Abstract
The cellular activity of Yondelis (trabectedin, Ecteinascidin 743, Et743) is known to depend on transcription-coupled nucleotide excision repair (TCR). However, the subsequent cellular effects of Et743 are not fully understood. Here we show that Et743 induces both transcription- and replication-coupled DNA double-strand breaks (DSBs) that are detectible by neutral COMET assay and as γ-H2AX foci that colocalize with 53BP1, Mre11, Ser1981-pATM, and Thr68-pChk2. The transcription coupled-DSBs (TC-DSBs) induced by Et743 depended both on TCR and Mre11-Rad50-Nbs1 (MRN) and were associated with DNA-PK–dependent γ-H2AX foci. In contrast to DNA-PK, ATM phosphorylated H2AX both in NER-proficient and -deficient cells, but its full activation was dependent on H2AX as well as DNA-PK, suggesting a positive feedback loop: DNA-PK-γ-H2AX-ATM. Knocking-out H2AX or inactivating DNA-PK reduced Et743's antiproliferative activity, whereas ATM and MRN tended to act as survival factors. Our results highlight the interplays between ATM and DNA-PK and their impacts on H2AX phosphorylation and cell survival. They also suggest that γ-H2AX may serve as a biomarker in patients treated with Et743 and that molecular profiling of tumors for TCR, MRN, ATM, and DNA-PK might be useful to anticipate tumor response to Et743 treatment.
INTRODUCTION
Natural products are a rich source for medicinal drugs with highly specific mechanisms for targeting biological systems (Pommier and Cherfils, 2005). Ecteinascidin 743 (Yondelis, Et743) was purified from extracts of the marine tunicate Ecteinascidia turbinata in 1990 (Rinehart et al., 1990) and was selected as an anticancer agent because of its extraordinary activities in experimental models (Rinehart et al., 1990; Sakai et al., 1992; Guan et al., 1993; Izbicka et al., 1998; Valoti et al., 1998). Et743 has recently been approved in the European Union for the treatment of soft tissue sarcomas and is in clinical trials for ovarian, breast, and prostate cancers and for pediatric sarcomas. Et743 has been granted Orphan Drug designation from the European Commission (EC) and the US Food and Drug Administration (FDA) for soft tissue sarcomas and ovarian cancer (http://www.pharmamar.com/en/pipeline/yondelis.cfm).
The mechanism of action of Et743 is unique (for reviews see Verschraegen and Glover, 2001; Aune et al., 2002; D'Incalci and Jimeno, 2003; Fayette et al., 2006). Et743 alkylates the N2-position of guanines in the DNA minor groove, binding preferentially to guanines that are 5′ from another guanine or a cytosine (Pommier et al., 1996; Zewail-Foote and Hurley, 1999) and bending the DNA toward the major groove (Pommier et al., 1996; Zewail-Foote and Hurley, 1999). Transcription is a major target for Et743, which sets it apart from many approved anticancer drugs whose activity tends to be restricted to dividing cells. Not only does Et743 interfere directly with transcription in a variety of cellular systems (Minuzzo et al., 2000; Synold et al., 2001; Friedman et al., 2002), but it also targets the transcription-coupled nucleotide excision repair (TCR; Erba et al., 2001; Takebayashi et al., 2001b).
Under normal conditions, TCR repairs bulky DNA adducts (such UV photoproducts, cisplatin, and carcinogenic base adducts) in transcribing genes using a set of factors that belong to the XP family of nucleotide excision repair (NER) factors (for review see de Laat et al., 1999; Sancar et al., 2004; Sugasawa, 2008). Nucleotide excision repair in the rest of the genome (i.e., the nontranscribing genome) is initiated by another set of XP factors that are specific for global genome repair (GGR). XP stands for xeroderma pigmentosum, a rare genetic disease with eight complementation groups. Among the XP factors that are relevant to the present study, XPC (along with XPE, specific for GGR), or CSB (Cockayne syndrome B; along with CSA, specific for TCR) detect the lesion, together with additional factors. The TCR and GGR lesion recognition subpathways then converge toward a common set of XP factors involved in excising and repairing the damaged DNA. Unwinding of the damaged DNA is mediated by the TFIIH helicases, XPD and XPB, whose combined action creates short stretches of single-stranded DNA around the lesions, which facilitates the recruitment of XPA and RPA. The association of XPA with RPA generates a sensor that detects, simultaneously, backbone and base-pair distortion of DNA (Missura et al., 2001). XPG and XPF are the two nucleases that incise the adducted strand before repair synthesis (de Laat et al., 1999; Sancar et al., 2004; Sugasawa, 2008).
The Et743 adducts have been proposed to trap XPG-DNA complexes (Takebayashi et al., 2001b; Herrero et al., 2006) and prevent further processing, thereby generating DNA single-strand breaks (Takebayashi et al., 2001a). As a consequence of the specific poisoning of TCR by Et743, TCR-deficient cells are resistant to Et743 (Erba et al., 2001; Takebayashi et al., 2001b; Zewail-Foote et al., 2001), and cells selected for resistance to Et743 have been shown to be deficient for XPG (Takebayashi et al., 2001b).
The outcome of TCR poisoning by Et743 is the focus of the present study, in which we investigated the mechanisms of double-strand break (DSB) formation by Et743. Recent studies have shown that Et743 induces replication-dependent DSBs (RC-DSBs; Guirouilh-Barbat and Pommier, 2006; Soares et al., 2007) that are repaired by homologous recombination (Soares et al., 2007). The present study is a further characterization of Et743-induced RC-DSBs using histone H2AX phosphorylated on serine 139 [γ-H2AX] as a marker of DSBs (Redon et al., 2002). We also demonstrate that Et743 induces transcription-coupled DSBs (TC-DSBs), which are replication-independent but are dependent on TCR and the Mre11-Rad50-Nbs1 (MRN) complex. We examined the role of ATM (ataxia telangiectasia–mutated gene) and DNA-PK, two protein kinases implicated in genome integrity and DNA damage sensing, in the responses to both RC-DSBs and TC-DSBs.
MATERIALS AND METHODS
Cells
All cell lines were maintained in DMEM containing 10% fetal calf serum. XPD and their stably complemented counterparts XPD-c were provided by Dr. Kenneth Kraemer (Basic Research Laboratory, National Institutes of Health, Bethesda, MD). Normal human fibroblasts (GM00637), XPA, XPC, XPF, and CSB cells were obtained from the Coriell cell repository (Camden, NJ). Colon carcinoma cell lines HCT116 and HT29 were obtained from the Developmental Therapeutics Program (National Cancer Institute, NIH). HCT116 cells complemented for Mre11 (HCT116-Mre11) were established in our laboratory (Takemura et al., 2006). Nbs1-deficient cells, NBS-1LBI, K1 and xrs6 (wild-type CHO and CHO deficient for KU80, respectively) were provided by Dr. Bernard S. Lopez (CEA, Fontenay aux Roses, France). Mre11-deficient ATLD2 and the counterpart complemented for wild-type Mre11 (ATLD2-Mre11) or for the nuclease inactive version of Mre11 (ATLD2-Mre11-3; Stracker et al., 2002; Uziel et al., 2003) were provided by Dr. Yosef Shiloh (Sackler School of Medicine, Tel Aviv, Israel) as well as the ATM-deficient cells and the counterparts complemented with ATM (pEBS7 and pEBS7-YZ5, respectively; Ziv et al., 1997). Mouse embryonic fibroblasts (MEFs) lacking H2AX (H2AX-KO) and their wild-type counterparts (H2AX-WT; Celeste et al., 2002) were obtained from Dr. William Bonner (Laboratory of Molecular Pharmacology, NIH, Bethesda, MD). The human peripheral lymphocytes were obtained from the Blood Bank at the NIH and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum.
Drugs and Antibodies
Et743 was a kind gift from Pharmamar (Madrid, Spain). Ten millimolar stock solutions in DMSO were stored at −20°C. Aphidicolin (APD) and DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) were purchased from Sigma Chemical Co. (St. Louis, MO). The ATM inhibitor (KU55933; Hickson et al., 2004) and the DNA-PK inhibitor (KU57788; Leahy et al., 2004) were obtained from Kudos Pharmaceuticals (Cambridge, United Kingdom). Two anti-γ-H2AX antibodies were used: a mouse mAb purchased from Upstate Biotechnology (Lake Placid, NY) and a rabbit polyclonal antibody obtained from Dr. William Bonner (Laboratory of Molecular Pharmacology, NIH). The mouse monoclonal anti-XPF and the mouse monoclonal anti-Ser1981-pATM antibodies were purchased from Neomarkers (Fremont, CA) and Cell Signaling (Danvers, MA), respectively. The rabbit polyclonal anti-Mre11 and the rabbit polyclonal anti-53BP1 were purchased from Novus Biologicals (Littleton, CO).
Immunocytochemistry and Confocal Microscopy
Cells used for microscopy studies were grown 1 day before drug treatment in Nunc chamber slides (Nalgene, Rochester, NY). After treatment, the medium was aspirated out, and cells were washed in phosphate-buffered saline (PBS). For XPF and Mre11 staining, soluble proteins were extracted with a 5-min incubation in 0.5% NP-40 on ice before fixation with 2% paraformaldehyde for 20 min at room temperature. Cells were then permeabilized with a 5-min incubation on ice in 0.5% Triton X-100. Otherwise, cells were immediately fixed and permeabilized by a 20-min incubation at room temperature with 2% paraformaldehyde and a 5-min incubation at room temperature with prechilled 70% ethanol. To block nonspecific binding, cells were incubated with 8% bovine serum albumin for 1 h at room temperature. Cells were stained for 1 h 30 min with the primary antibodies and tagged for 45 min with fluorescent secondary antibodies (Alexa-488 or Alexa-568, Molecular Probes, Carlsbad, CA, 1/800). All incubations were made in 1% bovine serum albumin at room temperature. The primary antibodies were diluted as follows: mouse γ-H2AX, 1/1000; rabbit γ-H2AX, 1/800; 53BP1: 1/500, XPF, 1/100; Mre11, 1/200; and Ser1981-pATM, 1/250. Nuclei were stained with propidium iodide (0.05 mg/ml) and RNase A (0.5 mg/ml) for 10 min at 37°C. Slides were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and visualized using a Nikon Eclipse TE-300 confocal laser scanning microscope system (Nikon, Inc., Augusta, GA). For each sample in each experiment, 50–200 cells were scored. Positive cells were defined as containing three or more nuclear foci. Quantification of staining intensity was done with Adobe Photoshop 7.0 and normalized to the number of analyzed cells. In every experiment, the intensity measured in the wild type or the complemented cells treated for 6 h with Et743 without inhibitors was taken as 1.
For the simultaneous detection of γ-H2AX and IdU, cells were treated with Et743 for 1 h and pulse-labeled with 100 mM IdU for the last 30 min. After treatment, the medium was aspirated out, and cells were washed in PBS. Cells were immediately fixed with 2% paraformaldehyde for 20 min at room temperature and permeabilized by a 5-min incubation at room temperature in prechilled 70% ethanol. We first performed the staining for γ-H2AX as described above. After the final PBS wash, the cells were again fixed with 4% paraformaldehyde for 5 min, followed by a 10-min incubation with 1.5 M HCl at 37°C to denature the DNA. Cells were washed again, incubated with 0.5% Tween 20 in PBS for 5 min, and incubated with normal goat serum for 20 min. IdU primary antibody (mouse anti-BrdU; BD Biosciences, San Jose, CA) was diluted in blocking buffer and incubated for 2 h. Cells were washed and incubated with a secondary fluorescent anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA) for 1 h, washed, and mounted by using Vectashield mounting medium. Images were visualized by using a Nikon Eclipse TE-300 confocal microscope.
Western Blotting
Cells were lysed at 4°C in buffer containing 1% SDS, 1 mM sodium vanadate, 10 mM Tris-HCl, pH 7.4, supplemented with protease inhibitors (Complete; Roche Diagnostics, Basel, Switzerland) and phosphatase inhibitors (Sigma). Viscosity of the samples was reduced by brief sonication, and 30 μg of protein was incubated in loading buffer (125 mM Tris-HCl, pH 6.8, 10% β-mercaptoethanol, 4.6% SDS, 20% glycerol, and 0.003% bromophenol blue), separated by SDS-polyacrylamide gel, and transferred to polyvinylidene difluoride membrane (Immobilon-P, Millipore, Bedford, MA). After blocking nonspecific binding sites for 1 h by 5% milk in PBS-T (PBS, Tween 20 0.5%), the membranes were incubated for 1 h with primary mouse anti-γ-H2AX antibody (1/2000). After three washes in PBS-T, the membrane was incubated with horseradish peroxidase–conjugated goat anti-mouse antibody (Amersham Biosciences, Amersham, Buckinghamshire, United Kingdom, 1/5000) for 1 h and then washed three times in PBS-T. Immunoblot was revealed using enhanced chemiluminescence detection kit (Pierce, Rockford, IL) by autoradiography.
COMET Assays
The neutral COMET assay (single-cell gel electrophoresis assay) was performed according to the manufacturer's instructions (Trevigen, Gaithersburg, MD) except that the electrophoresis was performed at 4°C. The quantification of tail length was made with Adobe Photoshop 7.0 measuring the number of pixels corresponding to the tail of the COMET.
Cell Survival (MTT) Assays
Cells were seeded on day 0 at a density of 1000 per well in 96-well microtiter plates. On day 1, Et743 was added and incubation was continued for 72 h. After 72 h, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to each well (0.5 mg/ml; Sigma Aldrich), and plates were maintained at 37°C for 4 h. The medium was then discarded, and DMSO was added to each well to lyse the cells. Absorbance was measured at 450 nm using a multiwell spectrophotometer (Emax, Molecular Devices, Sunnyvale, CA).
RESULTS
Et743 Induces DSBs That Rely on Functional TCR
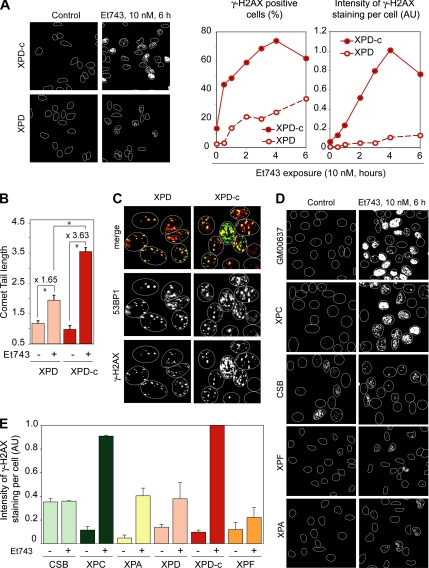
To investigate whether Et743 would induce DSBs in a TCR-dependent manner, we treated NER-deficient XPD cells and their NER-proficient complemented counterparts (XPD-c) with Et743 and analyzed the formation of γ-H2AX foci, a marker for DNA damage and DNA DSBs (Redon et al., 2002). Although Et743 induced γ-H2AX foci in both XPD and XPD-c cells, the induction was far greater in XPD-c than in XPD cells (Figure 1A). The percentage of positive cells that reached ∼70% in XPD-c cells was ∼25% in XPD cells (Figure 1A). Also, γ-H2AX–positive cells appeared at early times in XPD-c cells, whereas the induction was slower in XPD cells (Figure 1A). Western blotting analyses showed the lower phosphorylation of H2AX in XPD compared with XPD-c cells and confirmed the immunofluorescence microscopy results (Figure S1).
Figure 1.
Et743 induces DNA double-strand breaks TCR-dependent DSBs. (A–C) Reduced DSBs induction in XPD cells after Et743 treatment. (A) Left panels, representative pictures of γ-H2AX staining in XPD and XPD-c cells treated with 10 nM Et743 for 6 h. Right panels, percentage of γ-H2AX–positive cells (left plot) and intensity of γ-H2AX staining normalized to the number of cells analyzed (right plot; AU, arbitrary unit; see Materials and Methods) in XPD and XPD-c cells treated with 10 nM Et743 for the indicated times. (B) Lower induction of DSBs in XPD cells, measured by neutral COMET assays. XPD and XPD-c cells were treated with 10 nM Et743 for 6 h. Quantification of tail length (mean ± SD, 12–23 cells were analyzed per sample; *p <10−3, Student's t test). (C) XPD and XPD-c cells were treated with 10 nM Et743 for 6 h and costained for 53BP1 and γ-H2AX. (D and E) Reduced formation of γ-H2AX foci in CSB, XPA, XPD, and XPF cells compared with XPC, XPD complemented XPD-c cells, and wild-type GM00637 cells. (D) Representative pictures. (E) Intensity of γ-H2AX staining normalized to the number of cells analyzed (mean ± SD; n = 2–4; AU, arbitrary units; see Materials and Methods).
To investigate whether the differential induction of γ-H2AX foci in NER-proficient and -deficient cells was the hallmark of a differential induction of DSBs, we performed neutral COMET assays. As shown on Figure 1B, Et743 induced significantly more DSBs in NER-proficient XPD-c cells than in NER-deficient XPD cells. In parallel, we tested another marker of DSBs, 53BP1 (Schultz et al., 2000), and observed that Et743 induced 53BP1 foci with greater frequency in XPD-c cells than in XPD cells (Figure 1C). Moreover, 53BP1 colocalized with γ-H2AX, validating the use of γ-H2AX as a marker of DSBs in cells treated with Et743.
To confirm that Et743 induced γ-H2AX foci in a NER-dependent manner, we used two other NER-deficient fibroblast cell lines, XPA and XPF, and compared them to normal human fibroblasts, GM00637. Figure 1D shows reduced induction of γ-H2AX in the XPA- and XPF-deficient fibroblasts. In addition, γ-H2AX staining was consistently lower in the XPD, XPA, and XPF cells than in the XPD-c and GM00637 cells (Figure 1, A and D). Because XPD, XPA, and XPF are implicated in the common NER pathway elicited both by GGR and TCR, we wanted to determine whether the induction of γ-H2AX by Et743 was dependent on TCR or/and GGR. To that effect, we compared the γ-H2AX responses in TCR-deficient CSB fibroblasts and GGR-deficient XPC fibroblasts. Representative pictures (Figure 1D) and quantification of several independent experiments (Figure 1E) showed that Et743 did not induce more γ-H2AX in CSB cells than the basal level that was relatively high in those cells. By contrast, the induction of γ-H2AX in XPC cells treated with Et743 was very similar to the one observed in two NER-proficient cell lines, the XPD-complemented XPD-c cells and the wild-type fibroblasts GM00637 (Figure 1, D and E).
From these results we conclude that Et743 induces more DSBs in TCR-proficient than in TCR-deficient cells and that at least a fraction of Et743-induced DSBs are TCR-dependent.
The TCR-dependent DSBs Induced by Et743 Are Transcription-dependent
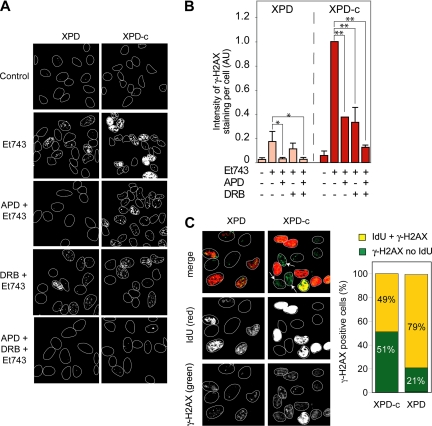
Next we investigated the contribution of DNA replication in the differential induction of γ-H2AX foci in XPD and XPD-c cells. For this purpose, we treated XPD and XPD-c cells with the DNA polymerase inhibitor, aphidicolin, which has been shown to effectively prevent the formation of replication-mediated DSBs (Furuta et al., 2003). Aphidicolin completely inhibited the appearance of γ-H2AX foci in XPD cells (Figure 2, A and B), which shows that in NER-deficient cells, γ-H2AX foci are primarily dependent on replication. In contrast, as shown on the representative pictures in Figure 2A, aphidicolin only partially inhibited γ-H2AX formation in XPD-c cells. The quantitation of several independent experiments (Figure 2B) showed a 62% reduction of Et743-induced γ-H2AX in XPD-c cells treated with aphidicolin. These results demonstrate that, in XPD-c cells, only a fraction of the γ-H2AX foci is dependent on replication. Thus, we conclude that, in NER-proficient cells, a fraction of the total γ-H2AX foci is replication-independent.
Figure 2.
NER-related DSBs are transcription-dependent. (A and B) Replication inhibition by aphidicolin (APD) is sufficient to inhibit γ-H2AX foci formation in XPD cells. However, concomitant inhibition of transcription (DRB) is necessary in XPD-c cells. XPD and XPD-c cells were preincubated with APD (1 μM, 15 min) and/or DRB (50 μM, 1 h) before the addition of 10 nM Et743 for the next 6 h. (A) Representative pictures of γ-H2AX foci in XPD and XPD-c cells. (B) Quantification of γ-H2AX staining intensity normalized to the number of cells analyzed (mean ± SD, n = 3; AU, arbitrary unit; see Materials and Methods, *p ≤ 0.05, **p ≤ 0.01, Student's t test). (C) Presence of γ-H2AX foci in XPD-c cells that do not incorporate iododeoxyuridine (IdU). XPD and XPD-c cells were treated with 10 nM Et743 for 1 h and pulse-labeled with 100 mM IdU for 30 min. Left panels, representative pictures. Right panel, quantitation of the number of γ-H2AX–positive cells that were negative or positive for IdU incorporation (n = 100 cells counted for XPD-c and n = 33 cells counted for XPD).
To confirm the occurrence of replication-independent γ-H2AX foci specifically in TCR-proficient cells, we pulse-labeled XPD and XPD-c cells with iododeoxyuridine (IdU), which is incorporated in replication foci selectively in cells going through S-phase (Conti et al., 2007; Seiler et al., 2007), and performed a costaining for γ-H2AX after treatment with Et743. As shown in Figure 2C, most (79%) of the XPD cells positive for γ-H2AX were also positive for IdU, which confirms that in NER-deficient cells, γ-H2AX foci are prominently in S-phase cells. In contrast, in XPD-c cells, approximately half of the cells positive for γ-H2AX showed no detectable IdU incorporation (indicated by arrows on the representative pictures on Figure 2C). In fact, among the XPD-c cells positive for γ-H2AX, only half of them were also positive for IdU (Figure 2C). Thus, in NER-proficient cells, a significant fraction of the Et743-induced γ-H2AX foci occurs independently of replication.
Because Et743 is known for its strong impact on transcription and specific poisoning of TCR (see Introduction), we tested the effect of the transcription inhibitor DRB on the induction of γ-H2AX foci by Et743. DRB had no effect on the induction of γ-H2AX foci by Et743 in XPD cells (Figure 2, A and B). By contrast, DRB inhibited ∼60% of γ-H2AX foci formation in XPD-c cells (Figure 2, A and B). Thus, in NER-proficient cells, a fraction of the γ-H2AX foci are transcription-dependent. Finally, the combination of aphidicolin and DRB resulted in the almost complete disappearance of γ-H2AX staining in XPD-c cells (Figure 2, A and B), indicating that, in those cells, Et743 induces both replication- and transcription-dependent DSBs.
Altogether our results show that Et743 induces TCR-dependent transcription-coupled γ-H2AX foci, which are independent from NER, and also replication-dependent γ-H2AX foci, which are independent of NER.
Induction of γ-H2AX foci by Et743 in Nonproliferative Human Lymphocytes and Colocalization of γ-H2AX with 53BP1, Mre11, and XPF
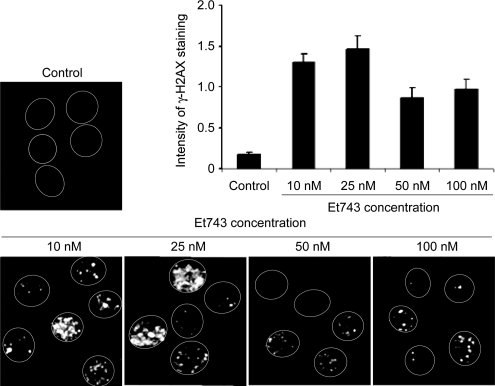
To gain further evidence that replication was not absolutely required for the induction of γ-H2AX foci by Et743, we exposed unstimulated normal human peripheral lymphocytes to various concentrations of Et743 for 3 h. Figure 3 shows that Et743 induced γ-H2AX foci in those nonreplicating cells at concentrations as low as 10 nM. Increasing drug concentration produced no further increase in the γ-H2AX signal in those cells.
Figure 3.
Induction of γ-H2AX foci by Et743 in nonproliferating human lymphocytes. Lymphocytes were treated with the indicated concentrations of Et743 for 3 h and analyzed for γ-H2AX foci induction by immunofluorescence. The histogram represents the quantitation of the intensity per cell measured on 80–100 cells (mean ± SD).
Because the NER pathway only induces DNA single-strand breaks (de Laat et al., 1999), our results raised the question of which factors were involved in the production of the TCR-dependent DSBs observed in response to Et743. We investigated Mre11 because of its known nuclease activities (D'Amours and Jackson, 2002) and first analyzed whether Et743 induced Mre11 foci that would colocalize with DSBs and/or NER proteins.
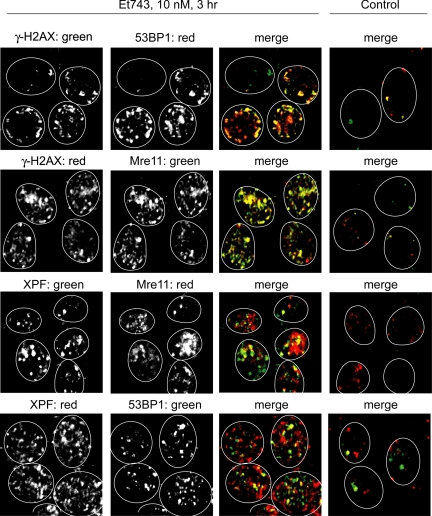
To visualize only the replication-independent γ-H2AX foci and avoid the normal Mre11 staining at replication forks (Mirzoeva and Petrini, 2003), we performed those experiments in unstimulated human lymphocytes. We first confirmed that the γ-H2AX foci induced by Et743 were consistent with the production of DSBs by staining the cells simultaneously with two markers of DSBs, γ-H2AX and 53BP1. As shown in Figure 4, both γ-H2AX and 53BP1 foci colocalized, confirming the occurrence of replication-independent DSBs in normal cells treated with Et743. Staining for Mre11 (Figure 4) showed the formation of Mre11 foci that also colocalized with γ-H2AX. This result is consistent with prior studies showing that Mre11 colocalizes with γ-H2AX at DSBs sites (Kobayashi et al., 2004). To investigate the potential direct implication of NER and Mre11 in the DSBs induced by Et743, we tested whether the Mre11 foci and 53BP1 foci colocalized with the NER protein XPF. As shown in Figure 4, Et743 induced the formation of XPF foci that partially colocalized with Mre11 and 53BP1. Together these results provide further evidence that Et743 induces NER-dependent DSBs and that Mre11 is present at those DSBs sites in nonreplicating cells.
Figure 4.
γ-H2AX foci induced by Et743 in nonproliferating human lymphocytes colocalize with 53BP1 and Mre11 foci and colocalize partially with XPF. Representative pictures of human peripheral lymphocytes treated with 10 nM Et743 for 3 h. Top panels, colocalization (yellow on the merge image) of γ-H2AX (green) and 53BP1 (red) foci. Top middle panels, colocalization (yellow on the merge image) of γ-H2AX (red) and Mre11 (green) foci. Bottom middle panels, partial colocalization (yellow on the merge image) of XPF (green) and Mre11 (red) foci. Bottom panels, partial colocalization (yellow on the merge image) of XPF (red) and 53BP1 (green) foci.
Dependency of Et743-induced DSBs on Mre11-Rad50-Nbs1
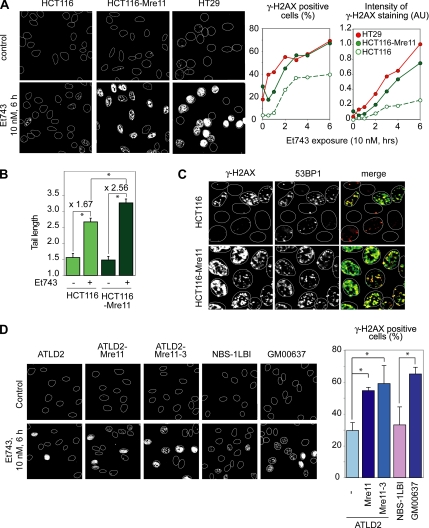
To determine the functional impact of Mre11 in Et743-induced DSBs, we analyzed the induction of γ-H2AX foci after Et743 treatment in three cell lines with different Mre11-Rad50-Nbs1 (MRN) expression: HCT116 cells, which are deficient for Mre11 (Giannini et al., 2002; Furuta et al., 2003), their counterpart complemented for Mre11, HCT116-Mre11 (Takemura et al., 2006), and another colon carcinoma cell line, HT29, wild-type for Mre11 (Giannini et al., 2002; Takemura et al., 2006). Representative pictures and quantitation of a time-course experiment (Figure 5A) showed that the two Mre11-proficient cell lines (HCT116-Mre11 and HT29) exhibited a faster and more intense induction of γ-H2AX than the Mre11-deficient HCT116 cells. The differential induction of γ-H2AX in HCT116-Mre11 and HCT116 was confirmed by Western blotting (Figure S1). Accordingly, neutral COMET assays and examination of 53BP1 foci showed greater induction of DSBs in HCT116-Mre11 cells compared with HCT116 cells (Figure 5, B and C). Again, the 53BP1 foci colocalized with the γ-H2AX foci in those cells. Together, these experiments demonstrate that Et743-induced DSBs are partially dependent on Mre11.
Figure 5.
The Mre11-Rad50-Nbs1 (MRN) complex is involved in the formation of DSBs and γ-H2AX foci after Et743 treatment. (A) HCT116 cells complemented for Mre11 (HCT116-Mre11) and HT29 (wild-type for Mre11) produce higher levels of γ-H2AX foci than Mre11-deficient HCT116 cells. Left panels, representative pictures of cells treated for 6 h with 10 nM Et743. Right panels, quantification of the percentage of γ-H2AX–positive cells (left plot) and of the intensity of the γ-H2AX staining normalized to the number of cells (right panel; AU, arbitrary unit; see Materials and Methods) in cells treated with 10 nM Et743 for the indicated times. (B) Increased induction of DSBs in HCT116 cells complemented for Mre11 (HCT116-Mre11) measured by neutral COMET assays. HCT116 and HCT116-Mre11 were treated with 10 nM Et743 for 6 h. Quantification of tail length (mean ± SD; 15–18 cells scored per sample; *p < 0.005, Student's t test). (C) The attenuated induction of γ-H2AX foci in HCT116 cells is coupled to the reduced formation of 53BP1 foci. HCT116 and HCT116-Mre11 cells were treated with 10 nM Et743 for 6 h and costained for 53BP1 (red) and γ-H2AX (green). (D) The MRN complex is involved in the creation of Et743 induced γ-H2AX foci, independently of Mre11 nuclease activities. Left panels, representative pictures of Mre11-deficient ATLD2 cells, their complemented counterparts ATLD2-Mre11, and their counterpart complemented with the nuclease inactive version of Mre11, Mre11-3 (Stracker et al., 2002; Arthur et al., 2004), Nbs1-deficient cells (NBS-1LBI) and wild-type fibroblasts GM00637. Right panel, quantification of the percentage of γ-H2AX–positive cells in response to 10 nM Et743 for 6 h (mean ± SD; n = 2–3; *p ≤ 0.05)
To confirm the implication of Mre11 in Et743-induced DSBs, we tested other cell lines deficient for the components of the MRN complex. The induction of γ-H2AX after Et743 treatment was attenuated in Mre11-deficient ATLD2 cells when compared with their Mre11-complemented counterpart ATLD2-Mre11 (Figure 5D). Because Nbs1 is an essential molecular partner for Mre11, we examined the γ-H2AX response of Nijmegen-breakage syndrome fibroblasts (NBS-1LBI) and compared them with wild-type fibroblasts, GM00637. Figure 5D shows reduced induction of γ-H2AX in NBS-1LBI compared with GM00637 fibroblasts.
To determine whether the Mre11 nuclease activities (for review see D'Amours and Jackson, 2002) were required for γ-H2AX and DSBs induction in response to Et743, we tested Mre11-deficient ATLD2 cells complemented with a nuclease-deficient mutant of Mre11, Mre11-3 (H129D/L130V; Stracker et al., 2002; Arthur et al., 2004). Figure 5D shows that those Mre11-nuclease–deficient cells were as proficient as the cells complemented with wild-type Mre11 for inducing full γ-H2AX response to Et743 (Figure 5D). Together, these results indicate that the MRN complex contributes to the production of DSBs and γ-H2AX response after Et743 treatment. However that contribution appears to be independent from Mre11's endonuclease activities.
The MRN Complex Is Implicated in the TC-DSBs Induced by Et743
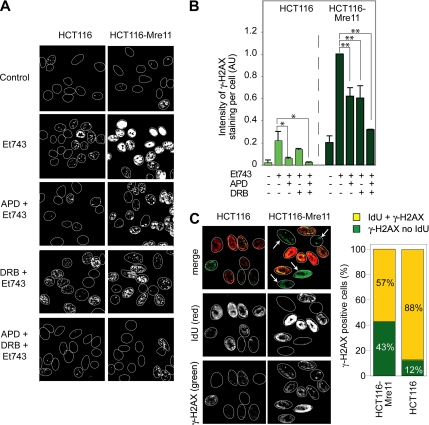
To investigate whether Mre11 is specifically involved in the TC-DSBs induced by Et743, we treated the Mre11-deficient (wild-type) HCT116 cells and their isogenic complemented counterpart (HCT116-Mre11) with Et743 in combination with aphidicolin and/or DRB.
Aphidicolin inhibited almost completely γ-H2AX induction in HCT116 cells (Figure 6, A and B), indicating that Mre11 is not implicated in the induction of replication-dependent DSBs by Et743. In contrast, in the Mre11-complemented HCT116-Mre11 cells, aphidicolin only had a partial effect (Figure 6, A and B), suggesting that in Mre11-proficient cells a fraction of the γ-H2AX foci is replication-independent. To gain further evidence that the Mre11-dependent γ-H2AX foci were formed independently of replication, we used the IdU pulse-labeling approach previously described in Figure 2C to identify replicating versus nonreplicating cells. In the Mre11-deficient HCT116 cells, almost all the γ-H2AX–positive cells (88%) were also positive for IdU (Figure 6C). By contrast, in the Mre11-complemented HCT116-Mre11 cells, a significant fraction of cells positive for γ-H2AX were negative for IdU (indicated by arrows on the representative pictures on Figure 6C). Quantitation of those results shows that, upon Mre11 complementation, 43% of the γ-H2AX–positive cells were negative for IdU (i.e., nonreplicating; Figure 6C). These results suggest that Mre11 enables the formation of the replication-independent DSBs induced by Et743.
Figure 6.
MRN-related DSBs are transcription-dependent. (A and B) Replication inhibition by aphidicolin (APD) was sufficient to inhibit γ-H2AX foci formation in HCT116 cells. However concomitant inhibition of transcription (DRB) was necessary in HCT116-Mre11 cells. HCT116 and HCT116-Mre11 cells were preincubated with APD (1 μM, 15 min) and/or DRB (50 μM, 1 h) before the addition of 10 nM Et743 for the next 6 h. (A) Representative pictures of γ-H2AX foci in HCT116 and HCT116-Mre11 cells. (B) Quantification of γ-H2AX staining intensity normalized to the number of cells analyzed (mean ± SD; n = 3; AU, arbitrary units; see Materials and Methods, *p ≤ 0.05, **p ≤ 0.01, Student's t test). (C) γ-H2AX foci were present in HCT116-Mre11 cells that did not incorporate iododeoxyuridine (IdU). HCT116 and HCT116-Mre11 cells were treated with 10 nM Et743 for 1 h and pulse-labeled with 100 mM IdU for 30 min. Left panels, representative pictures. Right panel, quantitation of the percentage of γ-H2AX–positive cells that were negative or positive for IdU incorporation (n = 77 cells counted for HCT116-Mre11 and n = 48 cells counted for HCT116).
We then investigated the implication of transcription in the production of those replication-independent γ-H2AX foci. The transcription inhibitor, DRB decreased the occurrence of γ-H2AX foci by 40% in the Mre11-complemented HCT116-Mre11 cells, whereas it had no significant effect in the Mre11-deficient HCT116 cells (Figure 6, A and B). These experiments indicate that Mre11 is implicated in the formation of transcription-dependent Et743-induced DSBs. In addition, the combination of aphidicolin and DRB resulted in the almost complete inhibition of γ-H2AX foci formation in HCT116-Mre11 treated with Et743 (Figure 6, A and B).
Together, the use of NER- and MRN-proficient/deficient cell lines coupled with inhibitors of replication and transcription reveal two mechanisms leading to DSBs after Et743 treatment. The first mechanism is replication-dependent and NER- and MRN-independent, whereas the second one is replication-independent, and transcription-, TCR-, and MRN-dependent.
Both ATM and DNA-PK Phosphorylate H2AX in Response to Et743
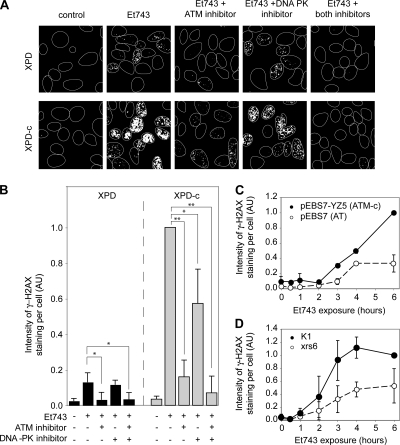
To identify the kinases leading to the phosphorylation of H2AX after Et743 treatment, we first used the specific inhibitors of ATM (KU55933; Hickson et al., 2004) and DNA-PK (KU57788; Leahy et al., 2004). Figures 7, A and B, show that the ATM inhibitor almost completely prevented the phosphorylation of H2AX in XPD cells and reduced γ-H2AX by ∼80% in XPD-c cells. Accordingly, experiments in AT cells (pEBS7) and their complemented counterpart (pEBS7-YZ5; Ziv et al., 1997) showed impaired formation of γ-H2AX in the ATM-deficient cells (Figure 7C). In those cells, ATM deficiency led to a decrease of γ-H2AX staining by 67% for a 6-h treatment with Et743, which is consistent with the decrease observed with the ATM inhibitor in XPD-c cells (see Figure 7B).
Figure 7.
Both ATM and DNA-PK phosphorylate H2AX in response to Et743 treatment. (A and B) Inhibition of Et743-induced formation of γ-H2AX foci by ATM- and DNA-PK-inhibitors (KU55933 and KU57788, respectively). XPD and XPD-c cells were preincubated with the ATM inhibitor (KU55933, 50 μM) and/or the DNA-PK inhibitor (KU57788, 20 μM) for 1 h before the addition of 10 nM Et743 for the next 6 h. (A) Representative pictures of γ-H2AX staining. (B) Quantification of the γ-H2AX staining intensity normalized to the number of cells analyzed (mean ± SD; n = 4; AU, arbitrary units; see Materials and Methods; *p ≤ 0.05, **p ≤ 0.01, Student's t test). (C) Reduced formation of γ-H2AX foci in the ATM-deficient cells (pEBS7) compared with their isogenic complemented counterparts (pEBS7-YZ5). Quantification of the intensity of the γ-H2AX staining normalized to the number of cells analyzed (mean ± SD, n = 3, AU, arbitrary units; see Materials and Methods). (D) Reduced γ-H2AX foci formation in DNA-PK–deficient xrs6 cells, compared with their wild-type counterpart (K1 cells). Quantification of the γ-H2AX staining intensity normalized to the number of analyzed cells (mean ± SD; n = 3; AU, arbitrary units; see Materials and Methods).
Noticeably, although the DNA-PK inhibitor had no effect in the XPD cells, it decreased the γ-H2AX signal by almost half in XPD-c cells (Figure 7, A and B). That result suggests that DNA-PK is primarily implicated in the TCR-dependent phosphorylation of H2AX. To confirm the implication of DNA-PK, we tested the γ-H2AX response of DNA-PK–deficient cells (Ku80-deficient xrs6). The xrs6 cells exhibited reduced γ-H2AX compared with the wild-type K1 cells (Figure 7D). After a 6-h treatment, γ-H2AX in xrs6 cells was reduced by ∼50% compared with K1 cells, which is comparable to the partial inhibition of γ-H2AX by the DNA-PK inhibitor in XPD-c cells (Figure 7B).
From those results we conclude that ATM phosphorylates H2AX in both NER-proficient and -deficient cells, whereas DNA-PK appears preferentially implicated in the TCR-dependent phosphorylation of H2AX.
Ser1981-pATM Foci Colocalize with Both TCR-MRN–dependent and –independent γ-H2AX Foci
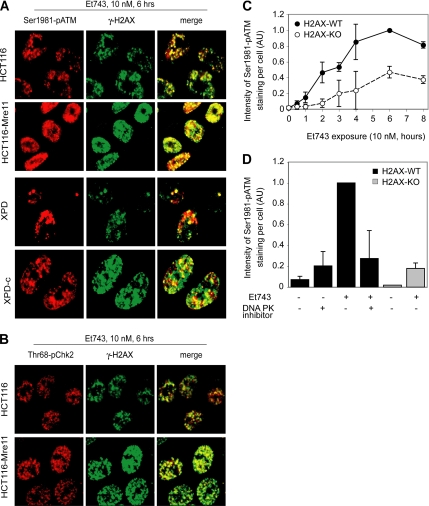
Next we examined the impact of Et743 treatment on ATM activation, which can be determined as ATM autophosphorylation on serine 1981 (Bakkenist and Kastan, 2003; Uziel et al., 2003). The representative pictures shown in Figure 8A illustrate the close relationship between the phosphorylations of ATM and H2AX in MRN-deficient and -proficient cells (HCT116 and HCT116-Mre11, respectively; Takemura et al., 2006). In both cell lines, 97% of the cells positive for Ser1981-pATM were also positive for γ-H2AX. Moreover, Et743-induced Ser1981-pATM foci colocalized with γ-H2AX foci, and, as for H2AX (see Figure 5), the fraction of Ser1981-pATM–positive cells after a 6-h treatment with Et743 was markedly higher in the Mre11-complemented than in the wild-type HCT116 cells (70 vs. 45%, respectively).
Figure 8.
Phosphorylation of H2AX is necessary for the full activation of ATM after Et743 treatment. (A) Colocalization (yellow in the merge image) of Ser1981-pATM (red) and γ-H2AX (green) foci. Representative pictures in HCT116, HCT116-Mre11, XPD, and XPD-c cells treated with 10 nM Et743 for 6 h. (B) Colocalization (yellow in the merge image) of the downstream target of ATM, Thr68-pChk2 (red), and γ-H2AX (green) foci. Representative pictures in HCT116 and HCT116-Mre11 cells treated with 10 nM Et743 for 6 h. (C) Reduced activation of ATM in embryonic fibroblasts (MEF) from H2AX knockout mice compared with normal MEF. Quantification of the intensity of the Ser1981-pATM staining normalized to the number of analyzed cells (mean ± SD; n = 5; AU, arbitrary units; see Materials and Methods). (D) The inhibition of DNA-PK in H2AX-WT MEF led to a reduced activation of ATM. Quantification of the intensity of the Ser1981-pATM staining normalized to the number of analyzed cells (mean ± SD; n = 2; AU, arbitrary units; see Materials end Methods) in H2AX-WT MEF incubated with the DNA-PK inhibitor (KU57788, 20 μM) for 1 h before the addition of 10 nM Et743 for the next 6 h and in H2AX-KO MEF incubated with 10 nM Et743.
ATM phosphorylation/activation was also correlated with the NER status of the cells. Phosphorylation of both ATM and H2AX was more intense in XPD-c than in XPD cells, and the two phosphorylated proteins colocalized (Figure 8A).
We also investigated ATM activation by the phosphorylation of its target, Chk2, on the threonine 68 (Thr68-pChk2). Thr68-pChk2 also colocalized with γ-H2AX and was activated to the same extent as H2AX and ATM in HCT116 and HCT116-Mre11 (Figure 8B).
From these experiments we conclude that Ser1981-pATM colocalizes with the Et743-induced γ-H2AX foci and that both are present at the sites of TCR/MRN-dependent and TCR/MRN-independent DSBs.
H2AX Is Necessary for the Full Activation of ATM
To determine whether γ-H2AX formation might promote ATM activation, in a positive-feedback loop (Stucki et al., 2005; Lou et al., 2006), we compared the kinetics of ATM activation in MEFs lacking H2AX (H2AX-KO) and their wild-type counterparts (H2AX-WT; Celeste et al., 2002). Figure 8C shows reduced ATM activation in the H2AX-KO cells, indicating that H2AX is necessary for the full activation of ATM after Et743 treatment. As we have shown above that DNA-PK phosphorylates H2AX at the site of NER-dependent DSBs, we hypothesized that not only H2AX by itself but rather the phosphorylation of H2AX by DNA-PK could lead to the activation of ATM in a positive feedback loop. We then measured the activation of ATM in H2AX-WT cells incubated with the DNA-PK inhibitor KU57788 and compared it with the activation of ATM in H2AX-KO cells. As shown on Figure 8D, the inhibition of DNA-PK led to a substantial reduction of the activation of ATM, bringing it down to the level of activation observed in H2AX-KO cells.
From these experiments we conclude that H2AX and its phosphorylation by DNA-PK are necessary for the full activation of ATM after Et743 treatment.
Opposite Impacts of DNA-PK-H2AX and MRN-ATM on the Antiproliferative Activity of Et743
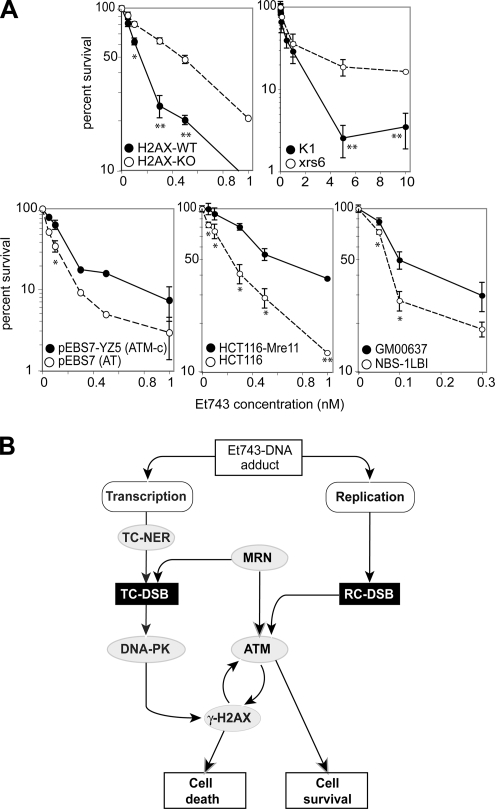
To determine the functional implication of H2AX, DNA-PK, MRN, and ATM, we performed cellular proliferation assays in genetically altered pairs of cell lines (deficient vs. wild-type or complemented) treated with Et743 (Figure 9A). H2AX-KO exhibited an unexpected ≈3-fold resistance. Consistently with that result and with the role of DNA-PK in the phosphorylation of H2AX, the DNA-PK–deficient, xrs6 cells were also more resistant to Et743 than their wild-type counterparts (K1). On the other hand, ATM-deficient cells showed approximately twofold greater sensitivity to Et743 (Damia et al., 2001). In the same manner, MRN deficiency confers a slight sensitivity (NBS-1LBI and HCT116 cells vs. GM00637 and HCT116-Mre11, respectively).
Figure 9.
Opposite roles for H2AX and ATM in the antiproliferative responses to Et743. (A) Cell proliferation assays. Cells were exposed to the indicated concentrations of Et743 for 72 h and analyzed by MTT assay. Open symbols correspond to the deficient cell lines (H2AX-KO, Ku80-deficient xrs6, ATM-deficient pEBS7, Mre11-deficient HCT116, and Nbs1-deficient NBS-1LBI), and filled symbols correspond to their wild-type (H2AX-WT, K1, and GM00637) or complemented (ATM-complemented pEBS7-YZ5 and HCT116-Mre11) counterparts. (B) Flowchart describing the proposed molecular pathways leading to Et743-induced DSBs and the subsequent cellular responses leading either to cell death (via DNA-PK/H2AX) or survival (via MRN/ATM).
Proposed Model Integrating NER, MRN, H2AX, ATM, and DNA-PK in the DSB Responses to Et743
Together our results suggest a model integrating H2AX, DNA-PK, ATM, MRN, and TCR in the cellular responses to Et743 (Figure 9B). We propose that two pathways lead to the formation of DSBs and refer to these two pathways/mechanisms as TC-DSBs and RC-DSBs. In the first pathway (left in Figure 9B), the Et743-DNA adducts trap the NER machinery in a transcription-dependent manner. However, those adducts cannot be properly excised by the NER machinery (Erba et al., 2001; Takebayashi et al., 2001b), probably because they interfere with the XPG endonuclease (Takebayashi et al., 2001b; Herrero et al., 2006). As a result, they lead to the stabilization of single-strand breaks (Takebayashi et al., 2001a,b) and to the trapping of NER complexes (Erba et al., 2001; Takebayashi et al., 2001b; Zewail-Foote et al., 2001). In a manner that remains to be elucidated, the MRN complex is recruited, which results in the creation of DSBs. Those DSBs are recognized by DNA-PK, which phosphorylates H2AX around each DSB site and promotes cell death. γ-H2AX also triggers the activation of ATM, which can phosphorylate H2AX in a positive feedback loop.
In the second mechanism, Et743 induces RC-DSBs and ATM phosphorylates H2AX at the sites of these RC-DSBs (right in Figure 9B). ATM and MRN both play a dual role: they participate to the phosphorylation of H2AX, thereby amplifying the death signal on one side, but they can also act as survival factors, probably by the activation of the checkpoints (Shiloh, 2006; Figure 8).
DISCUSSION
The present study shows that Et743 induces the formation of both transcription-coupled DNA TC-DSBs and RC-DSBs.
Although Et743-induced RC-DSBs have recently been reported (Guirouilh-Barbat and Pommier, 2006; Soares et al., 2007), here we provide further characterization of those RC-DSBs, and we demonstrate that those RC-DSBs are independent of TCR (Figure 2; Soares et al., 2007). RC-DSBs have been reported for camptothecins, as camptothecins trap topoisomerase I–mediated DNA single-strand breaks, which are converted into RC-DSBs by DNA polymerase “run-off” (Strumberg et al., 2000; Takemura et al., 2006). Because Et743-induced RC-DSBs are independent from TCR (present study and Soares et al., 2007), whereas the induction of DNA single-strand breaks by Et743 is dependent on TCR (Takebayashi et al., 2001b), it is unlikely that Et743-induced RC-DSBs are the consequence of DNA polymerase run-off. Thus, Et743-induced DSBs may involve “replication fork collapse” (Paulsen and Cimprich, 2007), a mechanism that has recently been reported for aminoflavone, because aminoflavone produces DNA–protein complexes that arrest replication forks (Meng et al., 2005). Abnormal DNA structures and helicase deficiencies have also been shown to produce RC-DSBs detected as γ-H2AX foci (Rao et al., 2007; Shimura et al., 2008).
The most novel aspect of our study is the induction of TCR- and MRN-dependent TC-DSBs. Although Soares et al. (2007) only discussed replication-dependent DSBs, their Supplementary Figure 7 is suggestive of reduced induction of DSBs by Et743 in XPG-deficient cells. However, they could not detect DSBs in nonproliferating lymphocytes by neutral COMET assay, whereas we did by immunodetection of nuclear foci containing γ-H2AX together with 53BP1 and Mre11 (Figures 3 and 4). This apparent discrepancy is probably due to the greater sensitivity of immunocytochemistry compared with neutral COMET assays. We also observed replication-independent γ-H2AX foci in cultured cells, outside of S-phase (IdU-negative; Figures 2 and 6).
The γ-H2AX response to Et743 (present study) is different from the γ-H2AX response to UVC irradiation (Marti et al., 2006), although both are dependent on NER and can occur in nonreplicating cells. UVC-induced γ-H2AX staining is diffuse (“pan-nuclear”) and does not colocalize with pNbs1 and 53BP1 (Marti et al., 2006), suggesting that it is not correlated with DSBs. In contrast, Et743 produced well-defined γ-H2AX foci. Their colocalization with DSBs markers, including Ser1981-pATM, Thr68-pChk2, and 53BP1 (Figures 1, 4, 5, and 8) is indicative of DSBs.
The replication-independent DSBs are likely to be transcription-dependent because we observed a significant reduction of γ-H2AX after inhibition of transcription with DRB (Figures 2 and 6). In fact, concomitant inhibition of both replication and transcription was required to block fully the formation of Et743-induced γ-H2AX foci in proliferative cells (Figures 2 and 6). The TC-DSBs are unlikely to result from two closely spaced TCR-dependent single-strand breaks on opposite strands (Takebayashi et al., 2001a; Takebayashi et al., 2001b). Indeed, a significant induction of γ-H2AX foci was observed in HCT116-Mre11 cells at concentrations as low as 0.05 nM Et743, which would be estimated to produce less than one adduct per 105 base pairs (Takebayashi et al., 2001b; Figure S2). Furthermore, in nonproliferating lymphocytes, increasing Et743 concentrations did not lead to an augmentation of γ-H2AX staining (Figure 3). Further investigations will be required to determine whether the NER endonucleases, XPF and XPG, may be responsible for the TC-DSBs. Indeed, it is possible that the Et743-DNA adducts alter the specificity of those endonucleases for the damaged strand (de Laat et al., 1998). Also, the stabilization of the NER repair intermediates (Zewail-Foote et al., 2001) may induce the recruitment of other nucleases that mediate the TC-DSBs.
Although Mre11 is endowed of known nuclease activities (D'Amours and Jackson, 2002), we found similar γ-H2AX induction in Mre11-deficient ATLD2 cells complemented with a nuclease inactive Mre11 (Arthur et al., 2004) or with wild-type Mre11, suggesting that Mre11 is not implicated in the direct production of TC-DSBs through its nuclease activities. Mre11 might promote the formation of TC-DSBs by tethering DNA damage response factors and/or nucleases, as recently demonstrated for C terminus Binding Protein-Interacting Protein (CtIP) (Limbo et al., 2007; Sartori et al., 2007), to the transcription damage sites. The defective response of Mre11-deficient cells to Et743 was more pronounced in the analysis of γ-H2AX foci than the neutral COMET assay (Figure 5, A and D, vs. B). This might result from the implication of Mre11 as a coactivator of ATM (Uziel et al., 2003; Horejsi et al., 2004; Falck et al., 2005; Lee and Paull, 2005; Dupre et al., 2006) and from the role of ATM in Et743-induced phosphorylation of H2AX (Figure 7).
Recent studies showed that H2AX is required for cell death after exposure to UVA (Lu et al., 2006) and that DNA-PK phosphorylates H2AX during apoptosis (Mukherjee et al., 2006). We found that DNA-PK phosphorylated H2AX in response to Et743-induced TC-DSBs (Figure 7). This could explain why cells defective for H2AX and DNA-PK were resistant to Et743 (Figure 9). DNA-PK has also been reported to trigger ionizing radiation-induced apoptosis in mouse thymocytes and fibroblasts (Wang et al., 2000; Woo et al., 2002) and the regulatory subunits of DNA-PK Ku70 and Ku80 have been identified as partners of Bax and Bcl-2 to regulate their apoptotic function (Sawada et al., 2003; Feki et al., 2005; Wang et al., 2008).
The enhanced sensitivity of ATM-deficient cells to Et743 (Damia et al., 2001; Figure 9) highlights the possible loops between ATM and H2AX. γ-H2AX can initiate pathways leading to cell death, but also promote the full activation of ATM (Figure 8), which may act as survival factor and activate checkpoint responses (Figure 8; Shiloh, 2006). The MRN complex could also play a dual role. It could be implicated in the creation of Et743-induced DSBs (Figure 5) and could act by enabling ATM-triggered checkpoints responses and allowing cell survival (Uziel et al., 2003; Horejsi et al., 2004; Falck et al., 2005; Lee and Paull, 2005; Dupre et al., 2006). Accordingly, we found that MRN-deficient cells were more sensitive to Et743 than their Mre11-proficient counterparts (Figure 9A).
Because γ-H2AX is readily formed in peripheral lymphocytes at drug concentrations within pharmacological range (10 nM and below; Puchalski et al., 2002; Garcia-Carbonero et al., 2004), our study suggests γ-H2AX as a pharmacodynamic marker in clinical studies with Et743. Our study also suggests that tumor profiling for TCR, MRN, ATM, and DNA-PK might be useful to anticipate patients' response to Et743 treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Chiara Conti, Andrew Jobson, Olivier Sordet, and Kurt W. Kohn for critical reading of the manuscript and suggestions during the course of these experiments. We also thank Dr. Roger Griffin, Department of Chemistry, University of Newcastle, and Dr. Ian Hickson, KuDos Pharmaceuticals. for generously providing the ATM and DNA-PK small molecule inhibitors. This work was supported by the National Cancer Institute (NCI) Intramural Program, Center for Cancer Research, NCI, National Institutes of Health.
Abbreviations used:
- APD
aphidicolin
- AT
ataxia telangiectasia
- ATM
ataxia telangiectasia–mutated gene
- CS
Cockayne syndrome
- DNA-PK
DNA-dependent protein kinase
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- DSB
DNA double-strand break
- Et743
ecteinascidin 743
- GGR
global genome repair
- γ-H2AX
histone H2AX phosphorylated on serine 139
- MRN
Mre11-Rad50-Nbs1
- NER
nucleotide excision repair
- RC-DSB
replication-coupled DSB
- RPA
replication protein A
- TC-DSB
transcription-coupled DSB
- TCR
transcription-coupled nucleotide excision repair
- XP
xeroderma pigmentosum.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0215) on July 16, 2008.
REFERENCES
- Arthur L. M., Gustausson K., Hopfner K. P., Carson C. T., Stracker T. H., Karcher A., Felton D., Weitzman M. D., Tainer J., Carney J. P. Structural and functional analysis of Mre11-3. Nucleic Acids Res. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune G. J., Furuta T., Pommier Y. Ecteinascidin 743, a novel anticancer drug with a unique mechanism of action. Anticancer Drugs. 2002;13:545–555. doi: 10.1097/00001813-200207000-00001. [DOI] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Celeste A., et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C., Seiler J. A., Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6:2760–2767. doi: 10.4161/cc.6.22.4932. [DOI] [PubMed] [Google Scholar]
- D'Amours D., Jackson S. P. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- D'Incalci M., Jimeno J. Preclinical and clinical results with the natural marine product ET-743. Expert Opin. Investig. Drugs. 2003;12:1843–1853. [PubMed] [Google Scholar]
- Damia G., Silvestri S., Carrassa L., Filiberti L., Faircloth G. T., Liberi G., Foiani M., D'Incalci M. Unique pattern of ET-743 activity in different cellular systems with defined deficiencies in DNA-repair pathways. Int. J. Cancer. 2001;92:583–588. doi: 10.1002/ijc.1221. [DOI] [PubMed] [Google Scholar]
- de Laat W. L., Appeldoorn E., Sugasawa K., Weterings E., Jaspers N. G., Hoeijmakers J. H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W. L., Jaspers N.G.J., Hoeijmakers J.H.J. Molecular mechanisms of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- Dupre A., Boyer-Chatenet L., Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- Erba E., Bergamaschi D., Bassano L., Damia G., Ronzoni S., Faircloth G. T., D'Incalci M. Ecteinascidin-743 (ET-743), a natural marine compound, with a unique mechanism of action. Eur. J. Cancer. 2001;37:97–105. doi: 10.1016/s0959-8049(00)00357-9. [DOI] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S. P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Fayette J., Coquard I. R., Alberti L., Boyle H., Meeus P., Decouvelaere A. V., Thiesse P., Sunyach M. P., Ranchere D., Blay J. Y. ET-743, a novel agent with activity in soft-tissue sarcomas. Curr. Opin. Oncol. 2006;18:347–353. doi: 10.1097/01.cco.0000228740.70379.3f. [DOI] [PubMed] [Google Scholar]
- Feki A., Jefford C. E., Berardi P., Wu J. Y., Cartier L., Krause K. H., Irminger-Finger I. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene. 2005;24:3726–3736. doi: 10.1038/sj.onc.1208491. [DOI] [PubMed] [Google Scholar]
- Friedman D., Hu Z., Kolb E. A., Gorfajn B., Scotto K. W. Ecteinascidin-743 inhibits activated but not constitutive transcription. Cancer Res. 2002;62:3377–3381. [PubMed] [Google Scholar]
- Furuta T., et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 2003;278:20303–20312. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
- Garcia-Carbonero R., et al. Phase II and pharmacokinetic study of ecteinascidin 743 in patients with progressive sarcomas of soft tissues refractory to chemotherapy. J. Clin. Oncol. 2004;22:1480–1490. doi: 10.1200/JCO.2004.02.098. [DOI] [PubMed] [Google Scholar]
- Giannini G., et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–254. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Sakai R., Rinehart K. L., Wang A. H. Molecular and crystal structures of ecteinascidins: potent antitumor compounds from the Caribbean tunicate Ecteinascidia turbinata. J. Biomol. Struct. Dyn. 1993;10:793–818. [PubMed] [Google Scholar]
- Guirouilh-Barbat J., Pommier Y. Et-743 induced DNA double strand breaks: a link between histone gamma-H2AX, transcription coupled nucleotide excision repair and Mre11 endonuclease (Abstract number 4642). 97th Annual Meeting of the American Association for Cancer Research; Washington, DC. 2006. p. 1090. [Google Scholar]
- Herrero A. B., Martin-Castellanos C., Marco E., Gago F., Moreno S. Cross-talk between nucleotide excision and homologous recombination DNA repair pathways in the mechanism of action of antitumor trabectedin. Cancer Res. 2006;66:8155–8162. doi: 10.1158/0008-5472.CAN-06-0179. [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Horejsi Z., Falck J., Bakkenist C. J., Kastan M. B., Lukas J., Bartek J. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene. 2004;23:3122–3127. doi: 10.1038/sj.onc.1207447. [DOI] [PubMed] [Google Scholar]
- Izbicka E., Lawrence R., Raymond E., Eckhardt G., Faircloth G., Jimeno J., Clark G., Von Hoff D. D. In vitro antitumor activity of the novel marine agent, ecteinascidin-743 (ET-743, NSC-648766) against human tumors explanted from patients. Ann. Oncol. 1998;9:981–987. doi: 10.1023/A:1008224322396. [DOI] [PubMed] [Google Scholar]
- Kobayashi J., Antoccia A., Tauchi H., Matsuura S., Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair. 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Leahy J. J., Golding B. T., Griffin R. J., Hardcastle I. R., Richardson C., Rigoreau L., Smith G. C. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z., et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Lu C., Zhu F., Cho Y. Y., Tang F., Zykova T., Ma W. Y., Bode A. M., Dong Z. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti T. M., Hefner E., Feeney L., Natale V., Cleaver J. E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Kohlhagen G., Sausville E. A., Pommier Y. DNA-protein crosslinks and replication-dependent histone H2AX phosphorylation induced by aminoflavone (NSC 686288), a novel anticancer agent active against breast cancer cells. Cancer Res. 2005;65:5337–5343. doi: 10.1158/0008-5472.CAN-05-0003. [DOI] [PubMed] [Google Scholar]
- Minuzzo M., Marchini S., Broggini M., Faircloth G., D'Incalci M., Mantovani R. Interference of transcriptional activation by the antineoplastic drug ecteinascidin-743. Proc. Natl. Acad. Sci. USA. 2000;97:6780–6784. doi: 10.1073/pnas.97.12.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva O. K., Petrini J. H. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- Missura M., Buterin T., Hindges R., Hubscher U., Kasparkova J., Brabec V., Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B., Kessinger C., Kobayashi J., Chen B. P., Chen D. J., Chatterjee A., Burma S. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair. 2006;5:575–590. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Paulsen R. D., Cimprich K. A. The ATR pathway: fine-tuning the fork. DNA Repair. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Cherfils J. Interfacial protein inhibition: a nature's paradigm for drug discovery. Trends Pharmacol. Sci. 2005;28:136–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Kohlhagen G., Bailly C., Waring M., Mazumder A., Kohn K. W. DNA sequence- and structure-selective alkylation of guanine N2 in the DNA minor groove by ecteinascidin 743, a potent antitumor compound from the Caribbean tunicate Ecteinascidia turbinata. Biochemistry. 1996;35:13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- Puchalski T. A., et al. Pharmacokinetics of ecteinascidin 743 administered as a 24-h continuous intravenous infusion to adult patients with soft tissue sarcomas: associations with clinical characteristics, pathophysiological variables and toxicity. Cancer Chemother. Pharmacol. 2002;50:309–319. doi: 10.1007/s00280-002-0498-3. [DOI] [PubMed] [Google Scholar]
- Rao V. A., Conti C., Guirouilh-Barbat J., Nakamura A., Miao Z. H., Davies S. L., Sacca B., Hickson I. D., Bensimon A., Pommier Y. Endogenous γ-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., Bonner W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- Rinehart K. L., et al. Bioactive compounds from aquatic and terrestrial sources. J. Nat. Prod. 1990;53:771–792. doi: 10.1021/np50070a001. [DOI] [PubMed] [Google Scholar]
- Sakai R., Rinehart K. L., Guan Y., Wang A. H. Additional antitumor ecteinascidins from a Caribbean tunicate: crystal structures and activities in vivo. Proc. Natl. Acad. Sci. USA. 1992;89:11456–11460. doi: 10.1073/pnas.89.23.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kaccmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada M., Sun W., Hayes P., Leskov K., Boothman D. A., Matsuyama S. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat. Cell Biol. 2003;5:320–329. doi: 10.1038/ncb950. [DOI] [PubMed] [Google Scholar]
- Schultz L. B., Chehab N. H., Malikzay A., Halazonetis T. D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler J. A., Conti C., Syed A., Aladjem M. I., Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell Biol. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Shimura T., Torres M. J., Martin M. M., Rao V. A., Pommier Y., Katsura M., Miyagawa K., Aladjem M. I. Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J. Mol. Biol. 2008;375:1152–1164. doi: 10.1016/j.jmb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares D. G., Escargueil A. E., Poindessous V., Sarasin A., de Gramont A., Bonatto D., Henriques J. A., Larsen A. K. From the cover: replication and homologous recombination repair regulate DNA double-strand break formation by the antitumor alkylator ecteinascidin 743. Proc. Natl. Acad. Sci. USA. 2007;104:13062–13067. doi: 10.1073/pnas.0609877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T. H., Carson C. T., Weitzman M. D. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- Strumberg D., Pilon A. A., Smith M., Hickey R., Malkas L., Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. Xeroderma pigmentosum genes: functions inside and outside DNA repair. Carcinogenesis. 2008;29:455–465. doi: 10.1093/carcin/bgm282. [DOI] [PubMed] [Google Scholar]
- Synold T. W., Dussault I., Forman B. M. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- Takebayashi Y., Goldwasser F., Urasaki Y., Kohlhagen G., Pommier Y. Ecteinascidin 743 induces protein-linked DNA breaks in human colon carcinoma HCT116 cells and is cytotoxic independently of topoisomerase I expression. Clin. Cancer Res. 2001a;7:185–191. [PubMed] [Google Scholar]
- Takebayashi Y., et al. Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair. Nat. Med. 2001b;7:961–966. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- Takemura H., Rao V. A., Sordet O., Furuta T., Miao Z. H., Meng L., Zhang H., Pommier Y. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J. Biol. Chem. 2006;281:30814–30823. doi: 10.1074/jbc.M603747200. [DOI] [PubMed] [Google Scholar]
- Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoti G., Nicoletti M. I., Pellegrino A., Jimeno J., Hendriks H., D'Incalci M., Faircloth G., Giavazzi R. Ecteinascidin-743, a new marine natural product with potent antitumor activity on human ovarian carcinoma xenografts. Clin. Cancer Res. 1998;4:1977–1983. [PubMed] [Google Scholar]
- Verschraegen C. F., Glover K. ET-743 (PharmaMar/NCI/Ortho Biotech) Curr. Opin. Investig. Drugs. 2001;2:1631–1638. [PubMed] [Google Scholar]
- Wang Q., Gao F., May W. S., Zhang Y., Flagg T., Deng X. Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol. Cell. 2008;29:488–498. doi: 10.1016/j.molcel.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Guo M., Ouyang H., Li X., Cordon-Cardo C., Kurimasa A., Chen D. J., Fuks Z., Ling C. C., Li G. C. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc. Natl. Acad. Sci. USA. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo R. A., Jack M. T., Xu Y., Burma S., Chen D. J., Lee P. W. DNA damage-induced apoptosis requires the DNA-dependent protein kinase, and is mediated by the latent population of p53. EMBO J. 2002;21:3000–3008. doi: 10.1093/emboj/cdf307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewail-Foote M., Hurley L. H. Ecteinascidin 743, a minor groove alkylator that bends DNA toward the major groove. J. Med. Chem. 1999;42:2493–2497. doi: 10.1021/jm990241l. [DOI] [PubMed] [Google Scholar]
- Zewail-Foote M., Li V. S., Kohn H., Bearss D., Guzman M., Hurley L. H. The inefficiency of incisions of ecteinascidin 743-DNA adducts by the UvrABC nuclease and the unique structural feature of the DNA adducts can be used to explain the repair-dependent toxicities of this antitumor agent. Chem. Biol. 2001;8:1033–1049. doi: 10.1016/s1074-5521(01)00071-0. [DOI] [PubMed] [Google Scholar]
- Ziv Y., Bar-Shira A., Pecker I., Russell P., Jorgensen T. J., Tsarfati I., Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.