Abstract
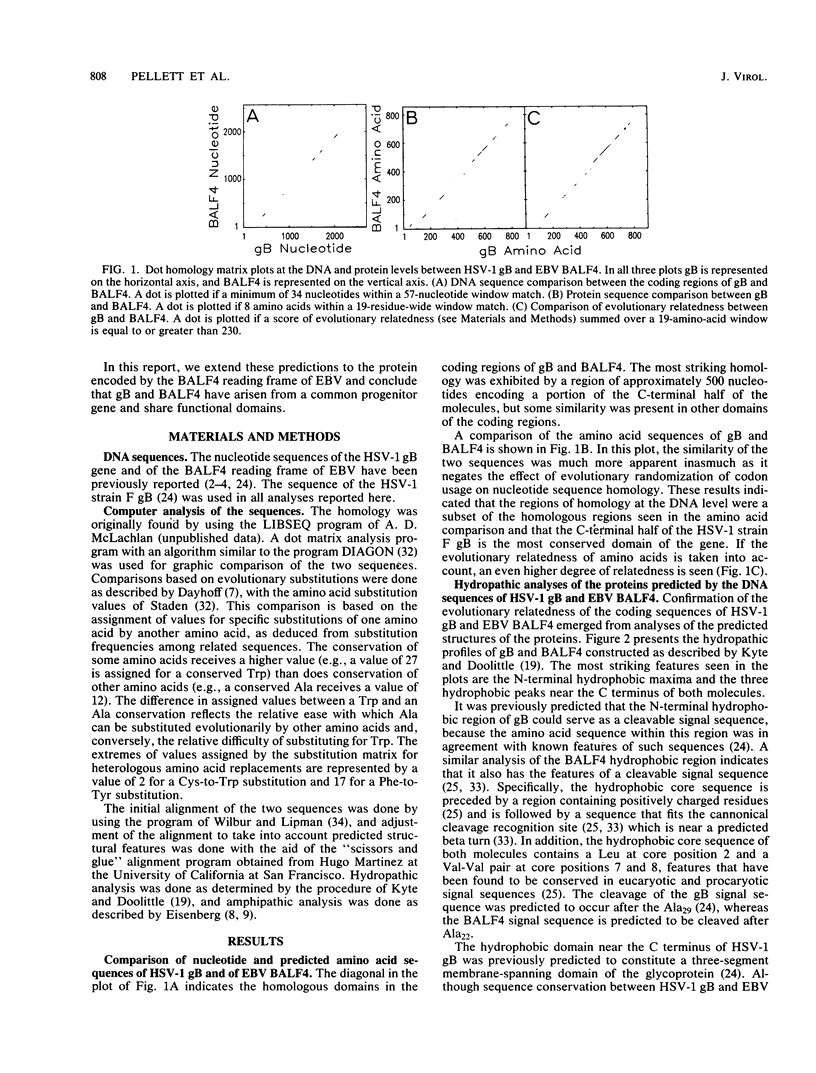
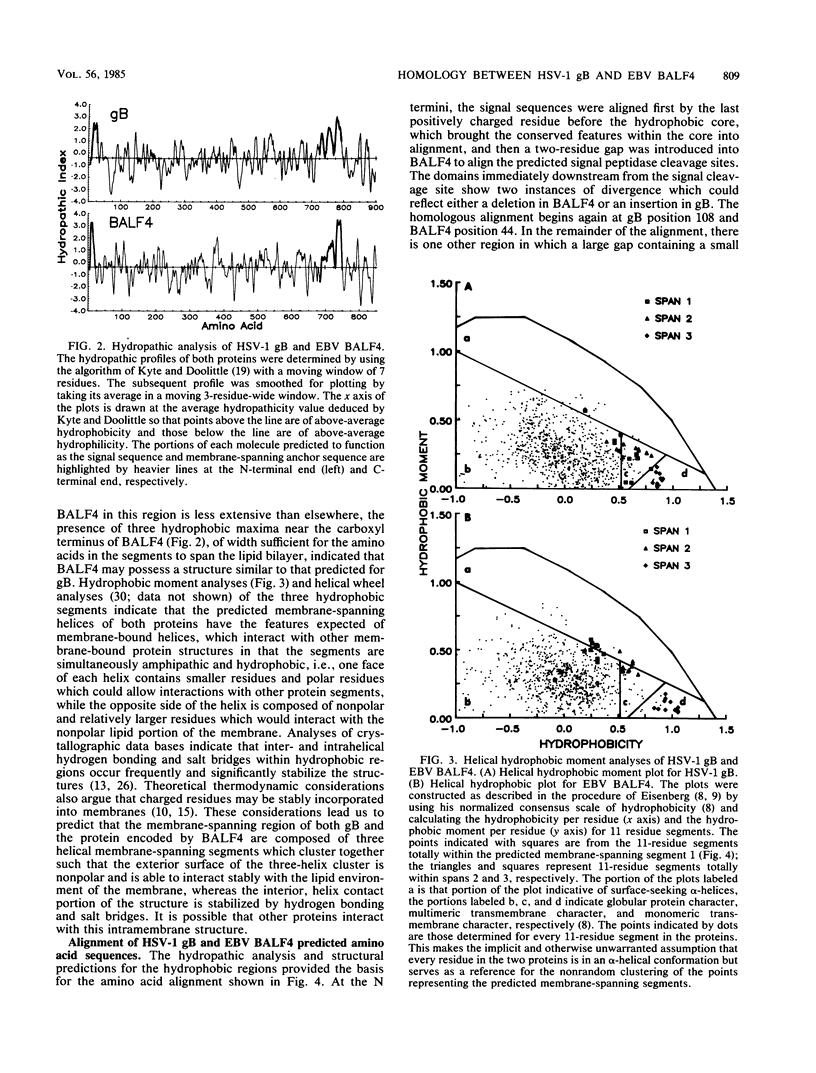
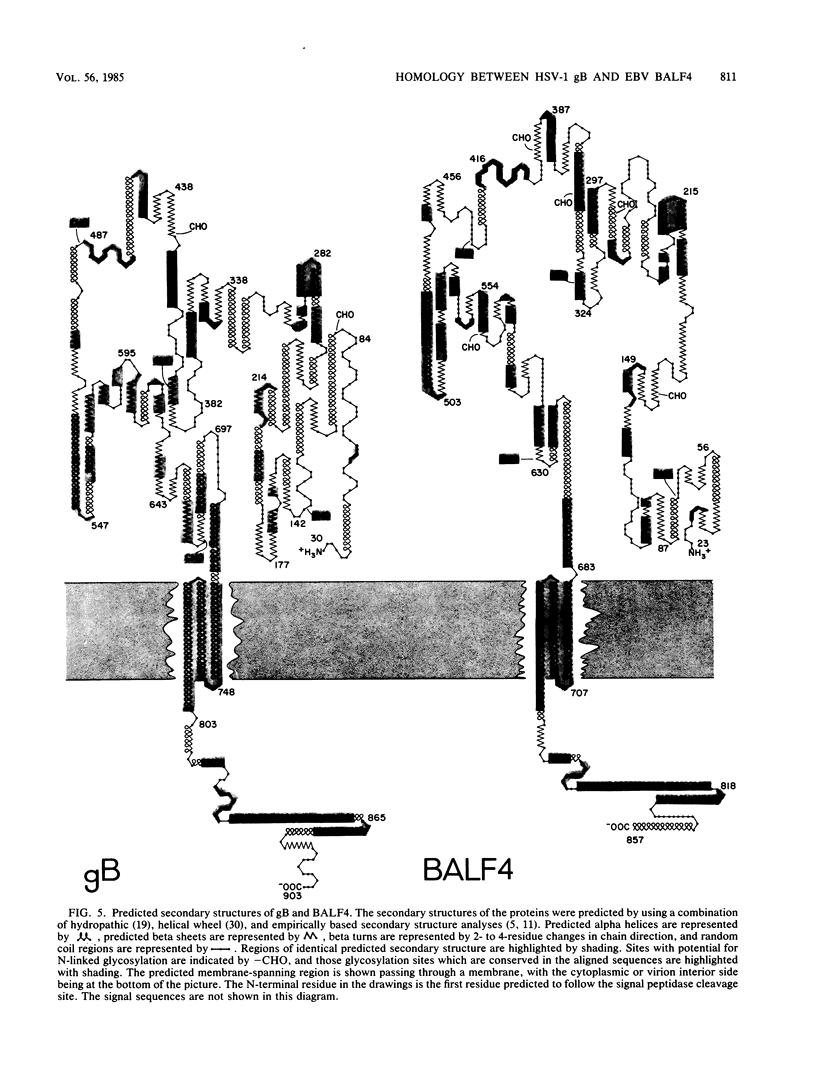
We report significant sequence and predicted secondary structure homology between the herpes simplex virus 1 glycoprotein B (gB) and a protein predicted to be encoded by the BALF4 reading frame of Epstein-Barr virus (EBV). Homology was detectable at the DNA level and was highly significant at the protein level and when evolutionary substitution frequencies of amino acids in related proteins were taken into account. Hydropathic analyses predicted that the two proteins possess conserved N-terminal and C-terminal hydrophobic domains. The N-terminal hydrophobic domains share features in common with known cleavable membrane insertion signal sequences. The amino acid sequences of the C-terminal hydrophobic domains predict three adjacent membrane-spanning segments as had been previously predicted for gB. In an alignment of the two amino acid sequences, 247 of 903 gB residues had a matched pair in the BALF4 sequence, and 247 of 854 BALF4 residues were found to have a matched pair in the gB sequence. In addition, all 10 cysteine residues located outside the predicted signal sequence of both proteins were conserved, as were four predicted N-linked glycosylation sites. In all, 43% of the residues in the aligned sequences are predicted to possess equivalent secondary structures. gB is a virion envelope glycoprotein required for virus entry into cells. The domain of gB determining the rate of entry into cells has been mapped; the predicted structure of this domain in gB and the predicted EBV protein are almost identical. Similarly, the cytoplasmic domain of gB postulated to interact with submembrane proteins was also nearly identical in predicted structure to that of the EBV protein. These results suggest that EBV encodes a protein similar in structure and function to the herpes simplex virus 1 gB.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bankier A. T., Deininger P. L., Farrell P. J., Barrell B. G. Sequence analysis of the 17,166 base-pair EcoRI fragment C of B95-8 Epstein-Barr virus. Mol Biol Med. 1983 Jul;1(1):21–45. [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984 Aug;137(1):185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984 Mar;133(2):301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Draper K. G., Kelly T. J., Wagner E. K. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J Virol. 1985 May;54(2):317–328. doi: 10.1128/jvi.54.2.317-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gibson T., Stockwell P., Ginsburg M., Barrell B. Homology between two EBV early genes and HSV ribonucleotide reductase and 38K genes. Nucleic Acids Res. 1984 Jun 25;12(12):5087–5099. doi: 10.1093/nar/12.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. M., Matthews B. W. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J Mol Biol. 1984 May 5;175(1):75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- Holland T. C., Marlin S. D., Levine M., Glorioso J. Antigenic variants of herpes simplex virus selected with glycoprotein-specific monoclonal antibodies. J Virol. 1983 Feb;45(2):672–682. doi: 10.1128/jvi.45.2.672-682.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B. H., Hubbell W. L. Stability of "salt bridges" in membrane proteins. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5412–5416. doi: 10.1073/pnas.81.17.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. How good are predictions of protein secondary structure? FEBS Lett. 1983 May 8;155(2):179–182. doi: 10.1016/0014-5793(82)80597-8. [DOI] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousoulas K. G., Pellett P. E., Pereira L., Roizman B. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology. 1984 Jun;135(2):379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Little S. P., Jofre J. T., Courtney R. J., Schaffer P. A. A virion-associated glycoprotein essential for infectivity of herpes simplex virus type 1. Virology. 1981 Nov;115(1):149–160. doi: 10.1016/0042-6822(81)90097-0. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K. Assessment of secondary-structure prediction of proteins. Comparison of computerized Chou-Fasman method with others. Biochim Biophys Acta. 1983 Oct 28;748(2):285–299. doi: 10.1016/0167-4838(83)90306-0. [DOI] [PubMed] [Google Scholar]
- Pellett P. E., Kousoulas K. G., Pereira L., Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985 Jan;53(1):243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Rashin A. A., Honig B. On the environment of ionizable groups in globular proteins. J Mol Biol. 1984 Mar 15;173(4):515–521. doi: 10.1016/0022-2836(84)90394-2. [DOI] [PubMed] [Google Scholar]
- Roizman B., Carmichael L. E., Deinhardt F., de-The G., Nahmias A. J., Plowright W., Rapp F., Sheldrick P., Takahashi M., Wolf K. Herpesviridae. Definition, provisional nomenclature, and taxonomy. The Herpesvirus Study Group, the International Committee on Taxonomy of Viruses. Intervirology. 1981;16(4):201–217. doi: 10.1159/000149269. [DOI] [PubMed] [Google Scholar]
- Ruyechan W. T., Morse L. S., Knipe D. M., Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979 Feb;29(2):677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento M., Haffey M., Spear P. G. Membrane proteins specified by herpes simplex viruses. III. Role of glycoprotein VP7(B2) in virion infectivity. J Virol. 1979 Mar;29(3):1149–1158. doi: 10.1128/jvi.29.3.1149-1158.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]


