Abstract
We have reported previously that the 27nt repeat polymorphism in endothelial nitric-oxide synthase (eNOS) intron 4—a source of 27nt small RNA—inhibits eNOS expression. In the current study, we have investigated how 27nt small RNA suppresses eNOS expression. Using a chromatin immunoprecipitation assay, we examined histone acetylation in the 27nt repeat element of eNOS intron 4, the promoter region up to −1486 bp, and the 5′ enhancer region (−4583/−4223bp) in human aortic endothelial cells (HAECs) treated with 27nt RNA duplex. 27nt RNA duplex induced hyperacetylation in H3 (lysine8, 12, and 23) and H4 (lysine 9 and 12) at the 27nt repeat element, which then interacted with nuclear actin, histone deacetylase 3 (HDAC3), and NonO proteins. In contrast, the histone H3 and H4 became hypoacetylated at the eNOS core promoter. HAECs treated with 27nt RNA duplex had reduced eNOS expression, but treatment with either HDAC3 small interfering RNA or NonO siRNA significantly attenuated the 27nt small RNA-induced suppression. We further found that 27nt small RNA induced DNA methylation in a region approximately 750nt upstream of the intron 4 repeats, and a methyltransferase inhibitor reversed the effect on methylation and eNOS expression. Our study demonstrates that 27nt small RNA may suppress eNOS expression by altering histone acetylation and DNA methylation in regions adjacent to the 27nt repeat element and core promoter.
INTRODUCTION
Endothelial-derived nitric oxide (NO), synthesized primarily by endothelial nitric-oxide synthase (eNOS), maintains the functional integrity of endothelial cells, regulates hemodynamic responses, and contributes to arterial wall remodeling and the establishment of collateral circulation (Gibbons and Dzau, 1994; Dimmeler and Zeiher, 1999). Adequate NO production is essential for antiatherogenic and antithrombotic processes in the normal arterial wall, and the reduced availability of NO promotes vascular diseases. Decreased basal NO release may predispose to hypertension, thrombosis, vasospasm, and atherosclerosis. In contrast, overproduction of NO can result in excessive oxidative stress and inflammation, both of which promote vascular diseases. Adequate eNOS expression and eNOS enzyme activity are central to maintaining appropriate physiological functions. Tremendous research efforts have recently focused on the regulation of eNOS enzyme activities, either by direct eNOS protein modification (e.g., phosphorylation) or availability of cofactors. However, progress in regulating eNOS expression at the transcriptional, posttranscriptional, and translational levels is relatively slow.
Despite extensive study of the association between eNOS polymorphisms and vascular diseases, the functional roles of variants of eNOS DNA are not clear. A meta-analysis of 12,990 subjects has shown a significant association between vascular diseases and the 27nt repeat polymorphism in eNOS intron 4 (Casas et al., 2004). Studies of ours and others suggest that the 27nt repeat polymorphism of the eNOS gene may regulate eNOS expression. Specifically, we have shown that the 27nt repeats may act as an enhancer/repressor in regulating eNOS expression; the regulatory effect is maintained during in vitro cell replication in ECs of different genotypes (Wang et al., 2002). Second, we have shown that the 27nt repeats in eNOS intron 4 produce 27nt small RNA, which seems to inhibit eNOS expression (Ou et al., 2005; Zhang et al., 2005). In the current study, we investigated the molecular mechanisms responsible for the 27nt small RNA-mediated eNOS suppression. Because 27nt small RNA is found exclusively in the nucleus and has a major effect on transcription, we focused our investigation on changes in histone acetylation and DNA methylation in the eNOS gene. We found that 27nt small RNA differentially altered histone acetylation in eNOS intron 4 and the promoter region. In addition, 27nt induced hypermethylation in the upstream region of the 27nt repeat element in eNOS intron 4.
MATERIALS AND METHODS
Small RNA Preparation
We designed target-specific small 27nt RNA duplexes according to sequences of the type AA(N27)UU (N, any nucleotide) from the eNOS genomic DNA. The sense is 5′-AAAAAUGUUCACCUACAUCUGCAACCACAUU-3′ and the antisense is 5′-AAUGUGGUUGCAGAUGUAGGUGAACAUUUUU-3′. We also designed a small 27nt RNA by using the luciferase gene coding region as the template to be used as a negative control. The sense is 5′-AAGUCUGACAGUUACCAAUGCUUAAUCAGUU-3′, and the antisense is 5′-AACUGAUUAAGCAUUGGUAACUGUCAGAC-3′. In evaluation of the roles of NonO and histone deacetylase 3 (HDAC3) in histone acetylation in eNOS genome, we used gene-specific siRNA to knockdown the expression of these two genes. We designed target-specific small 21nt RNA duplexes according to sequences of the type AA(N21)UU (N, any nucleotide) from the HDAC3 and NonO genes. The sequences for siRNAs are as follows. For NonO, the sense is 5′-AAUCAUACUCCAAGGAAGCAUUU-3′ and the anti-sense is 5′-AAAUGCUUCCUUGGAGUAUGAUU-3′. For HDAC3, the sense is 5′-AAAAAGGUGCAUGGUUCAGCAUCUU-3′ and the antisense is 5′-AAGAUGCUGAACCAUGCACCUUUUU-3′. We also designed a small 21nt RNA used as a negative control by using Luciferase gene coding region as the template. The sense is 5′-AACAGUUACCAAUGCUUAAUCAGUU-3′, and the antisense is 5′-AAAAGCAUUGGUAACUGUCAGAC-3′. All small RNAs were chemically synthesized by Integrated DNA Technologies (Coralville, IA). To examine the interaction between HDAC3 and 27nt small RNA, the 27nt RNA duplex was labeled with biotin at 5′ end and synthesized directly by Integrated DNA Technologies.
Cell Culture, Transfection, and Treatment
Human aortic endothelial cells (HAECs) were cultured in EGM-2 medium (Lonza Walkersville, Walkersville, MD) containing 3% fetal bovine serum (FBS). HeLa cells from American Type Culture Collection (Manassas, VA) were used in transfection assays as a control because HeLa cells do not constitutively express eNOS, but do have the necessary eNOS transcriptional machinery to drive the transfected eNOS constructs. The small RNA duplex or siRNA was transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). 5-Azacytidine (5-aza) (Sigma-Aldrich, St. Louis, MO) was used at a final concentration of 5 nM. Cells were treated every 48 h with 5-aza (a methyltransferase inhibitor) for 7 d. Trichostatin A (TSA) (Sigma-Aldrich), an HDAC inhibitor, was dissolved in 100% ethanol and used at a final concentration of 1 mM. Cells were treated for 24 h with either TSA or an equal amount of ethanol, which was used to dissolve TSA as a vehicle control.
mRNA Stability Assay
To assess the effect of the 27nt small RNA on eNOS mRNA stability, we cultured HAECs in six-well plates up to 90% confluence. We then transfected these cells with 27nt small RNA duplexes by using liposome. The transcription was blocked by adding actinomycin D (2 μg/ml). Treated HAECs were then harvested at 6, 12, 24, 48, and 72 h after the actinomycin D treatment. The relative levels of eNOS mRNA were determined by quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) adjusted by housekeeping gene β-actin.
Northern Blot Analysis
Total RNAs were extracted with TRIzol (Invitrogen). For eNOS mRNA detection, RNase protection assays (RPA) were performed by using the RPA III kit from Ambion (according to the manufacturer's instruction). Five to 20 μg of total RNA was used per reaction. Cyclophilin antisense control template from Ambion (Austin, TX) was labeled using T7 RNA polymerase as the internal control. 28S rRNA stained with ethidium bromide was used to control the amount of RNA loading.
Antibodies for Western Blot, Coimmunoprecipitation, and ChIP Assays
The ChIP assay kit was purchased from Millipore (Billerica, MA). Anti-acetyl histone H3 antibodies, anti-acetyl histone H4 antibodies, and anti-eNOS antibodies were purchased from Cell Signaling Technology (Danvers, MA). We purchased anti-HDAC3 antibody from Sigma-Aldrich, and the anti-NonO antibody from Abcam (Cambridge, MA).
Cellular Extracts and Western Blot Analysis
For total protein lysate preparation, the cell pellet was homogenized in lysis buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% Na desoxycholate, 0.1% SDS, 10% glycerol, 0.2 mM EDTA, 200 mg/ml leupeptin, 10 mg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaFluorure, and 1 mM Na othovanadate). The cell pellet was suspended in ice-cold lysis buffer before cells were disrupted by sonication. After centrifugation at 12,000 × g for 10 min at 4°C, an aliquot of the supernatant was used for protein determination using the Coomassie Blue Plus protein assay reagent (Pierce Chemical, Rockford, IL), as per manufacturer's instruction. The remaining protein solution was used for Western blot analysis. Electrophoresis was carried out in 10% SDS-polyacrylamide gel, which was transferred to a nitrocellulose membrane (Hybond-C Super; GE Healthcare, Chalfont St. Giles, United Kingdom). The membranes were then blocked in 5% milk and probed with the appropriate specific antibodies. Primary antibodies were incubated at 4°C overnight in 5% milk. Horseradish peroxidase-linked goat anti-rabbit or mouse immunoglobulin G (IgG) (H&L) antibodies (Cell Signaling Technology) were used as secondary antibodies, and detection was performed with the Pierce ECL Western Blotting Substrate solution (Pierce Chemical) according to manufacturer's instruction. Anti-β-actin antibody (Sigma-Aldrich) was used for loading normalization.
Immunofluorescence Confocal Microscope Analysis
HAECs were seeded on plates with coverslips in medium containing 10% FBS for 24 h. Cells were fixed in 4% paraformaldehyde and permeabilized in phosphate-buffered saline (PBS) containing 0.5% Triton X-100. Indirect immunofluorescence was performed using a monoclonal antibody (mAb) against HDAC3 (Sigma-Aldrich), nuclear actin (2G2 antinuclear actin, catalog no. 651132; Progen Biotechnik, Heidelberg, Germany) and NonO (Abcam). Goat anti-mouse cyanine (Cy)2 (green) and goat anti-rabbit Cy5 (red) secondary antibodies (Vector Laboratories, Burlingame, CA) were used to detect the locations of HDAC3, nuclear actin, and NonO in HAECs. Laser-scanning confocal microscopy was performed on an LSM410 (Carl Zeiss, Jena, Germany) with krypton-argon and helium-neon lasers; three-dimensional projections were generated with the accompanying software.
Coimmunoprecipitation
To study the interaction between HDAC3, nuclear actin, and NonO, HAECs were harvested in the lysis buffer for immunoprecipitation with a mouse anti-nuclear actin antibody (anti-actin 2G2) or mouse anti-HDAC3 mAb. The immune complexes were further analyzed by immunoblotting with the HDAC3 or NonO antibody.
Isolation of Nuclear Extracts and Streptavidin-Bead Precipitation Assay
We extracted nuclear protein from 1 × 106 cultured HAECs, according to the instructions of the NE-PER nuclear and cytoplasmic extraction reagents kit (Pierce Chemical). Protease inhibitor cocktail (1:100 dilution) (Sigma-Aldrich) was added to the extracted nuclear proteins, with the protein concentration being determined using a bicinchoninic acid protein assay kit with bovine serum albumin as a standard (Sigma-Aldrich). The 27nt RNA was 5′ labeled with biotin was synthesized by Integrated DNA Technologies Inc. (Houston, TX) mixed with 30 μl packed Streptavidin-beads for 3 h at 4°C, and incubated with 500 μg of nuclear protein extracts overnight at 4°C. After washing four times with ice-cold PBS containing 0.5% Triton X-100, 5 mM EDTA, proteins were eluted from the beads by the addition of 2× SDS loading buffer and boiled for 5 min. The eluted nuclear proteins were subjected to Western blot analysis using the anti-HDAC3 mAb.
ChIP Assay
We used the ChIP assay kit (Millipore) per the manufacturer's protocol. Briefly, ∼3 × 106 cells were used per ChIP assay. The cells were fixed in 1% formaldehyde solution for 10 min at 37°C. Sonication was performed on ice using a Sonics and Materials Vibra-Cell sonicator (Misonix, Farmingdale, NY) with a 3-mm tip set at 30% maximum power; chromatin fragments, ranging in size from 200 to 400 bp, were generated by using 5 × 10-s pulses with a 10-s interval between sonications. Samples were diluted 10-fold in ChIP dilution buffer, and a 20-μl aliquot (1% of total) was removed to serve as an input sample. Chromatin was precleared with 80 μl of a mixture of salmon sperm DNA/protein A/protein G at 4°C with rotation for 2 h, followed by the addition of 2 μg of anti-acetyl-histone H3, HDAC3, and NonO antibodies. Immunoprecipitation was performed at 4°C overnight with rotation. To collect immune complexes, 60 μl of a mixture of salmon sperm DNA/protein A/protein G was added and incubated at 4°C with rotation for 2 h. After washing immune complexes, formaldehyde cross-linking was reversed in immunoprecipitated samples and the input chromatin sample by the addition of 20 μl of 5 M or 2 μl of NaCl and incubation at 65°C for 4 h. After proteinase K treatment, phenol/chloroform extraction, and ethanol precipitation, the DNA was resuspended in 25 μl of water. We used yeast tRNA to aid the precipitation process. ChIP analyses were performed at least three times on three independent lots of HAECs. The primers for the ChIP assay are listed in Table 1.
Table 1.
ChIP assay primer sets for eNOS gene enhancer, promoter, and the 27nt repeats region of intron 4
| Primer pair | Sequence | Amplicon location |
|---|---|---|
| En-4917S | 5′-CTGTGCTGGGAAGGGAAGCCG-3′ | Enhancer |
| En-4626AS | 5′-GCACTCAGCATGAGTTACATCC-3′ | 4817 to −4626 |
| Pr-1421S | 5′-CTGTCTCAGGAGGACGACAAGGAG-3′ | Promoter |
| Pr-1304AS | 5′-GGATCCAGCCCCTACTTTTC-3′ | 1421 to −1304 |
| Pr-891S | 5′-TCAAGTGCCTGGAGAGTGCTGG-3′ | Promoter |
| Pr-796AS | 5′-GAAGAGCTTGATGCCCTGGTG G-3′ | 891 to −796 |
| Pr-483S | 5′-ACCAGGGGGTCATAAAGGTC-3′ | Promoter |
| Pr-312AS | 5′-GGGGAGGTGAAGGAGAGA-3′ | 483 to −312 |
| Pr-238S | 5′-AGCCCTGGCCTTTTCCTTAG-3′ | Promoter |
| Pr-13AS | 5′-GCTCCCACTTATCAGCCTCA-3′ | −238 to −13 |
| 27Re-5318S | 5′-AGTACTGGAGGGGAAAGAAGT-3′ | 27nt repeat region |
| 27Re-5464AS | 5′-AGTACTGAAGCTCCGGTCAGC-3′ | +5318 to +5464 |
Direct Sodium Bisulfite Sequencing for DNA Methylation Detection
Genomic DNA (5 μg) from the HAECs that were treated with the 27nt RNA duplex complementary to the exon 1 of the luciferase gene (siLuc) and the 27nt RNA duplex complementary to the 27nt repeat element of the eNOS intron 4 were subjected to sodium bisulfite treatment. Given that the nascent strand of replicating DNA is hemimethylated immediately after DNA replication, only quiescent postconfluent cells were used for DNA isolation. BamHI-digested DNA was denatured with 0.3 M NaOH for 15 min at 37°C in a volume of 20 μl and treated with 3.1 M sodium bisulfite and 0.5 mM hydroquinone. After being overlaid with mineral oil, the reaction solution was incubated at 55°C for 16 h in the dark. Free bisulfite was removed using a Wizard DNA clean-up desalting column (Promega, Madison, WI), and the solution was incubated with 0.3 M NaOH for 15 min at 37°C to denature and remove the -SO3 adduct from the uracil bases before PCR. Sample DNA was then neutralized with NH4OAc, precipitated, and stored at −20°C. An aliquot of the bisulfite-treated DNA (25–50 ng) was subjected to 35 cycles of PCR amplification, followed by another 35 cycles of nested PCR amplification. PCR primers in the eNOS promoter, enhancer, and 1.2 kb 5′ upstream of intron 4 27nt repeats were designed for the sodium bisulfite-modified template (Table 2). Oligonucleotides were generated using an Oligo 1000 DNA synthesizer (Beckman Coulter, Fullerton, CA). The final PCR products were subcloned using the TA cloning kit (Invitrogen) to yield 10–20 individual plasmid clones for sequencing.
Table 2.
Sodium bisulfite genomic sequencing primer sets
| Primer pair | Sequence | Amplicon location |
|---|---|---|
| En-4998S | 5′-GAATTTTTAGTATTTTAGAAGT-3′ | Enhancer |
| En-45944AS | 5′-CCCTACCTTCCCTCTCAA-3′ | −4998 to −4544 |
| En-4941S | 5′-TTT GTA ATT TGG AAG TTG AGT AA-3′ | |
| En-4554AS | 5′-CCTCTCAAACCAAAATACTCTACAA-3′ | −4941 to −4554 |
| Pr-1295S | 5′-GGTAGTAGGGTATATTAGTGTTTAGG-3′ | Promoter |
| Pr-860AS | 5′-AATACACCAACACTCTCCAAACA-3′ | −1295 to −860 |
| Pr-1211S | 5′-TGGTGTGATTTTAGAGTTTTGAGTAT-3′ | |
| Pr-880AS | 5′-AACACTTCAAACCAAAATACATAC-3′ | −1211 to −880 |
| Pr-965S | 5′-GATTAGATGTTTAGTTAGTGGTT-3′ | |
| Pr-583AS | 5′-TTATCCTTAAATCTAACATTAA-3′ | −965 to −583 |
| Pr-884S | 5′-AGTGTTTGGAGAGTGTTGGTGTATT-3′ | |
| Pr-617AS | 5′-CCTCCCAACCCCAATTTCCTAA-3′ | −884 to −617 |
| Pr-538S | 5′-AAGGATTTAAGGGTGGGGATT-3′ | |
| Pr-74AS | 5′-AACTCTCCAATACTAACCCTC-3′ | −538 to −74 |
| Pr-484S | 5′-GAGAGAATTTGTATGATTTTAGAGGTT-3′ | |
| Pr-145AS | 5′-AACTCTAAAACCTCAACTCCAC-3′ | −484 to −145 |
| Pr-346S | 5′-GTGTTATATTATAGAAGGATTTTTATG-3′ | |
| Pr-286S | 5′-TGTTTTAGTTTTTATGTTGTAGTTTTAG-3′ | |
| Pr+3AS | 5′-ACTACCTACTCCAACAAAACCCTAACC-3′ | −286 to +3 |
| +4381S | 5′-TCACCCATCCCACCCCTGCACCCTT-3′ | |
| +5066AS | 5′-GCGCCCGGGTCTCTCAGTCGCTGG-3′ | +4381 to +5066 |
Statistical Analyses
All experiments were performed at least three times, and representative results are presented here. Quantitative data are expressed as mean ± SEM of three separate experiments. Independent Student's t test was used for the comparisons of between group differences; and a one-way analysis of variance was applied for multiple group comparisons with post hoc Bonferroni correction for multiple between group comparisons. SPSS version 15.0 for Windows (SPSS, Chicago, IL) was used for the statistical analyses. Outcomes were considered statistically significant with two-tailed, p < 0.05.
RESULTS
Effect of 27nt RNA Duplex on Acetylation Status of Histone 3 and 4 in eNOS Gene
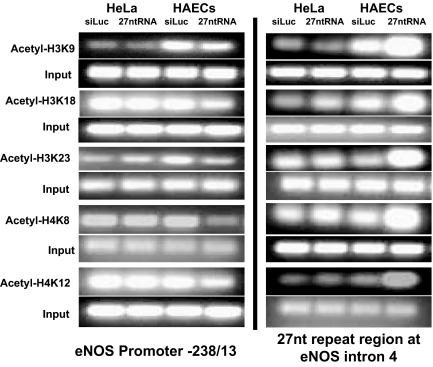
We reported previously that 27nt small RNA derived from the 27nt repeat elements in the eNOS intron 4 suppressed eNOS expression in endothelial cells (Zhang et al., 2005). Previous studies suggested that small RNA can regulate gene transcription by modifying acetylation or methylation of histones. To investigate whether 27nt small RNA affects histone acetylation, we used a quantitative ChIP technique to measure the acetylation status of histone H3 and H4 located in the 27nt repeat element (6340-6516 bp) in eNOS intron 4 and the eNOS promoter region (up to −4917 bp). Chromatin was isolated from HAECs treated with exogenous 27nt microRNA (miRNA) duplex for 24 h. ChIP was performed on soluble chromatin fragments that averaged 200–400 base pairs. We used antibodies against H3 lysine 9 (H3K9), H3 lysine 18 (H3K18), H3 lysine 23 (H3K23), H4 lysine 8 (H4K8), and H4 lysine 12 (H4K12) in the ChIP assay. As shown in Figure 1, levels of acetylated H3K9 and H3K18 in the 27nt repeat element were higher in eNOS-expressing HAECs than in non—eNOS-expressing HeLa cells. When these cells were treated with 27nt RNA, the acetylation status at all five residues increased in the 27nt repeat element. In contrast, H3K9 and H3K23 seemed to be more acetylated in the eNOS core promoter region (−238 to −13 bp; Figure 1) in HAECs than in HeLa cells. On treatment with 27nt RNA, the acetylation status was reduced in all five measured lysine residues in the core promoter. The acetylation changes in other regions of the promoter (−1421/−1304 bp, −891/−79 6bp, −483/−312 bp) and enhancer (−4917/−4626 bp) were not remarkable (data not shown).
Figure 1.
Effect of 27nt RNA duplex on histone acetylation binding with the eNOS core promoter −238/−13 and intron 4 regions. ChIP assay was performed with anti-acetylated H3 or H4 antibodies in HAECs treated with either siLuc small RNA (negative control) or 27nt RNA. HeLa cells were treated as a control. The eluted DNA was subjected to PCR for the eNOS promoter region (left) and the 27nt repeats in eNOS intron 4.
Effect of Nuclear β-Actin in 27nt RNA-induced Changes in eNOS Histones
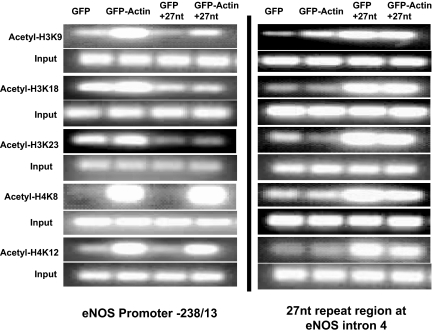
We have previously reported that 27nt small RNA duplex bound with nuclear β-actin and that the inhibitory effect of 27nt RNA on eNOS was attenuated by overexpression of β-actin (Ou et al., 2005). To study the effect of nuclear actin on the interaction between 27nt RNA and eNOS transcription, we measured histone acetylation status in 27nt RNA-treated HAECs with or without actin overexpression. HAECs were transfected with actin/EGFP plasmids for 24 h and then treated with the 27nt RNA duplex. A systematic ChIP analysis was carried out to examine the intron 4 and promoter region of the eNOS gene. As shown in Figure 2, actin overexpression in ECs did not affect the increased histone acetylation by 27nt RNA at the 27nt repeat element; however, a decreased trend for H4K8 and H4K12 was noted. In contrast, overexpression of β actin increased acetylation status at H3K9, H4K8 and H4K12 in the eNOS core promoter region (−238/−13 bp) in all cells with or without 27nt RNA treatment.
Figure 2.
Effect of nuclear actin overexpression on acetylation status of histones interacting with 27nt RNA duplex and eNOS promoter and intron 4 regions. ChIP assay shows that nuclear actin overexpression offset the influence of 27nt small RNA at the eNOS core promoter at −238/−13 region and the 27nt repeat region of the intron 4 in HAECs. Chromatin was prepared from HAECs transfected with GFP/actin plasmids and then treated with or without 27nt small RNA. These results were obtained in the same condition as for the ChIP assay described in Figure 1.
Protein–RNA and Protein–Protein Interaction
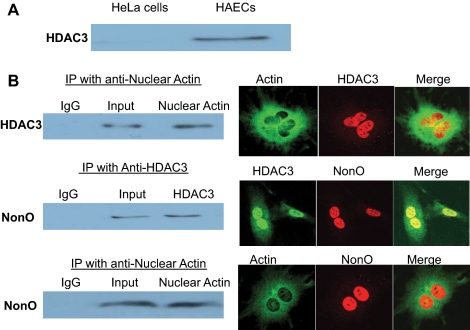
To understand how 27nt RNA modified histone acetylation, we next studied the interactions between 27nt RNA and HDACs. We used a streptavidin-bead precipitation assay to assess the in vitro binding of HDACs to 27nt RNA. Nuclear proteins were extracted from HAECs and then mixed with 5′-biotin labeled 27nt RNA duplex. We used the magnetic bead technique to isolate nuclear proteins bound to 27nt RNA; the eluted nuclear proteins were then subjected to Western blot analysis by using the anti-HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC6, and HDAC7 monoclonal antibodies. Nuclear extracts from HeLa cells were used as a control. Of all the HDACs studied, only HDAC3 bound the 27nt RNA (Figure 3A).
Figure 3.
Interaction among nuclear proteins associated with the 27nt repeat element in eNOS intron 4. (A) Streptavidin-bead precipitation assay for the assessment of the in vitro binding of HDACs to 27nt RNA. Nuclear proteins were extracted from HAECs and then mixed with 5′-biotin labeled 27nt RNA duplex. The pull-down proteins were analyzed by Western blot for the presence of HDACs (HDAC1, -2, -3, -4, -5, -6, and -7) and only HDAC3 bound with 27nt RNA. (B) HAECs lysates were immunoprecipitated (IP) with mAb against nuclear actin (top and boom), HDAC3 (middle), or IgG (as control). With the starting whole protein extracts at 300 μg, the immunoprecipitates (∼20 μg) were subjected to immunoblot analysis using antibody directed against HDAC3 (top) or NonO (middle and bottom). The nuclear protein interactions are also shown via confocal microscopy on the right panel with respective antibodies that illustrate their partial colocalization.
To search for nuclear proteins that may participate in 27nt RNA-mediated eNOS suppression, we first examined the interactions between nuclear proteins that bind to 27nt RNA or DNA. Using an immunoprecipitation assay, we detected interactive binding between nuclear actin and HDAC3 (Figure 3B, top left). We further detected interaction between HDAC3 and NonO protein (Figure 3B, middle left), and between nuclear actin and NonO (Figure 3B, bottom left). Our previous work showed that NonO protein, when recruited to the target genomic location, is frequently associated with suppression of transcription (Zhang et al., 2007). The recruitment of the NonO protein to the HDAC3 and nuclear actin complex in the 27nt repeat region could be responsible for the suppressive effects of the 27nt small RNA on eNOS transcription. Using confocal microscopy, we further illustrated the colocalization of HDAC3, actin, and NonO within the nucleus (Figure 3B, right). Most of the actin was present in the cytoplasm, but a small amount was visible within the nucleus (Figure 3B, top and bottom right). Most of the HDAC3 was found within the nucleus, but a trace amount was seen in the cytoplasm. NonO was found predominantly within the nucleus.
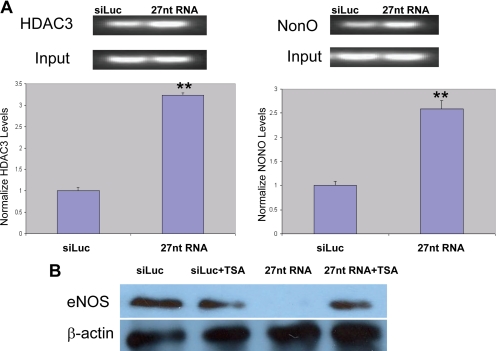
To determine whether the HDAC3 and NonO interaction occurred in the 27nt repeat element in eNOS intron 4, we performed a ChIP assay by using anti-HDAC3 or anti-NonO antibodies to pull down the nucleosomes extracted from HAECs treated with either 27nt RNA duplex or luciferase gene-specific siRNA as a control. We amplified the genomic DNA using the primers covering the 27nt repeat element. As shown in Figure 4A, HDAC3 and NonO did bind on the 27nt repeat element in eNOS intron 4. Treatment of HAECs with 27nt RNA significantly increased the binding between the HDAC3 or NonO and the 27nt repeat element (Figure 4A, p < 0.01). The role of HDAC in 27nt RNA induced eNOS suppression was further supported in cells treated with 1 mM TSA (Figure 4B).
Figure 4.
Interaction between HDAC3, NonO and 27nt repeats in eNOS intron 4. (A) ChIP assays using anti-HDAC3 or anti-NonO antibodies were performed to pull down the nucleosomes extracted from HAECs treated with either 27nt RNA duplex or luciferase gene-specific siRNA as a control. We amplified the genomic DNA by using the primers covering the 27nt repeat element. Bottom, band density in reference to the input band intensity, and an independent Student's t test was performed to compare the differences between siLuc siRNA and 27nt RNA treated cells (**p < 0.01). The quantitative data represent the results of three independent experiments. (B) HAECs were cultured and transfected with siLuc (control) siRNA or 27nt RNA duplex. The transfected HAECs were then treated with1 μM TSA for 24 h, and eNOS protein levels were determined by Western blot. The 27nt RNA duplex induced eNOS suppression was partly relieved by the TSA treatment.
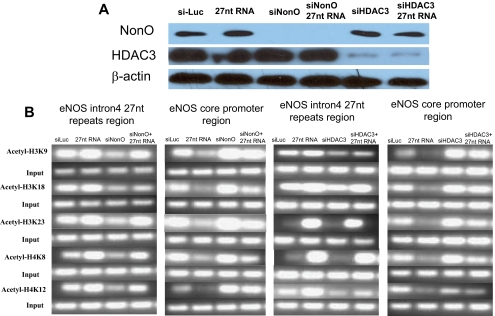
To further investigate the role of NonO in 27nt RNA-mediated eNOS regulation, we silenced the expression of these two genes in HAECs treated with or without 27nt RNA. When NonO was knocked down in cultured HAECs (Figure 5A), histones bound to the 27nt repeat region were hypoacetylated; histones bound to the core eNOS promoter region were generally hyperacetylated (Figure 5B). The effects on histone acetylation at 27nt repeat and eNOS core promoter regions were counteracted by the addition of the 27nt RNA duplex (Figure 5B). This is consistent with the involvement of NonO in 27nt RNA-mediated eNOS regulation.
Figure 5.
Involvement of HDAC3 and NonO in histone acetylation in eNOS gene. (A) Western blot of nuclear extracts from HAECs treated with NonO or HDAC3 specific siRNA. It shows effective reduction in the target protein levels in the knockdown cells. (B) ChIP assay covering the regions of 27nt repeats and core promoter (−238/13 bp) of the eNOS gene in HAECs treated with NonO or HDAC3 specific siRNAs or control siRNA (siLuc) with or without the 27nt RNA duplex treatment. Although acetylated histones bound the 27nt repeats were significantly reduced by the NonO knockdown, more acetylated histones bound to the core promoter region of the eNOS gene. Suppression of HDAC3 expression showed similar effects on the pattern of histone acetylation as that by the NonO knockdown.
We next examined whether HDAC inhibition would eliminate the 27nt RNA-mediated eNOS suppression. We used HDAC3-specific siRNA to suppress HDAC3 expression (Figure 5A). 27nt RNA-treated HAECs were cultured in EGM-2 medium for 24 h followed by HDAC inhibition for 24 h. Total protein extracts were obtained from the treated HAECs for Western blot analysis by using anti-eNOS antibody. When HAECs were treated with HDAC3-specific siRNA, more acetylated histones were bound to the eNOS core promoter (−238/13 bp; Figure 5B). The effects on the histone acetylation at the eNOS promoter were counteracted by the addition of the 27nt RNA duplex (Figure 5B).
Effects of 27nt RNA on eNOS DNA Methylation
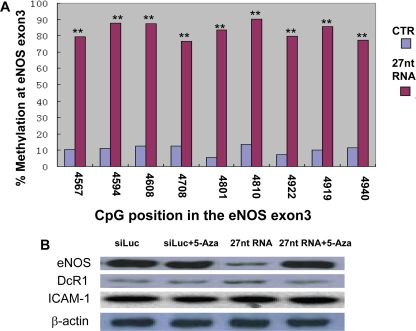
Because DNA methylation may be a mechanism for small RNA-induced transcription suppression (Klenov and Gvozdev, 2005; Klenov et al., 2007), we looked for changes in methylation status of the CpG islands located in the eNOS DNA sequence upstream from the 27nt repeat element. 27nt RNA treatment did not affect CpG methylation status in the eNOS promoter region (data not shown); however, HAECs treated with the 27nt RNA had significantly increased methylation of CpG immediately upstream from the 27nt repeat element in intron 4. Using sodium bisulfite genomic sequencing, we found that nine CpG dinucleotides located within the +4567/+4940 region were highly methylated by treatment with 27nt RNA (Figure 6A). The methylation rate at site 4567 increased from 10 to 80%, at site 4594 from 12 to 89%, at site 4608 from 12 to 92%, at site 4708 from 13 to 76%, at site 4801 from 8 to 83%, at site 4810 from 15 to 90%, at site 4922 from 7 to 81%, at site 4929 from 10 to 87%, and at site 4940 from 12 to 82%.
Figure 6.
Effect of 27nt RNA duplex on CpG island methylation in eNOS gene. (A) Methylation status in CpG islands in the genomic region immediate upstream of the eNOS intron 4. Sodium bisulfite genomic sequencing was used for the experiments in HAECs treated with 27 RNA duplexes or siLuc (negative control) for 24 h. The x-axis indicates the CpG island position in reference to the transcription starting site of the eNOS gene. Each column represents the percentage of the methylated CpG of a total of 10–15 individual clones. An independent Student's t test was performed to compare the differences between siLuc siRNA and 27nt RNA treated cells. (**p < 0.01). The quantitative data represent the results of three independent experiments. (B) Western blot of the eNOS, DcR1 and ICAM-1 proteins in HAECs treated with 27nt RNA duplex with or without the 5-aza–methyl transferase inhibitor. 5-aza seemed to recover the 27nt RNA duplex-induced eNOS depression in HAECs.
To assess whether the 27nt-induced DNA methylation was responsible for eNOS suppression, we treated HAECs transfected with 27nt RNA with 5 μM 5-aza, a methyltransferase inhibitor, for 72 h. Bisulfite genomic sequencing confirmed that 5-aza treatment resulted in demethylation in the eNOS region of +4567/+4940 (data not shown). The eNOS suppression was reversed by 5-aza treatment as shown using Western blot (Figure 6B). In contrast, neither 27nt RNA nor the 5-aza had any effect on the expression of DcR1 and intercellular adhesion molecule (ICAM)-1 from the cultured endothelial cells (Figure 6B).
Effects of 27nt RNA on eNOS mRNA Stability
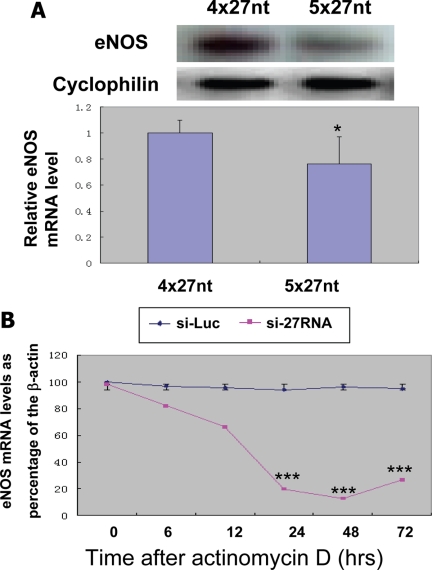
Our previous study has shown that more 27nt repeats the eNOS gene have the more 27nt small RNA is produced and the lower the eNOS expression levels (Zhang et al., 2008). Our current study further demonstrated that human endothelial cells with 5x27nt repeats produced less eNOS mRNA levels (Figure 7A). Although our experiments showed that the 27nt RNA could affect eNOS transcript at DNA level, we further examined whether 27nt RNA could also affect the eNOS mRNA stability. Using actinomycin D to block the eNOS transcription, our experiment showed that eNOS mRNA in endothelial cells treated with 27nt RNA degraded significantly faster than the endothelial cells treated with the nonspecific small RNA (Figure 7B). These findings suggest that the 27nt small RNA may also reduce the eNOS mRNA stability.
Figure 7.
Effect of 27nt RNA duplex on eNOS mRNA expression and stability. (A) Human umbilical venous endothelial cells (HUVECs) at fourth passage homozygous for 5x27nt repeats or homozygous for 4x27nt repeats in the eNOS intron 4 were cultured to 100% confluence before harvested for RNA extraction. The RNA samples were electrophoresized on 1% acrylamid/8 M urea gel and analyzed by Northern blot for eNOS mRNA using the exon4 specific mRNA probe (top lane). The cyclophilin RNA was used as the loading control. The bottom of bar graph represents average eNOS RNA levels from HUVECs of three individuals homozygous for 4x27nt three individuals homozygous for 5x27nt. *p < 0.05 by the Student test. (B) Effects of 27nt small RNA on eNOS mRNA stability in HAECs. HAECs were plated onto six-well plates and cultured up to 90% confluence. We then transfected 27nt RNA duplexes to these cells using liposome. The transcription was blocked by adding actinomycin D (2 μg/ml). Cells were then harvested for RNS extraction at 0, 6, 12, 24, 48 and 72 h after treatment. The relative levels of eNOS mRNA were determined by quantitative realtime RT-PCR adjusted by housekeeping gene β-actin. ***p < 0.001 by the Student's t test comparing to the levels at 0 and 6 h.
DISCUSSION
As the first to study eNOS-derived 27nt small RNA, we have shown that the 27nt repeat element-derived small RNA increased histone acetylation in the repeat region of the eNOS intron 4 through the complementary binding between the 27nt RNA and 27nt DNA. Acetylation of the histones resulted in recruitment of the NonO protein, which has been shown to have a suppressive effect on gene transcription (Zhang et al., 2007). Through protein–protein interaction, nuclear actin, which binds on the 27nt repeat element, recruits HDAC3 to the site. Then, HDAC3 induces histone deacetylation at the core promoter region of the eNOS, which represses eNOS transcriptional activity.
Currently, several types of small RNAs have been described in plants and animals: miRNA, piwiRNA (Aravin et al., 2006), siRNA, tiny noncoding RNA, heterochromatic siRNAs, and repeat associated small interfering RNAs (rasiRNAs) (Reinhart and Bartel, 2002; Ambros et al., 2003; Aravin et al., 2003). These small RNAs are usually ∼21–25 nt; rasiRNAs may be longer (Aravin et al., 2003). Although siRNAs usually arise from an exogenous source or during virus infection, miRNAs are mainly transcribed through designated genomic sequences (Cullen, 2004). Small RNAs usually inhibit gene expression by promoting the degradation of sequence-matched target mRNA in the cytoplasm via the RNA-induced silencing complex. The finding of endogenous miRNAs further indicates that miRNA can induce translational repression through imperfect match-binding at 3′ untranslated region (Cullen, 2004). In contrast, rasiRNAs have been reported to be mostly derived from centromere heterochromatic repeats and tend to be a few nucleotides longer than a typical mature miRNA or siRNA (Pelissier and Wassenegger, 2000; Cao et al., 2003; Matzke et al., 2004; Rana, 2007). One of the critical differences between rasiRNAs and miRNAs is that rasiRNAs regulate gene expression by modifying histones and DNA of the target regions (Aravin et al., 2001; Gvozdev et al., 2003; Lippman et al., 2004) (e.g., histone methylation and acetylation) (Volpe et al., 2002; Hall et al., 2003); this effect seems to be reversible with the deacetylase inhibitor TSA (Hall et al., 2002). The rasiRNAs in conjunction with one or more DNA methyltransferases and possibly chromatin-modifying factors are thought to trigger the de novo methylation and transcriptional silencing of the homologous promoter at the target locus (Aufsatz et al., 2002).
In comparison with the known classes of small RNAs, the 27nt small RNA could be either a new class of small RNA or an atypical form of the miRNA. Because the 27nt small RNA is transcribed from the host gene eNOS, we hypothesize that the intron 4-derived 27nt small RNA may function as a negative feedback regulator and maintain the stable transcription of eNOS. The more the host gene is transcribed, the more gene-specific repeat element derived-RNA will be produced, which forms a negative feedback regulatory loop to fine-tune gene expression in the host. Variable numbers of the repeats in the population may be responsible for quantitative interindividual differences in the expressed genes and disease susceptibility. More specifically, the 27nt repeats in the eNOS intron 4 can be converted to short intronic repeat RNA (sir-RNA) and can function as a negative feedback regulator of eNOS expression. Individuals with different numbers of 27nt repeats will have different levels of 27nt sir-RNA, which is responsible for between-individual differences in eNOS expression, and hence susceptibility to vascular diseases.
The current study is limited by the relatively preliminary nature of the evidence. Although our novel findings suggest that intron-derived 27nt small RNA suppresses the host gene via alteration of histone acetylation and DNA methylation at the transcriptional level, we do not have mechanistic data regarding the nuclear proteins involved in this process. Further experiments are necessary to completely understand how 27nt RNA induces hyperacetylation in histones located at the site of the 27nt repeat element and how the subsequent recruitment of HDAC3 causes histone deacetylation in the upstream promoter region rather than deacetylation of the histones in the 27nt repeat region. Furthermore, whether NonO directly inhibits eNOS transcription or merely facilitates changes in histone acetylation is worth investigating. The findings that 27nt RNA also decreases the eNOS mRNA stability suggest that the intron derived small RNA could also regulate the host gene posttranscriptionally. However, it is not clear whether this posttranscriptional regulation targets at the eNOS pre-mRNA or mature mRNA. Despite these unknowns, the finding that 27nt RNA site-specifically alters the eNOS histone acetylation and methylation status and eNOS mRNA stability is novel. Our findings may lead to the establishment of a new negative feedback regulatory mechanism at the transcriptional level—one that can more efficiently and quickly maintain the host gene within physiological range. This finding may have genome-wide implications for genes sharing the similar repeat structure.
In summary, we have established that the 27nt repeat element-derived small RNA can suppress transcription of the host gene by modifying histone acetylation and DNA methylation of the eNOS gene. Hyperacetylation at the repressor region (i.e., the 27nt repeat element) results in recruitment of a negative transcription factor, such as NonO. Hypoacetylation at the eNOS core promoter induced by the 27nt small RNA may inhibit eNOS suppression.
ACKNOWLEDGMENTS
We are grateful to Dr. Rebecca Bartow, medical editor at the Texas Heart Institute, for editorial assistance. This study is supported by National Institutes of Health/National Heart, Lung, and Blood Institute grants R01HL071608, R01HL066053, and P50HL083794 and the American Heart Association grants 0440001N and 0565134Y.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1186) on July 9, 2008.
REFERENCES
- Ambros V., Lee R. C., Lavanway A., Williams P. T., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Aravin A., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Naumova N. M., Tulin A. V., Vagin V. V., Rozovsky Y. M., Gvozdev V. A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aufsatz W., Mette M. F., van der Winden J., Matzke M., Matzke A. J. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M. F., Huang M. S., Matzke M., Jacobsen S. E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Casas J. P., Bautista L. E., Humphries S. E., Hingorani A. D. Endothelial nitric oxide synthase genotype and ischemic heart disease: meta-analysis of 26 studies involving 23028 subjects. Circulation. 2004;109:1359–1365. doi: 10.1161/01.CIR.0000121357.76910.A3. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Derivation and function of small interfering RNAs and microRNAs. Virus Res. 2004;102:3–9. doi: 10.1016/j.virusres.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Dimmeler S., Zeiher A. M. Nitric oxide–an endothelial cell survival factor. Cell Death Differ. 1999;6:964–968. doi: 10.1038/sj.cdd.4400581. [DOI] [PubMed] [Google Scholar]
- Gibbons G. H., Dzau V. J. The emerging concept of vascular remodeling. N. Engl. J. Med. 1994;330:1431–1438. doi: 10.1056/NEJM199405193302008. [DOI] [PubMed] [Google Scholar]
- Gvozdev V. A., Aravin A. A., Abramov Y. A., Klenov M. S., Kogan G. L., Lavrov S. A., Naumova N. M., Olenkina O. M., Tulin A. V., Vagin V. V. Stellate repeats: targets of silencing and modules causing cis-inactivation and trans-activation. Genetica. 2003;117:239–245. doi: 10.1023/a:1022952315467. [DOI] [PubMed] [Google Scholar]
- Hall I. M., Noma K., Grewal S. I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., Grewal S. I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Klenov M. S., Gvozdev V. A. Heterochromatin formation: role of short RNAs and DNA methylation. Biochemistry. 2005;70:1187–1198. doi: 10.1007/s10541-005-0247-4. [DOI] [PubMed] [Google Scholar]
- Klenov M. S., Lavrov S. A., Stolyarenko A. D., Ryazansky S. S., Aravin A. A., Tuschl T., Gvozdev V. A. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Matzke M., Aufsatz W., Kanno T., Daxinger L., Papp I., Mette M. F., Matzke A. J. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Ou H., Shen Y. H., Utama B., Wang J., Wang X., Coselli J., Wang X. L. Effect of nuclear actin on endothelial nitric oxide synthase expression. Arterioscler. Thromb. Vasc. Biol. 2005;25:2509–2514. doi: 10.1161/01.ATV.0000189306.99112.4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelissier T., Wassenegger M. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA. 2000;6:55–65. doi: 10.1017/s135583820099201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T. M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Bartel D. P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., Martienssen R. A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang J., Dudley D., Wang X. L. Haplotype-specific effects on endothelial NO synthase promoter efficiency: modifiable by cigarette smoking. 2002;22:e1–e4. doi: 10.1161/01.ATV.0000016248.51577.1F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., et al. TNF-alpha suppresses prolyl-4-hydroxylase alpha1 expression via the ASK1-JNK-NonO pathway. Arterioscler. Thromb. Vasc. Biol. 2007;27:1760–1767. doi: 10.1161/ATVBAHA.107.144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. X., Ou H., Shen Y. H., Wang J., Coselli J., Wang X. L. Regulation of endothelial nitric oxide synthase by small RNA. Proc. Natl. Acad. Sci. USA. 2005;102:16967–16972. doi: 10.1073/pnas.0503853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. X., Zhang C., Shen Y. H., Wang J., Li X. N., Zhang Y., Coselli J., Wang X. L. Biogenesis of short intronic repeat 27nt small RNA from endothelial nitric oxide synthase gene. J. Biol. Chem. 2008;283:14685–14693. doi: 10.1074/jbc.M801933200. [DOI] [PMC free article] [PubMed] [Google Scholar]