Abstract
Obscurin is an ∼800-kDa protein composed of structural and signaling domains that organizes contractile structures in striated muscle. We have studied the Rho-GEF domain of obscurin to understand its roles in morphogenesis and signaling. We used adenoviral overexpression of this domain, together with ultrastructural and immunofluorescence methods, to examine its effect on maturing myofibrils. We report that overexpression of the Rho-GEF domain specifically inhibits the incorporation of titin into developing Z-disks and disrupts the structure of the Z-disk and Z/I junction, and alters features of the A/I junction. The organization of other sarcomeric markers, including α-actinin, was not affected. We identified Ran binding protein 9 (RanBP9) as a novel ligand of the Rho-GEF domain and showed that binding is specific, with an apparent binding affinity of 1.9 μM. Overexpression of the binding region of RanBP9 also disrupted the incorporation of titin into developing Z-disks. Immunofluorescence localization during myofibrillogenesis indicated that the Rho-GEF domain assembles into sarcomeres before RanBP9, which first occurs in myonuclei and later in development translocates to the myoplasm, where it colocalizes with obscurin. Both the Rho-GEF domain and its binding region on RanBP9 bind directly to the N-terminal Ig domains of titin, which flank the Z-disk. Our results suggest that the Rho-GEF domain interacts with RanBP9 and that both can interact with the N-terminal region of titin to influence the formation of the Z-disk and A/I junction.
INTRODUCTION
Myofibrillogenesis is a complex process that requires the coordinated assembly and integration of many contractile, cytoskeletal, and signaling proteins into regular arrays, the sarcomeres. Two giant proteins, nebulin and titin, are responsible for organizing the thin and thick filaments of sarcomeres, respectively. Recently, another giant protein and member of the titin family was identified and named “obscurin” (Young et al., 2001). Unlike nebulin and titin, which are oriented longitudinally in each half sarcomere, obscurin surrounds the sarcomere (Kontrogianni-Konstantopoulos et al., 2003). Preliminary experiments suggest that it too plays an important role in organizing the contractile apparatus, as well as the sarcoplasmic reticulum.
Obscurin's ability to bind to a small form of ankyrin 1 (sAnk1;Ank1.5) is thought to provide a critical link between the contractile apparatus and the sarcoplasmic reticulum in striated muscle (Kontrogianni-Konstantopoulos et al., 2003, 2006b; Bagnato et al., 2003). As had been postulated previously for the obscurin homologue UNC-89 in Caenorhabditis elegans (Benian et al., 1996), obscurin also plays a critical role in the formation of the A-band and M-line during myofibrillogenesis in mammalian muscle (Kontrogianni-Konstantopoulos et al., 2004, 2006b; Borisov et al., 2006). Recently, changes in obscurin have been linked to hypertrophic cardiomyopathies (Borisov et al., 2003; Arimura et al., 2007).
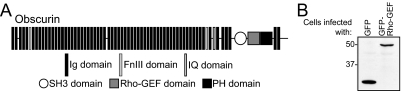
Although composed primarily of Ig domains, obscurin has several domains commonly used for intracellular signaling, including IQ, Src homology 3 (SH3), and tandem Rho-guanine nucleotide exchange factor (Rho-GEF), and pleckstrin homology (PH) domains (Young et al., 2001; Figure 1A). Because it surrounds the contractile apparatus at the Z-disks and M-bands, obscurin is well placed to detect alterations in muscle architecture during the contractile cycle and to transduce those signals to the surrounding myoplasm (Hoshijima, 2006). Elucidating the role of obscurin in myofibrillogenesis is complicated by the fact that it is expressed in several different isoforms, which are found at different positions along the sarcomeres of skeletal muscle. In particular, distinct isoforms containing the Rho-GEF domain are present at the level of the A/I junction and the M-band (Bowman et al., 2007).
Figure 1.
Overexpression of the Rho-GEF domain of obscurin in primary cultures of skeletal myotubes. (A) Structure of obscurin. The Rho-GEF domain was cloned into adenovirus as a GFP fusion protein. (B) P1 rat myotubes were infected 96 h after plating with adenovirus encoding GFP or GFP-Rho-GEF. After 48 h, cultures were collected and the proteins analyzed by SDS-PAGE and immunoblotting with an anti-GFP antibody. Both proteins are expressed at similar levels.
Rho-GEF domains and their best characterized substrates, Rho-GTPases, are known to play a critical role in myofibrillogenesis (Hoshijima et al., 1998; Schmidt and Hall, 2002; Bryan et al., 2005). Although the Rho-GEF/PH domains of obscurin have been implicated in pathologies (Borisov et al., 2003), their function is still unknown. We used adenoviral overexpression studies in developing muscle to investigate the possible roles of the Rho-GEF domain in myofibrillogenesis. We have also begun to characterize the interaction of obscurin's Rho-GEF with other proteins. Our studies of small GTPases are still in progress (Ford, Roche, and Bloch, unpublished). Here, we identify Ran binding protein 9 (RanBP9, also known as RanBPM) as a ligand of the Rho-GEF domain and study its role in myofibrillogenesis. Our results indicate that both the Rho-GEF domain and RanBP9 can interact with titin to regulate its incorporation into Z-disks in developing muscle cells.
MATERIALS AND METHODS
Plasmids
We used the Superscript first strand synthesis system (Invitrogen, Carlsbad, CA) in the reverse transcriptase PCR reaction to amplify the Rho-GEF, Rho-GEF/PH, and PH domains of obscurin from rat skeletal muscle poly A+ RNA (Clontech, Palo Alto, CA). The primers used are in Supplemental Table 1. The domain fragments were cloned into the EcoRI/BamHI sites of pGBKT7 (Clontech, Mountain View, CA). RanBP9 deletion constructs and full-length RanBP10 were generated by PCR, digested with SmaI/XhoI and EcoRI/XhoI, respectively, and ligated into the pGADT7 vector (Clontech). RanBP9108-729 was amplified via PCR and ligated into the following vectors: XhoI/HindIII sites of pRSETC (Invitrogen), the SmaI/NotI sites of pGEX-4T-1 (GE Healthcare, Piscataway, NJ), the XhoI/HindIII sites of pHcRed1-C1 (Clontech) and the XbaI/HindIII sites (after NheI/HindIII digestion) of pMAL-c2X (New England Biolabs, Ipswich, MA).
EGFPC2-Rho-GEF and EGFPC2-Rho-GEF/PH were created from the respective pGBKT7 plasmids and introduced into the EcoRI/BamHI sites of EGFPC2 (Clontech) and the EcoRI/XhoI sites of pGEX-4T-1. HcRedC1-RanBP91-729 (full length) was created by digesting the plasmid pGBKT7-RanBP9 (Wang et al., 2002) with Sal/HindIII and cloning into the Xho/HindIII sites of pHcRedC1.
Plasmid pDys246 (Amann et al., 1998) was kindly provided by Dr. J. Ervasti (University of Minnesota, Minneapolis, MN). Plasmids pEGFPC3-RanBP10 and pGBKT7-RanBP9 were the kind gift of Dr. G. Wu (University of Rochester Medical Center, Rochester, NY).
Yeast Two-Hybrid Screening and β-Galactosidase Assay
The Matchmaker two-hybrid system was used as described by the manufacturer (Clontech, Mountain View, CA). pGBKT7-Rho-GEF/PH was transformed into Saccharomyces cerevisiae strain AH109 and mated with yeast pretransformed with a cDNA library from human skeletal muscle. Mated yeast was plated on SD-His/-Ade/-Leu/-Trp plates and transformants on SD-His/-Ade/-Leu/-Trp plates with X-α-Gal. Positive plasmids were recovered by electroporation into Escherichia coli DH10B (Invitrogen) and sequenced.
Liquid β-galactosidase assays were performed following the Yeast Protocols Handbook (Clontech) by using chlorophenol red-β-d-galactopyranoside (CPRG). Three independent colonies were assayed for each construct. Results represent average values. Significance was determined with a two-tailed t test or a Mann–Whitney U-test (see text for details).
Purification of Recombinant Fusion Proteins
Expression of the recombinant 6X-His form of RanBP9108-729 and the actin binding domain of dystrophin by pDys246 (see above) was induced with 1.0 and 0.5 mM isopropyl β-d-thioglucopyranoside (IPTG), respectively, for 3 h. Proteins were purified on TALON resin (Clontech), following the manufacturer's instructions. The recombinant glutathione transferase (GST) form of Rho-GEF and RanBP9 were induced with 0.3 mM IPTG for 4 h, solubilized, and purified as described previously (Frangioni and Neel, 1993; Mercado-Pimentel et al., 2002; Bowman et al., 2007). Expression of recombinant fusion proteins of maltose binding protein (MBP) with RanBP9 (MBP-RanBP9) or the first two Ig domains of titin (MBP-TitinZIgI/II) (Kontrogianni-Konstantopoulos and Bloch, 2003) was induced with 0.2 mM IPTG for 3 h. The fusion proteins were affinity purified on amylose resin (New England Biolabs).
Antibodies
Rabbit anti-titin-M-x246 (against the M band portion of titin, 3 μg/ml; Centner et al., 2001) was a gift from Dr. S. Labeit (Universitätsklinikum Mannheim, Mannheim, Germany). Mouse antibodies to myomesin (B4, diluted 1:1; Grove et al., 1984) were a gift from Dr. J. C. Perriard (Institute of Cell Biology, ETH, Zurich, Switzerland).
GST-Rho-GEF and GST-RanBP9108-729 were used to generate antibodies in rabbits (Covance Research Products, Denver, PA). Antibodies were purified over affinity columns prepared with MBP-Rho-GEF and MBP-RanBP9108-729, respectively. Affinity-purified antibodies were used at 2 μg/ml for immunofluorescence and 100 ng/ml for immunoblotting. GST-Rho-GEF antibodies were also prepared in guinea pigs, and purified and used as described above. Immunodepletion was performed by incubating 2 μg/ml of each antibody with 200 μg/ml the corresponding antigen in phosphate-buffered saline (PBS)/bovine serum albumin (BSA)/normal goat serum (NGS) (PBS containing 3% BSA, 10 mM NaN3, and 5% normal goat serum; Jackson ImmunoResearch Laboratories, West Grove, PA) in a total volume of 1 ml for 16 h at 4°C and demonstrated that labeling was abolished after preadsorption. We also used: mouse anti-myosin II (clone MY-32, 1:400 dilution; Sigma-Aldrich, St. Louis, MO), mouse anti-α-actinin (clone EA53, 1:400 dilution; Sigma-Aldrich), mouse 9D10 to the PEVK region of titin (supernatant fraction, used at 1:1 dilution; Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit antibodies to the Z-disk region of titin (titin-Z; 3 μg/ml; Kontrogianni-Konstantopoulos et al., 2006b), goat anti-GST (GE Healthcare), mouse anti-MBP (New England Biolabs), and mouse anti-GFP (clone 11E5; Invitrogen).
Goat antibodies to mouse, guinea pig, and rabbit immunoglobulin Gs, linked to Alexa-488 or Alexa-568 (Invitrogen), were used as secondary antibodies at dilutions of 1:200.
Tissue Collection and Culture
Rats were killed under anesthesia by cardiac perfusion with buffered 2% paraformaldehyde, and muscles were collected, snap-frozen, and sectioned as described previously (Ursitti et al., 2004).
Primary cultures of rat myotubes were prepared as reported previously (De Deyne et al., 1998). C2C12, a mouse myogenic cell line (America Type Culture Collection, Manassas, VA), was cultured as specified by the supplier.
Fluorescent Immunolabeling and Confocal Microscopy
For immunofluorescence, myotubes were fixed, permeabilized, and incubated first in PBS/BSA/NGS to block nonspecific binding as described previously (Ursitti et al., 2004), then with the appropriate primary antibodies in PBS/BSA/NGS overnight at 4°C. Samples were washed, incubated with species-specific secondary antibodies in PBS/BSA/NGS, mounted in Vectashield, and observed with a Zeiss 410 confocal laser scanning microscope (Carl Zeiss, Tarrytown, NY). Frozen longitudinal and cross sections of rat tibialis anterior and quadriceps muscle were labeled as described previously (Kontrogianni-Konstantopoulos et al., 2003) except that the sections were not treated with detergents before labeling.
Pull-Down Assay
Homogenates of 8-d-old cultures of rat myotubes were prepared as described previously (Kontrogianni-Konstantopoulos et al., 2004) and supplemented with benzonase (Novagen, Madison, WI) at 50 U/ml. Equal amounts of recombinant His-Dystrophin1-246 and His-RanBP9108-729 were bound to TALON resin and incubated with 0.5 mg of homogenate protein at 4°C for 8 h. Beads were washed extensively at 4°C with 10 mM NaPO4, pH 7.2, 120 mM NaCl, 10 mM NaN3, 0.1% Tween 20, and heated for 30 min at 42°C in NuPAGE LDS sample buffer and reducing agent (Invitrogen). Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE); transferred to nitrocellulose; incubated for 6 h at room temperature (RT) in PBS, 10 mM NaN3, 0.5% Tween 20, and 3% dry milk; and probed with antibodies to the Rho-GEF domain of obscurin.
Experiments to assess binding of fusion proteins to the Z-disk region of titin were done similarly, except that glutathione resin bound to equivalent amounts of GST, GST-RanBP9108-729, or GST-Rho-GEF were incubated with equal amounts of MBP fused to the two N-terminal domains of titin, rather than with muscle homogenates, and blots were probed with anti-MBP antibodies.
Blot Overlay
Aliquots (2.5 μg of protein) of His-Dystrophin1-246, and His-RanBP9108-729 were separated on 4–12% Bis-Tris acrylamide gels and transferred to nitrocellulose. Blots were incubated in 50 mM Tris, pH 7.2, 120 mM NaCl, 3% BSA, 0.1% Tween 20, and 2 mM dithiothreitol for 3 h at RT and then with 1.5 μg/ml GST or GST-Rho-GEF polypeptides in the same buffer for 3 h at RT. After washing, blots were probed with anti-GST antibodies, as described above.
Surface Plasmon Resonance
Binding of RanBP9 to the Rho-GEF domain of obscurin was evaluated with a BIAcore 3000 biosensor (GE Healthcare) as described previously (Kontrogianni-Konstantopoulos et al., 2003). In brief, a CM5 sensor chip was charged with goat anti-GST, yielding ∼12,250 response units (RUs) per surface. Kinetic analysis was performed with similar amounts (measured in RUs) of control GST and GST-Rho-GEF immobilized in flow cell (FC) 1 and FC2 respectively, by binding to immobilized anti-GST antibody. MBP-RanBP9108-729 was then passed over both surfaces at concentrations from 0.9 to 2.6 μM. Binding of MBP to GST-Rho-GEF was analyzed as an additional control.
Generation of Recombinant Adenoviruses and Use with P1 Myotubes
EGFPC2-Rho-GEF and EGFPC2-Rho-GEF/PH were digested with Xba/BamH1, ligated into pAdlox (Hardy et al., 1997) and cotransfected with the adenoviral DNA partner ψ5 into human embryonic kidney 293 cells that stably express cre recombinase (CRE8). Purified recombinant products were selected after cell lysis by repeated passage in CRE8 cells and two rounds of plaque purification in agarose overlays. Virus was amplified and purified by banding in CsCl step gradients and the titer in plaque-forming units (pfu) was determined from agarose overlay (Graham and Prevec, 1995). Control EGFP virus was produced as described previously (Kontrogianni-Konstantopoulos et al., 2004).
Adenovirus expressing HcRed, HcRed-RanBP91-729, and HcRed-RanBP9108-729 were created with AdEasy (Stratagene, La Jolla, CA) following the manufacturer's instructions (see He et al., 1998), and amplified as described above.
Cultures of myotubes were infected with 3 × 107 pfu/ml DMEM at RT for 90 min. Cultures were supplemented with 1 ml of DMEM plus 20% fetal bovine serum and 40 μM cytosine arabinoside. After 48 h, cells were fixed, immunolabeled with antibodies to sarcomeric markers, and scored blindly. Approximately 50 myotubes each from at least three independent experiments were classified as having myofibrils that were completely disorganized, moderately disorganized, or organized normally. The percentages of fibers infected with GFP-Rho-GEF and HcRed-RanBP9108-729 that showed different levels of organization for each sarcomeric marker were normalized to the percentages obtained for fibers infected with GFP alone or HcRed-RanBP91-729, respectively, that showed the same morphology (completely disorganized, moderately disorganized, or organized normally). Significance was established with a one-sample t test.
Electron Microscopy
Myotube cultures prepared on glass coverslips and infected with either green fluorescent protein (GFP) or GFP-Rho-GEF virus were fixed with 2% paraformaldehyde for 30 min at room temperature and washed with PBS. The locations of infected cells, indicated by GFP fluorescence, were marked with a diamond-tipped scribing objective (Carl Zeiss). Coverslips were removed, and samples were fixed overnight in 2% glutaraldehyde, 5 mg/ml tannic acid, and 0.2 M cacodylate, pH 7.2. After washing with 0.2 M cacodylate buffer, pH 7.2, cultures were postfixed in 1% osmium tetroxide in 50 mM acetate buffer, pH 5.5, en bloc stained with 1% uranyl acetate in 65% ethanol, dehydrated, and embedded in Araldite-Embed 812 (Electron Microscopy Sciences, Fort Washington, PA). The glass coverslips were separated from the cells with hydrofluoric acid. Sections were cut at a thickness of ∼60–90 nm with an LKB MT5000 microtome (LKB-Pharmacia, Bromma, Sweden). Sections were picked up on 200 mesh copper grids, stained with uranyl acetate followed by lead citrate, and examined under a Philips 201 electron microscope (Philips, Eindhove, The Netherlands) or a Zeiss 10C (Carl Zeiss) electron microscope at a magnification of 30,000× and 31,500×, respectively. Pictures were taken on Kodak 4489 film and digitally scanned at 600 dpi.
Materials
Unless otherwise noted, all materials were purchased from Sigma-Aldrich and were the highest grade available.
RESULTS
Effects of Overexpressing the Rho-GEF Domain of Obscurin on Myofibrillogenesis
We postulated that the Rho-GEF domain of obscurin plays a significant role in the formation or stabilization of sarcomeres. To test this, we used adenoviral vectors to deliver and overexpress fluorescent fusion proteins of the Rho-GEF domain. Virus expressing GFP alone served as a control. Both viruses expressed the proteins they encoded at high levels, as shown by Western blotting (Figure 1B). Approximately 70% of myotubes in culture were infected with GFP-Rho-GEF virus, and slightly more with virus expressing GFP. We used antibodies to several different protein markers to examine the effects of expressing these fusion proteins on the development of myofibrils.
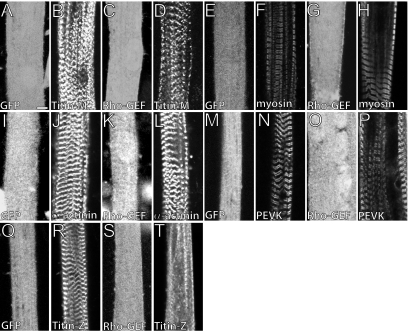
We found that the accumulation of α-actinin at the Z-disk (Figure 2, J and L), myosin at the A-band (Figure 2, F and H), and titin epitopes at the I-band (Figure 2, N and P), and M-band (Figure 2, B and D) assembled normally in cells that overexpress either GFP protein or the Rho-GEF domain of obscurin. By contrast, myotubes that overexpressed the Rho-GEF domain failed to incorporate the N-terminal epitopes of titin fully into developing Z-disks (Figure 2T). These epitopes incorporated into Z-disks normally in myotubes expressing GFP alone (Figure 2R). We ruled out the possibility that the titin molecule was being extensively proteolyzed by examining SDS-PAGE gels of treated and control myotubes stained with silver (Tatsumi and Hattori, 1995). Staining of an ∼3-MDa band, which we presumed to be titin, was unchanged in the experimental sample, suggesting that much of the titin remained intact (data not shown). Additionally, we ruled out the possibility that the effects of the Rho-GEF domain are due to masking of N-terminal epitopes of titin by overlaying MBP-TitinZIgI/II with GST-Rho-GEF and demonstrating no difference in antibody labeling compared with GST alone (Supplemental Figure 1). Thus, the Rho-GEF domain of obscurin is likely to play a specific role in integrating titin into the Z-disk.
Figure 2.
Overexpression of the Rho-GEF domain of obscurin disrupts titin organization specifically at the Z-disk. We infected P1 rat myotube cultures 96 h after plating with adenoviral constructs expressing GFP or GFP-Rho-GEF and assessed changes in myofibrillar morphology 48 h later. Cells were immunolabeled with antibodies to myosin or α-actinin, or with antibodies to particular regions of titin. GFP alone or GFP-Rho-GEF had no significant effect on the organization of titin at the M-band (A, B, and C, D), myosin at the A-band (E, F and G, H), α-actinin at the Z-disk (I, J and K, L), and titin at the I-band (M, N and O, P). GFP alone had no effect on the organization of titin at the Z-disk (Q and R). GFP-Rho-GEF, however, disrupted the incorporation of titin into developing Z-disks (S and T). Bar, 5 μm.
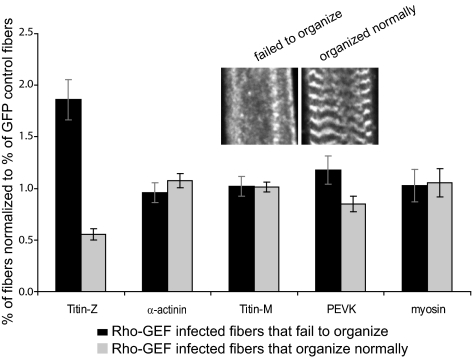
We quantitated our observations by categorizing the effects of overexpression on individual sarcomeric proteins as fully disorganized, moderately disorganized, or fully organized. The percentage of fibers in each category was then normalized to the percentage obtained for myotubes expressing GFP alone with the same respective morphology (fully disorganized, moderately disorganized, or fully organized), to correct for possible nonspecific effects of adenoviral infection and protein expression. Results for fibers that failed to organize and fibers that organized normally are shown in Figure 3. The association of the N-terminal region of titin with Z-disks was significantly inhibited in myotubes that overexpressed the Rho-GEF domain, compared with GFP alone (p ≤ 0.011 for fibers that fail to organize, and p ≤ 0.001 for fibers that organize normally; Figure 3). None of the other sarcomeric markers we examined, including α-actinin at Z-disks and epitopes of titin located at the M- or I-bands, were significantly altered by overexpression of the Rho-GEF domain (all p ≥ 0.15), indicating that its effect on the organization of titin at Z-disks was highly specific.
Figure 3.
Quantitation of the disruption of titin at the Z-disk by overexpression of obscurin's Rho-GEF domain, as illustrated in Figure 2. The data show that overexpression of GFP-Rho-GEF caused a significant increase (p ≤ 0.011) in fibers that fail to organize titin at the Z-disk and a significant decrease (p ≤ 0.001) in fibers in which titin organizes normally at the Z-disk, compared with fibers overexpressing GFP. No other morphological markers were altered to a significant extent (all p ≥ 0.15).
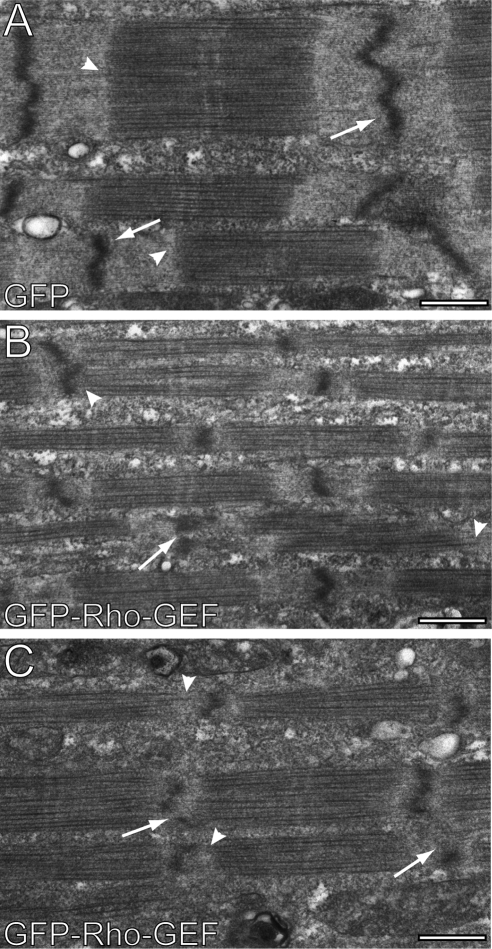
We used thin section electron microscopy to confirm the effect of overexpressing the Rho-GEF domain of obscurin on Z-disks. Consistent with our immunofluorescence results, the Z-disks were still present but were not as well developed and showed frequent interruptions (Figure 4, B and C, white arrows), compared with control samples, which overexpressed GFP (Figure 4A, white arrows). In addition, the A/I junctions were less organized (Figure 4, B and C, white arrowheads) than in control cells (Figure 4A, white arrowheads), and often showed regions at which thick filaments extended almost to the Z-disks. The apparent diameter of myofibrils also seemed to decrease, perhaps because the Z-disks are smaller. Although Figure 4B demonstrates a decrease in lateral alignment of myofibrils, this phenotype was also observable in controls and so was not specific to the effects of overexpression of the Rho-GEF domain. Two independent blinded observers analyzed 34 images and correctly identified 26 and 28 of the images, respectively, as control or treated samples (p < 0.001, chi-square test). Thus, the disruption of titin's integration at the Z-disk in developing skeletal myotubes by overexpression of the Rho-GEF domain leads to instability of the Z-disk and the A/I junction.
Figure 4.
Overexpression of the Rho-GEF domain of obscurin in developing myotubes disrupts Z-disk formation and organization of the A/I junction at the ultrastructural level. Myotubes were infected as in Figure 2 with adenovirus expressing GFP alone (A) or GFP-Rho-GEF (B and C) and processed for thin section electron microscopy. Cells expressing GFP-Rho-GEF show disrupted Z-disks (B and C, white arrows), with frequent interruptions, so that they extend over much shorter distances than in controls (A). Additionally, many of the A/I junctions in myotubes overexpressing GFP-Rho-GEF lack clear borders between the thick and thin filaments (B and C, white arrowheads), due to the presence of thick filaments very close to Z-disks. Bar, 0.5 μm.
Binding of the Rho-GEF Domain of Obscurin to RanBP9
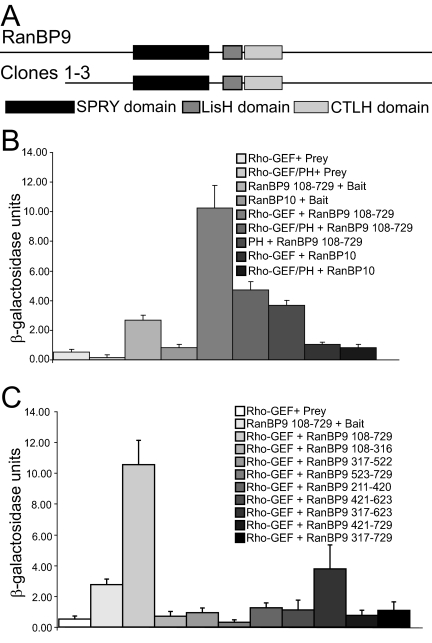
We used the yeast two-hybrid screen to identify other proteins that may modulate the effects of the Rho-GEF/PH domains of obscurin on myofibrillogenesis. With the Rho-GEF/PH domains as “bait,” we screened a human skeletal muscle cDNA library expressed in a “prey” vector that was pretransformed in yeast. After screening ∼8.8 × 105 transformants, we identified seven prey clones that activated of all four reporter genes (MEL1, ADE2, HIS3, and lacZ) and so met the most stringent criteria for specificity. DNA sequencing showed that three of them encoded amino acids 108–729 of RanBP9 (Figure 5A), containing all of its known functional domains (Nakamura et al., 1998).
Figure 5.
RanBP9 is a ligand of the Rho-GEF domain of obscurin. Yeast two-hybrid analysis identified RanBP9 as a ligand for the Rho-GEF/PH domains of obscurin. (A) The Rho-GEF/PH domains of obscurin were used as bait to screen a human skeletal muscle library. Three clones encoding amino acids 108–729 of RanBP9 were identified. We generated and expressed a series of constructs in the pGBKT7-bait and pGADT7-prey vectors, respectively, to identify the minimal sequences of obscurin and RanBP9 required for their interaction. We also examined the possible interaction between obscurin and RanBP10, a protein highly homologous to RanBP9. Averages of at least three independent samples are shown. (B) Assays of β-galactosidase activity indicated that the Rho-GEF domain of obscurin is sufficient for the interaction with RanBP9. The Rho-GEF domain does not interact with RanBP10. (C) Assays with smaller fragments of RanBP9 failed to show strong interactions with the Rho-GEF domain. The minimal binding domain on RanBP9 for the Rho-GEF domain of obscurin therefore seems to be composed of amino acids 108–729.
We further tested the role of the PH domain in binding of the Rho-GEF domain to RanBP9 with the yeast two-hybrid screen and quantitated our results with solution assays of β-galactosidase activity, with CPRG as the substrate. The Rho-GEF domain bound RanBP9 in the absence of the PH domain better than in its presence (p = 0.01). Consistent with this, the PH domain alone had no binding activity (p = 0.16; compared with the empty vector; Figure 5B). Significance was determined with a two-tailed t test (for PH, RanBP9 and Bait, RanBP9 interactions) or a Mann–Whitney U-test (for Rho-GEF, RanBP9 and Rho-GEF/PH, RanBP9 interactions). The Rho-GEF domain of obscurin failed to bind to RanBP10, another Ran binding protein that is also expressed in skeletal muscle (Wang et al., 2004). Although some Ran-binding proteins have been shown to be homologous to importins, this has not been demonstrated with RanBP9. Given their homology (68% amino acid identity), we selected RanBP10 as the most appropriate negative control (Wang et al., 2004; Murrin and Talbot, 2007; Schulze et al., 2008). Thus, the Rho-GEF domain binds RanBP9 specifically, without the participation of the PH domain.
We created deletion constructs of RanBP9 in the yeast two-hybrid prey vector and analyzed their interaction with the Rho-GEF domain of obscurin, to determine the regions required for binding. Binding of smaller fragments of RanBP9 to the Rho-GEF domain was not significantly different from controls (Figure 5C). These results suggest that much of the sequence of RanBP9 is required for binding to the Rho-GEF domain of obscurin in the yeast two-hybrid system. Based upon the yeast two-hybrid experiments, we used the Rho-GEF domain without the PH domain and amino acids 108–729 of RanBP9 for studies of binding in vitro.
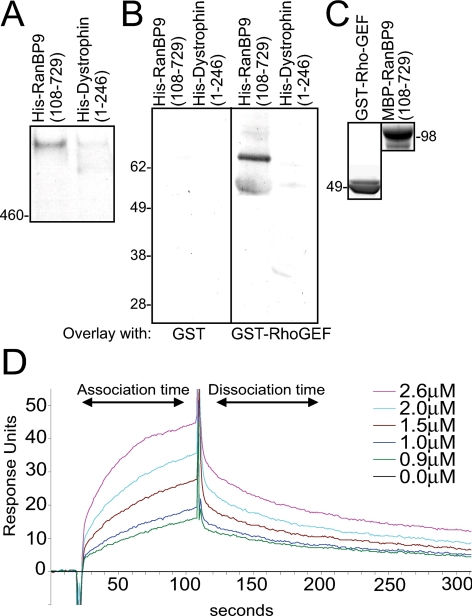
We used a “pull-down” assay to demonstrate that RanBP9 is able to bind native obscurin from homogenates of cultured rat myotubes, where much of the ∼800-kDa form of the protein is soluble (Kontrogianni-Konstantopoulos et al., 2003). A 6X-histidine (His)–tagged form of RanBP9 bound to TALON resin served as the adsorbent, and a similarly tagged N-terminal actin-binding domain of dystrophin (Amann et al., 1998) as control. RanBP9 but not the dystrophin domain bound the endogenous ∼800-kDa form of obscurin from myotubes (Figure 6A). RanBP9 was also able to interact with smaller isoforms of obscurin, which we are currently characterizing (our unpublished data).
Figure 6.
Binding of RanBP9 to the Rho-GEF domain of obscurin. We studied the interaction of the Rho-GEF domain of obscurin with RanBP9 in pull down, blot overlay, and surface plasmon resonance experiments. (A) We used bacterially expressed, affinity purified, RanBP9 and dystrophin carrying His tags, in a pull-down experiment to assay association with endogenous obscurin in extracts of cultured rat myotubes. The results show that His-tagged RanBP9108-729 associates with the ∼800-kDa isoform of obscurin, whereas a His-tagged control protein (the N-terminal domains of dystrophin) does not. (B) We assayed the ability of recombinant fusion proteins of RanBP9108-729 and the Rho-GEF domain of obscurin (shown after purification in C) to bind to each other directly, in blot overlay experiments. The Rho-GEF domain of obscurin binds to RanBP9108-729 but not to a control protein, the N-terminal region of dystrophin. Binding is therefore direct and specific. (C) SDS-PAGE analysis of the recombinant proteins used in blot overlay (B) and in surface plasmon resonance experiments (D). Proteins had the expected molecular masses, indicated. (D) We used surface plasmon resonance to assay direct binding of RanBP9108-729 to the Rho-GEF domain of obscurin. The results are consistent with specific binding in a 1:1 complex, with an affinity of 1.9 μM.
We next used “overlay” assays and surface plasmon resonance to assess direct binding between obscurin's Rho-GEF domain and RanBP9. Equivalent amounts of recombinant His-RanBP9108-729 and His-Dystrophin1-246 were separated by SDS-PAGE and transferred to nitrocellulose. Blots were incubated with either GST or a GST-Rho-GEF fusion protein. GST-Rho-GEF specifically bound to the His-tagged RanBP9108-729, but not to the His-tagged dystrophin fragment (Figure 6B, right). GST alone did not bind RanBP9 (Figure 6B, left). We also analyzed these interactions with a BIAcore 3000 biosensor (see Materials and Methods) and confirmed that binding of MBP-RanBP9108-729 to GST-Rho-GEF was specific. Real-time binding of MBP-RanBP9108-729 to GST-Rho-GEF was evaluated after subtraction of a reference control cell containing immobilized GST (Figure 6C). Fitting the curves with an assumed stoichiometry of 1:1 gave a dissociation constant, KD, of 1.9 μM (Figure 6D). Thus, the Rho-GEF domain of obscurin binds RanBP9 directly, with moderate affinity.
RanBP9 and Obscurin in Developing Muscle
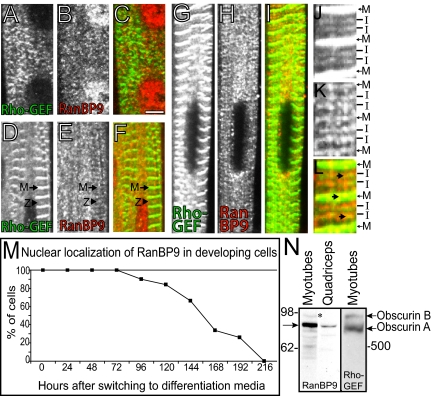
We studied the assembly of RanBP9 and the Rho-GEF domain of obscurin during myofibrillogenesis in primary rat skeletal myotubes in culture. Myoblasts fused into myotubes from 48 h to 72 h after plating. Cells were analyzed 72 h after plating and in 24-h increments thereafter (data from time points at 96, 120, and 168 h are shown). The Rho-GEF domain of obscurin began to occur in striations in most cells as early as 72 h and became more organized as development proceeded (Figure 7A, 96 h; D, 120 h; and G, 168 h), until it localized to the M-band and the I-band near the Z-disk. The organization of RanBP9 into striations was delayed compared with that of the Rho-GEF domain, but the two proteins began to colocalize by day 5 in culture (Figure 7, D–F). Although it is difficult to discern from our immunofluorescent experiments whether the Rho-GEF domain of obscurin and RanBP9 are present at Z-disks or adjacent I-band structures earlier in development, some images suggest that they are indeed at Z-disks (Figure 7, D–F, arrowheads). Given the effects of the Rho-GEF domain on titin, as well as its ability to bind to titin (see below), our immunofluorescent data and our previous results with antibodies to the C-terminal region of obscurin (Kontrogianni-Konstantopoulos et al., 2003), we propose that the Rho-GEF domain and RanBP9 both associate with Z-disks as they begin to form, although perhaps only transiently. At later times of development in vitro, the Rho-GEF domain and RanBP9 colocalized at the I-bands and M-bands (Figure 7, J–L, and insets), consistent with their ability to interact biochemically.
Figure 7.
Expression of obscurin and RanBP9 in developing P1 rat skeletal myotubes. Cultures of rat myotubes were collected between 72 and 168 h after initial plating and double immunolabeled with antibodies to the Rho-GEF domain of obscurin (A, D, G, and J) and to RanBP9 (B, E, H, and K). Overlap in labeling is shown in yellow in the color overlays (C, F, I, and L). (A–C) At 96 h, the Rho-GEF domain of obscurin (A and C, green) has begun to organize into striations. RanBP9 (B and C, red) occurs diffusely in the cytoplasm, as well as in the nucleus. (D–F) At 120 h, the Rho-GEF domain of obscurin is organized at the level of M-bands and Z-disks or nearby I-bands (D and F, green) and RanBP9 is beginning to organize into primitive striations at the level of the M-band (E and F, red). (G–I) At 168 h, both the Rho-GEF domain of obscurin (G and I, green) and RanBP9 (H and I, red) are organized in structures at both the M- and I-band. Colocalization can be seen at both the level of the M- and I-band. (J–L) Three-fold enlargements of the myotubes studied at 168 h after plating myotubes (G–I) are shown. Colocalization is indicated with arrows (D–F). Note that the nuclear staining of RanBP9 is only present at early times, and not in more mature myotubes. Bar, 5 μm. (M) Nuclear labeling of RanBP9 was quantified in developing P1 rat myotubes and in C2C12 cells. A representative quantitation of the translocation from the nucleus to cytoplasm of RanBP9 in C2C12 cells is shown. The results confirm the presence of RanBP9 in the nucleus at early stages and in the myoplasm at later stages of differentiation. (N) Immunoblotting of protein extracts from rat myotubes in culture and adult skeletal muscle. Affinity purified rabbit antibodies to RanBP9108–729 recognize an ∼90-kDa protein in blots of proteins separated from extracts of primary myotubes, and adult skeletal muscle (left two lanes). Affinity purified rabbit antibodies to the Rho-GEF domain recognize a predominant ∼800-kDa band in blots of proteins from primary myotubes. Molecular masses are indicated.
Affinity-purified rabbit antibodies to RanBP9 (Figure 7, B, E, and H) selectively labeled myonuclei at early stages of development, but this localization was lost with time in culture. Quantitation of the nuclear localization of RanBP9 in cultures of developing skeletal myotubes (C2C12, e.g., Figure 7M; neonatal rat, data not shown) showed a decrease in nuclear localization with development, with 50% loss occurring at ∼150 h in culture. As myonuclear labeling for RanBP9 diminished, its presence at sarcomeres became clearly discernible (Figure 7, E and H). Sequestration of RanBP9 in the nucleus may therefore restrict its ability to influence the early stages of myofibrillogenesis.
Expression of RanBP9 and Obscurin in Skeletal Muscle
Immunoblots with antibodies to RanBP9 of extracts of skeletal muscle, as well as myotubes in culture, showed a major band with an apparent molecular mass of ∼90 kDa (Figure 7N, arrow), consistent with the mass of the protein predicted from its amino acid sequence. Lower amounts of smaller polypeptides were also detected. Because RanBP9 is readily proteolyzed (our unpublished data), these are probably proteolytic fragments of the protein. A slightly larger isoform was also present in low amounts (Figure 7N, asterisk), which may represent phosphorylated or otherwise modified RanBP9. We have previously determined that antibodies to the Rho-GEF domain recognize obscurin A and B in adult skeletal muscle (Bowman et al., 2007). To determine whether both the immunolabeling of the M-band and I-band in developing myotubes represented both isoforms, we performed Western blot experiments with antibodies to the Rho-GEF domain. Both the ∼800- and ∼900-kDa isoforms of obscurin (A and B) were detected, suggesting that both of these isoforms are expressed in developing muscle (Figure 7N), consistent with our previous localization of these isoforms at M-bands (obscurin A) and near the A/I junction (obscurin B).
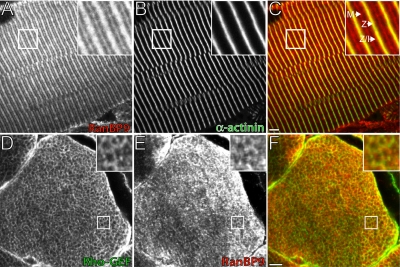
Because we used extracts of adult muscles in our immunoblotting studies, we also briefly investigated the localization of RanBP9 in quadriceps muscles of the adult rat. Consistent with our results with developing muscle cells, we observed RanBP9 at the level of M-bands (Figure 8, A and C, red) and in a single broad band overlying and flanking the Z-disks (Figure 8, A and C; Z-disks, yellow; Z/I junction, red). Careful comparison to labeling by antibodies to α-actinin, which marks the Z-disks (Figure 8, B and C, green), shown in an overlay image (Figure 8C), revealed that RanBP9 overlapped considerably with α-actinin at the Z-disk but also extended beyond the Z-disk into the Z-I junction (Figure 8C, arrows). Thus, RanBP9 colocalizes with α-actinin at Z-disks, but it is also present at the Z-I junctions and M-bands, where we have also localized the Rho-GEF domain of obscurin (Bowman et al., 2007). In cross-sections, RanBP9 and the Rho-GEF domain of obscurin also colocalized and occurred primarily in a reticular pattern (Figure 8, D–F), demonstrated previously for other C-terminal epitopes of obscurin (Kontrogianni-Konstantopoulos et al., 2003). Our results with adult skeletal muscle suggest that, like the Rho-GEF domain of obscurin, RanBP9 is present at M-bands, Z-disks, and Z-I junctions, primarily at the periphery of myofibrils.
Figure 8.
Localization of obscurin and RanBP9 in adult skeletal muscle. (A–C) RanBP9 (A and C, red) is present at the M-line and the Z-disk extending through the Z/I junction in quadriceps muscle, as determined by double immunostaining with antibodies to α-actinin (B and C, green). (D–F) Labeling of cross sections of quadriceps muscle with the guinea pig antibodies to the Rho-GEF domain of obscurin (D and F, green) and rabbit anti-RanBP9 (E and F, red) shows both proteins in a reticular pattern, suggesting that they surround myofibrils. Enlargements (top right corners [A–F]) indicate areas of colocalization. In all panels, inset images are threefold magnifications of the actual images. Bars, 5 μm.
Effects of Overexpressing RanBP9 on Developing Muscle
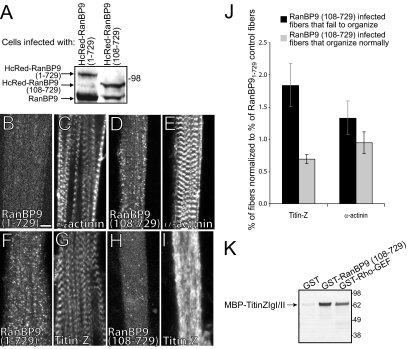
To determine whether RanBP9 could mediate the effects of the Rho-GEF domain of obscurin on myofibrillogenesis, we created adenoviral constructs encoding HcRed-RanBP9108-729, and HcRed-RanBP91-729 to infect myotubes in culture. We used HcRed-RanBP9108-729 as a dominant-negative construct, to learn whether it could exert an effect similar to the Rho-GEF domain, and HcRed-RanBP91-729, the full-length RanBP9, as control (HCRed alone seemed to be so highly expressed after adenoviral infection that it was toxic to the developing myotubes and thus could not be used as a control). Infected cells produced the fusion proteins at high levels, as shown by Western blotting (Figure 9A). Overexpression of the full-length RanBP91-729 did not disrupt the organization of either α-actinin (Figure 9C) or titin at the Z-disk (Figure 9G). By contrast, overexpression of the Rho-GEF binding fragment, RanBP9108-729, inhibited the organization of titin at the Z-disk (Figure 9I) without altering the organization of α-actinin there (Figure 9E). We scored these results as we did for overexpression of the Rho-GEF domain and confirmed that the effect of RanBP9108-729 on the assembly of titin at the Z-disk was specific and that the differences with RanBP91-729 were significant (p ≤ 0.04 for fibers that fail to organize; p = ≤0.004 for fibers that organize normally). The effects of overexpressing RanBP9108-729 were quantitatively similar to those obtained by overexpressing the Rho-GEF domain of obscurin (Figure 9J). These results suggest that the interaction between the Rho-GEF domain and RanBP9 specifically regulates the ability of the N-terminal region of titin to incorporate stably into the Z-disk of developing skeletal muscle.
Figure 9.
Overexpression of RanBP9108-729 disrupts the organization of titin at the Z-disk. (A) P1 rat myotubes were infected as above with adenovirus encoding HcRed-RanBP91-729 or HcRed-RanBP9108-729 and assayed 48 h later by immunoblotting with anti-RanBP9 antibodies. Endogenous RanBP9 (∼90 kDa) and the overexpressed forms of HcRed-RanBP9 (1–729 and 108–729, 108 and 96 kDa, respectively) are present at similar levels. Molecular masses are indicated. B-I. HcRed-RanBP91-729 has no effect on the organization of α-actinin (B and C) or titin (F and G) at the Z-disk. HcRed-RanBP9108-729 does not disrupt the ability of α-actinin (D and E) to organize normally at the Z-disk, but like GFP-Rho-GEF it disrupts the assembly of titin at Z-disks (H and I). (J) Quantitation of the effects of overexpressing RanBP9108-729, normalized to the effects of overexpressing RanBP91-729, shows selective disruption of titin organization at the Z-disk. The increase in fibers that fail to organize and the decrease in fibers that organize completely are both significant (p ≤ 0.04 and p ≤ 0.004, respectively). There was no significant effect on the organization of α-actinin in fibers that overexpressed HcRed-RanBP9108-729 (p > 0.3 and p > 0.8, respectively). (K) Equivalent amounts of bacterially expressed, affinity-purified GST, GST-RanBP9108-729, and GST-Rho-GEF were used in a pull-down experiment to assay association with purified MBP-TitinZIgI/II bound to amylose resin. Both GST fusion proteins, but not GST alone, bound directly to the ZIg domains of titin.
Interaction of the Rho-GEF Domain of Obscurin and RanBP9 with Titin
To investigate the mechanism by which the fusion proteins destabilize the association of the N-terminal region of titin with developing Z-disks, we tested their abilities to interact directly with the two N-terminal Ig domains of titin (ZIgI/II), which are located adjacent to the Z-disk (Gregorio et al., 1998). Glutathione resin bound to equivalent amounts of GST, GST-Rho-GEF, or GST-RanBP9108-729 was incubated with MBP-TitinZIgI/II and the bound proteins were analyzed by SDS-PAGE and immunoblotting. The N-terminal domains of titin bound to both the RanBP9108-729 fragment and the Rho-GEF domain of obscurin (Figure 9K), but not to GST alone. These results suggest that overexpression of these domains may affect titin's incorporation into Z-disks by binding directly to its N-terminal Ig domains.
DISCUSSION
Obscurin is the most recently discovered of three giant proteins that are thought to function as molecular templates in striated muscle and, like titin and nebulin, it has signaling domains that have the potential of linking contractile activity to the modulation of the structure, metabolism and gene expression of myofibers. Our results show that overexpression of the Rho-GEF domain of obscurin inhibits the incorporation of the N-terminal region of titin into the Z-disk and, to a lesser extent, disrupts the organization of the Z-disk and A/I junction. We also find that the Rho-GEF domain binds RanBP9 and that overexpression of the region of RanBP9 that binds obscurin's Rho-GEF domain, although not full-length RanBP9, has similar effects on the incorporation of the N-terminal region of titin into Z-disks. In developing myotubes, RanBP9 first concentrates in myonuclei and then translocates to the myoplasm, where it associates with the Rho-GEF domain of obscurin, already assembled in developing sarcomeres. Remarkably, both the Rho-GEF domain and its binding region on RanBP9 bind independently to the region of titin adjacent to the Z-disk. Our results suggest that the interaction between obscurin's Rho-GEF domain and RanBP9 modulate the association of titin with developing Z-disks as the contractile apparatus forms and matures.
Obscurin was shown previously to play a significant role in regulating the assembly of the A-band and M-band (Kontrogianni-Konstantopoulos et al., 2004, 2006b; Borisov et al., 2006). We were therefore surprised to find that its Rho-GEF domain interacted with N-terminal regions of titin, typically associated with the Z-disk, and that it had a selective effect on the association of this region of titin with developing Z-disks. The fact that RanBP9 associates with the same region of titin and overexpression of its binding region for obscurin's Rho-GEF domain has the same effect on the incorporation of titin into Z-disks, suggests that obscurin's Rho-GEF domain and RanBP9 are acting as a complex. The moderate binding affinity (1.9 μM) of RanBP9 and the Rho-GEF domain of obscurin suggests that their interaction might play a transient role in myotube development. The possible role in morphogenesis of small GTPases and their potential interactions with the Rho-GEF domain (Hall and Nobes, 2000; Schmidt and Hall, 2002; Jaffe and Hall, 2005), in the presence or absence of RanBP9 remain to be investigated.
RanBP9 was initially isolated as a binding partner of Ran-GTP, a signaling protein thought to play a role in nucleocytoplasmic transport, although it's unclear whether the full-length RanBP9 actually interacts with Ran-GTP (Mayans et al., 1998; Nakamura et al., 1998; Nishitani et al., 2001; Weis, 2003). RanBP9 is a 729-amino acid protein with a long stretch of prolines and glutamines at the N terminus, followed by an SPRY domain (found in the dual specificity kinase splA of Dictyostelium discoideum, and ryanodine receptors), a LisH domain (a Lissencephaly type-1-like homology motif), a CTLH domain (Rao et al., 2002), and a CRA motif, at the C terminus (Menon et al., 2004). Expressed in many tissues, RanBP9 is present at high levels in placenta, heart, and skeletal muscle (Wang et al., 2002). Characterized as a scaffolding protein based on the ability to interact with a variety of proteins in different subcellular locations, RanBP9 concentrates in the nucleus or the cytoplasm, depending upon the stage of the cell cycle (Jang et al., 2004), consistent with our results with developing muscle cells. RanBP9 has also been implicated in a variety of signaling cascades (reviewed in Murrin and Talbot, 2007). The association of RanBP9 with SOS promotes the phosphorylation of extracellular signal-regulated kinase (ERK) by Ras, and the downstream activation of mitogen-activated protein kinases (Wang et al., 2002). RanBP9 also binds Raf kinase directly (Johnson et al., 2006) and can modulate the ability of other proteins to phosphorylate ERK (Cheng et al., 2005). Recently, RanBP9 was found to be involved in sumoylation processes (Kabil et al., 2006). Thus, in addition to its interaction with the Rho-GEF domain of obscurin, RanBP9 is likely to interact with several important regulators in muscle cells.
Although RanBP9 is expressed at high levels in striated muscle, little is known about its subcellular distribution or function. Recent work has indicated that RanBP9 can inhibit the kinase and transcriptional coactivator activities of Mirk, which is known to activate factors critical for muscle differentiation, including myogenin, fast twitch troponin T, and muscle myosin heavy chain (Zou et al., 2003). Our results indicate that RanBP9 initially is found in myonuclei of newly formed myotubes but translocates to M- and I-band structures as these embryonic muscle cells develop in culture. In adult skeletal muscle, RanBP9 is found at the level of the I-band and Z-disk, as well as at the M-band (data not shown). The Rho-GEF domain of obscurin is present around mature myofibrils at several locations. Although our antibodies to the Rho-GEF domain do not clearly label the developing Z-disk, obscurin (or its fragments) has been localized there by several laboratories (Bang et al., 2001; Young et al., 2001; Kontrogianni-Konstantopoulos et al., 2003; Raeker et al., 2006; Arimura et al., 2007). Thus, it is possible that obscurin isoforms containing the Rho-GEF domain are present near the Z-disks of developing myofibrils but that the epitopes in the Rho-GEF domain are inaccessible to labeling by antibodies (Bowman et al., 2007). Alternatively, obscurin may associate with primordial Z-disk structures during development but dissociate from Z-disks once sarcomeres are fully formed.
Our morphological studies show that the Rho-GEF domain of obscurin and RanBP9 inhibit the incorporation of the N-terminal region of titin into developing Z-disks but does not alter the incorporation of α-actinin into Z-disks, or of titin into M-bands. Our ultrastructural studies, which show that Z-disks in myotubes that overexpress the Rho-GEF domain are present but are smaller than controls, is consistent with this observation and with the known role of the titin's N-terminal region in stabilizing Z-disks (Gregorio et al., 1998; Zou et al., 2006; Lee et al., 2006). Thus, the Rho-GEF domain of obscurin and RanBP9 can specifically regulate the incorporation of titin into Z-disks. Pull-down experiments clearly indicate that the Rho-GEF domain and RanBP9 bind directly to the N-terminal Ig domains of titin, which flank the Z-disk, suggesting that titin is unable to incorporate into Z-disks when its ZIg domains are bound to obscurin or RanBP9. Although we cannot rule out possible changes in signaling at this time, obscurin and RanBP9 are likely to alter titin's assembly into Z-disks by binding to its N-terminal region directly. This region of titin also binds telethonin/Tcap and disruption of this binding has pathological consequences (Gregorio et al., 1998) perhaps because it destabilizes Z-disks (Knoll et al., 2002; Lee et al., 2006; Pinotsis et al., 2006). Binding by the Rho-GEF domain of obscurin or RanBP9 may compete with binding of telethonin/Tcap, altering the association of titin with the Z-disk, consistent with the idea that Z-disk components are dynamic (Wang et al., 2005). Alternatively, full-length obscurin and RanBP9 may promote the early incorporation of titin into Z-disks, before telethonin/Tcap is expressed (Gregorio et al., 1998). Our results are most readily explained if, during development, the Rho-GEF domain binds to the N-terminal region of titin and promotes its incorporation into the Z-disk. As development proceeds and RanBP9 translocates from the nucleus to sarcomeric structures, it binds to the Rho-GEF domain of obscurin, the N-terminal Ig domains of titin, or both, to stabilize the preformed titin–obscurin complex.
It is perhaps paradoxical that the association of titin's N-terminal region with Z-disks is inhibited by the Rho-GEF domain of obscurin or by its binding domain on RanBP9, whereas the association of its C-terminal region in the M-band remains unaffected. Our results seem to be inconsistent with earlier models of myofibrillogenesis that propose that titin serves as a molecular template for key features of the sarcomere, beginning at the Z-disk through the I- and A-bands to the M-band, (Labeit and Kolmerer, 1995; Wang, 1996; van der Loop et al., 1996; Ehler et al., 1999). At the very least, our findings suggest that the stable association of titin with Z-disks is not a prerequisite for its stable association with M-bands and that M-bands can form during myofibrillogenesis even when titin is not stably incorporated into Z-disks. This is consistent with earlier results from our laboratory and others showing that the epitopes of titin that in adult muscle are separated by 0.5–1 μm are not clearly separated at early stages of myofibrillogenesis (Schultheiss et al., 1990; van der Ven and Furst, 1997; Kontrogianni-Konstantopoulos et al., 2006a). Thus, at the early stages of assembly of the contractile apparatus, titin may be anchored either at the Z-disk or at the M-band, but not both simultaneously. Our present results, as well as previous studies (Schultheiss et al., 1990; van der Ven and Furst, 1997), provide further data to support the theory that titin molecules assemble independently in primordial Z-disks and M-bands and align later in development. They confirm the need for a new model for titin's role in sarcomerogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. O'Neill, M. A. Borzok, W. Resneck and Drs. J. A. Ursitti, C. Antolik, R. Lovering, P. Reed, and D. W. Pumplin for useful discussions. This research was supported by National Institutes of Health grants NS-17282 and NS-43976 and the Muscular Dystrophy Association (to R.J.B.), National Institutes of Health grant R01 AR52768 and the Muscular Dystrophy Association (to A.K.K.), and by fellowships provided by training grants T32 GM-081818 (P.I., R.J.B.) and F30 NS048648 (to A.L.B.).
Abbreviations used:
- GEF
guanine nucleotide exchange factor
- PH
pleckstrin homology
- RanBP9
Ran binding protein 9.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0237) on June 25, 2008.
REFERENCES
- Amann K. J., Renley B. A., Ervasti J. M. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J. Biol. Chem. 1998;273:28419–28423. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- Arimura T., Matsumoto Y., Okazaki O., Hayashi T., Takahashi M., Inagaki N., Hinohara K., Ashizawa N., Yano K., Kimura A. Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2007;362:281–287. doi: 10.1016/j.bbrc.2007.07.183. [DOI] [PubMed] [Google Scholar]
- Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang M. L., et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Benian G. M., Tinley T. L., Tang X., Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov A. B., Raeker M. O., Kontrogianni-Konstantopoulos A., Yang K., Kurnit D. M., Bloch R. J., Russell M. W. Rapid response of cardiac obscurin gene cluster to aortic stenosis: differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem. Biophys. Res. Commun. 2003;310:910–918. doi: 10.1016/j.bbrc.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Borisov A. B., Sutter S. B., Kontrogianni-Konstantopoulos A., Bloch R. J., Westfall M. V., Russell M. W. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem. Cell Biol. 2006;125:227–238. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- Bowman A. L., Kontrogianni-Konstantopoulos A., Hirsch S. S., Geisler S. B., Gonzalez-Serratos H., Russell M. W., Bloch R. J. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan B. A., Li D., Wu X., Liu M. The Rho family of small GTPases: crucial regulators of skeletal myogenesis. Cell Mol. Life Sci. 2005;62:1547–1555. doi: 10.1007/s00018-005-5029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centner T., et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J. Mol. Biol. 2001;306:717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- Cheng L., Lemmon S., Lemmon V. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J. Neurochem. 2005;94:1102–1110. doi: 10.1111/j.1471-4159.2005.03254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Deyne P. G., O'Neill A., Resneck W. G., Dmytrenko G. M., Pumplin D. W., Bloch R. J. The vitronectin receptor associates with clathrin-coated membrane domains via the cytoplasmic domain of its beta5 subunit. J. Cell Sci. 1998;111:2729–2740. doi: 10.1242/jcs.111.18.2729. [DOI] [PubMed] [Google Scholar]
- Ehler E., Rothen B. M., Hammerle S. P., Komiyama M., Perriard J. C. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J. Cell Sci. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Frangioni J. V., Neel B. G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Prevec L. Methods for construction of adenovirus vectors. Mol. Biotechnol. 1995;3:207–220. doi: 10.1007/BF02789331. [DOI] [PubMed] [Google Scholar]
- Gregorio C. C., et al. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell Biol. 1998;143:1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove B. K., Kurer V., Lehner C., Doetschman T. C., Perriard J. C., Eppenberger H. M. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J. Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Nobes C. D. Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:965–970. doi: 10.1098/rstb.2000.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M. L. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima M., Sah V. P., Wang Y., Chien K. R., Brown J. H. The low molecular weight GTPase Rho regulates myofibril formation and organization in neonatal rat ventricular myocytes. Involvement of Rho kinase. J. Biol. Chem. 1998;273:7725–7730. doi: 10.1074/jbc.273.13.7725. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jang Y. J., Ji J. H., Ahn J. H., Hoe K. L., Won M., Im D. S., Chae S. K., Song S., Yoo H. S. Polo-box motif targets a centrosome regulator, RanGTPase. Biochem. Biophys. Res. Commun. 2004;325:257–264. doi: 10.1016/j.bbrc.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Johnson S. E., Winner D. G., Jr., Wang X. Ran binding protein 9 interacts with Raf kinase but does not contribute to downstream ERK1/2 activation in skeletal myoblasts. Biochem. Biophys. Res. Commun. 2006;340:409–416. doi: 10.1016/j.bbrc.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Kabil O., Zhou Y., Banerjee R. Human cystathionine beta-synthase is a target for sumoylation. Biochemistry. 2006;45:13528–13536. doi: 10.1021/bi0615644. [DOI] [PubMed] [Google Scholar]
- Knoll R., et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Bloch R. J. The hydrophilic domain of small ankyrin-1 interacts with the two N-terminal immunoglobulin domains of titin. J. Biol. Chem. 2003;278:3985–3991. doi: 10.1074/jbc.M209012200. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Bloch R. J. De novo myofibrillogenesis in C2C12 cells: evidence for the independent assembly of M bands and Z disks. Am. J. Physiol. Cell Physiol. 2006a;290:C626–C637. doi: 10.1152/ajpcell.00442.2005. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Randall W. R., Bloch R. J. Obscurin regulates the organization of myosin into A bands. Am. J. Physiol. Cell Physiol. 2004;287:C209–C217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Sutter S., Borisov A. B., Pumplin D. W., Russell M. W., Bloch R. J. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006b;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Jones E. M., Van Rossum D. B., Bloch R. J. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S., Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Lee E. H., Gao M., Pinotsis N., Wilmanns M., Schulten K. Mechanical strength of the titin Z1Z2-telethonin complex. Structure. 2006;14:497–509. doi: 10.1016/j.str.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Mayans O., van der Ven P. F., Wilm M., Mues A., Young P., Furst D. O., Wilmanns M., Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Menon R. P., Gibson T. J., Pastore A. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J. Mol. Biol. 2004;343:43–53. doi: 10.1016/j.jmb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Mercado-Pimentel M. E., Jordan N. C., Aisemberg G. O. Affinity purification of GST fusion proteins for immunohistochemical studies of gene expression. Protein Expr. Purif. 2002;26:260–265. doi: 10.1016/s1046-5928(02)00524-7. [DOI] [PubMed] [Google Scholar]
- Murrin L. C., Talbot J. N. RanBPM, a scaffolding protein in the immune and nervous systems. J. Neuroimmune Pharmacol. 2007;2:290–295. doi: 10.1007/s11481-007-9079-x. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Masuda H., Horii J., Kuma K., Yokoyama N., Ohba T., Nishitani H., Miyata T., Tanaka M., Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Hirose E., Uchimura Y., Nakamura M., Umeda M., Nishii K., Mori N., Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- Pinotsis N., Petoukhov M., Lange S., Svergun D., Zou P., Gautel M., Wilmanns M. Evidence for a dimeric assembly of two titin/telethonin complexes induced by the telethonin C-terminus. J. Struct. Biol. 2006;155:239–250. doi: 10.1016/j.jsb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Raeker M. O., Su F., Geisler S. B., Borisov A. B., Kontrogianni-Konstantopoulos A., Lyons S. E., Russell M. W. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev. Dyn. 2006;235:2018–2029. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- Rao M. A., Cheng H., Quayle A. N., Nishitani H., Nelson C. C., Rennie P. S. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J. Biol. Chem. 2002;277:48020–48027. doi: 10.1074/jbc.M209741200. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Schultheiss T., Lin Z. X., Lu M. H., Murray J., Fischman D. A., Weber K., Masaki T., Imamura M., Holtzer H. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J. Cell Biol. 1990;110:1159–1172. doi: 10.1083/jcb.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze H., Dose M., Korpal M., Meyer I., Italiano J. E., Jr., Shivdasani R. A. RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates non-centrosomal microtubules. J. Biol. Chem. 2008;283:14109–14119. doi: 10.1074/jbc.M709397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi R., Hattori A. Detection of giant myofibrillar proteins connectin and nebulin by electrophoresis in 2% polyacrylamide slab gels strengthened with agarose. Anal. Biochem. 1995;224:28–31. doi: 10.1006/abio.1995.1004. [DOI] [PubMed] [Google Scholar]
- Ursitti J. A., Lee P. C., Resneck W. G., McNally M. M., Bowman A. L., O'Neill A., Stone M. R., Bloch R. J. Cloning and characterization of cytokeratins 8 and 19 in adult rat striated muscle. Interaction with the dystrophin glycoprotein complex. J. Biol. Chem. 2004;279:41830–41838. doi: 10.1074/jbc.M400128200. [DOI] [PubMed] [Google Scholar]
- van der Loop F. T., van der Ven P. F., Furst D. O., Gautel M., van Eys G. J., Ramaekers F. C. Integration of titin into the sarcomeres of cultured differentiating human skeletal muscle cells. Eur. J. Cell Biol. 1996;69:301–307. [PubMed] [Google Scholar]
- van der Ven P. F., Furst D. O. Assembly of titin, myomesin and M-protein into the sarcomeric M band in differentiating human skeletal muscle cells in vitro. Cell Struct. Funct. 1997;22:163–171. doi: 10.1247/csf.22.163. [DOI] [PubMed] [Google Scholar]
- Wang D., Li Z., Messing E. M., Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- Wang D., Li Z., Schoen S. R., Messing E. M., Wu G. A novel MET-interacting protein shares high sequence similarity with RanBPM, but fails to stimulate MET-induced Ras/Erk signaling. Biochem. Biophys. Res. Commun. 2004;313:320–326. doi: 10.1016/j.bbrc.2003.11.124. [DOI] [PubMed] [Google Scholar]
- Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J. M., Sanger J. W. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskeleton. 2005;61:34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. Titin/connectin and nebulin: giant protein rulers of muscle structure and function. Adv. Biophys. 1996;33:123–134. [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Young P., Ehler E., Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P., Pinotsis N., Lange S., Song Y. H., Popov A., Mavridis I., Mayans O. M., Gautel M., Wilmanns M. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
- Zou Y., Lim S., Lee K., Deng X., Friedman E. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J. Biol. Chem. 2003;278:49573–49581. doi: 10.1074/jbc.M307556200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.