Abstract
Reduction in host-activated protein C levels and resultant microvascular thrombosis highlight the important functional role of protein C anticoagulant system in the pathogenesis of sepsis and septic shock. Thrombomodulin (TM) is a critical factor to activate protein C in mediating the anticoagulation and anti-inflammation effects. However, TM protein content is decreased in inflammation and sepsis, and the mechanism is still not well defined. In this report, we identified that the TM 5′ untranslated region (UTR) bearing the internal ribosome entry site (IRES) element controls TM protein expression. Using RNA probe pulldown assay, HuR was demonstrated to interact with the TM 5′UTR. Overexpression of HuR protein inhibited the activity of TM IRES, whereas on the other hand, reducing the HuR protein level reversed this effect. When cells were treated with IL-1β, the IRES activity was suppressed and accompanied by an increased interaction between HuR and TM 5′UTR. In the animal model of sepsis, we found the TM protein expression level to be decreased while concurrently observing the increased interaction between HuR and TM mRNA in liver tissue. In summary, HuR plays an important role in suppression of TM protein synthesis in IL-1β treatment and sepsis.
INTRODUCTION
There is ample evidence that inflammation and coagulation are intricately related processes, whereby inflammation not only leads to activation of coagulation, but coagulation also markedly affects inflammation activity (Esmon, 2005; Levi and Van der Poll, 2005). Inflammation-induced coagulation contributes to vascular thrombotic disease and is also the major consequence in the pathogenesis of microvascular failure and subsequent multiple organ failure in severe sepsis (Diehl and Borgel, 2005). Both preclinical and clinical studies have suggested that excessive microvascular thrombosis during sepsis results in part from depletion of endogenous anticoagulant systems, such as the heparin-antithrombin system, the protein C anticoagulant pathway, and the tissue factor pathway inhibitor system (Haley et al., 2004).
Thrombomodulin (TM) is an important anticoagulant protein present on the surface of vascular endothelial cells (Dittman and Majerus, 1990). TM forms a high-affinity complex with thrombin and results in approximately a 100-fold increase in the activation of protein C to execute anticoagulant effects (Esmon, 1993). Recent studies have shown that TM also plays an important role in attenuation of the inflammatory response (Van de Wouwer and Conway, 2004). One mechanism for TM's anti-inflammatory effect relates to the properties of activated protein C (APC). For example, APC has been found to inhibit endotoxin-induced production of tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, IL-6, and IL-8 in cultured monocytes/macrophages (Okajima, 2001). TM is also a critical cofactor for thrombin-mediated activation of the thrombin-activatable fibrinolysis inhibitor (TAFI), which is responsible for inactivation of complement factors C3a and C5a to protect against complement-mediated injury in the microvasculature (Campbell et al., 2002). Aside from TM acting as a cofactor to mediate anti-inflammatory effects, TM is also reported as playing a direct role in regulating the anti-inflammatory response. Using transgenic mice that lack the N-terminal lectin-like domain of TM (TMLeD/LeD), it was found that the lectin-like domain of TM provides the vascular endothelium with anti-inflammatory properties by interfering with neutrophil adhesion to endothelial cells (Conway et al., 2002). Another report indicates that the lectin-like domain of TM binds with a high-mobility group-B1 DNA-binding protein (HMGB1), a factor acting as an inflammatory mediator, thereby preventing its interaction with the receptor for advanced glycation end products (RAGE) and suppressing induction of proinflammatory events (Abeyama et al., 2005).
Though TM plays an important role in modulating inflammation, unfortunately it appears that TM expression is reduced in inflammation. In vitro studies have demonstrated that endothelial TM expression is potently inhibited by bacterial endotoxin and inflammatory cytokines such as IL-1β and TNF-α (Moore et al., 1987; Archipoff et al., 1991). Two kinds of mechanisms have been proposed to mediate the loss of TM function: the inhibition of transcription and stimulation of endocytosis (Conway and Rosenberg, 1988; Moore et al., 1989). However, in the study of transcriptional regulation of TM gene expression, Lentz et al. (1991) reported that the translational regulation of TM protein expression may occur under some conditions. Furthermore, it was observed that TM protein expression level was significantly decreased at 3 and 6 h, but recovered 12 h after lipopolysaccharide (LPS) treatment in rat sinusoidal endothelial cells. In contrast, the TM mRNA levels were reduced at 6 and 12 h yet slightly recovered 24 h after LPS treatment (Kume et al., 2003). Lack of correlation between protein and mRNA expression patterns indicates the translational regulation mechanism involved in TM protein expression. But, no further research has been conducted to study this mechanism.
In this study, we found the TM 5′ untranslated region (UTR) possesses the IRES activity, which controls the TM protein expression. The RNA-binding protein HuR interacts with TM 5′UTR and negatively regulates TM protein expression under IL-1β treatment is identified. Finally, we observed that this translational repression mechanism is functional in an animal model of sepsis.
MATERIALS AND METHODS
Materials
F-12K medium, fetal bovine serum (FBS), and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). Antibodies against HuR, actin, and thrombomodulin were sourced from Santa Cruz Biotechnology (Santa Cruz, CA). Human IL-1β and TNF-α were obtained from Calbiochem (San Diego, CA).
Cell Culture and Transient Transfection
A549 cells (human lung adenoma cell line) were grown in F-12K nutrient mixture (GIBCO, Rockville, MD) supplemented with 10% FBS, 100 μg/ml streptomycin, and 100 U/ml penicillin. Cells were propagated using standard culture techniques and maintained in a humidified 37°C, 5% CO2 environment. Transfection was performed using Lipofectamine 2000. All transfection assays were performed in triplicate on at least three independent experiments.
Plasmid Construction
The fragments of human TM promoter P1-Luc (nt −246 to + 150) and P2-Luc (nt −246 to + 2) were amplified from normal human genomic DNA. These PCR products were cloned into plasmid pGL3 vector (Promega, Madison, WI) using the HindIII and NcoI restriction sites. The full-length human TM 5′UTR (+169 to +1) fragment was amplified from normal human genomic DNA using primers TM-5′UTR-SpeI-F (5′-ACTAGTCATGTCAGAGGCTGCCTC GCAG-3′) and TM-5′UTR-NcoI-R (5′-CCATGGGTTACCCAGGCGCGCCGCGTG-3′). The PCR product was cloned into the pGL3 promoter vector digested by HindIII and NcoI restriction sites, and the resulting plasmids were designated pTM 5′UTR. Bicistronic reporter plasmids pRF and phpRF were a generous gift of A. E. Willis (The University of Nottingham, United Kingdom). Fragments of the TM 5′UTR were inserted into the SpeI and NcoI sites of these vectors and named as pRTMF and phpRTMF. The MS2hp vector was constructed by inserting the hairpin structure fragment, which bound with MS2 protein into the pGEM-T vector, and the MS2hp TM 5′UTR was created by cloning the TM 5′UTR fragment behind the MS2 hairpin fragment. The HuR protein expression vectors were constructed by amplifying the coding region of HuR and inserting them into the expression plasmid pcDNA3.1 V5 tag (Invitrogen) and pGEX6p-1.
Northern Blot Analysis
Twenty micrograms of total RNA per lane were separated in 1.2% (wt/vol) agarose/formaldehyde gels and transferred onto Hybond-N nylon membrane. Hybridization was performed in ExpressHyb hybridization solution (BD Bioscience, San Jose, CA) at 68°C for 2 h. The luciferase probe was generated by PCR amplification from luciferase gene by using forward primer: 5′-GGTTCCATCTGCCAGGTATCAGG-3′ and reverse primer: 5′-CGTCTTCGTCCCAGTAAGCTATG.-3′. The 300-bp probe was labeled by Rediprime II random prime labeling system (Amersham Bioscience, Piscataway, NJ). After hybridization and wash, the blots were exposed overnight at −80°C to x-ray film.
In Vitro RNA Synthesis and RNA Probe Pulldown Assay
The plasmids MS2hp, MS2hp TM 5′UTR and pTM 5′UTR were linearized to serve as templates to generate RNA probes for pulldown or translational assays. The RNAs were synthesized using the Riboprobe in vitro Transcription system (Promega). Briefly, in each 100 μl of reaction mixture, 1 μg of linearized DNA template was transcribed by T7 polymerase in the presence of 2.5 mM UTP, CTP, ATP, and GTP. After 2-h incubation at 37°C, the reaction was stopped by adding 2 U of RQ1 RNase-free DNaseI (Promega) for 15 min at 37°C. The RNA was purified with MicroSpin G-25 columns. For the RNA probe pulldown assay, 0.5 μg or 1 μg of Ms2hp TM 5′UTR RNA was incubated with A549 cytosol lysates for 1 h, respectively. After incubation, 10 μg of recombinant glutathione S-transferase (GST)-MS2 proteins or GST proteins were added to the reaction and incubated overnight at 4°C. Complexes were isolated using glutathione Sepharose 4B, and RNA-binding proteins in the pulldown material were analyzed by Western blot analysis using HuR antibodies.
RNA Immunoprecipitation Assay
Cytoplasmic extracts were incubated with 5 μg of HuR mouse mAb (Santa Cruz Biotechnology) in immunoprecipitation (IP) buffer (10 mM HEPES, pH 7.9, 100 mM NaCl, 1 mM MgCl2, 0.1% NP-40, 2% glycerol, 1 mM DTT, and 1× protease inhibitor) at room temperature (RT) for 2 h. For the control IP reaction, mouse IgG (5 μg) was used to perform this experiment, and then an equal volume of protein A Sepharose beads was added to the mixture and continually incubated for 2 h at RT. The protein A Sepharose beads were pelleted, washed with 1 ml of IP buffer, and suspended in 0.5 ml of TRI-Reagent (Invitrogen). The RNA was extracted, and RT-PCR was conducted with specific primers for TM or luciferase gene.
Ribosome Complex Pulldown
A549 cells were harvested in phosphate-buffered saline (PBS) and pelleted and suspended in lysis buffer (10 mM HEPES, pH 7.9, 40 mM KCl, 3 mM MgCl2, 0.5% NP-40, 5% glycerol, 2 mM DTT, and 1× protease inhibitor cocktail). Cells were kept on ice for 10 min, and lysates were centrifuged at 5000 rpm for 10 min, and the supernatant was saved as cytoplasmic lysate. 2 μg of ribosomal protein S6 antibody (Santa Cruz Biotechnology) was added to 800 μg cytoplasmic lysate and incubate at 4°C overnight. Protein A/G-agarose was added to the mixture to pull down the ribosome complex. The mRNAs bound with ribosome complex were extracted with TRI-reagent and analyzed the TM mRNA expression level by quantitative RT-PCR.
RNA Interference Assay
A chemically synthesized small interfering RNA (siRNA; 5′-AAGAGGCAAUUACCAGUUUCAtt-3′) targeted to the human HuR mRNA sequence was transiently transfected (final concentration 50 nM) into A549 cells (60% confluent in six-well plates) using Lipofectamine 2000. siRNA targeted to enhanced green fluorescent protein (EGFP) was used as a control. Forty-eight hours after transfection, whole cell lysates were collected for assessment of protein expression level or luciferase activity.
RT-PCR and Quantitative PCR
Total RNA was isolated using the TRI-Reagent, and 3 μg of RNA was subjected to reverse transcription with SuperScript II reverse transcriptase (Invitrogen). 1 μl of RT product was under 32 cycles of PCR analysis. The primers used are listed as follow: TM; forward primer: 5′-TGAGCGTTATTGGTCGGCAGCCT-3′ and reverse primer: 5′-CACAGGTAGGGTGACTCAGG-3′; Luciferase gene; forward primer: 5′-GGTTCCATCTGCCAGGTATCAGG-3′ and reverse primer: 5′-CGTCTTCGTCCCAGTAAGCTATG.-3′; and Renilla gene; forward primer: 5′-AAAGGTGAAGTTCGTCGTCCAAC-3′ and reverse primer: 5′-TTTGAGAACTCGCTCAACGAACG-3′. The quantitative PCR analysis was conducted on LightCycler 480 real-time PCR system by using LightCycler FastStart DNA Master SYBR Green I reagent. The primers used are listed as follow: human TM; forward primer: 5′-CCACTGCTACCCTAACTACG-3′ and reverse primer: 5′-TGTAGCCTTCAGGGCACTCA-3′; rat TM; forward primer: 5′-CGAATGCCTCACCAATGAA-3′ and reverse primer: 5′-TACCGTCGGATTGCTTGAT-3′; Luciferase gene; forward primer: 5′-GGATTACAAGATTCAAAGTGCG-3′ and reverse primer: 5′-TGATACCTGGCAGATGGAAC; and rat actin; forward primer: 5′-GGGTGTGATGGTGGGTAT-3′ and reverse primer: 5′-TTGTAGAAAGTGTGGTGCCAAA-3′.
Animal Model of Polymicrobial Sepsis
Male Sprague Dawley rats weighing 270–320 g were fasted overnight with free access to water. Sepsis was induced by cecal ligation and puncture (CLP) in accordance with a slightly modified previously described method (Wichterman et al., 1980). Under holthane anesthesia, a laparotomy was performed, and the cecum ligated with a 3-0 silk ligature and punctured twice with an 18-gauge needle. The cecum was then returned to the peritoneal cavity, and the abdomen was closed in two layers. In the sham-operated rats, a laparotomy was performed, and the cecum was manipulated but neither ligated nor punctured. All animals were individually resuscitated with 4 ml of isotonic sodium chloride solution per 100 g of body weight by sc injection at the completion of surgery and at 9 h after surgery (Hsieh et al., 2004). The study was conducted in accordance with National Institutes of Health's Guidelines for the use of experimental animals.
Immunofluorescence Microscopy
Before sacrifice, rats were perfused with 0.9% normal saline under anesthesia. Liver tissue was isolated and fixed by 4% paraformaldehyde overnight at 4°C. Then, the tissue was transferred to 30% sucrose in PBS and stored at 4°C for 3 d. Liver specimens were embedded in optimal cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA) cooled down with liquid nitrogen and then maintained at −80°C. The frozen OCT-embedded tissues were cut at 10-μm thickness and placed on silane-coated glass slides (DAKO, Carpinteria, CA). After blocking with 10% bovine serum albumin and 1% Triton X-100 in 1× PBS for 60 min at RT, sections were incubated with HuR mAb at 1:100 dilution or TM antibody at 1:100 dilution in 1× PBS containing 0.1% bovine serum albumin and 0.1% Triton X-100 for 2 h at RT and then overnight at 4°C and washed twice with PBS, followed by incubation with the secondary antibody conjugated with Alexa Fluor 568 (Molecular Probes at 1:200 dilution) for 1 h at RT. After washing three times with PBS, the nuclei were revealed by 4, 6-diamidino-2- phénylindole (DAPI; Sigma at 1:10,000). Confocal images were acquired on an Olympus IX71 confocal laser scanning system using a 100× immersion objective (Melville, NY).
RESULTS
TM 5′UTR Regulates TM Protein Expression
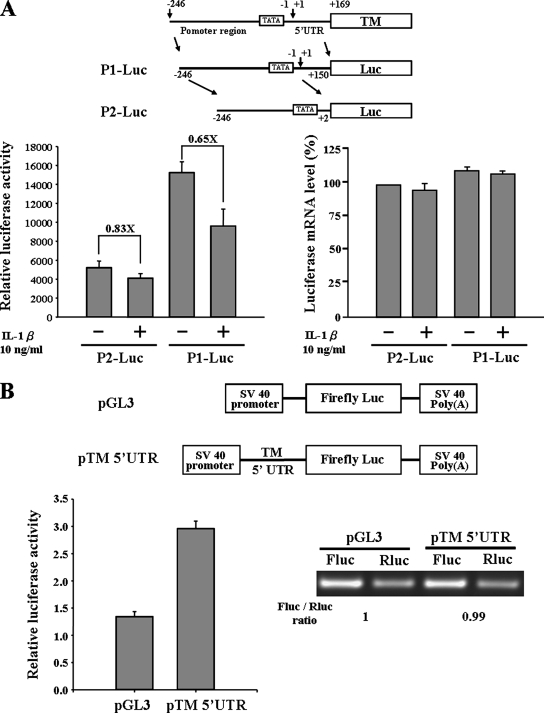
Two kinds of reporter constructs were created in studying the transcription regulation of TM promoter. The P1-Luc construct contained promoter and 5′UTR regions, and the P2-Luc construct only contained the promoter region. These constructs were transfected into A549 cells, and reporter assays were conducted. Unexpectedly, we found the reporter activity of P1-Luc construct to be higher than P2-luc construct (Figure 1A). After IL-1β treatment, the reporter activity of P1-Luc construct was reduced about 35% (0.65×), whereas the P2-Luc construct only decreased 17% (0.83×). This raises the possibility that the 5′UTR harbored a cryptic promoter element or other unknown elements that increased the total reporter activity, and this activity was repressed by IL-1β. However, the luciferase mRNA expression levels of P1-Luc and P2-Luc were similar. It indicated the transcription regulation was not the major reason to cause the difference in reporter activity. To further address the functional role of TM 5′UTR, the 5′UTR was cloned into the pGL3 promoter vector, and the reporter assay and RT-PCR were performed. From Figure 1B, we found the RNA expression level was equal for the pTM 5′UTR and pGL3 vector; however, the reporter activity was higher in the pTM 5′UTR construct. Because of the mRNA expression levels being the same, the higher reporter activity indicated the higher expression level of luciferase protein. From these results, it appears that TM 5′UTR contains an element that regulates the TM protein expression.
Figure 1.
TM 5′UTR contains cis-acting elements to regulate protein synthesis. (A) Two kinds of promoter constructs, P1-Luc and P2-Luc, were transfected into A549 cells. After 24 h, transfected cells were treated with or without 10 ng/ml IL-1β for 6 h. The cell lysate was collected for measuring luciferase activity. Total RNA was extracted, and the luciferase mRNA expression level was detected by quantitative RT-PCR. (B) Effect of TM 5′UTR in a monocistronic reporter assay. A549 cells were transfected with pGL3 or pTM 5′UTR vectors for 24 h. Cells were then lysed, and the luciferase activity was measured. In the right panel, total RNA was extracted, and the RT-PCRs were performed with specific primers targeted to firefly luciferase and Renilla luciferase. Error bars, SD as determined from at least three independent experiments performed in triplicate. The Renilla luciferase vector was cotransfected with the reporter constructs to normalize the transfection efficiency.
TM 5′UTR Contains IRES Element
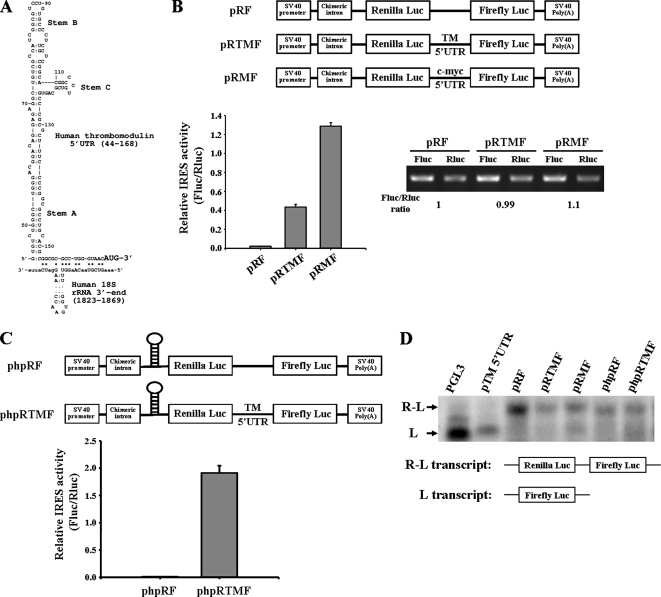
The 5′UTR of TM is 168 nt long and has a G+C content of 72%. Employing the computer algorithm of Zuker, which estimates the minimum free energy of a folded RNA molecule, the 5′UTR of TM was predicted to form a highly stable secondary structure with free energies (ΔG values) −80.2 kcal/mol (Mathews et al., 1999). It was reported that an RNA secondary structure with a free energy of −50 kcal/mol in 5′UTR was inhibitory to cap-dependent ribosomal scanning (Kozak, 1986). However, from our study we found that the TM 5′UTR increased the protein synthesis. This indicated the TM 5′UTR might contain an element that regulated the protein synthesis through a cap-independent translation mechanism. By using a computational approach, we found that TM 5′UTR contained a Y-shaped stem-loop structure (a three-way junction) followed by a short 18S rRNA-complementary sequence immediate to the initiator (Figure 2A). This structure is similar to the structure of cellular IRES, which initiated the cap-independent translation (Le and Maizel, 1997).
Figure 2.
TM 5′UTR contains IRES element. (A) The IRES element structure in the TM 5′UTR is predicted by computer software. The Y-shaped motif is denoted by Stems A, B, and C. The short 18S rRNA-complementary sequence is labeled by the asterisk (*). (B) A549 cells were transfected with bicistronic vectors pRF, pRTMF (contained TM 5′UTR), and pRMF (contained c-myc 5′UTR) for 24 h, and then cells were harvested for determination of firefly and Renilla luciferase activities. The IRES activity was expressed as a ratio of the downstream cistron to the upstream cistron (firefly/Renilla luciferase). Total RNAs from transfected cells were used for RT-PCR analysis. (C) A549 cells were transfected with phpRF and phpRTMF bearing a stable hairpin upstream of Renilla luciferase to block cap-dependent translation. After transfection for 24 h, firefly and Renilla luciferase activities were measured. Error bars, SD as determined from at least three independent experiments performed in triplicate. (D) Northern blot analysis of bicistronic mRNA expression. A549 cells were transfected with different kinds of bicistronic vectors that were used in this figure.
To further assess if the 5′UTR of the TM mRNA really has IRES activity, a bicistronic reporter construct named pRF was used (Stoneley et al., 1998). TM 5′UTR and c-myc 5′UTR were cloned into bicistronic pRF vector between the cDNA encoding Renilla and firefly luciferase genes. These plasmids were transiently transfected into A549 cells, and the activities of both luciferases (Renilla and firefly luciferase) were determined. The presence of the TM 5′UTR increased the expression of the downstream firefly luciferase relative to Renilla luciferase (Figure 2B, cf. pRTMF with pRF control). c-myc 5′UTR was reported to contain the IRES and showed as a positive control here. To clarify whether the increased activity was due to the elevated protein expression but not the mRNA expression, RT-PCR was conducted to determine the level of firefly luciferase mRNA and Renilla luciferase mRNA. As shown in Figure 2B, there were no differences in mRNA levels between pRF and pRTMF vectors, raising the question of whether the TM 5′UTR promoted the downstream cistron protein expression through a cap-independent mechanism. To address this question, the phpRF vector, which contains a palindromic sequence to form a stable mRNA hairpin (−55 Kcal/mol), located upstream of the Renilla luciferase cistron, was used to inhibit ribosome scanning (Figure 2C). Under this circumstance, the cap-dependent translation of Renilla luciferase cistron should be greatly diminished, whereas cap-independent IRES activity of the downstream firefly luciferase cistron should not be affected. Compared with phpRF, the phpRTMF which contained TM 5′UTR expressed a higher firefly luciferase activity (Figure 2C). This suggests that translation of firefly luciferase cistron is specifically driven by the TM 5′UTR, but is not dependent on ribosome scanning from cap-dependent translation. Furthermore, we demonstrated the integrity of the bicistronic transcript by Northern blot analysis. From Figure 2D, we found all the bicistronic transcripts were the full-length transcripts. No aberrant transcripts or monocistronic firefly luciferase transcripts were observed. Besides, RNase protection assay was performed to further confirm the integrity of the biocistronic transcripts (Supplementary Figure S1). Overall, these results demonstrate that TM 5′UTR possesses the IRES activity.
HuR Interacts with the TM 5′UTR
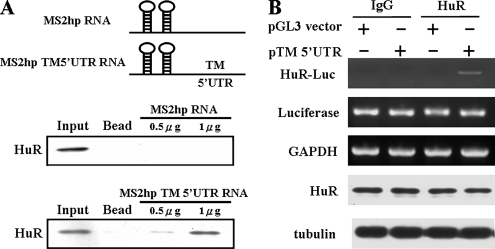
Internal initiation trans-acting factors (ITAFs), such as UNR, PTBP1, and PCBP1, were shown to modulate the IRES function (Stoneley and Willis, 2004). To investigate the molecular mechanism of how TM IRES regulates TM protein expression, we used the MS2 hairpin probe to perform a pulldown assay combined with the Western blot analysis to identify the possible trans-acting factors. The total cell lysates derived from A549 were incubated with MS2hp RNA or MS2hp TM 5′UTR RNA probes and then were pulled down by GST-MS2 fusion proteins. Using Western blot analysis with different ITAF antibodies, we found HuR bound with the TM 5′UTR in a dose-dependent manner (Figure 3A).
Figure 3.
HuR interacts with TM 5′UTR. (A) In vitro transcribed MS2hp and MS2hp-TM 5′UTR RNAs were incubated with cell lysates of A549 cells. After 1 h, the recombinant GST-MS2 proteins were added to the reaction mixture to perform a GST pulldown assay. The pulldown complexes were analyzed by SDS-PAGE and detected by HuR antibodies. (B) HuR binds with TM 5′UTR in A549 cells. Cells were transfected with pGL3 vector and pTM 5′UTR vector, and the cytosol extracts were immunoprecipitated with anti-HuR antibodies. The RNA associated with immunoprecipitated complex was extracted by Trizol reagent, and the level of HuR bound firefly luciferase mRNA (HuR-Luc) was detected by RT-PCR. The mRNA expression levels of Luciferase and GAPDH and the protein expression levels of HuR and tubulin were shown as an internal control.
To further demonstrate whether HuR binds with the TM 5′UTR in A549 cells, RNA-IP assays were performed. A549 cells were transfected with pTM 5′UTR and pGL3 promoter vectors, and the cytosol lysates were immunoprecipitated by HuR antibody. The HuR-interacted RNA was extracted, and RT-PCR was performed. As shown in Figure 3B, we found HuR specifically bound with the mRNA encoded by pTM 5′UTR construct, which bears TM 5′UTR before the luciferase gene, but not the pGL3 vector construct. To further rule out the increase binding between HuR and mRNA encoded by pTM5′UTR construct was due to the different expression level of luciferase mRNA or HuR protein under these conditions, the expression level of luciferase mRNA and HuR protein was examined. We observed no significantly differences existed between these conditions. The expression level of GAPDH mRNA and tubulin protein was detected as a loading control (Figure 3B). These results indicate that HuR specifically interacts with TM 5′UTR in a cultured cell system.
HuR Represses TM IRES-mediated Protein Translation
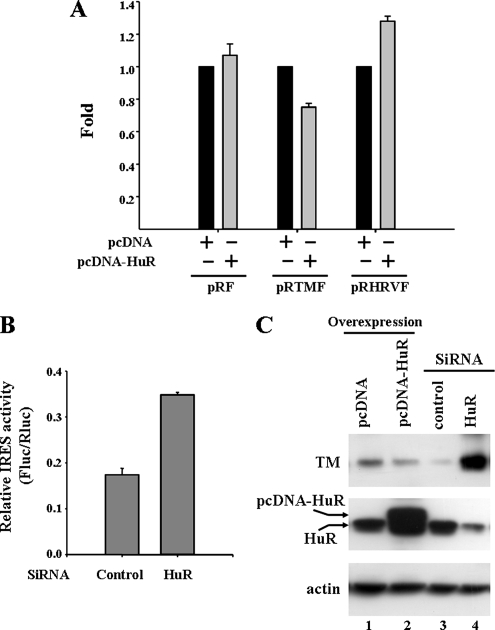
It is interesting to know the functional role of HuR in the TM 5′UTR. To address this question, the bicistronic constructs, pRF, pRTMF, and pRHRVF and the pcDNA-HuR expression plasmid were transiently cotransfected into the A549 cells. As shown in Figure 4A, HuR inhibited the TM IRES activity, but not the pRF and human rhinovirus IRES (pRHRVF) activity. On the other hand, under depletion of the HuR protein using siRNA technology, we observed the IRES activity increased when HuR protein expression was reduced (Figure 4B). Therefore, these results indicate HuR interacts with the TM 5′UTR and represses the cap-independent IRES activity.
Figure 4.
HuR represses the TM IRES-mediated translation. (A) HuR protein inhibits the TM IRES activity in A549 cells. The pRF, pRTMF, and pRHRVF (0.5 μg) vector was cotransfected with the pcDNA-HuR or pcDNA plasmid (0.5 μg) into A549 cells. After transfection for 24 h, the firefly and Renilla luciferase activities were measured. The IRES activities of HuR overexpression samples were normalized with pcDNA vector expression samples, and the values were presented as Fold to indicate the repression effect. (B) Reduced HuR protein expression increases the TM IRES activity. A549 cells were transfected with pRTMF together with siRNA against the HuR or scramble siRNA as a negative control. The cells were lysed 48 h later, and IRES activity was shown as FLuc/RLuc ratio. (C) The expression level of HuR protein regulates endogenous TM protein expression. A549 cells were transfected with siRNA targeted to the HuR mRNA or pcDNA-HuR expression vector. After 48 h, the cell lysates were collected, and HuR, TM, and actin protein levels were detected by Western blot analysis.
Furthermore, we also examined the endogenous TM protein expression under the condition of overexpressing or reducing HuR protein expression. We found that overexpression of HuR protein decreased the expression level of TM, and knockdown of the HuR protein expression by SiRNA dramatically increased the TM protein expression (Figure 4C). Taken together, HuR was demonstrated to negatively regulate the TM protein expression by repressing the IRES activity.
IL-1β Inhibits TM IRES Activity through Increasing the Interaction between HuR and TM 5′UTR
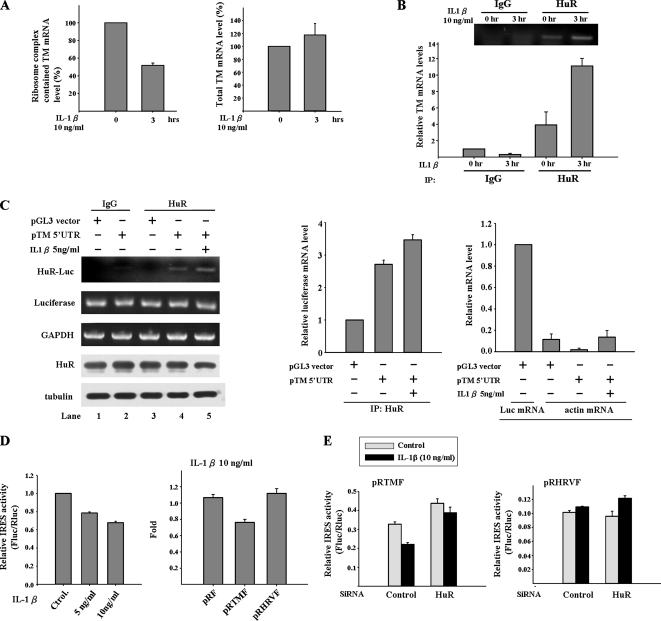
As shown in Figure 1A, the translational regulation mechanism may contribute to the inhibitory effect of IL-1β on the TM promoter construct P1-Luc. Whether IL-1β signal represses the TM protein translation in A549 cells is a key issue to be addressed. To answer this question, the ribosome complexes were pulled down by 40S ribosomal protein S6 antibody and then extracted the mRNAs bound with the ribosome complexes. TM mRNA expression level in ribosome complex was detected by quantitative RT-PCR and normalized with the level of ribosome bound GAPDH mRNA. Besides, the total TM mRNA level was also detected in A549 cell treated with or with IL-1β. From Figure 5A, we found the level of TM mRNA associated with ribosome complex was decreased after IL-1β treatment, whereas the TM mRNA expression level was slightly increased after IL-1β treatment for 3 h. It indicates the decrease in ribosome bound TM mRNA is not due to the decrease in total TM mRNA. On the other hand, IL-1β signal blocks the interaction of TM mRNA with ribosome complex and then shuts down TM protein synthesis. Furthermore, we found that the binding of HuR and TM mRNA was increased after IL-1β treatment by using RNA-IP method (Figure 5B). This result implies HuR may play a role in repressing TM protein synthesis under IL-1β treatment.
Figure 5.
IL-1β stimulates the binding of HuR to TM 5′UTR and represses TM IRES activity. (A) IL-1β treatment represses the TM protein synthesis in A549 cell. The ribosome complexes were pulled down by 40S ribosomal protein S6 antibody from A549 cells lysates treated with IL-1β. The mRNAs bound with ribosome complexes were extracted and analyzed by quantitative RT-PCR. TM mRNA expression level was detected and normalized with the mRNA expression level of GAPDH. In the right panel, the total RNA was also extracted from A549 cell with or without IL-1β treatment, and the TM mRNA level was detected as a positive control. (B) IL-1β treatment increases interaction between HuR and TM mRNA. A549 cells were treated with IL-1β for 3 h. The cytosol extracts were immunoprecipitated with anti-HuR antibodies. The mRNAs associated with immunoprecipitated complex were extracted and TM mRNA expression level was detected by quantitative RT-PCR. (C) IL-1β treatment increases interaction between HuR and TM 5′UTR. A549 cells were transfected with pGL3 and pTM 5′UTR vectors for 24 h and then treated with IL-1β for 3 h. The cytosol extracts were immunoprecipitated with anti-HuR antibodies. The RNA associated with immunoprecipitated complex was extracted by Trizol reagent, and the firefly luciferase mRNA level (HuR-Luc) was detected by RT-PCR. The mRNA expression levels of Luciferase and GAPDH and the protein expression levels of HuR and tubulin were shown as internal controls. The mRNA expression level was also detected by the quantity PCR with luciferase and actin primers to precisely measure their relative mRNA expression level. The actin expression level was compared with the TM mRNA expression level of sham control and is shown as a negative control. (D) IL-1β inhibits the TM IRES activity. A549 cells were treated with different amounts of IL-1β for 3 h or transfected with pRF, pRTMF, and pRHRVF vector for 24 h and then treated with IL-1β for 3 h. Cell lysates were collected and firefly and Renilla luciferase activities measured. In the left panel, the IRES activity was graphed relative to the untreated control, which was given a value of 1. In the right panel, the IRES activities of different constructs under IL-1β treatment were normalized with the IRES activities without IL-1β treatment, and the values were presented as Fold to indicate the repression effect. (E) Reduced the HuR expression prevents the decrease of TM IRES activity induced by IL-1β. A549 cells were transfected with pRTMF or pRHRVF together with siRNA against the HuR or scramble siRNA as a negative control. After 24 h, cells were treated with IL-1β for 3 h. Cell lysates were collected, and firefly and Renilla luciferase activities were measured.
From the results of Figure 4, HuR interacts with TM 5′UTR and represses TM IRES activity. It is interesting to know whether the increase interaction between HuR and TM mRNA under IL-1β treatment is due to HuR interacts with TM 5′UTR and then inhibits the IRES activity. To address this question, RNA-IP was performed in A549 cell transfected with pTM 5′UTR construct under normal and IL-1β–treated conditions. From Figure 5C, we found the pTM 5′UTR construct, which encoded the TM 5′UTR-luciferase mRNA was associated with HuR in normal conditions, but not the pGL3 control vector (cf. lane 3 and lane 4 of HuR-Luc). This result is consistent with that of Figure 3B. Under IL-1β treatment, HuR increased the interaction with TM 5′UTR-luciferase mRNA (Figure 5C, cf. lane 4 and lane 5 of HuR-Luc). The expression level of luciferase mRNA and HuR protein was also examined to rule out the possibility that this phenomenon was due to the higher expression of luciferase mRNA and HuR protein in these conditions. The mRNA expression levels were also quantified by quantitative PCR, and the expression levels of actin mRNA were shown as negative control. Moreover, repression of the TM IRES activity by IL-1β was also demonstrated. pRTMF vector was transfected to A549 cells, and cells were then treated with IL-1β for 3 h. Our data showed that TM IRES activity decreased 22% (5 ng/ml) and 38% (10 ng/ml) in a dose-dependent manner under IL-1β treatment (Figure 5D). Furthermore, when the HuR protein expression was reduced by SiRNA technology, we found the inhibitory effect on TM IRES activity by IL-1β signal was abolished (Figure 5E). Overall, our results suggest that IL-1β treatment increases the interaction between HuR and TM 5′UTR and results in the repression of the TM IRES activity followed by decreasing protein production.
Increases of Interaction between HuR and TM mRNA in the Liver Tissue of Septic Rat
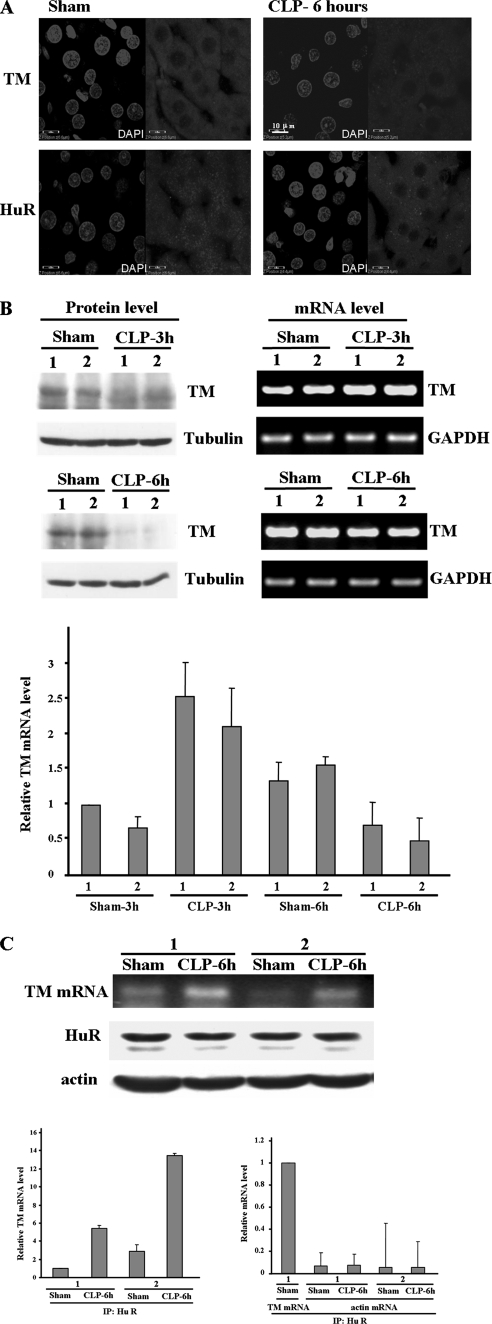
Sepsis is a systemic inflammatory condition and frequently is complicated by the development of pathological thrombosis. Unfortunately, the important anticoagulant TM protein is decreased in septic patients (Faust et al., 2001). This result prompts us to study whether HuR also represses TM protein synthesis in vivo during sepsis. To address this question, the technique of CLP was used to induce sepsis in rat model, and the liver tissue was used for the examination of intracellular distributions of both HuR and TM. From Figure 6A, we found the TM protein expression was reduced in the liver tissue of septic rat as other groups reported (Kume et al., 2003). In sham group, HuR was expressed in both cytosol and nucleus. However, the HuR was concentrated only in the cytoplasm region under sepsis conditions. TM protein expression in septic rat was also quantified by Western blot analysis. We found that TM protein expression was decreased after CLP for 3 h and became very low level at 6 h, but TM mRNA expression level detected by quantitative PCR was shown increase in CLP-3 h and minor decrease in CLP-6 h (Figure 6B). It indicated TM protein synthesis was blocked under CLP condition. To further address whether cytosolic HuR binds with TM mRNA to repress protein synthesis during sepsis, RNA-IP was conducted with HuR antibody followed by RT-PCR and quantitative RT-PCR to detect the TM mRNA expression level. The result showed that the interaction of HuR with TM mRNA was significantly increased under septic condition (Figure 6C). These results indicate the increased interaction between HuR and TM mRNA may play a role in the reduction of TM protein expression in sepsis.
Figure 6.
Interaction between HuR and TM mRNA is increased in septic rat. (A) Immunolocalization of TM and HuR in the liver tissue of septic rat. Rat liver section was prepared from normal rats or 6 h after cecal ligation and puncture (CLP) to induce sepsis. The intracellular localization patterns of HuR and TM were visualized by indirect immunofluorescence. The nuclei were identified after staining with DAPI. (B) TM protein expression was reduced in septic rat. Rat liver protein extract, and total RNA were prepared from normal rats or CLP induced sepsis for 3 and 6 h. Western blot and RT-PCR were performed to detect the TM protein and mRNA expression level. Also the quantitative PCR were conducted. The TM mRNA expression level was normalized with the mRNA level of GAPDH and showed as relative mRNA expression level compared with the sham-3 h control. (C) Increase of the interaction between HuR and TM mRNA. Rat liver extract was prepared from normal or CLP-6 h rats, and the protein extracts were immunoprecipitated with HuR antibody. The mRNAs bound with HuR were extracted, and TM mRNA expression levels were detected by RT-PCR. The protein expression levels of HuR and tubulin were shown as internal controls. The mRNA expression level was also detected by the quantitative PCR with TM and actin primers to precisely measure their relative mRNA expression level. The actin expression level was compared with the TM mRNA expression level of sham control and is shown as a negative control.
DISCUSSION
There are three major findings in the present study. First, TM 5′UTR possessed the IRES activity to regulate the TM protein synthesis. Second, the RNA-binding protein HuR, involved in the stabilization of mRNAs, played an important role in mediating the repression of TM protein expression in inflammation. Third, the increased interaction of HuR and TM mRNA to repress TM protein expression was observed in the liver tissue of the septic rat.
The importance of translation regulation is highlighted by the studies in yeast and mammalian cells that reported a striking lack of correlation between the steady-state levels of mRNAs, as determined using microarrays and the proteins (i.e., proteomes) encoded by those mRNAs (Gygi et al., 1999; Ideker et al., 2001). Protein translation occurs in three steps: initiation, elongation, and termination. In the initiation steps, two kinds of mechanisms are proposed: a cap-dependent scanning mechanism and an IRES-driven mechanism (Gray and Wickens, 1998). Cap-dependent scanning mechanism accounts for translation of the majority of cellular mRNA; however, nearly 10% of cellular mRNA can be translated by the IRES-driven mechanism (Pickering and Willis, 2005). Translation by internal ribosome entry was first identified in picornaviruses (Pelletier and Sonenberg, 1988), but a number of cellular mRNAs containing IRESs have subsequently been found, including basic fibroblast growth factor (Vanger et al., 1995), c-myc (Stoneley et al., 1998), p53 (Yang et al., 2006), and HIF-1α (Lang et al., 2002). Till now, about 80 cellular IRES elements have been identified, almost half found in genes encoding oncogenes, growth factors, and proteins involved in the regulation of apoptosis. This suggests that IRES-dependent translation is an important regulatory mechanism for the maintenance of cellular homeostasis, especially when cap-dependent initiation is inhibited (Baird et al., 2006). Here, we provide evidences that TM 5′UTR acts as an IRES by using a well-known bicistronic construct analysis (Stoneley et al., 1998). The quantities of firefly luciferase mRNA and Renilla luciferase mRNA were also examined to rule out whether the increased protein expression was due to the TM 5′UTR containing the cryptic promoter activity (Figure 2). TM is a multiple function protein involved in the anticoagulation, anti-inflammation, and cancer development processes (Weiler and Isermann, 2003). Therefore, the 5′UTR bearing IRES strengthened the important roles of TM in maintenance of homeostasis in both physiological and pathophisiological conditions.
Regulation of IRES activity was dependent on canonical translation initiation factors and noncanonical ITAFs for efficient initiation of translation (Stoneley and Willis, 2004). It has been proposed that the ITAFs interact with IRESs to maintain or to attain the correct 3D structure required for efficient assembly of the 48S ribosomal initiator complex (Pilipenko et al., 2000). Several ITAFs were identified to regulate IRES activity such as polypyrimidine tract binding protein, poly r(C) binding protein 1, upstream of N-ras (UNR), HuR and La protein, which are all cellular RNA-binding proteins with multiple function in cells (Vanger et al., 2001). In this study, we identified HuR as a negative regulator on TM IRES. Using in vitro translation assay and transiently transfected dicistronic construct assay, we found that overexpression of HuR inhibited the TM IRES activity and consequently the luciferase protein expression. This phenomenon was also observed in endogenous TM protein expression under overexpressed or reduced HuR protein expression (Figure 4). Overall, these results imply the IRES in the TM 5′UTR regulates TM protein expression and the IRES activity is suppressed by HuR.
HuR is a ubiquitously expressed RNA-binding protein belonging to the embryonic lethal abnormal vision (ELAV) family, which was originally identified in Drosophila melanogaster as essential for neural development, and the major functional role of HuR was demonstrated to interact with mRNAs bearing AU-rich sequences (AREs) and to stabilize these mRNAs (Brennan and Steitz, 2001). Numerous inflammation-related genes, such as TNF-α (Rajasingh et al., 2006), COX-2 (Sureban et al., 2007), inducible NO synthase (Linker et al., 2005), and eotaxin (Atasoy et al., 2003), have been demonstrated to bind with HuR. It highlights that HuR plays an important role in inflammation. HuR acting as a translational regulator through binding with 3′UTR or 5′UTR was also reported recently. For example, HuR bound with the highly conserved AU-rich sequence in 3′UTR inhibiting Wnt-5a mRNA translation (Leandersson et al., 2006), p53 mRNA binding to polysomes and the increased translation was also identified as being HuR-mediated (Galban et al., 2003), and HuR repressed p27 translation via an IRES element in the p27 5′UTR (Kullmann et al., 2002). However, the detailed mechanism of HuR-mediated translational suppression is still unclear. Meng et al. (2005) proposed that HuR blocked the activity of the IGF-IR IRES through arresting the IRES-associated translation preinitiation complex in an inactive state. Another study indicated HuR interacted with the translational silencer TIA-1 to reduce the translation of TNF-α and COX-2 (Katsanou et al., 2005). Therefore, characterization of the proteins involved in the HuR-TM IRES complex will give us more information about the HuR-mediated translational repression mechanism in TM protein synthesis.
Because HuR stabilizes numerous inflammatory-related mRNAs and blocks the TM protein expression, HuR plays as a proinflammatory factor. However, Katsanou et al. (2005) reported that HuR acts as a negative posttranscriptional modulator to reduce the translation of TNF mRNA in mouse macrophages after LPS challenge. This contradictory result may due to the different cell lines and animal model used for studying. Recently, the low-molecular-weight inhibitors for HuR were identified (Meisner et al., 2007). These compounds interfered with the formation of HuR dimers and consequently abolished the binding ability of HuR with RNA, and the cytokines expression levels were decreased in the activated primary human monocytes under these compounds treatment. Therefore, these compounds will be valuable tools to study the exactly functional role of HuR in inflammation.
To sum up, we reported a novel translational regulation mechanism in the suppression of TM protein expression by proinflammatory cytokines. Because TM plays an important role in anticoagulation and anti-inflammation response after severe inflammation, especially in severe sepsis, prevention of the decrease of TM protein could be a good way to increase the quantity of activated protein C and prevent the microvascular thrombosis. Therefore, blocking the interaction between HuR and TM 5′UTR through inhibiting the HuR activity will provide a potential new therapeutic strategy in treating severe sepsis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Anne Willis (School of Pharmacy, The University of Nottingham, United Kingdom) for kindly providing the pRF, pRMF, pRHRVF, and phpRF constructs. This work was supported by National Science Council Grant NSC 94-2311-B-006-004 and NSC 95-2320-B-006-066-MY3 of Taiwan, National Cheng-Kung University Project of Promoting Academic Excellence and Developing World Class Research Center, and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations used:
- CLP
cecal ligation and puncture
- IRES
internal ribosome entry site
- IL-1β
interleukin 1β
- TNF-α
tumor necrosis factor-α
- TM
thrombomodulin
- 5′UTR
5′ untranslated region.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0962) on June 25, 2008.
REFERENCES
- Abeyama K., et al. The N-terminal domain of thrombomodulin sequesters high-mobility group-B1 protein, a novel antiinflammatory mechanism. J. Clin. Invest. 2005;115:1267–1274. doi: 10.1172/JCI22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archipoff G., Beretz A., Freyssinet J. M., Klein-Soyer C., Brisson C., Cazenave J. P. Heterogeneous regulation of constitutive thrombomodulin or inducible tissue-factor activities on the surface of human saphenous-vein endothelial cells in culture following stimulation by interleukin-1, tumour necrosis factor, thrombin or phorbol ester. Biochem. J. 1991;273:679–684. doi: 10.1042/bj2730679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U., Curry S. L., Lopez de Silanes I., Shyu A. B., Casolaro V., Gorospe M., Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 2003;171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- Baird S. D., Turcotte M., Korneluk R. G., Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M., Steitz J. A. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W. D., Lazoura E., Okada N., Okada H. Inactivation of C3a and C5a octapeptides by carboxypeptidase R and carboxypeptidase N. Microbiol. Immunol. 2002;46:131–134. doi: 10.1111/j.1348-0421.2002.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Conway E. M., Rosenberg R. D. Tumor necrosis factor suppresses transcription of the thrombomodulin gene in endothelial cells. Mol. Cell. Biol. 1988;8:5588–5592. doi: 10.1128/mcb.8.12.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E. M., et al. The lectin-like domain of thrombomodulin confers protection from neutrophil-mediated tissue damage by suppressing adhesion molecule expression via nuclear factor kappaB and mitogen-activated protein kinase pathways. J. Exp. Med. 2002;196:565–577. doi: 10.1084/jem.20020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl J. L., Borgel D. Sepsis and coagulation. Curr. Opin. Crit. Care. 2005;11:454–460. doi: 10.1097/01.ccx.0000176692.03186.e7. [DOI] [PubMed] [Google Scholar]
- Dittman W. A., Majerus P. W. Structure and function of thrombomodulin: a natural anticoagulant. Blood. 1990;75:329–336. [PubMed] [Google Scholar]
- Esmon C. T. Molecular events that control the protein C anticoagulant pathway. Thromb. Haemost. 1993;70:29–35. [PubMed] [Google Scholar]
- Esmon C. T. The interactions between inflammation and coagulation. Br. J. Haematol. 2005;131:417–430. doi: 10.1111/j.1365-2141.2005.05753.x. [DOI] [PubMed] [Google Scholar]
- Faust S. N., Levin M., Harrison O. B., Goldin R. D., Lockhart M. S., Kondaveeti S., Laszik Z., Esmon C. T., Heyderman R. S. Dysfunction of endothelial protein C activation in severe meningococcal sepsis. N. Engl. J. Med. 2001;345:408–416. doi: 10.1056/NEJM200108093450603. [DOI] [PubMed] [Google Scholar]
- Galban S., Martindale J. L., Mazan-Mamczarz K., López de Silanes I., Fan J., Wang W., Decker J., Gorospe M. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol. Cell. Biol. 2003;23:7083–7095. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N., Wickens M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Gygi S. P., Rochon Y., Franza B. R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley M., Cui X., Minneci P. C., Deans K. J., Natanson C., Eichacker P. Q. Activated protein C in sepsis: emerging insights regarding its mechanism of action and clinical effectiveness. Curr. Opin. Infect. Dis. 2004;17:205–211. doi: 10.1097/00001432-200406000-00006. [DOI] [PubMed] [Google Scholar]
- Hsieh Y. C., Hsu C., Yang R. C., Lee P. Y., Hsu H. K., Sun Y. M. Isolation of bona fide differentially expressed genes in the 18-hour sepsis liver by suppression subtractive hybridization. Shock. 2004;21:549–555. doi: 10.1097/01.shk.0000126148.83935.6a. [DOI] [PubMed] [Google Scholar]
- Ideker T., Thorsson V., Ranish J. A. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Katsanou V., Papadaki O., Milatos S., Blackshear P. J., Anderson P., Kollias G., Kontoyiannis D. L. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M., Gopfert U., Siewe B., Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume M., Hayashi T., Yuasa H., Tanaka H., Nishioka J., Ido M., Gabazza E. C., Kawarada Y., Suzuki K. Bacterial lipopolysaccharide decreases thrombomodulin expression in the sinusoidal endothelial cells of rats—a possible mechanism of intrasinusoidal microthrombus formation and liver dysfunction. J. Hepatol. 2003;38:9–17. doi: 10.1016/s0168-8278(02)00324-0. [DOI] [PubMed] [Google Scholar]
- Lang K. J., Kappel A., Goodall G. J. Hypoxia-inducibile factor-1 alpha mRNA contains an internal ribosome entry site that allows efficiency translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Y., Maizel J. V. A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucl. Acid Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandersson K., Riesbeck K., Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz S. R., Tsiang M., Sadler J. E. Regulation of thrombomodulin by tumor necrosis factor-alpha: comparison of transcriptional and posttranscriptional mechanisms. Blood. 1991;77:542–550. [PubMed] [Google Scholar]
- Levi M., Van der Poll T. Two-way interactions between inflammation and coagulation. Trends Cardiovasc. Med. 2005;15:254–259. doi: 10.1016/j.tcm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Linker K., Pautz A., Fechir M., Hubrich T., Greeve J., Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D. H., Sabina J., Zuker M., Turner D. H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Meisner N. C., et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- Meng Z., King P. H., Nabors L. B., Jackson N. L., Chen C. Y., Emanuel P. D., Blume S. W. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. L., Andreoli S. P., Esmon N. L., Esmon C. T., Bang N. U. Endotoxin enhances tissue factor and suppresses thrombomodulin expression of human vascular endothelium in vitro. J. Clin. Invest. 1987;79:124–130. doi: 10.1172/JCI112772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. L., Esmon C. T., Esmon N. L. Tumor necrosis factor leads to the internalization and degradation of thrombomodulin from the surface of bovine aortic endothelial cells in culture. Blood. 1989;73:159–165. [PubMed] [Google Scholar]
- Okajima K. Regulation of inflammatory responses by natural anticoagulants. Immunol. Rev. 2001;184:258–274. doi: 10.1034/j.1600-065x.2001.1840123.x. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Pickering B. M., Willis A. E. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Pestova T. V., Kolupaeva V. G., Khitrina E. V., Poperechnaya A. N., Agol V. I., Hellen C. U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Rajasingh J., Bord E., Luedemann C., Asai J., Hamada H., Thorne T., Qin G., Goukassian D., Zhu Y., Losordo D. W., Kishore R. IL-10-induced TNF-alpha mRNA destabilization is mediated via IL-10 suppression of p38 MAP kinase activation and inhibition of HuR expression. FASEB J. 2006;20:2112–2114. doi: 10.1096/fj.06-6084fje. [DOI] [PubMed] [Google Scholar]
- Stoneley M., Paulin F. E., Le Quesne J. P., Chappell S. A., Willis A. E. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Stoneley M., Willis A. E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Sureban S. M., Murmu N., Rodriguez P., May R., Maheshwari R., Dieckgraefe B. K., Houchen C. W., Anant S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Van de Wouwer M., Conway E. M. Novel functions of thrombomodulin in inflammation. Crit. Care Med. 2004;32:S254–S261. doi: 10.1097/01.ccm.0000128036.64448.9e. [DOI] [PubMed] [Google Scholar]
- Vanger S., Galy B., Pyronnet S. Irresistible IRES. EMBO rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanger S., Gensac M. C., Maret A., Bayard F., Amalric F., Prats H., Prats A. C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler H., Isermann B. H. Thrombomodulin. J. Thromb. Haemost. 2003;1:1515–1524. doi: 10.1046/j.1538-7836.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- Wichterman K. A., Baue A. E., Chaudry I. H. Sepsis and septic shock—a review of laboratory models and a proposal. J. Surg. Res. 1980;29:189–201. doi: 10.1016/0022-4804(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Yang D. Q., Halaby M. J., Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.