Abstract
Telomere maintenance by telomerase is critical for the unlimited division potential of most human cancer cells. The two essential components of human telomerase, telomerase RNA (hTR) and telomerase reverse transcriptase (hTERT), are recruited from distinct subnuclear sites to telomeres during S phase. Throughout the remainder of the cell cycle hTR is found primarily in Cajal bodies. The localization of hTR to Cajal bodies and telomeres is specific to cancer cells where telomerase is active and is not observed in primary cells. Here we show that the trafficking of hTR to both telomeres and Cajal bodies depends on hTERT. RNA interference–mediated depletion of hTERT in cancer cells leads to loss of hTR from both Cajal bodies and telomeres without affecting hTR levels. In addition, expression of hTERT in telomerase-negative cells (including primary and ALT cancer cell lines) induces hTR to localize to both sites. Factors that did not stimulate hTR localization in our experiments include increased hTR RNA levels and Cajal body numbers, and expression of SV40 large T antigen and oncogenic Ras. Our findings suggest that the trafficking of telomerase to Cajal bodies and telomeres in cancer cells correlates with and depends on the assembly of the enzyme.
INTRODUCTION
Telomerase is a ribonucleoprotein (RNP) reverse transcriptase that utilizes a short motif within its integral RNA subunit (telomerase RNA or TR) to synthesize the DNA repeats that form the core of the telomere (Greider and Blackburn, 1987). In humans, the enzyme's activity is restricted to early prenatal development and the resulting telomeric repeats last throughout the normal lifespan of the organism (Wright et al., 1996; Cong et al., 2002). Most adult somatic tissues contain low or undetectable levels of the catalytic telomerase reverse transcriptase (hTERT) and telomerase activity, resulting in shortening of the telomeres and a limited replicative capacity (Harley et al., 1990; Allsopp et al., 1992; Kim et al., 1994; Cong et al., 2002; Masutomi et al., 2003; Shay and Wright, 2005). Although relatively inactive in the majority of adult somatic tissues, telomerase is reactivated in over 90% of human cancers, and telomere maintenance by telomerase is critical for the continued proliferation of these cells (Kim et al., 1994; Shay and Bacchetti, 1997; Shay and Wright, 2002).
Previously, our lab and that of Tamas Kiss reported cell cycle-regulated trafficking of hTR (and hTERT) in cancer cells that appears to reflect a cellular mechanism by which human telomerase activity is controlled (Jady et al., 2006; Tomlinson et al., 2006). The data indicate that access of hTR and hTERT to telomeres is restricted to S phase (Jady et al., 2006; Tomlinson et al., 2006) when telomere synthesis occurs (Ten Hagen et al., 1990; Wright et al., 1999). hTR is found in Cajal bodies in cancer cells during most of the cell cycle (Jady et al., 2006; Tomlinson et al., 2006). hTERT is similarly mobilized from separate nuclear foci to telomeres at S phase in human cancer cells (Jady et al., 2006; Tomlinson et al., 2006). In addition, during S phase near the time of telomere localization, both hTR and hTERT can be observed in foci directly associated with Cajal bodies (Jady et al., 2006; Tomlinson et al., 2006). Cajal bodies have been postulated to serve as sites of telomerase maturation and assembly and to deliver active telomerase to the telomere (Jady et al., 2004, 2006; Zhu et al., 2004; Matera and Shpargel, 2006; Tomlinson et al., 2006; Cristofari et al., 2007). Although hTR is expressed in primary (normal) human cells, it does not accumulate at Cajal bodies, telomeres, or any other intranuclear structures and is instead found diffusely distributed throughout the nucleoplasm (Zhu et al., 2004).
Here, we have taken advantage of the differences between normal and cancer cells to better understand the trafficking of hTR. We examined a variety of factors that differ between normal and cancer cells in order to identify factors that impact hTR localization. Our results indicate that hTERT is a key determinant in hTR trafficking and is essential for the localization of hTR to both Cajal bodies and telomeres.
MATERIALS AND METHODS
Cell Culture and Transfection
HeLa, VA13, VA13+hTR, VA13+hTR+hTERT, GM847, and GM847+hTERT cells were grown on coverslips in DMEM (Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (FCS; Mediatech, Manassas, VA). IIICF-T/C3 cells (Bryan et al., 1997) were grown in RPMI (American Type Culture Collection [ATCC], Manassas, VA) supplemented with 10% FCS. IMR90 and IMR90+hTERT primary fibroblasts were grown in minimum essential Eagle's medium (ATCC) with 10% FCS. MCF7 cells were cultured in minimum essential Eagle's media supplemented with 0.01 mg/ml bovine insulin (Sigma-Aldrich, St. Louis, MO) and 10% FCS. All human mammary epithelial (HME) cells (Elenbaas et al., 2001) were grown in MEGM media supplemented with bovine pituitary extract (BPE; Cambrex, Walkersville, MD). All cells were cultured at 37°C with 5% CO2. To increase Cajal body numbers in primary cells, IMR90 and HME cells were cultured at 32°C for 24 h before fixation as described (Carmo-Fonseca et al., 1993). Transfections were carried out using Fugene transfection reagent, according to the manufacturers protocol (Roche, Indianapolis, IN). For hTR overexpression, cells were transfected with construct encoding hTR under control of the U1 promoter (Li et al., 2004). For hTERT RNAi, cells were transfected with an shRNA against hTERT or empty vector control plasmid (Masutomi et al., 2003). Twenty-four hours after transfection, cells were selected with 1 μg/ml puromycin for 48 h before analysis.
hTR Fluorescence In Situ Hybridization
hTR fluorescence in situ hybridization (FISH) was performed using a combination of three cy3-conjugated DNA probes complementary to different regions of telomerase RNA (nts. 128–183, 331–383, and 393–449) essentially as described (Zhu et al., 2004; Tomlinson et al., 2006). Cells were grown on coverslips overnight and then washed once with 1× phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, and 1.4 mM KH2PO4, pH7.4) and fixed with 4% formaldehyde (Electron Microscope Sciences, Fort Washington, PA), 10% acetic acid, and 1× PBS for 10 min at room temperature. After two PBS washes, cells were permeablized in 70% ethanol overnight at 4°C. When hTR FISH was combined with visualization of green fluorescent protein (GFP), cells were fixed in 4% formaldehyde in 1× PBS for 10 min at room temperature. After two PBS washes, cells were permeablized in 0.2% Triton X-100 (Sigma-Aldrich) in 1× PBS for 5 min at 4°C. Cells were rinsed twice in 1× PBS and once in 50% formamide (Sigma-Aldrich), 2× SSC before FISH.
Immunofluorescence
After hTR FISH, cells were washed three times with 1× PBS and were incubated with either one or both of the following primary antibodies at the indicated dilution for 1 h at room temperature: mouse anti-p80 coilin to mark Cajal bodies (1:10,000; gift from G. Matera, Case Western Reserve University, Cleveland, OH), mouse anti-TRF2 (1:1000, Upstate/Millipore, Billerica, MA) or rabbit anti-TRF1 (1:100, gift from Susan Smith, Skirball Institute, New York, NY) to mark telomeres. Cells were washed three times in 1× PBS and then incubated with secondary antibody (1:100 of Cy2 conjugated goat anti-rabbit IgG (H+L), 1:100 cy2 conjugated goat anti-mouse IgG (H+L), and/or 1:100 Cy5 conjugated goat anti-mouse IgGγ) for 1 h at room temperature. hTERT immunofluorescence (IF) was performed essentially as described (Masutomi et al., 2003; Tomlinson et al., 2006) using a 1:5000 dilution of 2C4 monoclonal hTERT antibody (Abcam, Cambridge, MA) and a 1:100 dilution of cy2-conjugated goat anti-mouse IgM secondary antibody. All antibodies were diluted in 0.05% Tween-20 in PBS (PBST) and all secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Cells were then subjected to three final 1× PBS washes and mounted in either 90% glycerol, 1 mg/ml ρ-phenylenediamine, 1× PBS, and 0.1 μg/ml 4′6-diamidino-2-phenylindole (DAPI) or Prolong Gold (Molecular Probes/Invitrogen, Carlsbad, CA).
5-Bromodeoxyuridine Labeling
Before fixation, cells were incubated with 100 μM bromodeoxyuridine (BrdU; Sigma-Aldrich) for 30 min at 37°C. The cells were fixed as described above and denatured in 70% formamide (Sigma-Aldrich), and 2× SSC for 5 min at 80°C. After three PBS washes, BrdU was detected using a monoclonal BrdU antibody (1:1000, G3G4; Developmental Studies Hybridoma Bank, Iowa City, IA) and aminomethylcoumarin acetate (AMCA)-conjugated secondary antibody (1:100; Jackson ImmunoResearch) for 2 h at room temperature. Both antibodies were diluted in PBST. After three PBS washes, coverslips were mounted as described above. In cases where hTR FISH was to be performed, cells were fixed again in 4% formaldehyde in 1× PBS for 10 min at room temperature and washed twice in PBS.
S phase Synchronization
Synchronous populations of IMR90 and IMR90+TERT cells were obtained by double thymidine block. Cells were treated with 2 mM thymidine (Sigma-Aldrich) for 36 h. Cells were released by rinsing twice with 1× PBS and incubated in normal growth media for 12 h. Cells were retreated with 2 mM thymidine for another 36 h. At various time points after release, cells were fixed and analyzed by BrdU labeling and FISH.
Microscopy
Images were obtained with the Zeiss Axioskop 2 Mot Plus fluorescence microscope (Carl Zeiss Microimaging, Thornwood, NY) at 63× (Plan Apochromat objectives, NA 1.4) using a cooled charge-coupled device Retiga Exi Fast 1394 camera (Qimaging, Burnaby, BC, Canada) and IPLab Spectrum software (Scanalytics, Billerica, MA). Unless otherwise noted, the results reported are from analysis of at least 100 cells and at least two separate experiments.
RNase Protection Analysis
Total RNA samples were prepared from cells using Trizol according to the manufacturer's protocol (Invitrogen). RNase A/T1 protection was carried out as described (Goodall et al., 1990). 32P (UTP)-labeled, antisense U3 and U1 probes were synthesized from EcoRI linearized plasmids (Ganot et al., 1997; Lund, 1988) using SP6 polymerase. The template for the hTR probe was generated by PCR amplification of an hTR plasmid (Fu and Collins, 2003) using the following oligos: 5′-AGCCGCGAGAGTCAGCTTGG-3′ and 5′-ATTTAGGTGACACTATAGAGGTGACGGATGCGCACGATC-3′. The antisense hTR probe was generated by in vitro transcription of this PCR product using SP6 polymerase.
RESULTS
Identification of hTERT as a Factor in Nuclear Trafficking of hTR
To better understand the mechanisms governing trafficking of hTR to Cajal bodies and telomeres, we tested a series of factors that distinguish cancer cells (tumor-derived cancer lines) from normal cells (isolated primary strains), beginning with hTR expression levels.
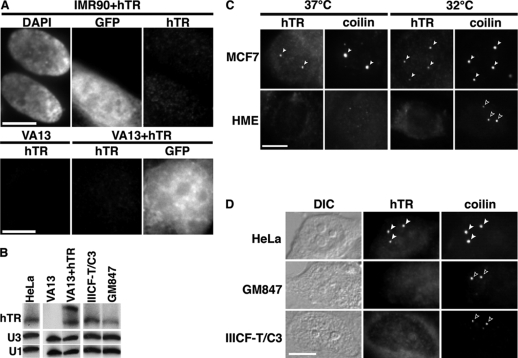
Although hTR is expressed in both normal and cancer cells, on average cancer cells contain twofold higher levels of hTR than normal cells (Yi et al., 1999, 2001). To determine whether expression of higher levels of hTR could account for hTR accumulation at intranuclear foci (like Cajal bodies and telomeres), we transiently expressed a construct encoding hTR (Li et al., 2004) in IMR90 primary lung fibroblasts. On this construct, hTR is expressed from a strong U1 snRNA promoter, which produces elevated levels of functional hTR (Li et al., 2004). The construct also encodes a GFP reporter gene, allowing us to determine which cells were transfected. After transfection, no hTR foci were observed in the GFP-positive cells; in fact, hTR exhibited the same diffuse localization pattern in transfected and untransfected cells (Figure 1A). Furthermore, expression of hTR from this same construct stably maintained in VA13 cells (an unusual cell line that lacks endogenous hTR transcripts; Bryan et al., 1997) produced higher hTR levels than are found in HeLa (cancer) cells (Figure 1B); however, no hTR foci were observed in the stable VA13+hTR lines (Figure 1A). The results indicate that increased hTR levels are not sufficient for localization of hTR into subnuclear structures.
Figure 1.
Localization of hTR to intranuclear foci is not stimulated by increased levels of hTR or Cajal bodies. (A) Ectopic overexpression of hTR does not lead to localization of hTR to nuclear foci. IMR90 primary fibroblasts transiently transfected with an hTR construct (Li et al., 2004), VA13, and VA13 cells stably expressing the same construct were analyzed by hTR FISH. GFP marks cells transfected with the hTR construct. Nuclei are stained with DAPI. (B) Comparison of hTR levels. RNase protection assay on total RNA from HeLa, IIICF-T/C3, GM847, VA13, and VA13+hTR cells. Note that hTR sometimes migrates as a doublet. U3 snoRNA and U1 snRNA were used as loading controls. (C) Increasing Cajal body numbers does not promote hTR localization. MCF7 breast cancer cells and human mammary epithelial (HME) primary cells were grown at 37°C (normal growth temperature) or 32°C (to increase Cajal body formation). Cells were costained by hTR FISH (hTR) and coilin IF (marker protein for Cajal bodies, coilin). Filled arrowheads, Cajal bodies that contain hTR; open arrowheads, Cajal bodies without hTR. (D) hTR does not localize to Cajal bodies in the ALT cell lines GM847 and IIICF-T/C3. HeLa, GM847, and IIICF-T/C3 cells were coanalyzed for hTR (detected by FISH, hTR) and coilin localization (detected by IF, coilin). Filled arrowheads denote Cajal bodies that contain hTR; open arrowheads indicate Cajal bodies without hTR. DIC, differential interference contrast. Scale bars, 10 μm.
In addition to lower hTR levels, primary cell lines generally contain fewer well-formed Cajal bodies. Less than 10% of primary cells in a given population organize typical Cajal body constituents such as coilin into distinct Cajal bodies (Spector et al., 1992). In contrast, over 90% of cancer cell lines have at least one Cajal body per nucleus (Spector et al., 1992). Therefore, we increased Cajal body numbers in HME cells and IMR90 cells by the previously established method of culturing the cells at 32°C (instead of 37°C) for 24 h (Carmo-Fonseca et al., 1993). Data for the HME cells is shown in Figure 1C. We confirmed that culturing at reduced temperature did not disrupt hTR localization in a cancer cell line (MCF7 breast cancer cells, Figure 1C). As expected, growing primary cells at 32°C produced detectable Cajal bodies (monitored by coilin IF) in over 40% of cells examined. However, no change in hTR localization was detectable in the HME and IMR90 cells grown at 32°C (Figure 1C and data not shown), indicating that the presence of well-formed Cajal bodies is also not sufficient to bring about hTR localization to Cajal bodies (or other subnuclear structures).
Finally, we addressed these two factors in combination in the ALT (alternative lengthening of telomeres) cell lines IIICF-T/C3 and GM847. ALT cells are immortalized cell lines that do not have active telomerase and instead maintain telomeres by a telomerase-independent mechanism (Henson et al., 2002). Importantly, these two cell lines express levels of hTR comparable to the commonly studied cancer line HeLa, and approximately one-third of these cells contain well-formed Cajal bodies (Figure 1B and Bryan et al., 1997). As shown in Figure 1D, hTR does not accumulate within Cajal bodies in either IIICF-T/C3 or GM847 cells. These results confirm that hTR levels and the presence of well-formed Cajal bodies do not account for the accumulation of hTR at Cajal bodies and telomeres in cancer cells.
We then examined a series of three gene products that is known to transform primary cells to an oncogenic state: large T antigen of SV40, hTERT, and oncogenic Ras (Hahn et al., 1999; Elenbaas et al., 2001). We examined these potential factors in a series of cell lines, each stably expressing one or more of the three proteins (Elenbaas et al., 2001). The lines were derived from HME primary cells, which do not accumulate hTR in detectable intranuclear foci (Figure 2A, HME). Expression of all three of these gene products in HME cells results in the appearance of hTR in nucleoplasmic foci in approximately one-third of cells examined (Figure 2A, HME + hTERT + large T + onc. Ras). However, we found that coexpression of large T antigen and oncogenic Ras (in the absence of hTERT) did not induce hTR localization (Figure 2A). At the same time, expression of hTERT alone was sufficient for the emergence of hTR foci (Figure 2, HME + hTERT), also producing hTR foci in one-third of cells examined. Finally, a similar pattern was observed in cells expressing both large T antigen and hTERT, whereas expression of large T antigen alone did not induce hTR localization (data not shown). Figure 2B shows that expression of hTERT in the HME cells did not produce a detectable increase in hTR levels. These results suggest that hTERT is important for the localization of hTR to specific nuclear foci and that this effect is not a consequence of increased hTR levels.
Figure 2.
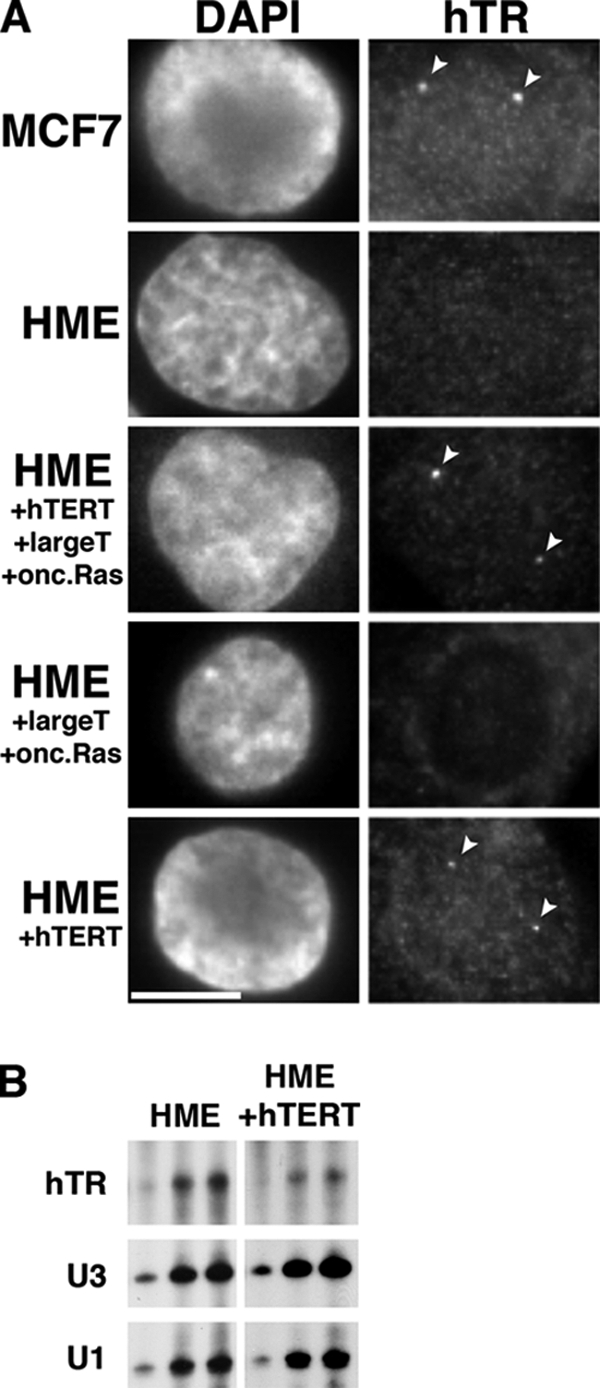
hTERT expression leads to accumulation of hTR within nuclear foci. (A) MCF7 breast cancer cells, human mammary epithelial (HME) normal cells, and HME cells expressing combinations of hTERT, large T antigen, and oncogenic (onc.) Ras were subjected to hTR FISH (hTR). Nuclei are stained with DAPI. Arrowheads denote hTR foci. Scale bar, 10 μm. (B) RNase protection assay on total RNA from HME and HME+hTERT cells. U3 snoRNA and U1 snRNA were used as loading controls.
hTERT Expression Is Essential for Localization of hTR to Cajal Bodies
To determine whether hTERT may play a role in hTR localization specifically to Cajal bodies, we examined the effect of hTERT expression in HME cell lines cultured at 32°C (to increase the frequency of readily detectable Cajal bodies). Although no accumulation of hTR in Cajal bodies was observed in HME cells cultured at 32°C in the absence of ectopic hTERT expression (Figure 1C), we found that approximately half of the Cajal bodies accumulated hTR in the HME cells expressing hTERT (Figure 3). Similar results were observed with IMR90+hTERT cells (Figure 3).
Figure 3.
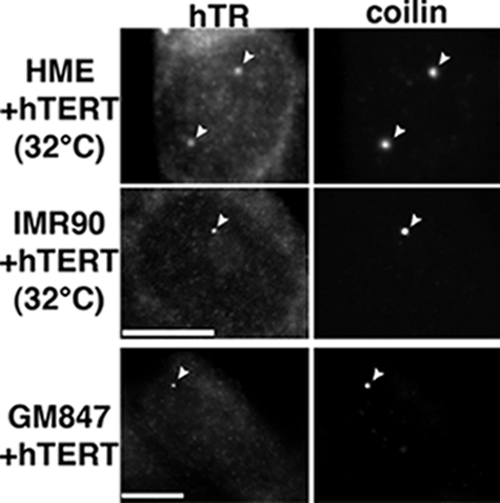
Expression of hTERT induces hTR localization to Cajal bodies. HME and IMR90 cells that ectopically express hTERT were cultured at 32°C to increase Cajal body numbers. These cells and GM847 (ALT) cells that stably express hTERT were analyzed by hTR FISH (hTR) and coilin IF (coilin). Arrowheads indicate hTR foci that colocalize with coilin (Cajal bodies). Scale bars, 10 μm.
Additionally, we tested the effect of hTERT expression on hTR localization in the ALT line GM847, where Cajal bodies are more prevalent but hTR is also not found in Cajal bodies (Figure 1D). Upon stable expression of hTERT in the GM847 cells, hTR was found in nearly every Cajal body observed (Figure 3). These results suggest that hTERT is important for the localization of hTR to Cajal bodies.
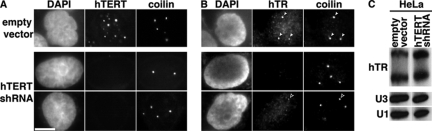
As a reciprocal approach, we knocked down expression of hTERT in HeLa and MCF7 cancer cells (where hTR is found in Cajal bodies) using RNA interference. Figure 4 shows data from the HeLa cell lines. We expressed a short hairpin RNA (shRNA) against the hTERT message and monitored hTERT knockdown efficiency by IF with an antibody against hTERT (Masutomi et al., 2003). In the HeLa cells transfected with an empty vector (and in untransfected cells, data not shown), 5–20 small hTERT foci were observed in over 90% of nuclei (Figure 4A, empty vector). Expression of the hTERT shRNA eliminated detectable hTERT foci in over 70% of cells and reduced hTERT signals in the remainder, indicating effective hTERT knockdown (Figure 4A, hTERT shRNA).
Figure 4.
hTERT expression is necessary for localization of hTR to Cajal bodies. (A and B) RNA interference mediated depletion of hTERT leads to a loss of hTR within Cajal bodies. HeLa cells were transfected with hTERT shRNA (middle and bottom rows) or an empty vector control (top row). (A) Cells were coanalyzed for hTERT and coilin localization (detected by IF); (B) hTR (detected by FISH) and coilin localization (detected by IF). Data in each set of panels were normalized relative to the empty vector control to allow direct visual comparison. Filled arrowheads, hTR in Cajal bodies; open arrowhead, a Cajal body with reduced level of hTR. DAPI was used to stain the DNA. Scale bar, 10 μm. (C) RNA interference mediated depletion of hTERT does not affect hTR levels. RNase protection assay on total RNA from HeLa cells transfected with hTERT shRNA or empty vector. Note that hTR sometimes migrates as a doublet. U3 snoRNA and U1 snRNA are used as loading controls.
We found that hTERT knockdown disrupted hTR localization to Cajal bodies in the cancer cell lines. We performed hTR FISH and coilin IF on cells expressing the hTERT shRNA and found no detectable hTR within Cajal bodies (or at any other nuclear sites) in over 70% of HeLa cells and reduced hTR signal intensity in other cells (Figure 4B, hTERT shRNA). By comparison, hTR was observed in Cajal bodies in over 80% of untransfected cells and cells transfected with empty vector (Figure 4B, empty vector; and data not shown). Importantly, RNase protection assays show that the effect on hTR localization to Cajal bodies is not attributable to a decline in hTR levels in the cells expressing the hTERT shRNA (Figure 4C). Similar results were seen with hTERT knockdown in MCF7 cancer cells where hTR was not observed in Cajal bodies in over 60% of cells (data not shown). Together, the findings presented here strongly indicate the importance of hTERT in the localization of hTR to Cajal bodies.
hTERT Promotes S phase–specific Recruitment of hTR to Telomeres
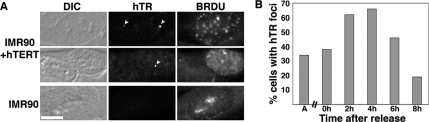
As shown in Figure 2, we found that hTERT induced the appearance of hTR foci in approximately one-third of normal cells in the absence of Cajal body induction, a pattern suggestive of hTERT-induced hTR localization to telomeres during S phase. First, to determine whether localization of hTR to these foci occurred during S phase, we performed hTR FISH in combination with BrdU labeling on normal cells ectopically expressing hTERT (IMR90+hTERT and HME+hTERT). Figure 5 shows data from the IMR90 cell lines. BrdU is incorporated into the DNA of cells undergoing DNA replication (i.e., cells in S phase) and distinct BrdU staining patterns are found in cells in early, mid, or late S phase (O'Keefe et al., 1992). Ectopic expression of hTERT in the normal cells resulted in the appearance of hTR foci as was previously observed (Figure 2), and we found that ∼70% of the cells that contained the hTR foci were in S phase (Figure 5A, IMR90+hTERT; and data not shown). No hTR foci were found in IMR90 or HME cells lacking ectopic hTERT expression in S phase or any other stage of the cell cycle (Figure 5A, IMR90; and data not shown).
Figure 5.
hTR foci in normal cells that express hTERT occur primarily in S phase. (A) hTR foci are predominantly found in S phase cells. IMR90 and IMR90+hTERT cells were subjected to hTR FISH (hTR) and BrdU labeling (BRDU). Arrowheads indicate hTR foci, which are present in BrdU-positive IMR90+TERT cells. Note that the BrdU-negative IMR90+hTERT cell in the middle row of panels does not contain hTR foci. DIC, differential interference contrast. Scale bar, 10 μm. (B) The percentage of cells with hTR foci increases in S phase the percentage of cells with hTR foci. The percentage of IMR90+hTERT cells with hTR foci is graphed relative to time after release from a double thymidine block. Data are compiled from over 100 cells and two independent experiments. A, asynchronous cells.
We quantified the relationship of the hTERT-induced hTR foci to the cell cycle in synchronized cells (Figure 5B). We synchronized IMR90 and IMR90+hTERT cells at the G1/S transition using a double thymidine block and analyzed cells at various time points after release by hTR FISH and BrdU labeling (to assess synchronization efficiency). The incidence of hTR foci in IMR90+hTERT cells peaks in mid-S phase (4 h after release from the block, assessed by BrdU staining, 79% of cells in S phase), where we found that 66% of cells contained hTR foci (Figure 5B). By 8 h after release (20% of cells still in S phase; majority of cells in G2), the frequency of cells with hTR foci had dropped to 18%. IMR90 cells do not display hTR foci in the absence of hTERT expression (Figure 5A) and the synchronization procedure did not induce the formation of hTR foci in IMR90 cells (data not shown). Together, the results indicate that hTR localization to the majority of the nuclear foci observed in hTERT-expressing normal cells occurs during S phase.
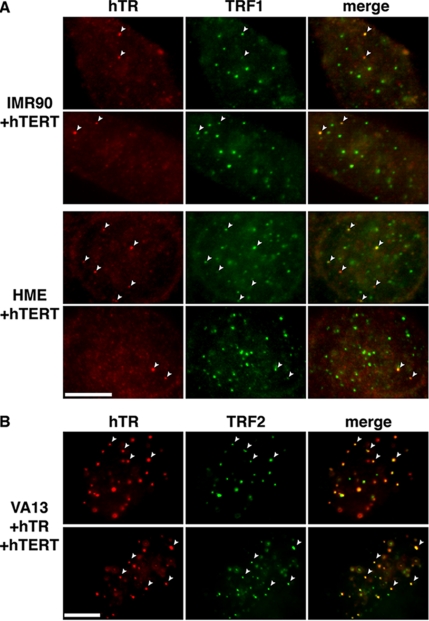
To determine whether the hTERT-induced hTR foci observed during S phase corresponded to telomeres, we performed hTR FISH in combination with IF using antibodies against the telomere-binding proteins TRF1 or TRF2 in IMR90+hTERT and HME+hTERT cells (Figure 6A). We found that hTR is present at telomeres in over 65% of the cells that have hTR foci. Localization of hTR to the telomere was only observed in BrdU-positive cells (data not shown), indicating that it is an S phase–specific event. In addition, we synchronized IMR90+TERT cells and found that greater than 90% of the hTR foci in S phase cells colocalize with telomeres (data not shown).
Figure 6.
hTERT expression is sufficient to stimulate localization of hTR to telomeres. (A) hTR localizes to a subset of telomeres in normal cells that ectopically express hTERT. IMR90+ hTERT (top panels) and HME+hTERT cells were analyzed by hTR FISH (hTR, red) and TRF1 IF (TRF1, green). Merge panels show superimposition of hTR and TRF1 panels; yellow indicates signal overlap. Some colocalizations of hTR and telomeres are denoted with arrowheads. (B) hTR localizes to most telomeres in VA13+hTR+hTERT cells. hTR FISH (hTR, red) and TRF2 IF (TRF2, green) were performed on VA13 cells that stably express both hTR and hTERT. Arrowheads point to representative examples of hTR and TRF2 colocalizations in each panel. Scale bars, 10 μm.
Finally, we examined hTR localization in VA13 ALT cells stably expressing both hTR and hTERT (VA13+hTR+hTERT). As described above, VA13 cell lines are normally devoid of both hTR and hTERT (Bryan et al., 1997) and expression of hTR alone in VA13 cells resulted in no hTR foci (see VA13+hTR, Figure 1A). In striking contrast, stable ectopic expression of hTR and hTERT in these cells results in numerous hTR foci, nearly all of which coincide with a telomere (as indicated by TRF2 staining; Figure 6B).
As shown in Figure 4, hTERT knockdown disrupted the accumulation of hTR in any detectable nuclear foci in cancer cells, indicating that hTERT is required for hTR localization to telomeres (as well as Cajal bodies). Taken together, our data suggest that hTERT expression is necessary and sufficient to enable localization of hTR to the telomere during S phase.
DISCUSSION
Current evidence suggests that the restriction of human telomerase activity to S phase of the cell cycle is accomplished by regulated trafficking of the enzyme. Access of both hTR and hTERT to the telomere is restricted to S phase (Jady et al., 2006; Tomlinson et al., 2006). Outside of S phase, the two components appear to be held primarily in separate intranuclear compartments: hTR in Cajal bodies and hTERT in other nuclear foci (“TERT foci”; Jady et al., 2006; Tomlinson et al., 2006). Because trafficking appears to be a mechanism for cell cycle–based regulation of the activity of telomerase, we undertook studies to further understand the trafficking of hTR. We found that the localization of hTR to both Cajal bodies and telomeres depends on hTERT. These findings explain the presence of hTR at Cajal bodies and telomeres in the majority of cancer cells but not normal cells and identify hTERT as the molecular basis for this difference. Moreover, our results suggest that the intranuclear trafficking of telomerase is coupled to telomerase biogenesis or activity as described below.
Getting to Telomeres during S phase
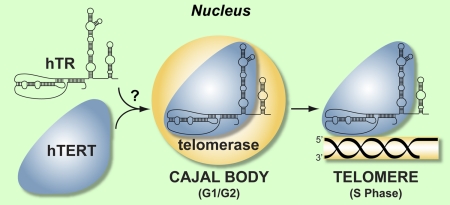
hTR and hTERT function together as integral components of the telomerase enzyme. Previously, we and others found that both hTR and hTERT are specifically recruited to subsets of telomeres in cancer cells during S phase (Jady et al., 2006; Tomlinson et al., 2006), the time when telomeres are replicated and presumably extended by telomerase (Ten Hagen et al., 1990; Wright et al., 1999; Marcand et al., 2000). The current studies revealed that the trafficking of hTR to telomeres during S phase depends on hTERT. The simplest model suggested by these findings is that hTR is only transported to telomeres with hTERT as an assembled complex (Figure 7). Consistent with this concept, we have also found that the localization of hTERT to telomeres depends on expression of telomerase RNA (manuscript in preparation). Moreover, biochemical and genetic studies in S. cerevisiae suggest that the localization of TERT (called Est2p in yeast) to telomeres also requires the expression of TR (TLC1 in yeast; Taggart et al., 2002; Fisher et al., 2004). This model suggests that telomerase (i.e., hTR and hTERT) assembly occurs before transport to telomeres. However, it is also possible that our results reflect more complicated scenarios wherein hTERT levels indirectly influence the trafficking of hTR or a transient interaction of the two components contributes to hTR localization.
Figure 7.
Localization of human telomerase RNA (hTR) to Cajal bodies and telomeres is linked to assembly with telomerase reverse transcriptase (hTERT). The simplest model that arises from the current data are that assembly of hTR and hTERT is a prerequisite for the trafficking of telomerase to Cajal bodies and telomeres. It is not clear whether hTR and hTERT assemble before or after localization to Cajal bodies (indicated by question mark). See Discussion for details.
Cajal Bodies and Telomerase
hTR is observed in Cajal bodies throughout most of the cell cycle in cancer cells (Jady et al., 2004; Zhu et al., 2004; Jady et al., 2006; Tomlinson et al., 2006) and in this study we found that this localization depends on hTERT. Because hTERT does not appear to be present at Cajal bodies outside of S phase (Tomlinson et al., 2006), it does not seem likely that cotransport to Cajal bodies as an assembled complex accounts for the hTERT dependence of hTR accumulation in Cajal bodies (though, it is possible that hTERT is present with hTR in Cajal bodies at concentrations below current detection limits). However, the observation that hTR accumulates in Cajal bodies only in the presence of hTERT points to a strong link between Cajal bodies and telomerase biogenesis. Although the mechanism by which hTERT promotes hTR trafficking is unclear, our results indicate that the localization of hTR to Cajal bodies is coordinated with ongoing or potential telomerase assembly (Figure 7).
Based initially on the observation that telomerase RNA accumulates in Cajal bodies, Cajal bodies have been implicated in various aspects of telomerase biogenesis (Lukowiak et al., 2001; Jady et al., 2004, 2006; Zhu et al., 2004; Tomlinson et al., 2006). Evidence indicates that these subnuclear structures are general sites of RNP assembly and RNA modification (Gall, 2000, 2003; Cioce and Lamond, 2005; Matera and Shpargel, 2006). Some evidence suggests that the assembly of telomerase may be mediated by the SMN (survival of motor neurons) complex, an RNP chaperone associated with Cajal bodies (Terns and Terns, 2001). SMN has been reported to interact with functional telomerase, and in particular with the telomerase-associated proteins Gar1 and hTERT (Bachand et al., 2002; Whitehead et al., 2002). Our new findings—that hTR does not accumulate in Cajal bodies in cancer cells in the absence of hTERT and that hTERT expression can induce hTR targeting to Cajal bodies in normal cells—strongly support the idea that Cajal body localization is intimately connected to telomerase biogenesis.
The mechanisms involved in the targeting and retention of hTR at Cajal bodies are not fully understood. Mutational analysis has demonstrated the importance of a motif called the CAB (Cajal body) box (ugAG) for localization of hTR to Cajal bodies (Jady et al., 2004; Cristofari et al., 2007; Theimer et al., 2007). The CAB box of hTR is located in a terminal loop in the CR7 domain of the molecule (Jady et al., 2004). Our finding that hTR fails to accumulate at Cajal bodies in cells that do not express hTERT indicates that the function of the CAB box of hTR depends (directly or indirectly) on the hTERT protein. Direct interaction of hTERT with hTR may activate the function of the CAB box. However, the CAB box is also present in a subset of functionally unrelated scaRNAs (small Cajal body RNAs), and it is unlikely that hTERT is required for the Cajal body association of this class of RNAs (Kiss et al., 2006; Terns and Terns, 2006).
Cajal bodies have also been proposed to function in the regulated delivery of telomerase to telomeres (Jady et al., 2006; Tomlinson et al., 2006; Cristofari et al., 2007). Cajal bodies are mobile within the nucleus and have been shown to interact with several chromosomal loci, including telomeres during S phase (Gall, 2000; Cioce and Lamond, 2005; Jady et al., 2006). Recently, we found that mutations in the CAB box element of hTR that reduce association of hTR with Cajal bodies (Jady et al., 2004) also reduce the frequency of association of hTR with telomeres and lead to shorter average telomere length (relative to cells expressing a wild-type copy of hTR; Cristofari et al., 2007). Because the CAB box mutations tested did not significantly affect hTR stability or ability to assemble into active enzyme in vivo, the results indicate that accumulation of hTR in Cajal bodies is critical in the localization of telomerase to telomeres.
The results of this study support the emerging paradigm of strong linkages between the physical trafficking pathways and biogenesis pathways of noncoding RNPs (see Matera et al., 2007).
ACKNOWLEDGMENTS
We are grateful to the following people for providing cell lines, plasmids, and antibodies: William Hahn (Dana-Farber Cancer Institute, Harvard Medical School; all HME cell lines, GM847+hTERT; Counter et al., 1998), Gregory Matera (Case Western Reserve University; HeLa cell line and anti-coilin antibody), Jerry Shay (UT Southwestern; IMR90+hTERT and GM847 cells); Roger Reddel (Children's Medical Research Institute, Sydney; IIICF-T/C3 cells), Michael Pierce (University of Georgia;MCF7 cells); Liz Blackburn (University of California, San Francisco; hTR plasmid for transfection), Kathy Collins (University of California, Berkley; hTR plasmid for RNase protection assay), Elsebet Lund (University of Wisconsin-Madison; antisense U1 plasmid), Tamas Kiss (Laboratoire de Biologie Moleculaire Eucaryote du CNRS; antisense U3 plasmid), and Susan Smith (Skirball Institute; anti-TRF1 antibodies). This work was supported by National Cancer Institute (NCI) Grant R01 CA104676 to M.P.T. and R.M.T, and R.L.T. was supported by a National Institutes of Health (NIH) training grant to the Department of Genetics at University of Georgia (GM07103) and a University of Georgia Dissertation Award. E.B.A. was supported by a University of Georgia GRO award and an NCI Diversity Supplement. H.L. was supported by the American Cancer Society (RSG-06-162-01-GMC). C.M.C. was supported by NIH Grant CA082481 and the Werner and Elaine Dannheisser Fund for Research in the Biology of Aging from the Lymphoma Foundation. C.M.C. is a Leukemia and Lymphoma Society Scholar.
Abbreviations used:
- hTR
human telomerase RNA
- hTERT
human telomerase reverse transcriptase
- ALT
alternative lengthening of telomeres.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0184) on July 2, 2008.
REFERENCES
- Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., Futcher A. B., Greider C. W., Harley C. B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand F., Boisvert F. M., Cote J., Richard S., Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol. Biol. Cell. 2002;13:3192–3202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T. M., Marusic L., Bacchetti S., Namba M., Reddel R. R. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum. Mol. Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Ferreira J., Lamond A. I. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis-evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M., Lamond A. I. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Cong Y. S., Wright W. E., Shay J. W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C. M., Meyerson M., Eaton E. N., Ellisen L. W., Caddle S. D., Haber D. A., Weinberg R. A. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Adolf E., Reichenbach P., Sikora K., Terns R. M., Terns M. P., Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Elenbaas B., et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher T. S., Taggart A. K., Zakian V. A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Fu D., Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- Gall J. G. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall J. G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer M., Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Wiebauer K., Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Futcher A. B., Greider C. W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Henson J. D., Neumann A. A., Yeager T. R., Reddel R. R. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Jady B. E., Bertrand E., Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady B. E., Richard P., Bertrand E., Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kiss T., Fayet E., Jady B. E., Richard P., Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Li S., Rosenberg J. E., Donjacour A. A., Botchkina I. L., Hom Y. K., Cunha G. R., Blackburn E. H. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- Lukowiak A. A., Narayanan A., Li Z. H., Terns R. M., Terns M. P. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA. 2001;7:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Lund E. Heterogeneity of human U1 snRNAs. Nucleic Acids Res. 1988;16:5813–5826. doi: 10.1093/nar/16.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Mann C., Gilson E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- Masutomi K., et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- Matera A. G., Shpargel K. B. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Matera A. G., Terns R. M., Terns M. P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- O'Keefe R. T., Henderson S. C., Spector D. L. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Shay J. W., Wright W. E. Telomerase: a target for cancer therapeutics. Cancer Cell. 2002;2:257–265. doi: 10.1016/s1535-6108(02)00159-9. [DOI] [PubMed] [Google Scholar]
- Shay J. W., Wright W. E. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- Spector D. L., Lark G., Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol. Biol. Cell. 1992;3:555–569. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart A. K., Teng S. C., Zakian V. A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Ten Hagen K. G., Gilbert D. M., Willard H. F., Cohen S. N. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M. P., Terns R. M. Macromolecular complexes: SMN-the master assembler. Curr. Biol. 2001;11:R862–864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- Terns M., Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb. Symp. Quant. Biol. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- Theimer C. A., Jady B. E., Chim N., Richard P., Breece K. E., Kiss T., Feigon J. Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol. Cell. 2007;27:869–881. doi: 10.1016/j.molcel.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Tomlinson R. L., Ziegler T. D., Supakorndej T., Terns R. M., Terns M. P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead S. E., Jones K. W., Zhang X., Cheng X., Terns R. M., Terns M. P. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 2002;277:48087–48093. doi: 10.1074/jbc.M204551200. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., Shay J. W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Tesmer V. M., Liao M. L., Shay J. W. Normal human telomeres are not late replicating. Exp. Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- Yi X., Tesmer V. M., Savre-Train I., Shay J. W., Wright W. E. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell Biol. 1999;19:3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Shay J. W., Wright W. E. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tomlinson R. L., Lukowiak A. A., Terns R. M., Terns M. P. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]