Abstract
Nuclear pore complexes (NPCs) are multisubunit protein entities embedded into the nuclear envelope (NE). Here, we examine the in vivo dynamics of the essential Drosophila nucleoporin Nup107 and several other NE-associated proteins during NE and NPCs disassembly and reassembly that take place within each mitosis. During both the rapid mitosis of syncytial embryos and the more conventional mitosis of larval neuroblasts, Nup107 is gradually released from the NE, but it remains partially confined to the nuclear (spindle) region up to late prometaphase, in contrast to nucleoporins detected by wheat germ agglutinin and lamins. We provide evidence that in all Drosophila cells, a structure derived from the NE persists throughout metaphase and early anaphase. Finally, we examined the dynamics of the spindle checkpoint proteins Mad2 and Mad1. During mitotic exit, Mad2 and Mad1 are actively imported back from the cytoplasm into the nucleus after the NE and NPCs have reformed, but they reassociate with the NE only later in G1, concomitantly with the recruitment of the basket nucleoporin Mtor (the Drosophila orthologue of vertebrate Tpr). Surprisingly, Drosophila Nup107 shows no evidence of localization to kinetochores, despite the demonstrated importance of this association in mammalian cells.
INTRODUCTION
In eukaryotes, the nuclear envelope (NE) defines the limits between the nucleus and the cytoplasm. The outer NE membrane is considered to be structurally and functionally part of the endoplasmic reticulum network, whereas the inner membrane, with its distinct protein composition, provides anchoring points for the chromatin and nuclear lamina. Nuclear pore complexes (NPCs) are embedded at the points of fusion between the inner and outer NE membrane and represent the sole channels of transport across the NE. NPCs are composed of multiple copies of ∼30 different proteins termed nucleoporins (Nups), most of which are organized into subcomplexes that associate with each other to build up the mature NPCs (for reviews, see Hetzer et al., 2005; Schwartz, 2005; Lim and Fahrenkrog, 2006; Tran and Wente, 2006).
During cell division, the NE and NPCs are subjected to major rearrangements. However, the extent to which the NE and NPCs disassemble at mitotic entry varies among organisms (for reviews, see Margalit et al., 2005; Prunuske and Ullman, 2006). Unlike in most yeast and fungi, characterized by a “closed mitosis,” NE disassembly is required in animal cells to allow spindle microtubule access to chromosomes. In vertebrates, cell division leads to complete NE breakdown at the prophase–prometaphase transition. During this “open mitosis,” integral membrane proteins of the NE and the soluble subcomplexes of the NPCs redistribute throughout the endoplasmic reticulum and the mitotic cytoplasm (for reviews, see Hetzer et al., 2005; Margalit et al., 2005; Prunuske and Ullman, 2006). In Drosophila and Caenorhabditis elegans embryos, however, the NE only partially disassembles near spindle poles in early mitosis. NPCs disassemble during prometaphase (or even after metaphase in early C. elegans embryos), thus leaving behind a fenestrated or leaky nuclear envelope (Stafstrom and Staehelin, 1984; Lee et al., 2000; Kiseleva et al., 2001). In Drosophila embryos, the spindles of syncytial mitoses remain confined within a membranous structure partly derived from the NE and previously termed spindle envelope (SE) (Stafstrom and Staehelin, 1984; Harel et al., 1989, and references therein). Accordingly, the term “semi-closed” has been used to describe Drosophila mitosis. In all metazoans, NE and NPC reassembly at mitotic exit is initiated around the two sets of chromosomes in late anaphase, and it continues until late telophase/early G1, leading to the reformation of two daughter nuclei (for reviews, see Hetzer et al., 2005; Margalit et al., 2005; Prunuske and Ullman, 2006; Gorjanacz et al., 2007).
These various aspects of NE and NPC remodeling during mitosis need to be spatially and temporally coordinated with other cell cycle events. Key regulators that contribute to this spatio-temporal coordination are mitotic kinases and phosphatases as well as the small GTPase Ran and the transport receptors of the karyopherin/importin β family (for reviews, see Dasso, 2002; Harel and Forbes, 2004; Hetzer et al., 2005; Margalit et al., 2005; see also Onischenko et al., 2005). In addition, there is growing evidence that NPCs functionally participate in mitosis. Indeed, several kinetochore constituents, including the checkpoint proteins Mad1 and Mad2, are localized to NPCs in interphase; conversely, a growing number of nucleoporins have been found on the mitotic spindle or at kinetochores during mitosis (for reviews, see Stukenberg and Macara, 2003; Hetzer et al., 2005; see also Babu et al., 2003, Liu et al., 2003; Arnaoutov et al., 2005; Galy et al., 2006; Fernandez and Piano, 2006; Rasala et al., 2006).
Among them, the vertebrate Nup107-160 subcomplex, which is composed in vertebrates of nine subunits (Belgareh et al., 2001; Vasu et al., 2001; Loiodice et al., 2004), contributes to various aspects of cell cycle progression. Besides playing crucial role at a very early stage of NPC reassembly (Harel et al., 2003; Walther et al., 2003a), a fraction of human Nup107-160 subcomplex and of C. elegans Nup107p localizes at kinetochores during mitosis (Belgareh et al., 2001, Harel et al., 2003; Loiodice et al., 2004; Galy et al., 2006) and was recently demonstrated to contribute to proper kinetochore functions in human cells (Zuccolo et al., 2007). In addition, this complex was reported to localize to spindle poles and proximal spindle fibers in prometaphase mammalian cells and throughout reconstituted spindles in Xenopus egg extracts (Orjalo et al., 2006).
Here, we have examined the in vivo dynamics of the Nup107 nucleoporin during mitosis in Drosophila, directly comparing it with the dynamics of other fluorescently tagged proteins of the nuclear envelope and with the checkpoint protein Mad2. We studied both the rapid mitosis of late syncytial embryos (cycles 10 to 13), in which synchronous nuclear divisions take place in a common cytoplasm close to the embryo cortex (Foe and Alberts, 1983; Stafstrom and Staehelin, 1984) and also the more typical mitosis of larval-stage neuroblasts or cellularized embryos, about which less is known concerning NPC and nuclear envelope dynamics. We have refined the temporal resolution of the order of events at the molecular level that take place in this organism during nuclear disassembly and reassembly. This study revealed both similarities and unique differences with other metazoan systems.
MATERIALS AND METHODS
Drosophila Stocks
Fly lines used in this study are listed in Table 1.
Table 1.
Fly lines used in this study
| Line | Genotype | Origin |
|---|---|---|
| nup107EP2403 | P[EP]Nup107EP2403/CyO | Szeged Stock Centre (Szeged, Hungary)/EP(2)2403 |
| nup107E8 | yw67; nup107E8/CyO; MKRS,Sb/TM2,y | This study |
| mRFP-Nup107 rescued line | yw67; nup107E8; P[w+, mRFP-nup107]7.1/TM2 | This study |
| GFP-Nup107 | yw67; Sp/CyO; P[w+, GFP-nup107]49.1.2/TM2 | This study |
| GFP-Histone | P[w+, His2AvDGFP] | Clarkson and Saint (1999) |
| GFP-LaminC/G00158 | y1w1118; P[w+]LamC G00158 ttvG00158/CyO | Schulze et al. (2005) |
| GFP-Mad2 | w1118; P[w+, GFP-mad2]9a | Buffin et al. (2005) |
| GFP-Mad2 rescued line | P[w+, GFP-mad2]/Y; +; mad2P/mad2P | Buffin et al. (2007) |
| ketelGFP | y; ketelGFP/y+CyO | Gift from A. Debec (Institut Jacques Monod, UMR 7592, Paris, France) |
| mRFP-Nup107, GFP-Histone | w1118; nup107E8; P[w+, mRFP-nup107] P[w+, GFP-His 2Av]/TM2 | This study (obtained by genetic crosses with above-described lines) |
| mRFP-Nup107, GFP-Mad2 | w1118; nup107E8; P[w+, mRFP-nup107] P[w+, GFP-mad2]/TM2 | |
| mRFP-Nup107, GFP-LaminC | w1118; nup107E8 P[GFP-LamC]/CyO;P[w+, mRFP-nup107]/TM2 | |
| mRFP-Nup107, ketelGFP | w1118; ketelGFP/CyO; P[w+, mRFP-nup107]/TM2 |
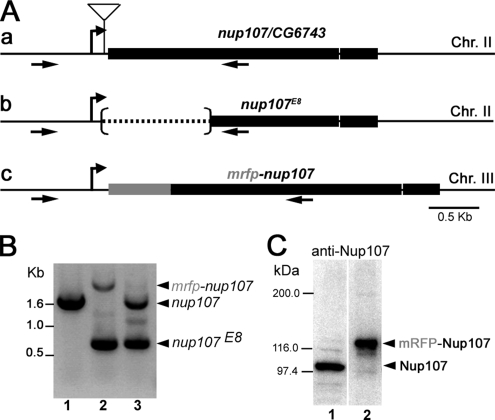
The nup107E8 deletion mutant was generated by imprecise excision of the P element from the nup107EP2403 allele (Szeged Stock Center, Szeged, Hungary) by using standard methods (Ashburner et al., 2005). The nup107E8 allele lacks 976-base pairs of the nup107 open reading frame (ORF), including the starting ATG, as confirmed by polymerase chain reaction (PCR) and sequencing (Figure 1A).
Figure 1.
Structure of Drosophila nup107, mutants, and transgene. (A) a, schematic representation of the nup107 genomic locus on chromosome II. The thick line corresponds to the nup107 ORF, interrupted by a unique intron. The insertion site of the P element, located in the 5′UTR, 64 base pairs from the start codon of nup107 is indicated. b, imprecise excision of the P element generated the nup107E8 deletion allele lacking 976 nt from the translated region. c, the mRFP-nup107 transgene comprises ∼4.9 kb of the nup107 genomic locus and the mRFP ORF (inserted 18 base pairs upstream of the starting ATG of nup107). The transgene used in this study was integrated on chromosome III. Arrows indicate the position of the two primers used for the PCR in B. (B) PCR analysis of wild type (lane 1), the rescued line w−; nup107E8/nup107E8; P[mRFP-nup107] (lane 2), and nup107E8/+ heterozygotes (lane 3). (C) Total protein extracts of 0- to 3-h-old embryos from wild type (lane 1) and the mRFP-Nup107 rescued line flies (lane 2) were analyzed by Western blot using a polyclonal anti-Drosophila Nup107 antibody (see Materials and Methods). Endogenous Drosophila Nup107 (theoretical molecular mass of 97 kDa) is absent from the rescued flies (lane 2), in which mRFP-nup107 is the only source of Nup107. Molecular mass markers are indicated on the left. The original Western blot also including extracts from larvae and adult flies and the corresponding anti-tubulin staining of the membrane are provided in Supplemental Figure 1.
For transgenic mRFP-Nup107 and GFP-Nup107 flies, a 7.7-kb fragment of the bacterial artificial chromosome (BAC) clone BACR19N18 (base pairs 4,540–12,268), containing the nup107/CG6743 gene (Fbgn0027868) was first subcloned into pBluescriptSK−. Next, a 4.2-kb XhoI–NsiI fragment, including 1.5 kb upstream and 0.8 kb downstream of nup107 ORF was introduced into the P transformation vector CasPer (Pirrotta, 1988). PCR-amplified monomeric red fluorescent protein 1 (mRFP) cDNA (Campbell et al., 2002) was inserted in-frame at a unique AgeI site located 18 base pairs upstream of the starting ATG of nup107. For the GFP-Nup107 transgene, the green fluorescent protein (GFP) DNA fragment (from the pEGFPC2 vector; Clontech, Mountain View, CA) was inserted in-frame as AgeI-XmaII (12 base pairs upstream of the starting ATG of nup107) in the above-described nup107-CasPer construct. The integrity of the transgenes was checked by sequencing. The mRFP-Nup107 and GFP-Nup107 transgenes were introduced into the germline of yw67 flies by standard methods (Ashburner et al., 2005). Two independent mRFP-nup107 insertion lines were tested for rescue of the lethality of the null nup107E8 allele, of which the mRFP-nup1077.1 rescued the nup107E8 mutation in a single copy. The stock w−; nup107E8; mRFP-nup1077.1/TM2 where the transgene is the only source of Nup107 (referred to as the rescued line in the text) was used for studying Nup107 dynamics.
Live Embryo Imaging
For live imaging, embryos were hand-dechorionated on double-sided adhesive tape and aligned on a coverslip covered with embryo glue in a Ludin chamber (Johansen and Johansen, 2004). Imaging was performed at 24–26°C by using an inverted confocal microscope equipped with an LSM5 META laser confocal imaging system, using a 40× oil/1.3 numerical aperture (N.A.) objective (Carl Zeiss MicroImaging, Jena, Germany). A 488-nm argon laser line with a 505- to 530-nm emission filter for GFP, and a 543-nm HeNe laser line with a 560- to 615-nm emission filter for rhodamine were used. The z-stacks (usually 2–4 planes, 0.5–0.8 μm apart) were acquired at zoom 6 or 10 every 10–30 s. Photobleaching was achieved after 1 or 2 prebleach image acquisitions, with 30 iterations of 60–85% 488-nm laser intensity within regions of interest of varying areas (as indicated on the figures). After acquisition, images were analyzed with MetaMorph software (Molecular Devices, Sunnyvale, CA) and converted to 8-bit images before being imported to Adobe Photoshop software 7.0 (Adobe Systems, Mountain View, CA). A median filter (MetaMorph) was applied to most images. Quantitative analyses were performed using MetaMorph software as described in the corresponding supplemental figure legends, and graphs were generated using Excel (Microsoft, Redmond, WA).
Microinjection of Embryos
For microinjections followed by imaging, 1- to 2-h embryo collections were hand-dechorionated, aligned in the center of an IWAKI plate coated with embryo glue, and covered with halocarbon oil (Voltalef 10S; Prolabo, Lutterworth, Leicestershire, United Kingdom) to avoid desiccation. To determine the embryo age, fluorescent and differential interference contrast images of the whole embryo were acquired before injection. Injections were performed with a manual microinjector (TransferMan NK2; Eppendorf, Hamburg, Germany) on a standard inverted microscope using a 10× objective. Colchicine (catalog no. 27620; Sigma-Aldrich, St. Louis, MO) was used at 1 mM in phosphate-buffered saline (PBS) or H20, and Alexa488-conjugated wheat germ agglutinin (WGA) (Invitrogen, Carlsbad, CA) at 0.1 mg/ml in PBS.
Larval Neuroblast Imaging
Larval neuroblasts were prepared and imaged as described previously (Buffin et al., 2005), using an Olympus IX-70 inverted microscope equipped with a focused xenon lamp and an OrcaER camera (Hammamatsu, Bridgewater, NJ), piloted by Cell-R hardware and software system (Olympus). Depending on the fly line, image acquisition times were 100–400 ms for GFP and 250–400 ms for red fluorescent protein (RFP). Images were usually collected at 15 s intervals (with the exception of the Lamin C/Nup107 movie, 30 s).
Antibodies, Western Blot Analysis, and Immunostaining
The anti-dmNup107 polyclonal rabbit serum was generated against the full-length Drosophila protein (cDNA LD18761 cloned in pBluescript and obtained from Invitrogen) fused to glutathione-S-transferase. For Western blot analysis, 5 μl of dechorionated embryos, two third-instar larvae, and two adult flies were homogenized in 1× SDS-Laemmli buffer. Samples were boiled for 5 min and centrifuged for 15 min at 14,000 rpm. Proteins were then separated on 8% SDS-polyacrylamide gel electrophoresis and transferred on nitrocellulose membrane. The membrane was probed with the anti-Drosophila Nup107 polyclonal antibody and mouse anti-tubulin α antibody (mouse DM1A; Sigma-Aldrich).
Drosophila Mad1 (CG2072) is listed in Flybase as TXBP181-like. The full-length Mad1 cDNA GM14169 (cloned in vector pOT2) was obtained from the Drosophila Genomics Resource Center (Indiana University, Bloomington, IN). A BglII–XhoI restriction fragment of this plasmid encoding the last 334 amino acids of Mad1 was cloned into the BamI and SalI sites of pET28b expression vector. The protein was expressed and His-tag purified and used to inoculate mice (Kernov Antibody Services, St. Etienne en Cogles, France). A polyclonal serum (mouse no. 1) was used in this study.
Fixation and immunostaining of 1- to 3-h-old embryos were performed essentially as described previously (Johansen and Johansen, 2004). Incubations were performed overnight at 4°C for primary antibodies and for 2–3 h at room temperature for secondary antibodies. Primary antibodies and dilutions used were rabbit anti-dmNup153 (1/500) and rat anti-Mtor (1/750) (Mendjan et al., 2006), rabbit anti-dmNup107 (1/1000), and mouse anti-Mad1 (1/50). After a 5-min incubation with 4,6-diamidino-2-phenylindole (DAPI), embryos were mounted with Mowiol and imaged using either the above-described Zeiss confocal system or (for Figure 9) a CSU10 spinning head (Yokogawa Electric, Tokyo, Japan) coupled with four solids diodes (405, 491, 561, and 635 nm) adapted on a inverted DMIRB microscope (Leica, Wetzlar, Germany). For confocal spinning imaging, selection of fluorophores was realized using an acousto-optic tunable filter on excitation and a filter wheel on emission (Roper-Sutter-Errol). The whole setup was driven with MetaMorph 7 software (Molecular Devices). Acquisitions were performed using an oil immersion objective100× PL APO HCX, N.A. 1.4 mounted on a piezo-electric motor (Physik Instrument, Irvine, CA) and a CoolSnap HQ camera (Photometrics). Image stacks were acquired with a binning of 2, with a plane spacing of 0.5 μm.
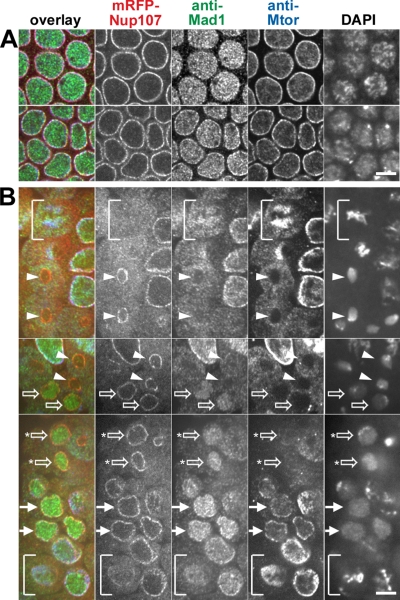
Figure 9.
Localization of mRFP-Nup107, Mad1, and the basket nucleoporin Mtor during embryonic mitotic divisions. Spinning disk confocal images of fixed syncytial and cellularized mRFP-Nup107 Drosophila embryos labeled with anti-Mad1 (green), anti Mtor (blue), and DAPI (not included in the overlay). (A) Unlike mRFP-Nup107 and Mtor, Mad1 is mainly localized in the nucleoplasm in syncytial embryos (top), and its NE localization becomes clearly detectable in cellularized embryos (bottom). (B) Successive steps of Mad1 and Mtor recruitment in the nucleus and at the NE during mitotic divisions of a cellularized embryo. The three panels arise from distinct areas and focal planes within the same embryo. The bracket in the bottom panel points to a prometaphase cell in which a fraction of Nup107 is still detectable at the NE, whereas Mad2 and Mtor are localized within the spindle area. The bracket in the top panel shows a metaphase cell revealing the typical localization of Mtor at the spindle matrix. Note that a fraction of Mad1 also accumulates within the spindle area whereas mRFP-Nup107 is more diffuse. Arrowheads point to cells in telophase in which Mad1 is still in the cytoplasm. Open arrows indicate cells with a nuclear accumulation of Mad1 and a cytoplasmic localization of Mtor. Some Mtor begins to be detectable at the NE in cells with an asterisk. Solid arrows indicate G1 cells in which Mtor and a fraction of Mad1 are localized to the NE. Bars, 5 μm.
RESULTS
Dynamics of Drosophila Nup107, an Essential Nuclear Pore Protein, during the Mitotic Cycles of Syncytial Embryos
The Drosophila nup107 gene (CG6743, mislabeled in Flybase as Nup170) was identified by BLASTN/P alignments by using the human Nup107 sequence. The nup107EP2403 allele, resulting from a P element insertion in the 5′ untranslated region (UTR) of the nup107 locus, was reported to be homozygous lethal. Imprecise excision of the P element yielded the nup107E8 allele, lacking ∼1 kb of the 5′ coding region of the nup107 gene, which is also recessive lethal (Figure 1, A and B). Further characterization of this phenotype will be reported elsewhere.
We generated transgenes encoding Drosophila Nup107 fused to GFP or mRFP1 (Campbell et al., 2002) under the control of the nup107 promoter (see Materials and Methods). The mRFP-nup107 transgene rescued in a single copy the lethal phenotype of nup107E8 flies (Figure 1, B and C, and Supplemental Figure 1), proving both that nup107 is an essential gene and that the transgene is fully functional. As anticipated, mRFP-Nup107 localized to the nuclear envelope in all Drosophila tissues and cell types examined (Figures 2 and 6; our unpublished data). Except when indicated, all of the studies described here used a rescued line in which all of Nup107 is derived from the mRFP-nup107 transgene.
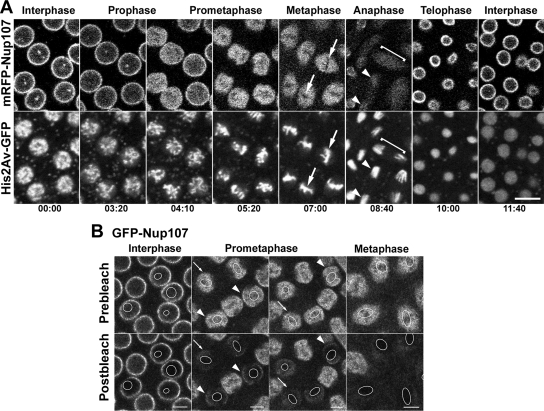
Figure 2.
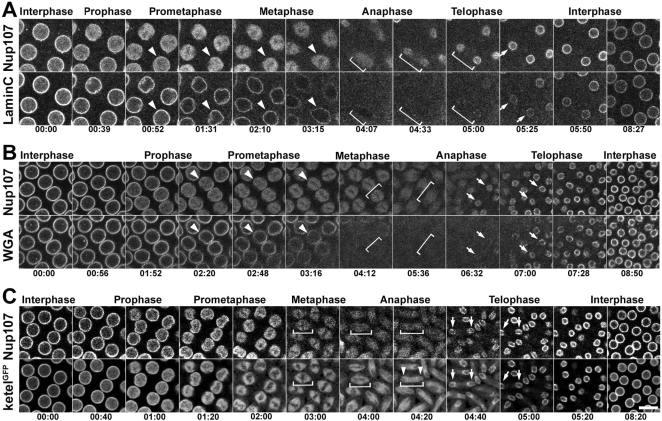
Dynamics of mRFP-Nup107 throughout the mitotic cycle in Drosophila syncytial embryos. (A) Selected frames of TLSCM acquisitions revealing the dynamics of mRFP-Nup107 together with His2Av-GFP during a complete embryonic cleavage cycle. A representative focal plane from a 3.6-μm z-stack acquisition is shown for each time point. Note the decreased NE localization and concomitant nuclear accumulation of mRFP-Nup107 in prometaphase and its persistence within the spindle area in metaphase. Arrows point to the chromatin-occupied (i.e., GFP-histone-labeled) areas from which mRFP-Nup107 is excluded. In anaphase, the signal fades within the spindle area (brackets) and redistributes over the segregated chromatids (arrowheads). The first rims reappear on the decondensing chromatin in late anaphase/early telophase. Time is in minutes:seconds. Bar, 10 μm. (B) Photobleaching analysis of GFP-Nup107 in embryonic nuclei. Small regions (outlined in white in the frames) within nuclei of GFP-Nup107 Drosophila embryos at various stages of the cell cycle were bleached. Acquisitions before and just after photobleaching are shown. Bars, 5 μm. Note the stable signal at the NE in interphase nuclei and the persistence of a minor fraction of GFP-Nup107 stably associated with the NE in early (arrowheads) but not late (arrows) prometaphase stages.
Figure 6.
NPC and NE dynamics in dividing larval neuroblasts. (A) Selected frames of wide-field time-lapse acquisition of dividing larval neuroblasts expressing mRFP-Nup107 and ketelGFP. Two representative series describing the prophase to metaphase (top) and the metaphase to telophase stages (bottom) are shown (see also Supplemental Movies 7 and 8). Note that ketelGFP is released from the prophase NE earlier than mRFP-Nup107 (top, 3:30 and 4:00 min). Both proteins are enriched within the area defined by the spindle envelope in early metaphase, and a fraction of ketelGFP but not mRFP-Nup107 persists within the spindle area in anaphase (brackets). Note that at these stages, both Nup107 and ketelGFP are always excluded from the chromatin area (arrowheads). The proteins are recruited simultaneously to the reforming nuclear envelope in early telophase (arrows). (B) Selected frames of wide-field time-lapse acquisition of a dividing larval neuroblast expressing mRFP-Nup107 and GFP-Lamin C. Note the persistence of the GFP-Lamin C staining at the spindle envelope until late anaphase (arrowheads). Arrows point to the reforming NE and NPCs in telophase. Bars, 5 μm. Time is in minutes:seconds.
To examine the dynamics of Nup107 during mitosis, we first followed by time-lapse scanning confocal microscopy (TLSCM) the last three syncytial mitotic cycles (cycles 10–13) of early Drosophila embryos expressing mRFP-Nup107 together with the GFP-tagged histone H2A variant His2AvD (Clarkson and Saint, 1999). Representative panels from one cleavage cycle (cycle 12) are shown in Figure 2A (Supplemental Movie 1). After entry into mitosis, the NE staining of mRFP-Nup107 persisted until prometaphase. At that stage, as the SE developed (Stafstrom and Staehelin, 1984, and references therein), the mRFP-Nup107 signal decreased at the rim and accumulated in the region delimited by the SE. Photobleaching analyses performed at this stage of the cell cycle by using the GFP-Nup107 line revealed that only a minor fraction of Nup107 remains stably associated with the NE in early prometaphase, whereas no NE-associated signal could be detected from late prometaphase on (Figure 2B). In metaphase, mRFP-Nup107 remained concentrated in the spindle region, but it was excluded from the area occupied by the chromosomes. This homogeneous, diffuse mRFP-Nup107 signal in the spindle region was independent of microtubules (see below, Figure 10A). During late anaphase, the signal faded within the spindle area, and Nup107 then began to accumulate on the two sets of separating chromatids. Finally, a rim-like staining reappeared at the periphery of the decondensing chromatin during telophase. This behavior was confirmed using polyclonal antibodies directed against dmNup107 in fixed wild-type syncytial embryos (Supplemental Figure 2).
Figure 10.
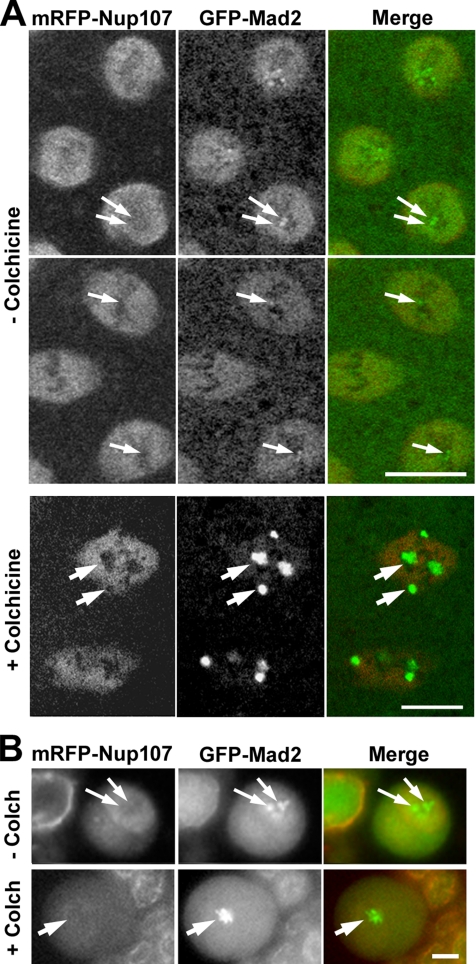
Nup107 is not detectable at kinetochores during Drosophila mitosis. (A) Confocal images of syncytial stage embryos expressing mRFP-Nup107 and GFP-Mad2 either untreated (−colchicine) and recorded in prometaphase and metaphase (30 s between the two frames), or imaged ∼5–10 min after microinjection with colchicine (1 mM). (B) Wide-field images of live larval neuroblasts expressing mRFP-Nup107 and GFP-Mad2 either untreated (−Colch) or imaged after an ∼10-min incubation with colchicine (10 μM). There is no detectable mRFP-Nup107 at kinetochores (arrows) in either cell type, even upon colchicine treatment, which leads to substantial accumulation of GFP-Mad2 on these structures. Note also that colchicine treatment does not affect the accumulation of Nup107 in the spindle region (in A, +colchicine). Bars, 5 μm.
NE and NPC Dynamics during Drosophila Embryonic Mitosis
To get a more general view of the respective dynamics of NPC and NE constituents during the various stages of mitosis, we next compared the localization of mRFP-Nup107 with that of other stably or transiently associated NE or NPC constituents (Figures 3–5).
Figure 3.
NE and NPC dynamics during syncytial mitotic divisions. Selected frames of TLSC acquisitions revealing the dynamics of mRFP-Nup107 in combination with markers for nuclear lamina (GFP-Lamin C) (A), the WGA-Nup (Alexa488-WGA) (B), and ketelGFP (C) during a complete embryonic cleavage cycle. A representative single focal plane from z-stack acquisitions is shown for each time point. (A) Note the persistence of the GFP-Lamin C signal at the SE (arrowheads) that surrounds the diffuse mRFP-Nup107 signal in prometaphase and metaphase, its even distribution in anaphase and early telophase (brackets), and its accumulation at the NE only once a full rim of Nup107 signal is clearly detected in late telophase (arrows). (B) Unlike mRFP-Nup107, WGA-Nup remains associated with the NE up to prometaphase (arrowheads), and it is evenly distributed from metaphase to anaphase (brackets). Also note that accumulation of WGA-Nup occurs slightly later than Nup107 in telophase (arrows). (C) Note the persistence of mRFP-Nup107 and ketelGFP within the spindle regions throughout metaphase and anaphase (brackets); ketelGFP seems to be excluded from the chromatin-occupied areas in anaphase (arrowheads). In telophase, the recruitment of mRFP-Nup107 and ketelGFP at the reforming NE is indistinguishable (arrows). Time is in minutes:seconds. Bar, 10 μm.
Figure 4.
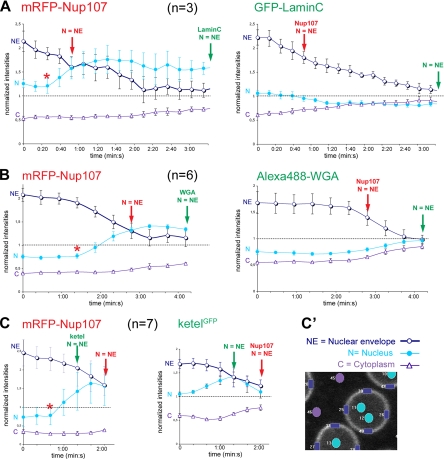
Quantitative analysis of the behavior of GFP-Lamin C, Alexa488-WGA, and ketelGFP compared with mRFP-Nup107 during the NE/NPC disassembly process. For each time point, the average intensity of small regions within the NE (4 rectangles) and nuclear interior (N; 2–3 circles) of n = 3–7 nuclei (as indicated in each graph) and within the cytoplasm (C; 6 circles) were measured. Typical regions are depicted in C′ for ketelGFP. Regions were identified based on the marker with the longest lasting signal at the NE (namely, GFP-Lamin C in A, Alexa488-WGA in B, and mRFP-Nup107 in C) and subsequently transferred to the other fluorescent channel. To allow comparison among markers, these values were normalized at each time point to the mean intensity within the entire field (so that a normalized intensity of one reflects a homogeneous distribution of the marker between the nucleus and the cytoplasm). Graphs represent the mean normalized intensities at the NE (dark blue circles and curves), within the nucleus (cyan dots and curves) and cytoplasm (purple triangles and curves) for mRFP-Nup107 (red, left) and either GFP-Lamin C (A), Alexa488-WGA (B), or ketelGFP (C) (green, right). Error bars are SD. Time points at which the NE can no longer be discriminated for the intranuclear signal (N = NE) are indicated for both markers. The stars on the mRFP-Nup107 disassembly curves indicate the time point at which the intranuclear signal begins to increase. Quantifications were performed on the movies shown in Figure 3, A–C (Supplemental Movies 2–4), and qualitatively similar results were obtained upon quantification of a distinct movie (data not shown).
Figure 5.
Quantitative analysis of the nuclear recruitment of GFP-Lamin C, Alexa488-WGA, and ketelGFP compared with mRFP-Nup107 during mitotic exit. To quantify the nuclear recruitment of these NE- or NPC-associated markers, circles of constant size broadly encompassing the area of five to eight reforming nuclei were manually identified based on the mRFP-Nup107 staining (red circles in Aa′–b′) and subsequently transferred to the corresponding GFP channels (green circles; GFP-Lamin C, Alexa488-WGA, or ketelGFP). For each time point and channel, the mean intensity in the cytoplasm (black circles) was subtracted from the mean intensity within the nuclear area. To allow comparison among markers, this value was then normalized to the maximal mean intensity reached for each marker at the last time point of the series. Graphs represent the mean of n = 5–8 nuclei as indicated. Error bars are SD. (A) Nuclear recruitment of GFP-Lamin C and mRFP-Nup107 at mitotic exit. A movie encompassing two successive cycles of the same embryo (a–a′ and b–b′) was used for these quantifications (not enough nuclei were in the correct focal plane in the Supplemental Movie 2). Black dots in the graphs indicate representative stages of the reassembly process illustrated on the right panels (a′ and b′), in which the areas used for quantifications are circled. Note that despite the difference in the rate of mRFP-Nup107 recruitment between these two cycles, GFP-Lamin C reaches its half recruitment intensity (dashed line at 0.5) significantly later than mRFP-Nup107 (1:50 and 1:20 min, respectively). (B and C) Kinetics of nuclear recruitment of mRFP-Nup107 and either WGA-Nup (8 nuclei from Supplemental Movie 3; note that time 0:00 in this graph corresponds to t = 06:04 in the movie and corresponding Figure 3B) (B) or KetelGFP (6 nuclei, from Supplemental Movie 4; note that time 0:00 in this graph corresponds to t = 4:20 in the movie and corresponding Figure 3C) (C). For each time point and nucleus, the focal plane with best rim staining for mRFP-Nup107 was used. As for GFP-Lamin C, qualitatively similar results were obtained upon quantification of a distinct movie (data not shown).
Live imaging of embryos expressing mRFP-Nup107 and GFP-Lamin C (from a protein trap transgenic line described in Schulze et al., 2005) (Figure 3A and Supplemental Movie 2) revealed that the GFP-Lamin C signal at the NE declined more progressively in prometaphase than mRFP-Nup107 (see quantifications in Figure 4A). GFP-Lamin C persisted at the SE into metaphase, where it surrounded the diffuse mRFP-Nup107 signal. In anaphase, however, GFP-Lamin C disappeared from the SE whereas Nup107 remained enriched within the spindle region (Figure 3A). Finally, GFP-Lamin C accumulated at the NE in telophase only once a full Nup107 rim signal was detected (a stage at which mRFP-Nup107 reached its half maximal intensity at the NE) and its recruitment was completed more than a minute later than Nup107 (see quantifications in Figure 5A). The dynamics of GFP-Lamin C during mitosis, in particular its persistence in the spindle envelope until late metaphase, are consistent with earlier observations of lamin behavior based on microinjection of embryos with fluorescently labeled anti-lamin Fab fragments (Paddy et al., 1996).
We next compared the behavior of mRFP-Nup107 with that of a widely used NPC marker WGA, a lectin that associates with O-linked N-acetyl-D-glucosamine-modified nucleoporins (Holt et al., 1987). In many species, the lectin WGA recognizes several Nups containing phenylalanine-glycine (FG)-repeated motifs, but in Drosophila it was demonstrated previously to reflect the localization of one FG-nucleoporin, Nup58 (Onischenko et al., 2004). For simplicity, we refer to the signal detected by WGA as “WGA-Nup.” Microinjection of Alexa488-labeled WGA into mRFP-Nup107 embryos (Figure 3B and Supplemental Movie 3) revealed that the gradual release of mRFP-Nup107 from the NE and its concomitant appearance in the nucleoplasm in prometaphase occurred before the decline of the WGA-Nup at the NE (Figure 4B). The WGA-Nup persisted at the NE into late prometaphase, longer than Nup107. As reported previously (Onischenko et al., 2005), and in marked contrast to Nup107, the WGA-Nup became evenly distributed in the cytoplasm and showed no enrichment in the spindle area during metaphase and anaphase. In telophase, the first reappearance of both markers at the NE was nearly coincident, although mRFP-Nup107 reached its half-maximal intensity at the NE slightly earlier than the WGA-Nup (Figure 5B).
We also investigated the localization of another FG-repeat nucleoporin, Nup153, in fixed mRFP–Nup107-expressing embryos by using specific anti-Nup153 antibodies (Mendjan et al., 2006). This analysis revealed that unlike WGA, Nup153 localized together with Nup107 within the spindle area during metaphase (Supplemental Figure 3). In this respect, the behavior of Nup153 is similar to that reported for the RL1 antibody (Snow et al., 1987) that recognizes FG nucleoporins in a variety of species and labels four nucleoporins (including an ∼150-kDa protein) in Drosophila (Onischenko et al., 2004). In addition, Nup153 seemed to be released from the NE before Nup107 in prophase, and it gave rise to a weaker signal than Nup107 on the reforming NE during telophase. This suggests that Nup153 and Nup107, although they both remain concentrated in the spindle area in metaphase, have slightly different behaviors during mitotic entry and exit.
Importin β, a key component of the nuclear import machinery, also plays essential roles in the NE, NPCs, and spindle assembly in vertebrates (for reviews, see Harel and Forbes, 2004). Additionally, it has been shown to interact with several nucleoporins, including Nup107 and Nup153 in Xenopus mitotic extracts (Walther et al., 2003b). In Drosophila, importin β is encoded by the ketel gene (Lippai et al., 2000). Here, we used an exon trap allele of ketel called ketelGFP in which a GFP exon is inserted between amino acids 18 and 19 of ketel within its Ran binding domain (Morin et al., 2001, Villanyi et al., 2008). Previous studies performed in vertebrates demonstrated that deletion of the first 10 or 32 residues of importin β impairs its interaction with RanGTP and leads to its increased residency at NPCs, probably at the import termination sites (i.e., the nuclear side of NPCs) (Kutay et al., 1997, and references therein). Consistent with this, fluorescence recovery after photobleaching analyses (Supplemental Figure 4) revealed that fluorescent recovery of ketelGFP at the NE and in the nucleus seemed to be slower compared with full-length GFP-human importin β expressed in HeLa cells (Rabut et al., 2004). Despite the reduced function of the ketelGFP allele compared with wild-type Ketel in the fly (Villanyi et al., 2008), its expression in one copy does not impair mitotic progression in the syncytial embryos (Figure 3C and Supplemental Movie 4). Live imaging of embryos expressing ketelGFP and mRFP-Nup107 further revealed their similar mitotic dynamics (Figure 3C and Supplemental Movie 4). Both proteins persisted within the spindle area throughout mitosis, although ketelGFP showed an increased persistence in the spindle area in anaphase, which may reflect the previously reported interaction between ketel and microtubules (Tirian et al., 2003) (Figure 3C; frame 4:20 min). In addition, ketelGFP was released from the NE slightly earlier than Nup107 (∼20–30 s) at mitotic entry and reached its half-maximal intensity at the NE slightly later than mRFP-Nup107 in telophase (Figures 4C and 5C and Supplemental Figure 5). The dynamics of ketelGFP differed from the dynamics described previously for wild-type Ketel (Trieselmann and Wilde, 2002) and rather reproduced the behavior of Nup153 (compare Supplemental Figure 3 and Figure 3C). Because importin β has been reported to have higher affinity for Nup153 compared with other vertebrate FG-repeat nucleoporins (Ben-Efraim and Gerace, 2001), the increased residency of the ketelGFP fusion at NPCs, and potentially its dynamics in mitosis, may thus reflect its binding to Nup153. Accordingly, although the ketelGFP allele does not seem to be an appropriate tool to study the dynamics of wild-type importin β in flies, it nevertheless provides a valuable marker to follow some features of NPCs dynamics in Drosophila.
NPC and NE Dynamics in Dividing Larval Neuroblasts
The rapid (10–12 min) cell cycles of the syncytial embryo are very unusual. To examine the behavior of Nup107 in a more conventional Drosophila mitosis, we recorded divisions in third-instar larval neuroblasts, another well characterized Drosophila mitotic cell type whose asymmetric division generates a large daughter neuroblast and a small ganglion mother cell (Knoblich, 2001; Chia and Yang, 2002).
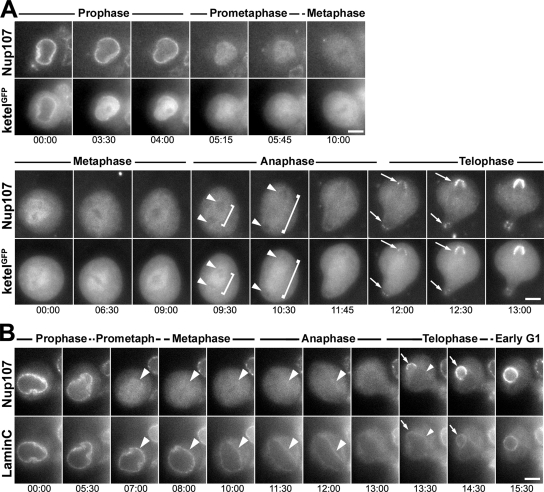
Time-lapse analysis of living neuroblasts expressing mRFP-Nup107 and ketelGFP confirmed an overall behavior for both proteins similar to that seen in syncytial embryos (Figure 6A and Supplemental Movies 5 and 6). In particular, we observed that 1) in prophase ketelGFP is released from the NE before Nup107 (Figure 6A, top, frames 3:30 and 4:00 min); 2) both proteins are enriched within the spindle area in prometaphase (albeit to a lesser extent than in syncytial embryos); 3) ketelGFP persists in the spindle area in late metaphase and anaphase, whereas Nup107 becomes gradually fainter and may even be slightly excluded from the spindle area (Figure 6A, bottom, frames 6:30–11:45 min); and 4) both proteins, although always excluded from chromatin before this stage, are simultaneously recruited to decondensing chromatin in early telophase (Figure 6A, bottom, frames 12:00–13:00 min).
In neuroblasts expressing GFP-Lamin C and mRFP-Nup107, the GFP-Lamin C signal again was found in a spindle envelope-like structure separating the spindle region from the cytoplasm and defining initially the limits of the diffuse Nup107 signal in prometaphase (Figure 6B, arrowheads, and Supplemental Movie 7). This Lamin C signal persisted throughout anaphase, although it became progressively less distinct, concomitant with the progressive cytoplasmic diffusion of Nup107. In late anaphase/telophase, Nup107 was again first seen accumulating on decondensing chromatin at the side proximal to the former spindle pole. At this stage, faintly labeled GFP-Lamin C structures lacking detectable mRFP-Nup107 staining were observed (Figure 6B, Supplemental Figure 6A, and Supplemental Movies 7 and 8).
The observed enrichment of ketelGFP and to some extent of mRFP-Nup107 within the spindle region of metaphase neuroblasts, together with the persistence of the GFP–Lamin C-labeled structure, suggested the existence of a spindle envelope in these cells. Although neuroblasts are well-studied cells, little is known concerning the dynamics of their NE in mitosis. In particular, it is unknown whether they undergo a semi-closed mitosis as reported for syncytial embryos (Stafstrom and Staehelin, 1984) and embryonic-derived cells in culture (Debec and Marcaillou, 1997), in which the mitotic spindles are enclosed in membranous structures that become permeable to 70-kDa molecules in prophase (Paddy et al., 1996). To further address this question, we examined dividing neuroblasts expressing GFP-protein disulfide isomerase (PDI), a component of the endoplasmic reticulum lumen (Bobinnec et al., 2003). This analysis revealed the presence of GFP–PDI-labeled membranous structures surrounding the mitotic spindle that persisted into anaphase (Supplemental Figure 6B).
In summary, our study has determined that the order of events governing the disassembly and reassembly of the NE and NPCs are quite similar in neuroblasts and syncytial embryos, despite differences in the mitotic kinetics of these two developmental stages. In particular, neuroblasts seem to undergo a semi-closed mitosis, similar to the mitosis reported previously for the embryonic tissues. The higher cytoplasmic signal of both ketelGFP and mRFP-Nup107 observed in neuroblasts (compared with syncytial embryos) could be due either to an increased permeability of the spindle envelope in differentiated tissues, or to the longer duration of their mitoses, allowing the progressive diffusion of large complexes.
The Association of Mad2 with NPCs at the End of Mitosis Is a Two-Step Process
Although best characterized with respect to its checkpoint function and kinetochore localization, Mad2 associates with NPCs during interphase in both human and yeasts (Campbell et al., 2001; Iouk et al., 2002). In Drosophila neuroblasts, Mad2 is primarily in the nucleoplasm, but a fraction of it is associated with the NE (Buffin et al., 2005). To further explore Mad2's association with the NE, we directly compared the behavior of GFP-Mad2 with that of mRFP-Nup107 during mitotic entry and exit.
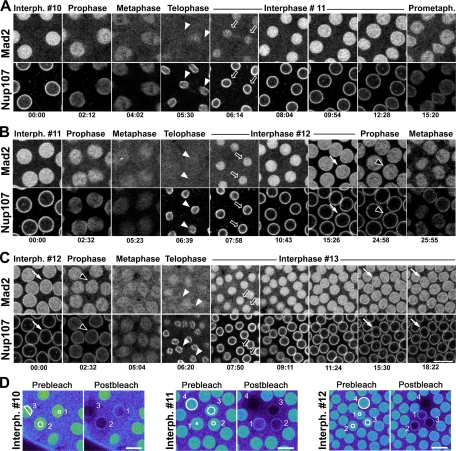
Somewhat surprisingly, NE-associated Mad2 was only obvious in the later interphases (cycles 12 and 13) of syncytial embryos. Mad2 localization at the NPCs of interphase nuclei 10 and 11 was almost undetectable compared with the strong intranuclear fluorescence (Figure 7and Supplemental Movies 9–11). One explanation for the almost undetectable NE-associated Mad2 signal in interphases 10 and 11 was that the intense nucleoplasmic GFP-Mad2 signal was masking a weaker signal at the NE. To test this, we performed photobleaching of small regions (corresponding to areas of 1–16 μm2) on interphase syncytial embryos whose only source of Mad2 is GFP-Mad2 (Buffin et al., 2007) (Figure 7D). The intranuclear pool of Mad2 seemed to be freely diffusible, because even a small photobleached spot (marked as nucleus 1 of the various cycles in Figure 7D) resulted in a nearly uniform 30–50% drop of fluorescence throughout the nucleoplasm. More extensive photobleaching of Mad2 to background levels revealed that Mad2 is also present in the cytoplasm in these syncytial embryos (compare the fluorescence intensities outside vs. inside the nuclei marked 2–4). Most importantly, these photobleaching experiments revealed that a fraction of Mad2 did in fact associate with the NE of cycles 10 and 11. This indicates that Mad2 is present at the NE as well as in the nucleus in all syncytial embryonic interphases, but the NE pool is masked by the intensity of the intranuclear Mad2 signal in cycles 10 and 11.
Figure 7.
Dynamics of GFP-Mad2 in syncytial embryonic mitoses. (A–C) Selected frames of TLSC acquisitions describing the behavior of GFP-Mad2 and mRFP-Nup107 during three consecutive embryonic cleavage cycles: interphase #10 to prometaphase #11 (A), interphase #11 to metaphase #12 (B), and interphase #12 to interphase #13 (C). Best focal plane of a 4.0 μm z-stack is shown for each time frame. Bar, 10 μm. Time is in minutes:seconds. In each cycle, accumulation of Mad2 in the reforming nuclei begins only once a continuous perinuclear Nup107 staining is observed (arrowheads and open arrows). Note the presence of a fraction of GFP-Mad2 in between the two reforming nuclei (seen in A and C). Arrows point to the NE localization of GFP-Mad2 in interphase nuclei of cycles #12 (B and C), and #13 (C), and black arrowheads to prophase cells, revealing its dissociation from the NE before mRFP-Nup107. (D) Photobleaching analysis of GFP-Mad2 in embryonic nuclei. Small regions (outlined in white in the first frames) within three or four nuclei of interphases #10, #11, or #12 were bleached. Images are in pseudocolor to facilitate the visualization of differences in fluorescence intensity. Bars, 10 μm. Note that the fluorescence signal at the NE of interphase nuclei of cycles #10, #11, and #12 becomes clearly detectable after bleaching the intranuclear pool (postbleach) and is of similar intensity in all three interphase cycles (compare nuclei 2 in the three series).
In prophase of cycle 12 embryos, GFP-Mad2 seemed to dissociate from the NE before mRFP-Nup107 (black arrowheads in Figure 7, B and C). In prometaphase and metaphase, GFP-Mad2 labels kinetochores and was also partly confined within the spindle envelope (Figure 7, A–C; see also Figure 10A for kinetochore staining). During telophase, at the time of the initial Nup107 recruitment on the reforming nuclei, there was no concomitant nuclear accumulation of Mad2 (white arrowheads in Figure 7, A–C). Only once the nucleus was entirely framed by a ring of mRFP-Nup107 did GFP-Mad2 begin to stream into the nucleus (open arrows in Figure 7, A–C). Interestingly, however, Mad2 did not immediately associate with the NE during its importation and became detectable at the NE of cycle 12 or cycle 13 embryos only several minutes later (Figure 7, B and C, and Movies 10 and 11).
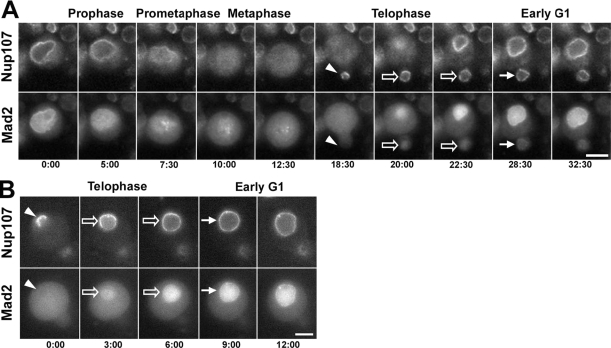
Both the early dissociation of GFP-Mad2 and the late recruitment of Mad2 to the newly formed NE were also seen in the more conventional mitoses of larval neuroblasts (Figure 8). As already observed for Nup107 and ketelGFP (Figure 6A), GFP-Mad2 was, however, more broadly diffused throughout the cytoplasm during prometaphase and metaphase in neuroblasts compared with syncytial embryos (compare Figure 8A with Figure 7, A–C). As in syncytial embryos, no nuclear accumulation of Mad2 could be detected in neuroblasts during telophase (arrowheads in Figure 8). GFP-Mad2 began to stream into the nucleus only ∼3 min later (open arrows) and NE-associated Mad2 again became evident after six to seven additional minutes, in early G1 (arrows). Thus, both in embryos and neuroblasts, Mad2 seems to be first imported into the nucleoplasm of the reforming nucleus, and only subsequently does a fraction of this checkpoint protein associate with the NE.
Figure 8.
Association of Mad2 with the NPCs at the end of mitosis is a two-step process. Selected frames of wide-field time-lapse acquisitions of dividing larval neuroblasts expressing mRFP-Nup107 and GFP-Mad2. In A, a complete mitosis is shown, with the smaller daughter cell in sharp focus. Note that GFP-Mad2 is released from the NE during prophase, whereas mRFP-Nup107 remains associated with the NE/SE until prometaphase (A, frames 5:00 and 7:30 min) and that Mad2, but not Nup107, associates with kinetochores. (B) The final steps of mitosis are depicted for a different neuroblast in which the nucleus of the large daughter cell (neuroblast) is in sharp focus. In A and B, note the diffuse cytoplasmic distribution of GFP-Mad2 in telophase, whereas mRFP-Nup107 is already being recruited at the periphery of the decondensing chromosomes (arrowheads). The progressive nuclear accumulation of GFP-Mad2 begins once a full mRFP-Nup107 rim has formed (open arrows) and its subsequent enrichment at the NE only occurs several minutes later, during early G1 (arrows). Time in minutes:seconds. Bars, 5 μm.
Nuclear Import of Mad2 and Mad1 Precedes the Recruitment of Mtor at NPCs in Telophase
In addition to Mad2, the checkpoint protein Mad1, which interacts with Mad2 throughout the cell cycle (Chen et al., 1998; Campbell et al., 2001; Luo et al., 2002) is also localized on the nuclear side of the NPCs in vertebrates and yeasts (Campbell et al., 2001). In budding and fission yeasts, the NE localization of Mad2 relies on Mad1 (Iouk et al., 2002; Ikui et al., 2002) and in Saccharomyces cerevisiae, the basket nucleoporins Mlp1/2 were further demonstrated to be major binding sites for Mad1 at NPCs (Scott et al., 2005). This prompted us to analyze the behavior of Drosophila Mad1 (CG2072, listed in Flybase as TXBP181-like) and Mtor (Megator, the Drosophila orthologue of S. cerevisiae Mlp1/2 and vertebrate Tpr; Qi et al., 2004) in fixed mRFP-Nup107 Drosophila embryos. As observed for GFP-Mad2, and unlike mRFP-Nup107 or Mtor, Mad1 was mainly nuclear in early embryos, and its NE localization only became apparent in late preblastoderm or cellularized embryos (Figure 9A, compare top and bottom panels, and Figure 9B). We therefore further investigated its behavior during the asynchronous mitotic divisions on cellularized embryos, in which Mad1 is best detected at the NE (Figure 9B). In prometaphase, a fraction of Nup107 was still detectable at the NE, whereas Mad1 and Mtor were localized within the spindle area (Figure 9B, bottom, brackets). In metaphase, Mtor displayed its typical localization at the spindle matrix (Qi et al., 2004), and a fraction of Mad1 also persisted within the spindle area, whereas mRFP-Nup107 had a more diffuse localization throughout the cell (Figure 9B, top, brackets). On NE reassembly, Mad1 was first excluded from the reforming nuclei, even from those already ringed with an apparently continuous mRFP-Nup107 signal (arrowheads in Figure 9B). In a subsequent stage, nuclear accumulation of Mad1 was observed, whereas Mtor was still exclusively localized in the cytoplasm (open arrows). Still later, some Mad1 was detected at the NE, and this was always coincident with NE localization of Mtor (arrows in Figure 9B).
Together, this analysis has revealed the shared behavior of Mad1 and Mad2 during all stages of mitosis, including their nuclear import before their NE anchoring at the end of mitosis. Moreover, their recruitment to the NE in early G1, coincident with NPC recruitment of Mtor, suggests that, as in yeast, Mtor may serve as the NPC anchoring determinant of these two checkpoint proteins in Drosophila (see Discussion).
Nup107 Is Undetectable at Drosophila Kinetochores
Another feature of metazoan mitosis is the relocalization of several NPC constituents to mitotic structures. In both human cells and C. elegans embryos, a fraction of the Nup107-160 NPC-subcomplex localizes to kinetochores during mitosis (Belgareh et al., 2001; Galy et al., 2006). Surprisingly, none of the mRFP-Nup107 movies acquired in fly embryos or neuroblasts revealed any kinetochore association (Figures 2–3 and 6–8). To further explore this issue, we compared the mitotic behavior of mRFP-Nup107 with that of GFP-tagged Mad2, which localizes to unattached kinetochores in mitosis (Buffin et al., 2005). Analysis of live embryos expressing both fusion proteins did not reveal any colocalization between mRFP-Nup107 and GFP-Mad2 at kinetochores (Figure 10A; also see Figure 7 and Supplemental Movies 9–11). Because mammalian Nup107 is enriched at kinetochores lacking microtubules (Orjalo et al., 2006; Zuccolo et al., 2007), we next examined embryos after injection of the microtubule-depolymerizing drug colchicine (Figure 10A). Under these conditions, which cause metaphase arrest and a notable accumulation of Mad2 at kinetochores, mRFP-Nup107 still showed no kinetochore association, and rather seemed to be excluded from the Mad2-labeled kinetochore area (Figure 10A). It is, however, noteworthy that the mRFP-Nup107 signal persisted within the area defined by the spindle envelope, indicating that this localization is independent of microtubules.
As in syncytial embryos, mRFP-Nup107 staining could not be detected at kinetochores in control or colchicine-treated neuroblasts (Figure 10B; also see Figure 8 and Supplemental Movie 12), indicating that the absence of detectable Nup107 at kinetochores is a general feature of fly mitosis. Similarly, no signal could be detected at kinetochores upon immunostaining of syncytial embryos by using specific anti-Nup107 antibodies (Supplemental Figure 2; our unpublished data). Thus, our data indicate that, unlike in vertebrates and worms, Drosophila Nup107 shows no evidence of association with kinetochores.
DISCUSSION
We have monitored the in vivo distribution of a functional fluorescently tagged Nup107 nucleoporin in Drosophila syncytial embryos and larval neuroblasts, and compared its behavior with that of other NPC and NE markers. In doing so, we have refined our understanding of the dynamics of NE and NPC components during Drosophila mitosis.
NPC Disassembly and Reassembly in Drosophila Mitosis
During prophase and prometaphase, Nup107 and the other NPC-associated proteins examined were progressively released from the NE, most likely reflecting the loose synchrony of NPC disassembly (Stafstrom and Staehelin, 1984; Kiseleva et al., 2001). Drosophila NPC disassembly was completed by metaphase and preceded lamina depolymerization, a property shared by C. elegans embryos (Lee et al., 2000). Compared with Nup107, ketelGFP (which, as discussed above, likely interacts with peripheral FG-Nups), as well as Nup153 and Mtor (the Drosophila orthologue of Tpr; Qi et al., 2004), two asymmetrically localized nuclear basket nucleoporins, were released slightly earlier, in prophase. Previous studies performed in Drosophila embryos, starfish oocytes, and in vitro assembled Xenopus nuclei (Kiseleva et al., 2001; Lenart et al., 2003; Cotter et al., 2007), also found an earlier release of peripheral NPC structures.
In contrast, the WGA signal, which was demonstrated previously to reflect the localization of Drosophila Nup58 (Onischenko et al., 2004), persisted longer than mRFP-Nup107 at the NE periphery. Although Nup58 forms part of the central channel of the NPC in mammals (Guan et al., 1995), a previous electron microscopy study suggested that the release of the central transporter or central material occurs at an early stage of NPC disassembly in Drosophila embryos (Kiseleva et al., 2001). Our unexpected result was however corroborated by a study that came out while this manuscript was under revision, revealing that human Nup58 remains longer than Nup133 (another constituent of the Nup107 complex) in fragments of the NE during disassembly (Dultz et al., 2008). During the closed mitosis of the fungus Aspergillus nidulans, however, FG-nups, including those making up the central channel, disperse throughout the cell, whereas constituents of the Nup107 subcomplex persist at NPCs (De Souza et al., 2004; Osmani et al., 2006). Accordingly, the fate of NPC components in Drosophila and human cells, in which all NPC constituents disassemble, seems to be distinct from that occurring during the closed mitosis of A. nidulans.
The various steps of NPC reassembly at the end of mitosis have been extensively investigated previously both in vivo and in vitro (for reviews, see Hetzer et al., 2005; Prunuske and Ullman, 2006). Our study, revealing the early recruitment of Nup107 to the reforming NE in late anaphase, the slightly delayed recruitment of ketelGFP and WGA-Nup, and the late recruitment of Mtor, is consistent with previous studies in other organisms (Bodoor et al., 1999; Burke and Ellenberg, 2002; Dultz et al., 2008). Along with data recently gathered in human cells (Dultz et al., 2008), it further indicates that NPC reassembly is not simply reversing the sequence of its disassembly.
Mad1 and Mad2 during Drosophila Mitosis
We found that a fraction of Drosophila Mad1 and Mad2 associates with the NE in syncytial embryos, which probably reflects their binding to the nuclear side of the NPCs as demonstrated in yeast and vertebrates (Campbell et al., 2001; Iouk et al., 2002). The release of GFP-Mad2 at an early stage of Drosophila NPC disassembly in prophase and the late recruitment of Mad1 and Mad2 at the NE corroborate previous observations in Xenopus and human cells (Chen et al., 1996; Shah et al., 2004). However, we show in addition that relocalization of Drosophila Mad1 and Mad2 to the NE occurs in two steps, with their nuclear import preceding their NE association. This shared behavior suggests that both proteins might be reimported into the nucleus as a complex.
The coincident NPC recruitment of Mad1 and Mad2 with Mtor indicates that Drosophila Megator likely represents their NPC-anchoring determinant as are the yeast Mtor orthologues (Mlp1/2) (Scott et al., 2005). This seems to be an evolutionarily conserved interaction, because small interfering RNA-induced depletion of Tpr (the vertebrate orthologue of yeast Mlp1/2-Drosophila Mtor) in HeLa cells significantly reduces Mad2 labeling at the NE (our unpublished data). Our in vivo study further revealed that in syncytial embryos, a fraction of GFP-Mad2 is found within the spindle area in metaphase and subsequently localizes between the two reforming nuclei during telophase (Figure 7), localizations similar to that of Mtor (Qi et al., 2004). It is thus conceivable that an interaction between Mad1-Mad2 and Mtor may also occur at this stage of mitosis.
Specific Features of Drosophila Mitosis
Although a spindle envelope was initially identified in syncytial Drosophila embryos (Stafstrom and Staehelin, 1984; Harel et al., 1989; Bobinnec et al., 2003; Frescas et al., 2006; Wagner et al., 2006) and embryo-derived Drosophila cell lines (Debec and Marcaillou, 1997; Maiato et al., 2006), it was unclear whether all Drosophila cells had one. In the S2R+ cell line for example, NE remnants were observed in cells fixed shortly after NE breakdown, but not at later stages of metaphase (Maiato et al., 2006). Our in vivo studies of larval neuroblasts, revealing the presence of a membrane-like structure stained by GFP-PDI and the persistence of a fraction of lamins around the mitotic spindle up to anaphase, indicate that the spindle envelope is not restricted to embryonic cells and thus seems to be a general feature of Drosophila mitosis. By confining the prometaphase chromosomes as well as a subset of proteins to a small region of the cell, the persistence of an envelope and/or the Mtor-defined spindle matrix (Qi et al., 2004) might explain the remarkable efficiency of fly mitosis, in which kinetochores capture spindle fibers so rapidly that chromosomes can segregate properly even in the absence of the spindle checkpoint (Buffin et al., 2007).
Another specific feature of Drosophila mitosis uncovered by our study was the absence of detectable Nup107 at kinetochores. In embryos and to a lesser extent larval neuroblasts, however, a fraction of Nup107 remained confined within the area defined by the spindle envelope up to late metaphase. Unlike in Xenopus extracts, in which the localization of the Nup107-160 complex throughout the mitotic spindle requires microtubules (Orjalo et al., 2006), this signal persists upon colchicine treatment in Drosophila embryos (Figure 10A). Accordingly, it may reflect either a weak interaction of the Nup107-160 complex with the Mtor-defined spindle, or the confinement of this huge complex within the spindle envelope.
Because the Nup107 complex is also absent from kinetochores in fission yeast, which undergoes a closed mitosis (Bai et al., 2004), the lack of Nup107 at kinetochores could potentially be related to the presence of a spindle envelope in flies. However, the localization of Nup107 at kinetochores of early C. elegans embryos in which NPCs and NE disassembly only take place in anaphase does not support this hypothesis (Lee et al., 2000; Galy et al., 2006). Moreover, Nup107 was not found at kinetochores during the open mitosis of the fungus Ustilago maydis in which Nup107 is first dispersed throughout the cytoplasm before being recruited to chromatin in metaphase (Theisen et al., 2008). Understanding how various spatial and temporal localization of the Nup107-160 complex occurring in distinct organisms may underlie species-specific properties of mitotic spindle assembly and chromosome segregation will be a fruitful avenue of future studies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Isabelle Loïodice for initial characterization of the Drosophila anti-Nup107 antibodies; Asifa Akhtar, Alain Debec, Arnaud Echard, and B. Chia's laboratory for providing fly lines and/or antibodies; János Szabad for sharing results before publication; T. Piolot and X. Baudin (imaging platform; Institut Jacques Monod Paris) for help with spinning disk confocal imaging; and the Curie imaging staff and members of our laboratories for constant support and valuable comments during the course of this work. This research was supported by the Centre National de la Recherche Scientifique; the Institut Curie; the Ligue Nationale contre le Cancer (équipe labelisée 2006, to V. D.); the Association pour la Recherche contre le Cancer (to V. D. and R. K.); and the Ministère de l'Education Nationale, de la Recherche et de l'Enseignement Supérieur (A.C.I. “Jeunes chercheurs,” to V. D.). K. K. was supported by postdoctoral fellowships from the Human Frontier Science Program, the Centre National de la Recherche Scientifique, and the Institut Curie.
Abbreviations used:
- NE
nuclear envelope
- NPC
nuclear pore complex
- Nup
nucleoporin
- SE
spindle envelope
- TLSCM
time-lapse scanning confocal microscopy
- WGA
wheat germ agglutinin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1162) on June 18, 2008.
REFERENCES
- Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Golic K. G., Hawley R. S. Drosophila: A Laboratory Handbook. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Babu J. R., Jeganathan K. B., Baker D. J., Wu X., Kang-Decker N., van Deursen J. M. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S. W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I., Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Marcaillou C., Morin X., Debec A. Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil. Cytoskeleton. 2003;54:217–225. doi: 10.1002/cm.10094. [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W. H., Bastos R., Lohka M., Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 1999;112:2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Buffin E., Emre D., Karess R. E. Flies without a spindle checkpoint. Nat. Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M. E., Karess R. E. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- Burke B., Ellenberg J. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Campbell M. S., Chan G. K., Yen T. J. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Shevchenko A., Mann M., Murray A. W. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Waters J. C., Salmon E. D., Murray A. W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chia W., Yang X. Asymmetric division of Drosophila neural progenitors. Curr. Opin. Genet. Dev. 2002;12:459–464. doi: 10.1016/s0959-437x(02)00326-x. [DOI] [PubMed] [Google Scholar]
- Clarkson M., Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- Cotter L., Allen T. D., Kiseleva E., Goldberg M. W. Nuclear membrane disassembly and rupture. J. Mol. Biol. 2007;369:683–695. doi: 10.1016/j.jmb.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Dasso M. The Ran GTPase: theme and variations. Curr. Biol. 2002;12:R502–R508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- Debec A., Marcaillou C. Structural alterations of the mitotic apparatus induced by the heat shock response in Drosophila cells. Biol. Cell. 1997;89:67–78. doi: 10.1016/s0248-4900(99)80082-3. [DOI] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. G., Piano F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr. Biol. 2006;16:1757–1763. doi: 10.1016/j.cub.2006.07.071. [DOI] [PubMed] [Google Scholar]
- Foe V. E., Alberts B. M. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Frescas D., Mavrakis M., Lorenz H., Delotto R., Lippincott-Schwartz J. The secretory membrane system in the Drosophila syncytial blastoderm embryo exists as functionally compartmentalized units around individual nuclei. J. Cell Biol. 2006;173:219–230. doi: 10.1083/jcb.200601156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Askjaer P., Franz C., Lopez-Iglesias C., Mattaj I. W. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr. Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- Gorjanacz M., Jaedicke A., Mattaj I. W. What can Caenorhabditis elegans tell us about the nuclear envelope? FEBS Lett. 2007;581:2794–2801. doi: 10.1016/j.febslet.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Guan T., Muller S., Klier G., Pante N., Blevitt J. M., Haner M., Paschal B., Aebi U., Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Forbes D. J. Importin beta: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D. J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Harel A., Zlotkin E., Nainudel-Epszteyn S., Feinstein N., Fisher P. A., Gruenbaum Y. Persistence of major nuclear envelope antigens in an envelope-like structure during mitosis in Drosophila melanogaster embryos. J. Cell Sci. 1989;94:463–470. doi: 10.1242/jcs.94.3.463. [DOI] [PubMed] [Google Scholar]
- Hetzer M. W., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Holt G. D., Snow C. M., Senior A., Haltiwanger R. S., Gerace L., Hart G. W. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J. Cell Biol. 1987;104:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui A. E., Furuya K., Yanagida M., Matsumoto T. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 2002;115:1603–1610. doi: 10.1242/jcs.115.8.1603. [DOI] [PubMed] [Google Scholar]
- Iouk T., Kerscher O., Scott R. J., Basrai M. A., Wozniak R. W. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen K. R., Johansen J. Studying nuclear organization in embryos using antibody tools. In: Henderson D. S., editor. Methods in Molecular Biology, Vol. 247: Drosophila Cytogenetics Protocols. Totowa, NJ: Humana Press; 2004. pp. 215–234. [DOI] [PubMed] [Google Scholar]
- Kiseleva E., Rutherford S., Cotter L. M., Allen T. D., Goldberg M. W. Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J. Cell Sci. 2001;114:3607–3618. doi: 10.1242/jcs.114.20.3607. [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. Asymmetric cell division during animal development. Nat. Rev. Mol. Cell Biol. 2001;2:11–20. doi: 10.1038/35048085. [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde E., Bischoff F. R., Mattaj I. W., Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. K., Gruenbaum Y., Spann P., Liu J., Wilson K. L. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P., Rabut G., Daigle N., Hand A. R., Terasaki M., Ellenberg J. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J. Cell Biol. 2003;160:1055–1068. doi: 10.1083/jcb.200211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R. Y., Fahrenkrog B. The nuclear pore complex up close. Curr. Opin. Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Lippai M., et al. The Ketel gene encodes a Drosophila homologue of importin-beta. Genetics. 2000;156:1889–1900. doi: 10.1093/genetics/156.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. T., Chan G. K., Hittle J. C., Fujii G., Lees E., Yen T. J. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Maiato H., Hergert P. J., Moutinho-Pereira S., Dong Y., Vandenbeldt K. J., Rieder C. L., McEwen B. F. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A., Vlcek S., Gruenbaum Y., Foisner R. Breaking and making of the nuclear envelope. J. Cell Biochem. 2005;95:454–465. doi: 10.1002/jcb.20433. [DOI] [PubMed] [Google Scholar]
- Mendjan S., et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E. A., Gubanova N. V., Kieselbach T., Kiseleva E. V., Hallberg E. Annulate lamellae play only a minor role in the storage of excess nucleoporins in Drosophila embryos. Traffic. 2004;5:152–164. doi: 10.1111/j.1600-0854.2004.0166.x. [DOI] [PubMed] [Google Scholar]
- Onischenko E. A., Gubanova N. V., Kiseleva E. V., Hallberg E. Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol. Biol. Cell. 2005;16:5152–5162. doi: 10.1091/mbc.E05-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo A. V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S. G., Fontoura B., Briggs S., Dasso M., Forbes D. J. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. H., Davies J., Liu H. L., Osmani S. A. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddy M. R., Saumweber H., Agard D. A., Sedat J. W. Time-resolved, in vivo studies of mitotic spindle formation and nuclear lamina breakdown in Drosophila early embryos. J. Cell Sci. 1996;109:591–607. doi: 10.1242/jcs.109.3.591. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Prunuske A. J., Ullman K. S. The nuclear envelope: form and reformation. Curr. Opin. Cell Biol. 2006;18:108–116. doi: 10.1016/j.ceb.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rasala B. A., Orjalo A. V., Shen Z., Briggs S., Forbes D. J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S. R., Curio-Penny B., Li Y., Imani R. A., Rydberg L., Geyer P. K., Wallrath L. L. Molecular genetic analysis of the nested Drosophila melanogaster lamin C gene. Genetics. 2005;171:185–196. doi: 10.1534/genetics.105.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T. U. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Scott R. J., Lusk C. P., Dilworth D. J., Aitchison J. D., Wozniak R. W. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D. W. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Snow C. M., Senior A., Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom J. P., Staehelin L. A. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur. J. Cell Biol. 1984;34:179–189. [PubMed] [Google Scholar]
- Stukenberg P. T., Macara I. G. The kinetochore NUPtials. Nat. Cell Biol. 2003;5:945–947. doi: 10.1038/ncb1103-945. [DOI] [PubMed] [Google Scholar]
- Theisen U., Straube A., Steinberg G. Dynamic rearrangement of nucleoporins during fungal “open” mitosis. Mol. Biol. Cell. 2008;19:1230–1240. doi: 10.1091/mbc.E07-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirian L., Timinszky G., Szabad J. P446L-importin-beta inhibits nuclear envelope assembly by sequestering nuclear envelope assembly factors to the microtubules. Eur. J. Cell Biol. 2003;82:351–359. doi: 10.1078/0171-9335-00324. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Trieselmann N., Wilde A. Ran localizes around the microtubule spindle in vivo during mitosis in Drosophila embryos. Curr. Biol. 2002;12:1124–1129. doi: 10.1016/s0960-9822(02)00934-x. [DOI] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W. H., Forbes D. J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanyi Z., Debec A., Timinszky G., Tirian L., Szabad J. Long persistence of importin-beta explains extended survival of cells and zygotes that lack the encoding gene. Mech. Dev. 2008;125:196–206. doi: 10.1016/j.mod.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wagner N., Kagermeier B., Loserth S., Krohne G. The Drosophila melanogaster LEM-domain protein MAN1. Eur. J. Cell Biol. 2006;85:91–105. doi: 10.1016/j.ejcb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Walther T. C., et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003a;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I. W., Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003b;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Zuccolo M., et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.