Abstract
To determine whether the expression of cardiac genes changes in a graded manner or by on/off switching when cardiac myocytes change genetic programs in living animals, we have studied two indicator genes that change their expression oppositely in mouse binucleate ventricular cardiomyocytes during development and in response to cardiac hypertrophy. One is a single-copy transgene controlled by an α-myosin heavy chain (aMHC) promoter and coding for CFP. The other is the endogenous β-myosin heavy chain (bMHC) gene modified to code for a YFP–bMHC fusion protein. Using high-resolution confocal microscopy, we determined the expression of the two indicator genes in individual cardiomyocytes perinatally and after inducing cardiac hypertrophy by transverse aortic constriction. Our results provide strong evidence that the cardiac genes respond by switching their expression in an on/off rather than graded manner, and that responding genes within a single cell and within the two nuclei of cardiomyocytes do not necessarily switch concordantly.
Keywords: gene switching, myosin heavy chain
Cells in different organ systems such as the heart commonly respond during development and after stress in adults by changing their gene expression programs. The changes involve multiple genes and can be in either direction. Substantial work in the past decade has focused on the pathways governing gene regulation and has uncovered molecular mechanisms that integrate the actions of multiple cell surface receptors (1–3), signal transduction pathways (4–6), and nuclear chromatin remodeling (7–10) into changes in gene expression profiles (11). However, important questions remain, including how expression varies in different cardiac genes within the same cardiac myocyte and in different myocytes within the heart. For example, it is not known whether individual myocytes change their expression levels in a graded manner or via an on/off-type switch (12). It is also not known whether the expressions of responding genes change coordinately or independently within individual myocytes.
A close to ideal system in which to answer these questions is provided by the cardiac α-myosin heavy chain (aMHC) and β- myosin heavy chain (bMHC) genes. These two isoforms are normally expressed in a developmental stage-specific manner in the mouse ventricle. Expression of bMHC predominates during embryonic and fetal stages, at which stage the cells are mononucleate. Expression of aMHC predominates soon after birth (13), at which stage the cells are binucleate in the mouse (14, 15). Reversal of this developmental change in expression occurs after experimental cardiac hypertrophy, during which aMHC is down-regulated, and bMHC is up-regulated. The response of the cardiac ventricle to stress is accompanied by an increase in the size of its myocytes without any appreciable increase in their numbers (16). The hypertrophy-associated changes in gene expression include many genes in addition to the MHC genes and affect a variety of structural and functional features of the myocytes, including their contractile efficiency, metabolic state, and current-modulating ion channels (17–19).
To take advantage of this behavior of the MHC genes to answer our questions, we used mice expressing a YFP-bMHC fusion protein from a modified allele at the bMHC locus and a CFP from a single-copy transgene at the hypoxanthine phosphoribosyltransferase locus (Hprt). We followed the expression patterns of these two indicator genes in individual cells after the induction of hypertrophy. Our results provide strong evidence that changes in protein expression of cardiac genes are mediated by switching on/off with the probability that a gene will switch increasing in relation to local fibrosis, although the switching of responding genes within a single cell and within the nuclei of binucleated cardiomyocytes are not necessarily concordant events. Our data are not consistent with expression changing only in a graded manner.
Results
Generation of Transgenic Animals.
To generate an indicator gene (aMHC-CFP) that would change its expression during development and in response to hypertrophy in a manner similar to that of an endogenous gene, we generated mice with a single-copy transgene targeted to the X-linked Hprt locus. The transgene consists of a 5.5-kb aMHC promoter driving expression of sequences encoding CFP. This 5.5-kb promoter has been used extensively to direct ventricle-specific expression of genes and is known to respond appropriately to developmental and hypertrophic stimuli (20, 21). (The GFP transgene upstream of the CFP transgene and separated from the aMHC-CFP transgene by two chicken β-globin HS4 insulators was intended for use in future experiments. The GFP transgene is under the control of a 5.6-kb bMHC promoter, but proved not to be expressed in the adult cardiac myocyte under any conditions.) Single-copy CFP transgenic mice were generated by gene targeting as illustrated in Fig. 1. We chose to target the transgene to the Hprt locus because it is neutral for transgene expression, as judged by previous work showing no notable stimulatory or suppressive effects on transgene expression (22). Because the transgene is a single copy, difficulties associated with multicopy transgenes are avoided. Hemizygous males and homozygous females with this X-linked CFP transgene were used in the present study; they are phenotypically normal.
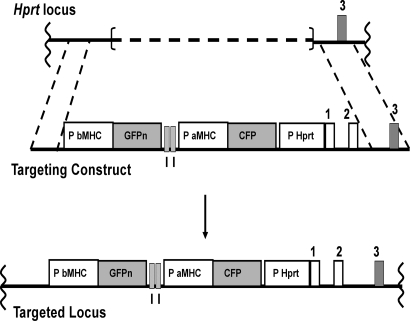
Fig. 1.
Generation of CFP transgenic animals. (Top) Schematic of the partially deleted Hprt locus of BK4 cells (42). The dashed line represents the deleted region. (Middle) Schematic of the targeting construct. (Bottom) Schematic of the targeted locus. An aMHC promoter (P aMHC)-driven CFP gene is upstream of a fragment of the human Hprt gene that includes its promoter (P Hprt) and exons 1 and 2 (coding exons are gray, noncoding exons and promoters are white). Details of the targeting construct have been described (42). GFPn represents the coding region for GFP with a nuclear-locating sequence under control of a 5.5-kb bMHC promoter sequence (P bMHC). The boxes labeled I represent a 1.2-kb chicken β-globin insulator. The schematics are not to scale.
The CFP Transgene Is Regulated in Vivo During Development Like the Endogenous aMHC Gene.
To determine whether our single-copy CFP transgene at the Hprt locus is appropriately regulated in vivo, we analyzed its expression pattern during neonatal development. High-resolution confocal microscopy was used to assess CFP fluorescence in the ventricles of neonatal pups of ages 1 and 10 days. Supporting information (SI) Fig. S1A shows the expression pattern of CFP in thick ventricular sections. Some, but not all, cells in the day 1 pups express CFP at very low levels, as indicated by the faint and patchy cyan fluorescence. By neonatal day 10, CFP is expressed at high levels in virtually all cells (Fig. S1B), a pattern that continues throughout adult age. This pattern mirrors the reported expression pattern of the endogenous aMHC gene (13), indicating that the CFP transgene changes its expression in vivo during neonatal development like the endogenous aMHC gene.
The CFP Transgene Responds to Hypertrophy Like the Endogenous aMHC Gene.
To investigate the response to hypertrophic stimuli of genes of interest, we induced hypertrophy in 6-month-old animals by using transverse aortic constriction (TAC). This surgical procedure causes chronic pressure overload, stimulates renin production by the kidneys, and leads to left ventricular hypertrophy accompanied by a shift from the adult to the fetal MHC isoforms. Controls received sham treatment (similar surgery without TAC). The animals in the TAC group responded with a significant increase in their heart weight (HW)/body weight (BW) ratio (P < 0.005) (Fig. 2A), with all of the echocardiographic hallmarks of decompensated cardiac hypertrophy, including decreases in fractional shortening and increases in left ventricular mass (Table S1).
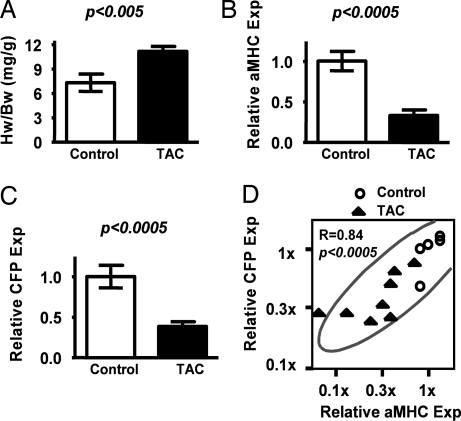
Fig. 2.
Induction of aMHC-CFP transgene expression during hypertrophy. (A–C) Comparisons of HW/BW ratios (A) and mRNA expressions relative to mean of control (Relative Exp) for aMHC (B) and CFP (C) from hearts of TAC (filled bars) and control animals (open bars). Quantitative gene expressions were determined by Taqman Real-Time PCR. (D) Bivariate analyses between mRNA levels of the relative expression of CFP (Relative CFP Exp) and aMHC (Relative aMHC Exp). Circles denote control animals (n = 6), and solid triangles represent TAC animals (4-week treatment, n = 9). The scales of both axes are logarithmic.
To test whether the CFP transgene responds to hypertrophic stress like the endogenous aMHC gene, we determined the expression of the two genes at the mRNA levels by using RT-PCR. As expected from the hypertrophic state of the hearts, the average expression levels of aMHC were significantly reduced (to ≈40% of controls, P < 0.0005) in the ventricles of animals in the TAC group (Fig. 2B). Expression of the CFP transgene was reduced comparably (to ≈40% of controls, P < 0.0005), showing that the transgene is responsive to the hypertrophic stimuli (Fig. 2C).
To test whether the CFP mRNA expression levels parallel the expression levels of the native aMHC gene, we conducted bivariate analyses of mRNA expression levels of CFP and aMHC in the ventricles of individual animals. Fig. 2D illustrates the resulting data as a scatterplot matrix of expression levels of CFP and aMHC. Each point corresponds to an individual animal. The plot demonstrates that expression of CFP at the organ level is highly correlated (r = 0.84, P < 0.0005) with expression of aMHC. Together, these data show that the CFP transgene responds to hypertrophy like the endogenous aMHC gene.
The CFP Transgene Is Down-Regulated in Individual Cells Surrounding Fibrosis.
We have previously generated mice expressing a YFP-bMHC fusion protein and have shown that the YFP-bMHC allele, a fusion of YFP coding sequences to the endogenous bMHC gene, faithfully tracks bMHC expression during development and hypertrophy (23). This work also revealed that a modest increase in bMHC expression occurs during aging. Both the marked increase that occurs with hypertrophy and the modest aging effect are correlated with fibrosis.
To test whether individual cells in which the CFP transgene is down-regulated are also associated with increased fibrosis, we counterstained ventricular sections with a fluorescently labeled (Tritc) lectin, wheat germ agglutinnin (WGA). This lectin binds to collagen and serves as an indicator of fibrosis (23, 24). Fig. 3 A and C shows CFP fluorescence at low power (Fig. 3A) and high power (Fig. 3C), while Fig. 3 B and D show WGA fluorescence in the same ventricular sections. Fig. 3 illustrates that the down-regulation of the CFP transgene (solid ovals in Fig. 3C), like the up-regulation of the YFP-bMHC gene, occurs in cells close to or surrounded by regions with increased collagen (solid ovals in Fig. 3D). In contrast, cells showing high levels of CFP expression (dashed ovals in Fig. 3C) are surrounded by normal levels of collagen (dashed ovals in Fig. 3D).
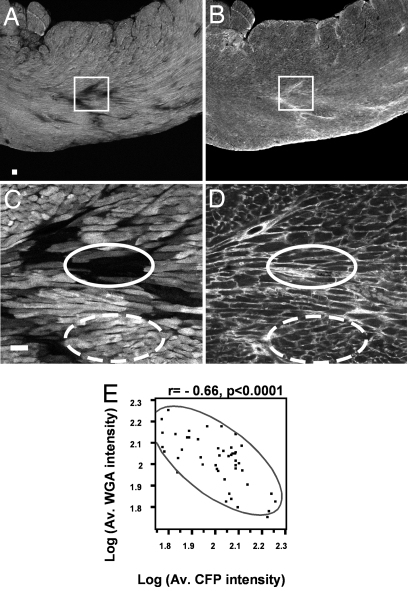
Fig. 3.
aMHC down-regulation is associated with fibrosis. (A–D) Confocal images of a single vibratome transverse section from a transgenic TAC (4-week treatment) heart imaged with a CFP filter (A and C) and a Tritc filter (B and D) for WGA lectin, using ×4 (A and B) or ×20 (C and D) oil immersion objectives. In A and B, squares indicate the regions that were imaged in C and D. In C and D, solid ovals delineate cells with low levels of CFP and dashed ovals delineate cells with high levels of CFP. (E) Bivariate analysis between log10 of mean CFP pixel intensity [Log(Av. CFP intensity)] and log10 of mean WGA pixel intensity [Log(Av. WGA intensity)]. (Scale bar: 50 microns.)
To quantitate these findings, confocal images of the left ventricles from hypertrophic animals were scored for fibrotic and nonfibrotic areas and for CFP fluorescence in discrete areas across sections of the ventricles. Fig. 3E illustrates the resulting data in the form of a scatterplot matrix of mean WGA pixel intensity per area against mean CFP pixel intensity per area and demonstrates a highly significant inverse correlation between WGA staining and CFP expression (r = −0.66, P < 0.0001). To exclude the possibility that cells with undetectable levels of CFP are dead, we counterstained ventricular sections with fluorescent phalloidin, which binds to actin, to determine the cytoarchitecture of the cells. As shown in Fig. S2, cells that express undetectable levels of CFP (* in Fig. S2A) show the architecture expected for living cells (* in Fig. S2B). Thus, cells in which CFP levels change, like cells in which YFP-bMHC levels change, occur in regions of the ventricles where there is increased collagen deposition.
Expression of the Two MHC Indicator Genes Is Independently Controlled in Individual Cells.
To test whether our two myosin-related indicator genes are coordinately regulated in individual cells, we mated the CFP transgenic mice with mice expressing the YFP-bMHC gene. Animals heterozygous for the YFP-bMHC gene and hemizygous (if male) or homozygous (if female) for the X-linked CFP transgene were submitted to TAC (4 and 8 weeks). We then used quantitative confocal microscopy to visualize the expression of the two indicator genes. To quantitate the protein expression in individual cells from hypertrophic and control hearts, we determined the average pixel intensities of CFP and YFP in single cells. Approximately seven images spanning the left ventricular free walls of six hypertrophic and three untreated double heterozygote animals were acquired, and the average fluorescence intensities per pixel of YFP and CFP in individual cells (≈19,000 cells total; 3,000 cells per TAC heart) were determined by using National Institutes of Health program ImageJ.
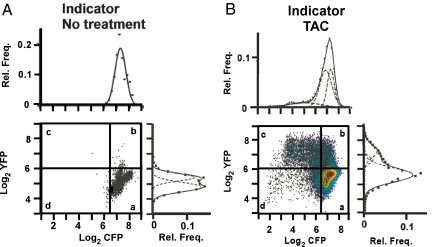
Fig. 4 illustrates the resulting data as a scatterplot matrix of log2 of average YFP fluorescence (vertical axis) and log2 of average CFP intensity (horizontal axis) of individual cells from untreated transgenic animals (Fig. 4A) and transgenic animals with TAC treatment (Fig. 4B). The crossed vertical and horizontal lines in Fig. 4 are drawn, respectively, two standard deviations to the left and above the mean log2 CFP and mean log2 YFP intensities of the control cells; 99% of the control cell populations (Fig. 4A) lie within the lower right quadrant (a) formed by these lines. Fig. 4B shows that after TAC the expression of the YFP fusion gene increases in some, but not all, myocytes (38% of the points are now above the horizontal line). Accompanying these increases in YFP expression, the expression of CFP decreased (25% of the points are now left of the vertical line). These data consequently demonstrate that, although most of the cells in the ventricles of TAC-treated animals are hypertrophic (23), the expression levels of the indicator genes change in only some of them, and the two indicator genes do not always change concordantly. More cells increase YFP expression than decrease CFP expression.
Fig. 4.
MHC isoform switching. Bivariate scatterplots representing log2 of average pixel intensity per cell of CFP (x axis) and YFP (y axis) of ventricular cells in transgenic animals with no treatment (A) and in transgenic mice treated with TAC (B). The crossed vertical and horizontal lines are drawn, respectively, two standard deviations to the left and above the mean Log2 CFP and Log2 YFP intensities of the control cells. Contour lines in the data for TAC animals present one-10th quantile values. Relative frequency (Rel. Freq.) of average CFP pixel intensity per cell (Upper) and average YFP pixel intensity intensity per cell (Lower) are presented alongside each bivariate scatterplot. Solid lines represent the best-fit curves, and dashed lines represent the parameters of the constituent populations determined by nonlinear regression analyses. Data represent ≈19,000 cells from six TAC hearts (4- and 8-week treatments, 3,000 cells per heart) and three control hearts (800 cells per heart). The data for the nine individual animals that are included are presented in Fig. S3 and Fig. S4. The bimodal distribution of YFP pixel intensity for the control animals is not caused by the presence of two populations in each animal, but is caused by small differences in YFP expression between the different control animals that each show a unimodal distribution (Fig. S3).
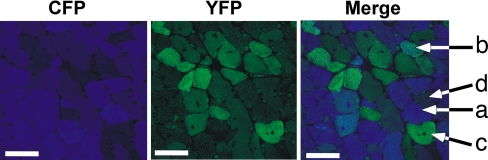
Fig. 5 illustrates cells, close to areas of fibrosis, that correspond to the four types of cells represented by the quadrants in Fig. 4B: high CFP, low YFP (a); high CFP, high YFP (b); low CFP, high YFP (c); and low CFP, low YFP (d). The lack of concordance is further documented by noting that the fraction of cells in quadrants d and b (31%) with only one of the genes changing expression is approximately twice the fraction in quadrant c (16%) with both genes changing expression.
Fig. 5.
Confocal images of a single saggital vibratome section from a transgenic TAC heart imaged with filters for CFP and YFP using a ×60 objective. Merge image represents the merging of CFP and YFP. Arrows point to cells that express CFP only (a), both CFP and YFP (b), YFP only (c), and neither CFP nor YFP (d) and represent cells corresponding to each of the four quadrants defined in Fig. 4. (Scale bars: 50 microns.)
To confirm our finding of a lack of concordance between CFP and YFP expression, we conducted regression analysis of CFP fluorescence against YFP fluorescence at the level of individual cells (≈19,000), and the results show only a weak correlation (Spearman rank test ρ = −0.22). This test indicates that the changes in expression of CFP and YFP are largely independent in individual cells. We conclude that TAC induces tissue-wide changes in expression of the two indicator genes that are inversely correlated at the level of individual ventricles, but at the level of individual cells the changes in expression levels of the two genes are not obligatorily coupled.
Changes in Expression of the CFP Indicator Gene Do Not Follow a Graded Model.
We next tested whether the CFP indicator gene responds to the stresses of TAC in a graded manner or by on/off switching. To make these tests we analyzed the fluorescence intensity data to determine statistically whether the distributions are unimodal or bimodal. If, after TAC, the indicator genes change expression in a graded manner in all cells, the distribution of CFP fluorescence intensities should shift downward and that of YFP should shift upward but each distribution should remain unimodal. If the changes are on/off, the distributions should become bimodal (one population remaining unchanged, and the other changed). Nonlinear regression analyses (25) enable comparisons between models of varying complexity. Such analyses compare the statistical merits of the better fit that always results from complexity (i.e., more parameters) versus the statistical penalty (more degrees of freedom) imposed by the additional parameters. We applied tests of this type to the data presented in Fig. 4 without constraining any of the parameters. The tests show that the CFP fluorescence intensities in untreated mice are well described by a single Gaussian distribution, with the high mean intensity expected for a gene that is strongly expressed (best-fit curve above the bivariate scatterplot matrix in Fig. 4A). In contrast, the tests show that the CFP intensities for the TAC-treated animals (Fig. 4B) are best described by a trimodal distribution (P for bimodal versus trimodal < 0.0001; P for unimodal versus trimodal < 0.0001). Thus the intensities of CFP fluorescence in cardiomyocytes after TAC are best represented by three populations that differ in a stepwise manner, rather than by one or two populations with decreased mean levels of CFP expression.
Table 1 and Figs. S3 and S4 present the results of the nonlinear regression analyses for the nine animals whose data were pooled to generate Fig. 4. The best-fit averages from the distributions of cells in individual animals and the best-fit distributions of the cell populations in the individual animals are close to those of the pooled data. Table 1 also shows that the likelihood of finding one or more populations of cells with changed CFP expression is correlated with the HW/BW ratio (a measure of cardiac stress) of the individual mice. We used two tests to assess whether this correlation between the detection of more than one population and the HW/BW ratio is significant. In one test, we considered the number of populations (one, two, or three) detected in a given animal as a continuous variable and regressed the number of populations against the HW/BW ratio by standard linear regression. In the other test, we considered the number of populations as an ordinal number and assessed the correlation by ordinal logistic regression. The linear regression gave R2 = 0.6, P = 0.01; the logistic regression gave P = 0.004. Thus both tests show that the likelihood of detecting a population of cells with altered CFP expression in a given animal is correlated with the degree of stress (HW/BW ratio) to its heart.
Table 1.
Log2 of Mean fluorescence Intensities of the populations identified in individual animals
| Animal description | Log2 CFP mean fluorescence intensity ± SD |
Log2 YFP mean fluorescence intensity ± SD |
||||||
|---|---|---|---|---|---|---|---|---|
| Low pop. | Inter. pop. | High pop. | HW/BW | Low pop. | Inter. pop. | High pop. | HW/BW | |
| 1 No Ind., No Treat. | 2.5 ± 0.4 | 5.4 | 4.9 ± 0.3 | 5.4 | ||||
| 2 No Ind., No Treat. | 2.3 ± 0.2, 2.8 ± 0.3 | 5.7 | 5.2 ± 0.3 | 5.7 | ||||
| 3 Ind., No Treat. | 7.9 ± 0.2 | 4.8 | 5.4 ± 0.2 | 4.8 | ||||
| 4 Ind., No Treat. | 7.2 ± 0.2, 6.9 ± 0.3 | 5.5 | 4.7 ± 0.3 | 5.5 | ||||
| 5 Ind., No Treat. | 7.3 ± 0.2 | 5.8 | 5.2 ± 0.5 | 5.8 | ||||
| 6 Ind., TAC−4 Weeks | 7.1 ± 0.3 | 9.3 | 5.3 ± 0.4 | 6.2 ± 0.8 | 9.3 | |||
| 7 Ind., TAC−4 Weeks | 5.0 ± 1.8 | 6.6 ± 0.4 | 7.0 ± 0.2 | 14.7 | 4.9 ± 0.3 | 5.3 ± 0.9 | 14.7 | |
| 8 Ind., TAC−8 Weeks | 7.0 ± 0.4 | 7.5 ± 0.3 | 7.6 | 5.6 ± 0.2 | 6.4 ± 0.5 | 7.6 | ||
| 9 Ind., TAC−8 Weeks | 5.4 ± 1.3 | 6.6 ± 0.4 | 9.2 | 5.3 ± 0.3 | 6.3 ± 0.5 | 9.2 | ||
| 10 Ind., TAC−8 Weeks | 3.9 ± 0.3 | 5.6 ± 0.9 | 6.7 ± 0.4 | 10.5 | * | 6.7 ± 0.6 | 7.5 ± 0.4 | 10.5 |
| 11 Ind., TAC−8 Weeks | 6.7 ± 0.2 | 7.4 ± 0.6 | 10.6 | 5.8 ± 0.2 | 6.6 ± 0.8 | 10.6 | ||
| ALL TAC cells | 5.0 ± 1.3 | 6.8 ± 0.5 | 7.3 ± 0.3 | 5.6 ± 0.5 | 6.7 ± 0.2 | 7.3 ± 0.4 | ||
Indicator (Ind.) refers to animals having one copy of the YFP fusion gene and one copy (if males) or two copies (if females) of the X-linked CFP transgene. No Indicator (No. Ind.) refers to WT animals with no fluorescence genes. Mean fluorescence intensities and SDs of each population were acquired from the parameters of best-fit curves for each distribution as determined from nonlinear regression analyses. HW/BW indicates the HW to BW ratio (mg/g). Animals are arranged in the increasing order of HW/BW ratios within their groups. The occurrence of two subpopulations of cells with different CFP intensities (animals 2, No. Ind., No Treat., and 4, No Ind., No Treat.) are due to subtle changes in expression levels (see Fig. S3).
Table 1 shows, in addition, that the mean intensity of fluorescence of the intermediate population of cells is approximately midway between the mean intensities of the high expression cells (equivalent to cells from the untreated animals) and the cells with the lowest level of CFP expression (most responsive cells); the means of the three populations are significantly different (P < 0.001).
Equivalent analyses for YFP expression in the cells of TAC hearts show that the pooled data (Fig. 4B and Table 1) and the data from the single animals (Fig. S4 and Table 1) can be described by either a bimodal or trimodal distribution, but not by a unimodal distribution (P < 0.0001). The bimodal and trimodal distributions both identify one population (the majority of cells) with a mean intensity identical to that of the cells from the control animals and animals with no transgenes (indicating a complete shutdown of YFP gene). The bimodal and trimodal distributions identify two other populations, with means greater than the mean of the control population. As in the case of CFP expression, one population has a mean expression level approximately midway between the high and the low expressing populations (Fig. 4B and Table 1). Individual animals exhibit low, low plus intermediate, or intermediate plus high populations. The mean intensities of the three populations are consistent across the 11 animals and differ significantly (P < 0.01). Together, the CFP and YFP data reveal three discrete levels for both indicator genes and show that expression changes in a stepwise rather than a graded manner.
Discussion
In this article we describe experiments aimed at uncovering the manner in which individual cardiac myocytes change the expression levels of different genes in response to external stimuli. We chose to follow the changes in expression in cardiomyocytes of two indicator genes related to the aMHC and bMHC genes because of the well documented response of the two MHC genes to chronic pressure overload on the heart. One of the two indicator genes is a CFP transgene driven by the aMHC promoter and inserted as a single copy into the Hprt locus. The other is a modified form of the natural bMHC gene that produces a YFP-bMHC fusion protein; this gene is at its normal chromosomal locus and has all of its normal introns and control elements intact. Like the aMHC and bMHC genes, the two indicator genes respond antithetically to the stress induced by TAC.
Our investigations reveal that the CFP and YFP indicator genes are regulated discordantly in individual cells in vivo. Thus, we find that myocytes with decreased CFP levels do not necessarily have increased YFP levels, and vice versa, although we find a weak negative correlation between the expression of the CFP and YFP in individual cells. These observations are consistent with previous suggestions that the MHC genes are independently controlled (26–29). A recent report (30) suggests that re-expression of bMHC is the indirect effect of down-regulation of aMHC mediated by a microRNA, mir 208a, transcribed from an intron of the aMHC gene. However, our aMHC CFP indicator transgene lacks this intron, so that the weak correlation that we find between the expressions of our indicator genes cannot be caused by this microRNA. We think it to be more likely because the expressions of both genes are influenced by circumstances that promote fibrosis (23).
The most striking result of our study is the evidence it provides that the changes in expression of indicator genes in the heart in response to stress are mediated in an on/off rather than in a graded fashion, and that each indicator exhibits a trimodal pattern of expression. Thus in the hearts of animals subjected to TAC, we find for each indicator gene a major population of hypertrophic cells that have expression levels indistinguishable from the myocytes from untreated hearts that have high CFP and low YFP expression. A second population has expression levels midway between the first population and those of a third population, which have low levels of expression for CFP, and high levels of expression for YFP. This trimodal distribution of fluorescence intensities is not compatible with graded changes in the expression of the indicator genes. It is, however, readily understood if genes in the binucleate cardiomyocytes switch in an on/off manner that is not necessarily concordant. Under this interpretation, the population with high levels of expression corresponds to cells with the indicator gene on in both nuclei. The second population with intermediate levels of expression corresponds to cells with the indicator gene on in one nucleus and off in the other. The third population with undetectable levels of expression corresponds to cells with indicator genes off in both nuclei. Previous studies (15) have shown that the vast majority (>91%) of the adult mouse cardiac myocytes are binucleate, and Engelmannn et al. (31) have shown that multinucleate myocytes in the adult rat heart are least frequent in the left ventricle compared with the right ventricle and septum. Because our analyses were done exclusively in the left ventricle of the hearts, the relative contributions of the few mononucleate or multinucleate myocytes that may be present in the left ventricle are unlikely to have a major effect on our overall analyses and conclusions.
There has been substantial theoretical discussion (12, 32, 33) and experimental modeling (34–37) regarding the possibility that individual genes are regulated in an on/off or in a graded manner in individual cells. Our study provides evidence that on/off switching occurs in cardiac myocytes and confirms other observations that individual genes change expression in an on/off rather than graded manner (38–40). Our inference that genes within an individual cardiac myocyte nuclei can switch independently is consistent with previous work demonstrating that not all nuclei within multinucleate myotubes are transcriptionally active at a given time and that different loci are regulated independently (41). Our working hypothesis is that the switching we observe represents a change at the level of chromatin that follows when all of several, perhaps many, constantly associating and dissociating factors are transiently in the correct state of modification (phosphorylation, acetylation, etc.) and all are present on the relevant control regions at the same time. There may be more than one transient constellation of bound factors that is competent to induce the change in chromatin configuration. If a competent constellation of bound factors occurs only infrequently, then responding genes will appear to switch stochastically. And in a binucleate cell the gene in one nucleus will switch independently of the same gene in the other nucleus.
In summary, our results provide strong evidence that the changes in expression of the indicator genes that we have studied are accomplished by on/off switching, with the probability that a gene will switch increasing in relation to stress, but with the switching of responding genes within a single myocyte and within the nuclei of binucleated cardiomyocytes being independent events.
Experimental Methods
Generation of Transgenic Animals.
A 5.6-kb murine aMHC promoter and 5.5-kb murine bMHC promoter (kind gifts of Jeffrey Robbins, University of Cincinnati, Cincinnati) were used to drive the coding sequence of CFP and GFPn, respectively. The reporter transgene was cloned into the Hprt targeting vector, a modified version of pSKB1 (42). Cell culture, electroporation, selection, and microinjection of ES cells were as described (42). Transgenic animals from chimeras were maintained in mixed 129Svev/C57BL/6 background. Generation of YFP-bMHC has been described (23). The mice were maintained in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International according to Institutional Animal Care and Use Committee-approved guidelines.
Morphometrical Analyses.
To quantitatively analyze gene expression levels in individual cells, hearts from WT (no transgene), control, and TAC (double heterozygote) animals were sectioned sagitally by using a vibratome and stained with WGA lectin labeled with Alexa Fluor633 (23). Approximately seven images each from the left ventricular free walls were acquired from an Olympus FV500 confocal microscope with a ×20 objective. Cell boundaries were traced based on WGA staining, and the average pixel intensities of YFP and CFP in individual cells (≈3,000 cells per heart and ≈19,000 cells total for TAC; ≈800 cells per heart and 2,500 total for control) were acquired by using ImageJ.
Statistical Analyses.
Average CFP and YFP pixel intensities of cells were analyzed by nonlinear regression. Unimodal, bimodal, and trimodal populations were fitted by using a modified version of JMP software (SAS Institute), and the three models were compared by using F test with Graphpad Prizm software. The parameters of each curve (means, standard deviations, and amplitudes) were derived from the best-fit curves for the distributions determined by the nonlinear regression analyses. Statistical calculations were performed with JMP software. Linear and ordinal logistic regressions were carried out by considering the number of CFP populations, the dependent variable, either as a continuous or ordinal numerical variable, and the HW/BW ratio as a continuous, independent variable.
TAC, echocardiography, gene expression analyses, and confocal image analyses were conducted as described (23, 43). Animals of both genders were used, and no differences between them were observed. Six-month-old mice were used to minimize the effects of age-related changes in MHC expression. For the data presented in Fig. 3E ≈15 areas each from three hearts were analyzed; each individual area represents ≈1/50th ventricle.
Supplementary Material
Acknowledgments.
We thank D. Moore, B. Qaqish, M. Edgell, T. Doetschman, B. Ballou, R. Bagnell, A. Pendse, and J. Arbones-Mainar for discussions and helpful suggestions and J. Robbins for mouse MHC promoters. This work was supported by National Institutes of Health Grants HL71266 and HL49277 (to O.S.) and American Heart Association Grants 0325655U and 0525594U (to K.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805120105/DCSupplemental.
References
- 1.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 2.Koelle MR. Heterotrimeric G protein signaling: Getting inside the cell. Cell. 2006;126:25–27. doi: 10.1016/j.cell.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger J, Lemmon MA. Nuclear signaling by receptor tyrosine kinases: The first robin of spring. Cell. 2006;127:45–48. doi: 10.1016/j.cell.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: The role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–80. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 5.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Vivekanand P, Rebay I. Intersection of signal transduction pathways and development. Annu Rev Genet. 2006;40:139–157. doi: 10.1146/annurev.genet.40.110405.090555. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Schulze SR, Wallrath LL. Gene regulation by chromatin structure: Paradigms established in Drosophila melanogaster. Annu Rev Entomol. 2007;52:171–192. doi: 10.1146/annurev.ento.51.110104.151007. [DOI] [PubMed] [Google Scholar]
- 10.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 11.Lodish H, Berk A, Zipursky S, Matsudaira P, Baltimore D, Darnell J. Regulation of transcription initiation. In: Tenny S, editor. Molecular and Cellular Biology. New York: Freeman; 2000. pp. 342–404. [Google Scholar]
- 12.Zhang Q, Andersen ME, Conolly RB. Binary gene induction and protein expression in individual cells. Theor Biol Med Model. 2006;3:18. doi: 10.1186/1742-4682-3-18. Available at http://www.tbiomed.com/content/3/1/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss A, Leinwand LA. The mammalian myosin heavy chain gene family. Annu Rev Cell Dev Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
- 14.Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 15.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 16.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79:215–262. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 17.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 18.Chien KR, Olson EN. Converging pathways and principles in heart development and disease: CV@CSH. Cell. 2002;110:153–162. doi: 10.1016/s0092-8674(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 19.Copper GT. Basic determinants of myocardial hypertrophy: A review of molecular mechanisms. Annu Rev Med. 1997;48:13–23. doi: 10.1146/annurev.med.48.1.13. [DOI] [PubMed] [Google Scholar]
- 20.Heine HL, Leong HS, Rossi FM, McManus BM, Podor TJ. Strategies of conditional gene expression in myocardium: An overview. Methods Mol Med. 2005;112:109–154. doi: 10.1007/978-1-59259-879-3_8. [DOI] [PubMed] [Google Scholar]
- 21.Gulick J, Subramaniam A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 22.Hatada S, Kuziel W, Smithies O, Maeda N. The influence of chromosomal location on the expression of two transgenes in mice. J Biol Chem. 1999;274:948–955. doi: 10.1074/jbc.274.2.948. [DOI] [PubMed] [Google Scholar]
- 23.Pandya K, Kim HS, Smithies O. Fibrosis, not cell size, delineates β-myosin heavy chain reexpression during cardiac hypertrophy and normal aging in vivo. Proc Natl Acad Sci USA. 2006;103:16864–16869. doi: 10.1073/pnas.0607700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soderstrom KO. Lectin binding to collagen strands in histologic tissue sections. Histochemistry. 1987;87:557–560. doi: 10.1007/BF00492470. [DOI] [PubMed] [Google Scholar]
- 25.Motulsky HJ, Christopoulos A. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. San Diego: Graphpad Software Inc.; 2003. [Google Scholar]
- 26.Mariner PD, Luckey SW, Long CS, Sucharov CC, Leinwand LA. Yin Yang 1 represses α-myosin heavy chain gene expression in pathologic cardiac hypertrophy. Biochem Biophys Res Commun. 2005;326:79–86. doi: 10.1016/j.bbrc.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Sucharov CC, et al. The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses α-myosin heavy-chain gene expression. Mol Cell Biol. 2004;24:8705–8715. doi: 10.1128/MCB.24.19.8705-8715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sucharov CC, Mariner P, Long C, Bristow M, Leinwand L. Yin Yang 1 is increased in human heart failure and represses the activity of the human α-myosin heavy chain promoter. J Biol Chem. 2003;278:31233–31239. doi: 10.1074/jbc.M301917200. [DOI] [PubMed] [Google Scholar]
- 29.Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 31.Engelmann GL, Vitullo JC, Gerrity RG. Age-related changes in ploidy levels and biochemical parameters in cardiac myocytes isolated from spontaneously hypertensive rats. Circ Res. 1986;58:137–147. doi: 10.1161/01.res.58.1.137. [DOI] [PubMed] [Google Scholar]
- 32.Andersen ME, Yang RS, French CT, Chubb LS, Dennison JE. Molecular circuits, biological switches, and nonlinear dose-response relationships. Environ Health Perspect. 2002;110(Suppl 6):971–978. doi: 10.1289/ehp.02110s6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hume DA. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood. 2000;96:2323–2328. [PubMed] [Google Scholar]
- 34.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 35.Karttunen J, Shastri N. Measurement of ligand-induced activation in single viable T cells using the lacZ reporter gene. Proc Natl Acad Sci USA. 1991;88:3972–3976. doi: 10.1073/pnas.88.9.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM. Transcriptional control: Rheostat converted to on/off switch. Mol Cell. 2000;6:723–728. doi: 10.1016/s1097-2765(00)00070-8. [DOI] [PubMed] [Google Scholar]
- 37.Walters MC, et al. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 39.Ko MS, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newlands S, et al. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bronson SK, et al. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rockman HA, et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci USA. 1991;88:8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.