplasmic reticulum. This dual localization may reflect a means of regulating sterol synthesis and trafficking in growing cells. The dual localization of squalene epoxidase and the functional interaction between lipid particles and the endoplasmic reticulum are discussed.
MATERIALS AND METHODS
Strains and Culture Conditions
Yeast strains used in this study, their relevant genotypes, and the respective sources are listed in Table 1. Wild-type yeast strains were cultivated at 30°C on a rotary shaker with vigorous aeration in YPD medium containing 1% yeast extract, 2% peptone, and 2% glucose. Transformants harboring the recombinant plasmid pAF22 with the ERG1 gene (Jandrositz et al., 1991) were grown in minimal medium (Sherman, 1991) supplemented with the respective amino acids except leucine. For treatment with terbinafine, cells were grown to the exponential phase in YPD medium, transferred to fresh sterol-free medium (Hirsch and Henry, 1986) or Sabouraud medium (Merck, Darmstadt, Germany) containing 30 μg/ml terbinafine, and incubated at 30°C for 4 h. Terbinafine was added from a stock solution in dimethyl sulfoxide or ethanol.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| KLN | MATα, erg1::URA3, ura3, leu2, trp1 | Landl et al. (1996) |
| KLO | MATα, ura3, leu2, trp1 | Landl et al. (1996) |
| A2 | MATα, leu2, his3, can1 | Wild-type strain |
| W303 | MATα, leu2, ura3, his3, ade2, trp1 | Wild-type strain |
| W303D | Matα/MATa, leu2/leu2, ura3/ura3, his3/his3, ade2/ade2, trp1/trp1 | Diploid wild-type strain |
| Tetraploid wild-type strain | Kadowaki et al. (1995) | |
| X2180-1A | MATa, SUC2, mal, gal2, CUP1 | Wild-type strain |
The erg1 disruptant strain KLN and the isogenic wild-type strain KLO were grown anaerobically in YPD medium supplemented with 0.5% Tween 80 and 12 μg/ml ergosterol. Ergosterol was dissolved in Tween 80:ethanol (1:1, vol/vol). Agar plates and small liquid cultures were incubated in an anaerobic jar in the presence of Anaerocult A (Merck). For large cultures, 2 l of the above mentioned medium were inoculated with 10 ml of a stationary-phase yeast culture. Oxygen was removed by bubbling nitrogen through the culture. Incubation was carried out at 30°C for 60–70 h.
Isolation of Yeast Subcellular Fractions
Lipid particles (Leber et al., 1994), microsomes (Zinser et al., 1991), and vacuoles (Uchida et al., 1988) were isolated from late exponential phase cultures by following published procedures. The purity of subcellular fractions was judged by measuring marker enzyme activities or by Western blot analysis (see below) as summarized by Zinser and Daum (1995).
Protein Analysis
Proteins were quantified either by the method of Lowry et al. (1951) or Bradford (Ausubel et al., 1994) by using bovine serum albumin as the standard. Before proteins were quantified, lipid particles were delipidated by extracting the nonpolar lipids with 2 volumes of diethyl ether. Proteins were precipitated with trichloroacetic acid (10%, final concentration; wt/vol) and solubilized in 0.1% SDS and 0.1 M NaOH.
SDS-PAGE was performed by the method of Laemmli (1970). Western blot analysis was carried out after separating proteins on a 10% SDS-polyacrylamide gel and transferring to nitrocellulose filters (Hybond-C; Amersham, Arlington Heights, IL) or polyvinylidene difluoride membranes (Immobilon P; Millipore, Bedford, MA); (Haid and Suissa, 1983). Proteins were detected with polyclonal rabbit antiserum against the respective antigens and alkaline phosphatase- or peroxidase-conjugated goat anti-rabbit secondary antibodies, by the manufacturer’s instructions. Antiserum against Erg1p was raised in rabbits as described below. Antibodies against Sec61p and BiP were a gift from R. Schekman (University of California, Berkeley), and antibodies against Erg6p were obtained as described elsewhere (Leber et al., 1994). Immunoreactive proteins on blots were quantified densitometrically at 600 nm by using a Shimadzu CS 930 TLC scanner.
For amino acid sequencing, the excised Immobilon P membrane (Millipore) with the protein bound was soaked in 100 μl of 0.1 M Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl), pH 8.5, and 5% acetonitrile and incubated with 5 μl of a solution containing 1 mg/ml trypsin (sequencing grade, Boehringer Mannheim, Mannheim, Germany) at 37°C for 24 h. After 6 h of incubation, another 5-μl aliquot of the trypsin-containing solution was added. Separation of peptides was achieved on a Merck Supersphere C18 column (4 × 125, 100Å, 4 μm) at a flow rate of 1 ml/min with a column temperature of 30°C. Solvent A was 0.1% trifluoroacetic acid in water, and solvent B was 0.1% trifluoroacetic acid in acetonitrile. The linear gradient from 0% to 70% solvent B was run for 70 min. The elution profile was monitored at 214 nm and recorded on a Waters Maxima 820 system. Fractions containing the peptides were manually collected from the high-pressure liquid chromatography (HPLC), dried in a Speed Vac concentrator, and stored at 4°C. Automated amino acid sequence analysis was carried out by using an Applied Biosystems model 477A pulsed liquid-phase sequencer with online analysis on a 120 ABI HPLC system.
Antisera against Squalene Epoxidase
Squalene epoxidase was purified from lipid particles by SDS-PAGE and eluted from gel slices by using an Electro Eluter model 422 (Bio-Rad, Richmond, CA) according to the manufacturer’s instructions. This protein solution was injected into a rabbit for the production of antibodies.
In a second approach, a β-galactosidase-Erg1p fusion protein was expressed in Escherichia coli by using the MoBiTec pAX4a+ vector (MoBiTec, Göttingen, Germany), in which the ERG1 gene was cloned in-frame behind the lacZ gene by following standard procedures (Ausubel et al., 1994). Expression was induced by 1 mM isopropyl β-d-thiogalactoside for 6 h, and the 170-kDa fusion protein was isolated from cell extracts by SDS-PAGE. The fusion protein was used to immunize rabbits without prior elution of the protein from the gel. Immunization of rabbits was performed at the Sandoz Forschungsinstitut (Vienna, Austria).
Immunofluorescence Microscopy
Preparation of cells for immunofluorescence microscopy was as described by Pringle et al. (1989). Cells of a tetraploid wild-type strain (Table 1) were cultivated aerobically in 5 ml of YPD medium at 30°C. The erg1 disruptant KLN was grown under anaerobic conditions in the presence of ergosterol. Cells were fixed by adding 0.5 ml of 37% formaldehyde to the growth medium and incubation for 2 h at growth temperature, washed twice with 100 mM potassium phosphate (KPi) buffer, pH 7.5, and resuspended in 0.9 ml of 100 mM KPi buffer containing 1.2 M sorbitol. After addition of 50 μl of glucuronidase (Boehringer Mannheim) and 5 μl of 2-mercaptoethanol, cells were incubated for 20 min at 37°C. Cells were further spheroplasted by addition of 25 μl of zymolyase 20,000 (2 mg/ml in KPi buffer; Seikagaku Corp., Tokyo, Japan) for 7–15 min at 37°C. Spheroplasts were washed twice with phosphate-buffered saline (PBS), 1% bovine serum albumin (BSA), blocked with PBS/1% BSA for 1 h, and placed on polyethylenimine-coated multiwell slides. Immunolabeling was carried out by using polyclonal antibodies against Erg1p (diluted 1:100 in PBS/1% BSA), Erg6p (1:200 dilution), and Kar2p (binding protein; 1:300 dilution) overnight at 4°C. Antiserum against squalene epoxidase was pretreated with fixed and spheroplasted erg1-disruptant cells to remove unspecifically binding components. Cells were washed with PBS and 1% BSA and incubated with the secondary antibody, fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (1:100 dilution), in PBS/1% BSA for 1 h at room temperature. After washing once with PBS/1% BSA and twice with PBS, cells were mounted in 90% glycerol containing 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) for DNA staining.
For double immunofluorescence, cells were prepared and immunodecorated with polyclonal antibodies against Erg6p (1:200 dilution) or binding protein (1:300 dilution) as described above. After three washes with PBS/1% BSA cells were incubated with FITC-conjugated anti-rabbit IgG (1:100 dilution) in PBS/1% BSA for 1 h at room temperature. Cells were washed with PBS/1% BSA and incubated with Cy5-labeled Erg1p antibodies (1:5 dilution) for 2 h at 4°C. Labeling of the IgG fraction of the Erg1p antiserum with Cy5 was carried out by following the instructions of the manufacturer (Amersham). Then, cells were washed once with PBS/1% BSA and twice with PBS prior to mounting in 90% glycerol containing 1 μg/ml DAPI.
Fluorescence microscopy was carried out on a Leica TCS 4D confocal microscope, equipped with an Ar/Kr laser and set up with the appropriate filter sets. FITC emission detection at 515–535 nm and Cy5 emission detection at 690 nm (long pass filter) ensured complete optical separation of the respective fluorescence dyes. Differences in fluorescence intensities were adjusted by using an acousto-optical tunable filter for individual excitation line modulation. Both simultaneous scanning for FITC and Cy5 channels and sequential scanning (first Cy5 followed by FITC) to avoid bleaching of the more-sensitive red dye were performed and led to the same results. DAPI fluorescence was visualized by UV-epifluorescence and recorded with a Hamamatsu video system.
Squalene Epoxidase Assay
Squalene epoxidase activity was measured as described by Satoh et al. (1993). The standard assay mixture contained 0.35–0.7 mg of microsomal protein or/and 3.5–75 μg of lipid particle protein, 100 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.1 mM FAD, 3 mM NADPH, 0.1 mM squalene 2,3-epoxide cyclase inhibitor U18666A (Cenedella, 1980), and 32 μM [3H]squalene dispersed in 0.005% Tween 80, in a total volume of 0.5 ml. Assays were performed in screw-cap glass tubes at 30°C. After preincubation for 10 min, the reaction was started by adding squalene and terminated after 15, 30, or 45 min by lipid extraction with 3 ml of chloroform:methanol (2:1, vol/vol) and 750 μl of 0.035% MgCl2 according to Folch et al. (1957). Lipids in the organic phase were dried under a stream of nitrogen and dissolved in 0.5 ml of chloroform:methanol (2:1, vol/vol). For TLC analysis, aliquots of the samples were applied on silica gel 60 plates (0.2 mm; Merck) by using a CAMAG Linomat IV applicator (Muttenz, Switzerland) and developed with chloroform as a solvent. The positions of [3H]2,3-oxidosqualene and [3H]squalene were identified by using an automatic TLC linear analyzer (Berthold, Bad Wildbad, Germany). Radioactive spots were scraped off the plates, and radioactivity was quantified by liquid scintillation counting in Safety Scintillation Cocktail (Baker, Deventer, the Netherlands) containing 5% water. To identify the positions of the substrate and the product of the squalene epoxidase reaction on thin-layer plates, 14C-labeled squalene and [14C]2,3-oxidosqualene were prepared by labeling cell extracts with [14C]mevalonate (specific activity, 58 Ci/mol) in the presence of 0.1 mM of the squalene 2,3-epoxide cyclase inhibitor U18666A (stock solution in propyleneglycol:water, 1:1, vol/vol). The assay conditions were as described by Jandrositz et al. (1991).
Lipid Analysis
Neutral lipids of yeast homogenate and isolated subcellular fractions were extracted according to Folch et al. (1957). Quantification of ergosterol, ergosteryl esters, and triacylglycerols was carried out as described by Leber et al. (1995). In brief, lipid extracts were applied to silica gel 60 plates with the aid of a sample applicator, and chromatograms were developed in an ascending manner by using a two-step solvent system with light benzene:diethyl ether (1:1, vol/vol) and light benzene:diethyl ether (98:2, vol/vol; Leber et al., 1995). Ergosterol and ergosteryl esters were quantified after chromatographic separation by direct densitometry on thin-layer plates at 275 nm by using a Shimadzu CS 930 TLC scanner with ergosterol as a standard. Triacylglycerols were visualized by postchromatographic staining. Plates were dipped for 8 s into a developing reagent consisting of 0.63 g of MnCl2.4 H2O, 60 ml of water, 60 ml of methanol, and 4 ml of concentrated sulfuric acid by using a chromatogram immersion device (CAMAG), briefly dried, and heated to 120°C for 15 min. Quantification of triacylglycerols was carried out by densitometric scanning at 400 nm with triolein as a standard.
RESULTS
Identification of Erg1p as a Component of Lipid Particles
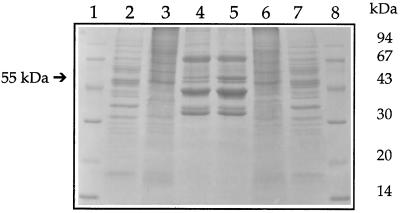
Lipid particles contain only small amounts of protein, and the pattern is rather simple (Leber et al., 1994). Major protein components with apparent molecular masses of 72, 43, 35, and 32 kDa, and other polypeptides with apparent molecular masses of 62, 57, 55, and 52 kDa are present in the purified lipid particle fraction (Figure 1).
Figure 1.
Protein pattern of yeast lipid particles grown in the presence or absence of the squalene epoxidase inhibitor terbinafine. Wild-type yeast cells were grown in the presence (30 μg/ml) or absence of terbinafine as described in MATERIALS AND METHODS. Lipid particles were prepared according to Leber et al. (1994), and 20 μg of total protein were applied to each lane of a 12.5% SDS gel and stained with Coomassie brilliant blue after electrophoretic separation. Lanes 1 and 8, molecular weight standards; lane 2, yeast homogenate (without terbinafine); lane 3, vacuoles (without terbinafine); lane 4, lipid particles (without terbinafine); lane 5, lipid particles (with terbinafine); lane 6, vacuoles (with terbinafine); lane 7, homogenate (with terbinafine).
Incubation of yeast cells for 4 h with the squalene epoxidase inhibitor terbinafine led to some fivefold accumulation of the 55-kDa protein in lipid particles. This increase was paralleled by a slight induction of a 43-kDa protein that had previously been identified as sterol Δ24-methyltransferase, Erg6p (see Figure 1). The 55-kDa protein was purified by SDS-PAGE, electroeluted from the gel, and subjected to amino acid sequencing. After tryptic digestion of the N-terminally blocked protein, two peptide fragments were isolated by reversed-phase HPLC. The amino acid sequence of both fragments unequivocally identified the 55-kDa protein as squalene epoxidase, the product of the ERG1 gene (Jandrositz et al., 1991; Figure 2).
Figure 2.
Amino acid sequence of squalene epoxidase (Erg1p). Two fragments (overlined with brackets) of a 55-kDa protein isolated from yeast lipid particles were identified as partial sequences of squalene epoxidase (Erg1p) by amino acid sequence analysis. Erg1p contains two potential hydrophobic membrane spanning regions (boxed) and one FAD binding site (indicated with asterisks).
Erg1p Localizes to Both Lipid Particles and Endoplasmic Reticulum
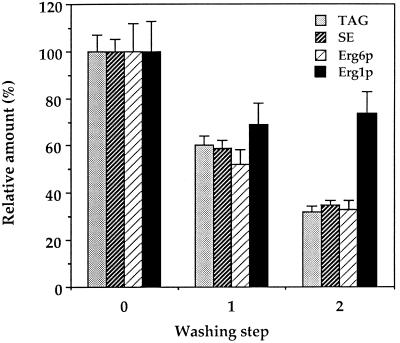
Polyclonal antibodies raised against the 55-kDa lipid particle protein or a β-galactosidase-Erg1 fusion protein (see MATERIALS AND METHODS) were used to determine the distribution of Erg1p in subcellular fractions. The reactivity of the antibody raised against the ERG1 gene product expressed in E. coli as a fusion protein with the 55-kDa lipid particle protein of yeast provided further proof to the identity of this protein. As expected, a significant amount of Erg1p was detected by Western blot analysis in lipid particles of late exponential cells. Moreover, a marked amount of squalene epoxidase was also present in the 30,000 × g microsomal fraction consisting mainly of endoplasmic reticulum (Table 2). To test whether the presence of Erg1p in the endoplasmic reticulum was due to contamination of this fraction with lipid particles, the 30,000 × g microsomal membrane pellet was isolated from the wild-type strain W303 and subjected to two sequential washes with 10 mM Tris-HCl, pH 7.5. After each washing step, microsomes were analyzed with respect to their content of neutral lipids, Erg1p, and Erg6p. The amount of immunoreactive Erg1p in “washed” microsomes was decreased after the first washing step but could not be reduced below 70% by further washes (Figure 3), indicating that a significant amount of Erg1p remained tightly associated with the microsomal fraction. Enzymatically active squalene epoxidase was only detectable in microsomal pellets but not in the supernatants of washed microsomes. In contrast, repeated washing of crude microsomes resulted in a stepwise decrease of the lipid particle-specific components, triacylglycerols, steryl esters, and Erg6p (Leber et al., 1994), to as low as 35% of the starting material. These results indicate that under standard preparation conditions microsomes contain loosely associated lipid particles. Thus, further localization experiments were performed with “washed” microsomes.
Table 2.
Distribution of components between the endoplasmic reticulum and lipid particles
| Marker | Enrichment (fold)
|
Relative recovery (%)a
|
||
|---|---|---|---|---|
| Endoplasmic reticulum | Lipid particles | Endoplasmic reticulum | Lipid particles | |
| Sec61pb | 3.9 ± 0.5 | 4.2 ± 0.6 | 98 | 2 |
| BiPb | 1.8 ± 0.3 | 2.7 ± 0.4 | 97 | 3 |
| Steryl estersc | 1.1 ± <0.1 | 860 ± 43 | 6 | 94 |
| Triacylglycerolsc | 1.9 ± 0.2 | 740 ± 59 | 1 | 99 |
| Erg6pb | 2.5 ± 0.3 | 700 ± 91 | 20 | 80 |
| Erg1pb | 4.0 ± 0.5 | 100 ± 12 | 62 | 38 |
Distribution of components was determined based on the relative recovery of components in isolated organelles (see Zinser and Daum, 1995) from cells harvested in the late logarithmic growth phase. Data are mean values from three independent experiments.
Equivalent amounts of protein of each fraction were separated by SDS-PAGE and subjected to Western blot analysis using the respective antiserum. The intensity of the immunological signal in the homogenate was set at 1, and the intensities of signals measured in the endoplasmic reticulum and lipid particles were set in relation. Data were obtained from at least three independent experiments.
Amount of triacylglycerols and steryl esters (mg) per mg of protein in the homogenate was set at 1, and the enrichment in the respective fraction was calculated from the corresponding values. Data were obtained from three independent experiments.
Figure 3.
Presence of Erg1p in the endoplasmic reticulum was not due to a contamination with lipid particles. Endoplasmic reticulum (30,000 × g microsomes) was prepared as described in MATERIALS AND METHODS and subjected to two additional washes with 10 mM Tris-HCl, pH 7.5. Triacylglycerols (TAG) and steryl esters (SE) were quantified after thin-layer chromatographic separation of lipids (see MATERIALS AND METHODS), and Erg6p and Erg1p in the membrane pellet were quantified before and after the respective washing steps by Western blot analysis. Mean values of three independent experiments are shown.
To clarify the localization of squalene epoxidase in late exponential cells, the enrichment over the homogenate of Erg1p and of markers of the endoplasmic reticulum (Sec61p and BiP) and lipid particles (steryl esters, triacylglycerols, and Erg6p) was measured in both fractions. These experiments revealed that Erg1p is not exclusively located in either lipid particles or the endoplasmic reticulum: squalene epoxidase was enriched only 100-fold in the lipid particle fraction, whereas typical lipid particle components such as steryl esters, triacylglycerols, and Erg6p were enriched 700- to 850-fold over the homogenate (Table 2). The latter components were also detectable in the endoplasmic reticulum, but the enrichment was rather low compared with lipid particles. The enrichment of Erg1p in the “washed” microsomal fraction was comparable to Sec61p and BiP. To calculate the distribution of marker proteins and marker lipids between the endoplasmic reticulum and lipid particles, we considered the relative recovery of these components in the two isolated fractions. These calculations demonstrated that 62% of cellular Erg1p is present in the endoplasmic reticulum and 38% is present in the lipid particle fraction (Table 2). In contrast, lipid particles harbor 80% and the endoplasmic reticulum harbors 20% of total cellular Erg6p.
Disruption and Overexpression of ERG1 Affect Squalene Epoxidase Levels in Both Lipid Particles and the Endoplasmic Reticulum
When present on the high-copy-number plasmid pAF22, the ERG1 gene is overexpressed resulting in a fivefold accumulation of squalene epoxidase in lipid particles and in microsomes (our unpublished observations). The amount of Erg1p present in these compartments of the overproducing strain is comparable to that of a wild-type strain grown in the presence of terbinafine. The relative distribution also remained unaltered in wild-type strain KLO grown under anaerobic conditions. These data indicate that neither overproduction of Erg1p nor anaerobiosis leads to an altered subcellular localization of the protein. As expected, disruption of the ERG1 gene in strain KLN leads to the absence of the gene product in both lipid particles and the endoplasmic reticulum.
Immunofluorescence Microscopy
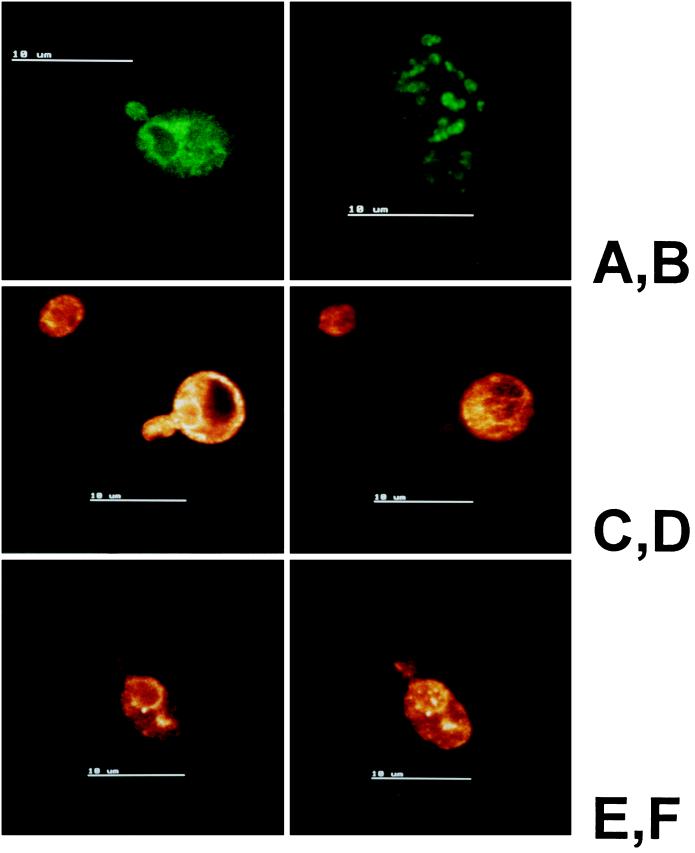
In addition to analysis by cell fractionation, Erg1p localization was determined by immunofluorescence microscopy. In a first set of experiments, we studied Erg1p localization in wild-type yeast cells (W303D) in different stages of growth (Figure 4). BiP (Kar2p) was used as a marker for the endoplasmic reticulum (Figure 4A), and sterol Δ24-methyltransferase (Erg6p) was used as a control for a typical lipid particle protein (Leber et al., 1994; Figure 4B). The distribution of Erg1p varied depending on the growth phase of the cells. In the early logarithmic phase, a distribution of the protein resembling endoplasmic reticulum was predominant (Figure 4, C and D), whereas in later stages of growth a more particulate distribution was observed (Figure 4, E and F). These microscopic observations are in agreement with our results obtained by Western blot analysis of organelles isolated from yeast cells harvested at different growth stages. In the early logarithmic phase, cells harbor 81% of cellular Erg1p in the microsomal fraction and 19% in lipid particles, whereas in the late logarithmic phase, 62% of cellular Erg1p was detected in microsomes and 38% was in lipid particles.
Figure 4.
Indirect immunofluorescence microscopy of Erg1p in yeast at various growth stages. Cells of the diploid strain Saccharomyces cerevisiae W303D were prepared as described in MATERIALS AND METHODS, and immunoreactive proteins were detected with FITC-conjugated anti-rabbit IgG. Primary antibodies against the endoplasmic reticulum marker BiP (Kar2p) (A), the lipid particle marker sterol Δ24-methyltransferase (Erg6p) (B), and Erg1p (C—F) were used. (C and D) Localization of Erg1p in two sections of the same cell in the logarithmic phase. (E) Localization of Erg1p in a single optical section of a late logarithmic-phase cell. (F) Extended focus image of the same cell.
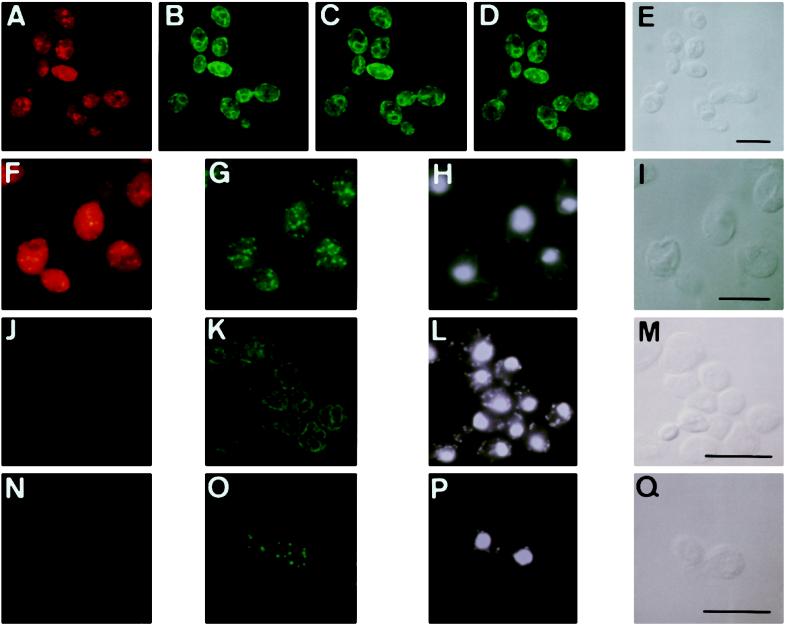
To determine Erg1p distribution relative to the marker proteins BiP (endoplasmic reticulum) and Erg6p (lipid particles) in greater detail, late-logarithmic-phase cells of a tetraploid wild-type strain (Kadowaki et al., 1995) were prepared for double immunofluorescence by using Erg1p antibody directly labeled with Cy5, and BiP or Erg6p antibodies visualized by reaction with FITC-conjugated anti-rabbit IgG (Figure 5). Because of the distant excitation and emission characteristics, no optical cross-talk between FITC- and Cy5-labeled probes was observed.
Figure 5.
Localization of Erg1p in the endoplasmic reticulum and a subpopulation of lipid particles. Late-logarithmic cells of a tetraploid wild-type strain (A–I) and the erg1 disruptant strain KLN (J–Q) were prepared for double immunofluorescence microscopy as described in MATERIALS AND METHODS. (A–E) Localization of Erg1p (A) and BiP (B–D; three different optical sections) in the same wild-type cells. (E) Transmission image. (F–I) Localization of Erg1p (F) and Erg6p (G) in the same wild-type cells. (H) DAPI staining. (I) Transmission image. (J–Q) erg1 disruptant strain KLN lacks a distinct signal with Cy5-labeled Erg1p antibody (J and N). Localization of BiP (K) and Erg6p (O), with DAPI staining (L and P) and transmission images (M and Q) of the respective cells of the strain KLN. Bar, 10 μm.
BiP-specific antibody detected the typical nuclear-rim structure of the endoplasmic reticulum (Koning et al., 1996; Figure 5, B–D), whereas Erg1p displayed a rather punctate staining close to the endoplasmic reticulum (Figure 5, A and F). The punctate Erg1p-staining completely overlapped the population of lipid particles recognized by the Erg6p antiserum (Figure 5G). Lipid particles stained with Erg1p antibodies are located close to the endoplasmic reticulum in proximity to the nucleus, which was visualized by DAPI staining (Figure 5H). Erg6p distribution, on the other hand, appeared more distinct and was mainly associated with lipid particle structures present throughout the cell (Figure 5G). The intensity distribution of Erg6p within the population of lipid particles was rather homogeneous, whereas the Erg1p fluorescence signal appeared brightly only in a few particles (Figure 5F). Conclusively, Erg6p as a typical lipid particle protein seems to be present in all lipid particles of the cell, whereas Erg1p with its higher tendency to associate with the endoplasmic reticulum is found only in a subset of lipid particles. Different subpopulations of lipid particles may exist, but it is not known how differential targeting/redistribution of proteins occurs. More detailed information about the biogenesis of lipid particles (see also DISCUSSION) will be required to address this question.
The erg1 disruptant KLN lacked a significant signal with the Cy5-labeled Erg1p antibody (Figure 5, J and N), whereas the BiP antibody recognized the typical structure of the endoplasmic reticulum (Figure 5K) and the Erg6p antibody detected lipid particles (Figure 5O) of the mutant. Thus, the antibody against Erg1p used in this study does not recognize putative Erg1p-related proteins. This result is in line with the fact that no Erg1p homologues were found by database searches of the entire yeast genome. The observation that an erg1 disruption strain is strictly auxotroph for sterols supports the view that other enzymes cannot functionally replace Erg1p.
Enzymatic Activity of Squalene Epoxidase In Vitro
Erg1p present in the endoplasmic reticulum of wild-type strain X2180 exhibited squalene epoxidase activity in vitro, as measured by the conversion of [3H]squalene to [3H]2,3-oxidosqualene (Table 3). In lipid particles of this strain, however, squalene epoxidase activity could not be detected in vitro, although a marked enrichment of Erg1p was found in this fraction (see Table 2). When lipid particles of the wild-type strain X2180 were added to 30,000 × g microsomes of X2180, the specific activity did not increase (Table 3).
Table 3.
Enzymatic activity of squalene epoxidase in yeast microsomes and lipid particles
| Yeast strain | Subcellular fraction | Specific activity (pmol per min per mg) |
|---|---|---|
| X2180-1A | Microsomes | 48.1 |
| X2180-1A | Lipid particles | ND |
| X2180-1Aa | Lipid particles X2180-1A + microsomes X2180-1A | 44.3 |
| W303 | Microsomes | 14.4 |
| W303 [pAF22] | Microsomes | 75.4 |
| W303 [pAF22] | Lipid particles | ND |
| KLO (anaerobic growth) | Microsomes | 49.2 |
| KLN (anaerobic growth) | Microsomes | ND |
| W303 [pAF22] + KLNa | Lipid particles W303 [pAF22] + microsomes KLN | 25.3 |
Results are mean values of at least three independent measurements (SD ± 10%). ND, not detectable.
In mixing experiments, the ratio of lipid particle proteins to microsomal proteins was 1:10.
Microsomes isolated from strain W303 [pAF22], which overproduces Erg1p, exhibited squalene epoxidase activity approximately fivefold higher than the corresponding wild-type strain W303 (Table 3). Lipid particles of W303 [pAF22], however, were devoid of squalene epoxidase activity. In microsomes prepared from the disruptant KLN (erg1::URA3), no enzymatic activity of squalene epoxidase could be detected in vitro. This result is in agreement with the lack of Erg1p in this strain and further supports the notion that the enzyme encoded by the ERG1 gene represents the only squalene epoxidase in yeast. When lipid particles isolated from W303 [pAF22] were mixed with 30,000 × g microsomes from the erg1 disruptant strain KLN, squalene epoxidase activity in vitro was partially restored. Thus, microsomal component(s) are required for the enzymatic reaction of squalene epoxidase and can activate Erg1p in highly purified lipid particles. The amount of the putative cofactor(s) present in the endoplasmic reticulum appears to be limiting.
DISCUSSION
Genes involved in sterol biosynthesis of the yeast Saccharomyces cerevisiae have been cloned and characterized during the last few years (for recent reviews, see Lees et al., 1995; Bard et al., 1996; Skaggs et al., 1996). The complex pathway of sterol biosynthesis in this unicellular eukaryote is reasonably well understood, but the localization of the enzymes involved is still a matter of discussion. Knowledge about the subcellular distribution of Erg proteins is of great importance for the understanding of sterol precursor and ergosterol traffic between organelles, the regulation and coordination of the pathway, and possible metabolic malfunction.
Herein we present evidence that one of the key enzymes of sterol biosynthesis, squalene epoxidase, is located in two subcellular structures in yeast. Western blot analysis and immunofluorescence microscopy revealed that Erg1p is enriched in the lipid particle fraction and is also associated with the endoplasmic reticulum. Trivial cross-contamination of the endoplasmic reticulum with lipid particles and vice versa is unlikely, because components characteristic for lipid particles such as steryl esters, triacylglycerols, and Erg6p, could be largely removed from 30,000 × g microsomes by repeated washing steps. In contrast, a significant portion of Erg1p remained associated with the endoplasmic reticulum. Two potential membrane-spanning hydrophobic domains are present in Erg1p at the C terminus and may be responsible for its membrane association (Jandrositz et al., 1991). Erg6p from yeast (Gaber et al., 1989) does not contain predictable transmembrane domains and has probably no such strong affinity to a membrane lipid bilayer.
The dual localization of Erg1p in the endoplasmic reticulum and lipid particles led us to speculate about a structural and functional relationship between these two compartments. Lipid particles are regarded as a depot for membrane lipid components, such as fatty acids and sterols, which are incorporated into triacylglycerols and steryl esters by enzymes located in the endoplasmic reticulum. Droplets containing neutral lipids at high concentration and a special subset of proteins may emerge from the endoplasmic reticulum, as has been suggested for storage oil bodies of plants (Murphy, 1993). This view is in good agreement with results presented recently by Lum and Wright (1995). These authors suggested that lipid particles are formed as a depot of membrane lipid components upon degradation of so-called karmellae, a compartment that is induced by the overproduction of 3-hydroxy-3-methylglutaryl coenzyme A reductase and resembles stacks of endoplasmic reticulum membranes. Association of lipid particles with endoplasmic reticulum-like structures was observed during the process of karmellae degradation, suggesting that lipid particle biogenesis can be regarded as a budding process from the endoplasmic reticulum. The idea that lipid particles may also serve as an end-stage degradation compartment is supported by the observations that the number of lipid particles increases during prolonged growth and that this fraction contains a substantial amount of Erg1p. Furthermore, the relative distribution of Erg1p and possibly other proteins may change during growth from the endoplasmic reticulum toward lipid particles. Detection of Erg1p in distinct isolated fractions may reflect the “extremes” of the localization of the protein. In vivo, a continuous blending of endoplasmic reticulum, pre-lipid particles associated with the endoplasmic reticulum, and removable mature lipid particles can be postulated, as suggested by fluorescence microscopy. Thus, deposition of certain proteins in lipid particles might also be regarded as “mistargeting” of proteins originally located to the endoplasmic reticulum. This may occur under conditions when synthesis of certain components in the endoplasmic reticulum, namely, neutral lipids and/or a specific set of proteins, is still ongoing, whereas proliferation of membranes ceases, e.g., at the onset of the stationary growth phase.
Lipid particles may also contribute actively to cellular lipid biosynthesis, as shown by the presence of in vitro enzymatically active sterol Δ24-methyltransferase (Leber et al., 1994) and glycerol-3-phosphate acyltransferase (Zinser et al., 1991). A close interaction between lipid particles and the endoplasmic reticulum may be required for a concerted action of these two compartments in sterol and phospholipid biosynthesis. According to this model, permanent or temporary surface contact would allow exchange of sterol precursors between enzymes in juxtaposition to each other. Isolation of lipid particles by centrifugation may destroy the association with the endoplasmic reticulum, thus, interrupting the potential biosynthetic interactions between these two compartments. This hypothesis would also explain the lack of measurable squalene epoxidase activity in isolated lipid particles. Squalene epoxidase is a mixed-function oxygenase and requires a reductase for its activity (Ryder, 1990). This reductase, which has not yet been identified in yeast or multicellular eukaryotes, may be present in the endoplasmic reticulum and could be the missing and reaction-limiting component for the activity of the enzyme in isolated lipid particles. Indeed, mixing of lipid particles of a wild-type strain, which lack squalene epoxidase activity, with microsomes of an erg1 disruption strain partially restored the activity of the enzyme (see Table 3). This result argues against inactivation of Erg1p in lipid particles during the isolation procedure but supports the view that a cofactor present only in the endoplasmic reticulum is required for squalene epoxidase activity. However, other factors required for squalene epoxidase activity have also been discussed. In mammalian cells, several components, e.g., a soluble protein factor, are necessary for squalene epoxidation (Bai and Prestwich, 1992). Gavey et al. (1978) proposed that a sterol carrier protein facilitates the movement of squalene on or within the microsomal membrane of mammalian cells, thus, activating its further metabolic conversion. Yeast squalene epoxidase, however, does not appear to depend on such soluble proteins in vitro (M’Baya and Karst, 1987). The existence of two isoforms of the enzyme, one that is enzymatically active and localizes to the endoplasmic reticulum and the other that is inactive and associates with lipid particles, is rather unlikely, because single disruption of the ERG1 gene results in the complete lack of Erg1p both in the endoplasmic reticulum and in lipid particles (see RESULTS). Alternatively, a modification of the protein could be the reason for localization of Erg1p in one or the other compartment and/or affect the enzymatic activity. Preliminary results, however, suggest that Erg1p is not modified by phosphorylation, palmitoylation, or glycosylation (Pump and Daum, unpublished data). Extensive proteolytic processing can also be ruled out, because the protein isolated from both compartments shows the same electrophoretic mobility as determined by SDS-PAGE and Western blot analysis. The enzyme associated either with the endoplasmic reticulum or lipid particles could have different binding affinities for FAD due to the different environments, but this has not been experimentally proven. Finally, localization of Erg1p in lipid particles may be explained by its structural properties. Although the majority of Erg1p is located in the endoplasmic reticulum, a certain amount of the protein may be deposited on the surface of the lipid particles during interaction with the endoplasmic reticulum. Hydrophobic domains of Erg1p may play an important role in that respect. Preliminary experiments (Leber and Turnowsky, unpublished results) indicate that truncated species of Erg1p lacking parts of the C-terminal hydrophobic stretches are retained in the endoplasmic reticulum.
Lange and Steck (1985) proposed that in mammalian cells a coordinate pathway of cholesterol biosynthesis and its movement to the plasma membrane exists. The authors suggested that squalene epoxide and other early membrane-bound sterol precursors are synthesized in the smooth endoplasmic reticulum and then translocated to specialized membranes where cholesterol synthesis is completed. As the last step in this process, the fusion of the cholesterol-rich precursor membranes with the plasma membrane was proposed. If such precursor membranes are small vesicles, they may fractionate with the endoplasmic reticulum. The result of this combined synthesis/translocation sequence would be that all precursor sterols are metabolized to cholesterol before incorporation into the plasma membrane. This model very much parallels the situation in yeast with lipid particles as a compartment that not only contains acyl esters of ergosterol and its late precursors but also harbors enzymes of sterol biosynthesis. Thus, certain late steps of sterol biosynthesis may be coordinated with the transport of sterols to the plasma membrane via lipid particles.
ACKNOWLEDGMENTS
We express our thanks to F. Paltauf and G. Högenauer for critically reading the manuscript; A. Ivessa for his help in microscopic analysis; R. Schekman, University of California, Berkeley, for providing antisera; A. Stütz and N. Ryder, Sandot Forschungs Institut Vienna, for samples of terbinafine; R. Cenedella, Kirksville, MO for providing the inhibitor U18666; and the SFI Vienna for the production of antisera. This work was financially supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (projects S-5811 and 11491 to G.D., projects S-5812 and F 706 to S.D.K., and project S-5814 to F.T.).
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1994. [Google Scholar]
- Bai M, Prestwich GD. Inhibition and activation of porcine squalene epoxidase. Arch Biochem Biophys. 1992;293:305–313. doi: 10.1016/0003-9861(92)90400-q. [DOI] [PubMed] [Google Scholar]
- Bard M, Brunner DA, Pierson CA, Lees ND, Biermann B, Freye L, Koegel C, Baruch R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci USA. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure B, Fegueur M, Servouse M, Karst F. Regulation of squalene synthase and squalene epoxidase activities in Saccharomyces cerevisiae. Lipids. 1989;24:1020–1023. doi: 10.1007/BF02544072. [DOI] [PubMed] [Google Scholar]
- Cenedella RJ. Concentration-dependent effects of AY-9944 and U18666A on sterol synthesis in brain. Biochem Pharmacol. 1980;29:2751–2754. doi: 10.1016/0006-2952(80)90006-4. [DOI] [PubMed] [Google Scholar]
- Clausen MK, Christiansen K, Jensen PK, Behnke O. Isolation of lipid particles from baker’s yeast. FEBS Lett. 1974;43:176–179. doi: 10.1016/0014-5793(74)80994-4. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavey KL, Scallen TJ. Studies on the conversion of enzymatically generated microsome-bound squalene to sterol. J Biol Chem. 1978;10:5176–5183. [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Hirsch PJ, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandrositz A, Turnowsky F, Högenauer G. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene. 1991;107:155–160. doi: 10.1016/0378-1119(91)90310-8. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Schneiter R, Hitomi M, Tartakoff AM. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isoenzymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell. 1996;7:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landl K, Klösch B, Turnowsky F. ERG1, encoding squalene epoxidase, is located on the right arm of chromosome VII of Saccharomyces cerevisiae. Yeast. 1996;12:609–613. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C609::AID-YEA949%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lange Y, Steck TL. Cholesterol-rich intracellular membranes: a precursor to the plasma membrane. J Biol Chem. 1985;260:15592–15597. [PubMed] [Google Scholar]
- Lange Y, Swaisgood MH, Ramos BV, Steck TL. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J Biol Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- Leber R, Zinser E, Hrastnik C, Paltauf F, Daum G. Export of steryl esters from lipid particles and release of free sterols in the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1234:119–126. doi: 10.1016/0005-2736(94)00270-y. [DOI] [PubMed] [Google Scholar]
- Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 1994;10:1421–1428. doi: 10.1002/yea.320101105. [DOI] [PubMed] [Google Scholar]
- Lees ND, Skaggs B, Kirsch DR, Bard M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae —A review. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lum PL, Wright R. Degradation of HMG-CoA reductase-induced membranes in the fission yeast, Schizosaccharomyces pombe. J Cell Biol. 1995;131:81–94. doi: 10.1083/jcb.131.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M’Baya B, Karst F. In vitro assay of squalene epoxidase of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1987;147:556–564. doi: 10.1016/0006-291x(87)90967-3. [DOI] [PubMed] [Google Scholar]
- Mercer EI. The biosynthesis of ergosterol. Pestic Sci. 1984;15:133–155. [Google Scholar]
- Murphy DJ. Structure, function and biogenesis of storage lipid bodies and oleosins in plants. Prog Lipid Res. 1993;32:247–280. doi: 10.1016/0163-7827(93)90009-l. [DOI] [PubMed] [Google Scholar]
- Paltauf F, Daum G, Zuder G, Högenauer G, Schulz G, Seidl G. Squalene and ergosterol biosynthesis in fungi treated with Naftifine, a new antimycotic agent. Biochim Biophys Acta. 1982;712:268–273. [Google Scholar]
- Paltauf F, Kohlwein SD, Henry SA. Regulation and compartimentalization of lipid synthesis in yeast. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 415–500. [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin D, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Reinhart MP, Billheimer JT, Faust JR, Gaylor JL. Subcellular localization of the enzymes of cholesterol biosynthesis and metabolism in rat liver. J Biol Chem. 1987;262:9649–9655. [PubMed] [Google Scholar]
- Ryder NS. Squalene epoxidase-enzymology and inhibition. In: Kuhn PJ, Trinci APJ, Jung MJ, Goosey MW, Copping LG, editors. Biochemistry of Cell Walls and Membranes in Fungi. Berlin: Springer; 1990. pp. 189–203. [Google Scholar]
- Ryder NS. Squalene epoxidase as a target for the allylamines. Biochem Soc Trans. 1991;19:774–777. doi: 10.1042/bst0190774. [DOI] [PubMed] [Google Scholar]
- Satoh T, Horie M, Watanabe H, Tsuchiya Y, Kamei K. Enzymatic properties of squalene epoxidase from Saccharomyces cerevisiae. Biol Pharm Bull. 1993;16:349–352. doi: 10.1248/bpb.16.349. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Skaggs BA, Alexander JF, Pierson CA, Schweitzer KS, Chun KT, Koegel C, Barbuch R, Bard M. Cloning and characterisation of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene. 1996;169:105–109. doi: 10.1016/0378-1119(95)00770-9. [DOI] [PubMed] [Google Scholar]
- Uchida E, Ohsumi Y, Anraku Y. Purification of yeast vacuolar membrane H+-ATPase and enzymological discrimination of three ATP-driven proton pumps in Saccharomyces cerevisiae. Methods Enzymol. 1988;157:544–562. doi: 10.1016/0076-6879(88)57103-3. [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CDM, Fasch E-V, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eucaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]