Abstract
The highly conserved LC8/DYNLL family proteins were originally identified in axonemal dyneins and subsequently found to function in multiple enzyme systems. Genomic analysis uncovered a third member (LC10) of this protein class in Chlamydomonas. The LC10 protein is extracted from flagellar axonemes with 0.6 M NaCl and cofractionates with the outer dynein arm in sucrose density gradients. Furthermore, LC10 is specifically missing only from axonemes of those strains that fail to assemble outer dynein arms. Previously, the oda12-1 insertional allele was shown to lack the Tctex2-related dynein light chain LC2. The LC10 gene is located ∼2 kb from that of LC2 and is also completely missing from this mutant but not from oda12-2, which lacks only the 3′ end of the LC2 gene. Although oda12-1 cells assemble outer arms that lack only LC2 and LC10, this strain exhibits a flagellar beat frequency that is consistently less than that observed for strains that fail to assemble the entire outer arm and docking complex (e.g., oda1). These results support a key regulatory role for the intermediate chain/light chain complex that is an integral and highly conserved feature of all oligomeric dynein motors.
INTRODUCTION
The LC8 protein (now termed DYNLL1 and DYNLL2 in mammals; Pfister et al., 2005) was originally identified using biochemical methods as a component of the Chlamydomonas outer dynein arm (Piperno and Luck, 1979; Pfister et al., 1982). Molecular cloning revealed that it defines a very highly conserved family of proteins sharing ∼90% sequence identity that are present in a wide variety of eukaryotes ranging from algae to mammals, including those that lack motile cilia and flagella (King and Patel-King, 1995). Subsequent studies demonstrated that LC8/DYNLL proteins are also integral components of, or are associated with, many distinct cellular systems including cytoplasmic dynein (King et al., 1996), the dynein that powers retrograde intraflagellar transport (Pazour et al., 1998; Rompolas et al., 2007), myosin V (Espindola et al., 2000), neuronal nitric oxide synthase (Jaffrey and Snyder, 1996), flagellar radial spokes (Yang et al., 2001), the Bim proapoptotic factor (Puthalakath et al., 1999), Rabies virus P protein (Raux et al., 2000; Poisson et al., 2001), Drosophila swallow (Schnorrer et al., 2000), and many others. More distant homologues are present in higher plants (King and Patel-King, 1995) and have been found to associate with sireviruses (Havecker et al., 2005). LC8/DYNLL proteins form symmetric dimers with two identical grooves into which short segments of the target proteins bind (Liang et al., 1999; Fan et al., 2001); the monomer-dimer transition can be modulated by altering the pH and by phosphorylation (Liang et al., 1999; Nyarko et al., 2005; Song et al., 2008).
In Drosophila, complete lack of LC8 (ddlc1) function is embryonic lethal due to the induction of apoptosis, whereas partial loss-of-function alleles show pleiotropic defects in bristle and wing development, female sterility, and altered neuronal development (Dick et al., 1996a; Phillis et al., 1996). The Aspergillus LC8/DYNLL orthologue (NudG) is required for nuclear distribution along hyphae presumably due to its role in cytoplasmic dynein activity (Beckwith et al., 1998). However, in both Saccharomyces cerevisiae (Dick et al., 1996b) and Schizosaccharomyces pombe (Miki et al., 2002) null alleles have only very minor phenotypes. Similarly, a null mutant for LC8 in Chlamydomonas grows at wild-type rates; however, it does exhibit marked defects in retrograde intraflagellar transport and forms only short flagellar stubs that are deficient for both outer and inner dynein arms, radial spokes, and the projections within the B-tubules of the axonemal outer doublet microtubules (Pazour et al., 1998).
In the Chlamydomonas outer dynein arm, LC8 is a component of the intermediate chain/light chain (IC/LC) complex that is located at the base of the soluble dynein particle and is involved in attachment of the motor to its target site within the flagellar axoneme (King et al., 1991; Wilkerson et al., 1995; and see King and Kamiya, 2008 for recent review). In addition, a LC8-related protein termed LC6 that shares 41% sequence identity is also present (Pfister et al., 1982; King and Patel-King, 1995); intriguingly, orthologues of LC6 have yet to be identified in other organisms, suggesting that its role in outer arm function may be Chlamydomonas-specific. Lack of LC6 leads to a subtle decrease in flagellar beat frequency but does not affect outer arm dynein assembly within the axonemal superstructure (Pazour and Witman, 2000; DiBella et al., 2005).
Here we demonstrate that Chlamydomonas expresses a third member of the LC8 family (termed LC10) that is an integral component of the outer dynein arm. We find that LC10 is necessary for wild-type motor function but is not required for dynein assembly. These observations suggest there are fundamental differences in the roles played by the various members of this ubiquitous class of proteins. Proteins closely related to LC10 are found across a broad phylogenetic spectrum in organisms with motile cilia, and we also present evidence to support the assignment of DNAL4 as the mammalian orthologue of Chlamydomonas LC10.
MATERIALS AND METHODS
Strains and Beat Frequency Analysis
The Chlamydomonas reinhardtii strain 1132D was used as wild type. The mutant strains used were fla14, ida1, ida4, oda1, oda2, oda3, oda4, oda4-s7, oda4-s7 oda11, oda5, oda6, oda6-r75, oda6-r88, oda7, oda8, oda9, oda12-1, oda12-2, oda13, and pf28 pf30 ssh1 (also known as the WS4 isolate). All strains except oda6-r75, oda6-r88, and the oda4-s7 oda11 double mutant may be obtained from the Chlamydomonas Center (http://www.chlamy.org/). Chlamydomonas were grown in R medium with continuous bubbling with 5% CO2 95% air under a 15 h/9 h light/dark cycle (Witman, 1986). Beat frequency was determined using the population-based fast Fourier transform method of (Kamiya, 2000) as described previously (Wakabayashi and King, 2006).
Flagellar Isolation and Dynein Purification
Flagella were detached from Chlamydomonas strains by treatment with dibucaine and were isolated using standard methods (King, 1995). Subsequently, the membrane and matrix components were solubilized with 1% IGEPAL CA-630 in 30 mM HEPES, pH 7.4, 5 mM MgSO4, 0.5 mM EDTA, 25 mM KCl, and the resulting axonemes were treated with 0.6 M NaCl to extract the dynein arms. Dyneins were then purified by sedimentation in 5–20% sucrose density gradients using a Beckman SW-55 rotor (Fullerton, CA).
Molecular Cloning of LC10
The LC10 coding sequence was first assembled from three expressed sequence tags (ESTs) and a probe encompassing the protein coding sequence obtained from first strand cDNA using the PCR. Subsequently a full-length cDNA was isolated from a λZapII library made from mRNA derived from wild-type Chlamydomonas that were actively regenerating their flagella (Wilkerson et al., 1995) using our standard methods (King and Patel-King, 1995).
Fusion Proteins and Antibody Production
The LC10 coding region was obtained using the PCR and subcloned into the pMAL-c2 vector across the XmnI/XbaI restriction sites. This resulted in the fusion of LC10 to the C-terminus of maltose-binding protein (MBP) via a hydrophilic linker that incorporated a Factor Xa proteolytic cleavage site. The full-length fusion protein was used as the immunogen for antibody production in rabbit CT247, and specific antibody was subsequently obtained from serum by blot purification with recombinant LC10. The MBP-LC6 and MBP-LC8 fusion proteins were described previously (King and Patel-King, 1995). Using a similar protocol, we also raised antibody CT254 against a MBP fusion with the murine LC10-related protein DNAL4; the DNAL4 coding sequence was obtained using the PCR from expressed sequence tag BQ895415.
Electrophoresis and Immunoblotting
All samples were separated by electrophoresis in 5–15% acrylamide gradient gels and were either stained with Coomassie blue, or blotted to nitrocellulose for immunostaining using standard methods. Nitrocellulose blots were first stained for total protein using 0.05% Reactive Brown 10 (aq.) before incubation with 5% nonfat dry milk 0.05% Tween in Tris-buffered saline. Primary antibody reactivity was detected with a peroxidase-conjugated goat anti-rabbit secondary antibody and enhanced chemiluminescence (ECL; GE Healthcare, Little Chalfont, United Kingdom).
Preparation of Cytoplasmic Extracts
Five hundred-milliliter cultures of Chlamydomonas were grown to late-log phase and harvested by low speed centrifugation to yield 1.5–2 ml of packed cells. Cells were resuspended in 6–10 ml of 10 mM HEPES, pH 7.4, containing protease inhibitor cocktail (P8340; Sigma, St. Louis, MO), 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride and deflagellated. After three washes with 10 mM HEPES, pH 7.4, cells (∼15 ml) were then disrupted by passage three times through a French press to ensure complete lysis. Subsequently, the lysate was subject to ultracentrifugation using a TLA100.2 rotor at 33,000 rpm for 2 h at 4°C. After centrifugation, the lysate supernatant was removed and concentrated to ∼400 μl using an Amicon Ultra (10,000 mol. wt. cutoff) ultrafiltration unit (Beverly, MA). The upper 200 μl of the concentrate was layered onto a 5-ml 5–20% sucrose gradient made in 30 mM HEPES, pH 7.4, 5 mM MgSO4, 0.5 mM EDTA, and 25 mM K acetate. The sample was spun for 10 h at 30,000 rpm in a SW55Ti rotor and fractionated. Note that the lower ∼200 μl of the concentrate was discarded because it contained a dense pigment that caused the sample to sink into the sucrose gradient.
Histology and In Situ Hybridization
Sections of murine lung and testis were stained for the presence of DNAL4 and counterstained with hematoxylin by the University of Connecticut Health Center (UCHC) histology core facility using standard methods. Images of murine embryos and brain probed for expression of various dynein components by in situ hybridization were obtained from the BGEM database, which is freely available for use (Magdaleno et al., 2006; http://www.stjudebgem.org).
Computational Methods
Analysis of the Chlamydomonas genome was performed at http://genome.jgi-psf.org/Chlre3/Chlre3.home.html. Searches of the nonredundant databases were made using BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Protein sequences were aligned using CLUSTALW and the output processed with BOXSHADE. The residues that are not conserved between Chlamydomonas LC8 and LC10 were mapped onto the rat DYNLL1 structure (pdb accession 1F3C) and molecular surface using MOLMOL (Koradi et al., 1996).
RESULTS
Molecular Analysis of the LC2-LC10 Genomic Region
Analysis of the genomic region surrounding the LC2 gene that is deleted in the oda12-1 mutant revealed a second potential dynein gene (here termed LC10) located within ∼2 kb (Figure 1a). The LC10 coding sequence was initially assembled from overlapping ESTs BI722005, BE337027, and BI719072. Subsequently, the entire 736-base pair cDNA was obtained from a λZapII library using a portion of the coding region obtained by PCR from first-strand cDNA as a probe. Several full-length clones were sequenced to confirm the EST assembly (this sequence is available under accession no. EU448319). Analysis of the Chlamydomonas genome (ver. 3.0) and Southern blotting (Figure 1b) demonstrated that there is one LC10 gene in Chlamydomonas. Northern analysis (Figure 1c) revealed a single message of ∼1.3 kb that was greatly up-regulated after deflagellation as is characteristic of flagellar proteins.
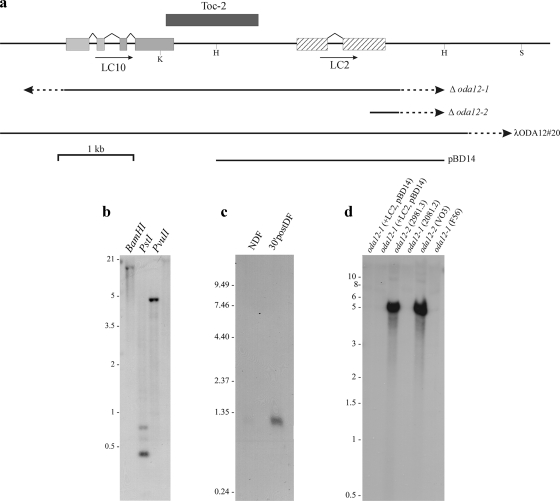
Figure 1.
Molecular analysis of LC10. (a) Map of the LC2/LC10 genomic region indicating the locations of both LC genes, the Toc-2 transposon, the regions missing in the oda12-1 and oda12-2 deletion mutants, and the λODA12#20 clone and HindIII fragment (pBD14) used previously by Pazour et al. (1999) to rescue the oda12-1 phenotype. Restriction sites: H, HindIII; K, KpnI; S, SalI. (b) Southern blot analysis of wild-type genomic DNA restricted with BamHI, PstI, and PvuII and probed with the LC10 cDNA indicates that there is a single gene for this LC in Chlamydomonas. (c) Northern analysis of RNA samples from nondeflagellated cells (NDF) and from cells that had been deflagellated by pH shock and allowed to undergo flagellar regeneration for 30 min (30′ post-DF). A single message of ∼1.3 kb was detected only in the deflagellated sample. (d) Southern blot of PvuII-digested genomic DNA from oda12-1 and oda12-2 mutant strains and oda12-1 rescued with pBD14, probed with the LC10 cDNA. The LC10 gene is present only in oda12-2 and is not rescued by transformation with pBD14.
Toc 2 is a ∼1.2-kb repeated sequence in the Chlamydomonas genome that contains imperfect terminal repeats of 14 base pairs and is related to class II transposable elements (Day, 1995). One copy of this element, in reverse orientation to the published sequence (X84663), starts at base pairs 670 of the LC10 gene. One of the LC10 cDNA clones we obtained contained this entire element at the 3′ end of the LC10 coding region. However, we presume this transcriptional read-through represents a rare event, as we did not observe a second message by Northern blot analysis (Figure 1c).
Previously, two insertional alleles at the ODA12 locus that are defective for LC2 were described (Pazour et al., 1999). The oda12-1 allele lacks the entire LC2 gene, and Southern blotting demonstrated that this strain is also a complete null for LC10 (Figure 1d). In contrast, oda12-2 lacks only the 3′ end of the LC2 gene and retains the entire LC10 gene. The oda12-1 motility phenotype may be essentially completely rescued by transformation with a λ clone (λODA12#20) that is now known to contain both LC2 and LC10 genes; use of an ∼3-kb HindIII fragment (pBD14) containing only the LC2 gene (Figure 1, a and d) also complemented the mutation (Pazour et al., 1999).
Properties of the LC10 Protein
The full-length LC10 clone encodes a protein of 103 residues with a mass of 12,086 Da and a calculated pI of 5.28. There is one in-frame stop codon located within the 5′ untranslated region and a perfect copy of the Chlamydomonas polyadenylation signal within the 3′ untranslated region. The LC10 protein shares 33% identity (45% similarity) with Chlamydomonas LC8 and 45% identity (61% similarity) with the sea urchin (Anthocidaris crassispina) outer arm dynein LC4 protein. A CLUSTALW alignment and phylogenetic analysis of various members of this protein family revealed three distinct groupings (Figure 2,a and b). Chlamydomonas LC10, sea urchin LC4, and the human DNAL4 protein form a subgroup that is distinct from both canonical LC8/DYNLL proteins and the related Chlamydomonas outer arm component LC6. In this analysis, the Tctex1-related outer arm dynein protein LC9, which is structurally related to LC8 although it shares little sequence homology (DiBella et al., 2005; Wu et al., 2005), was used as the out-group.
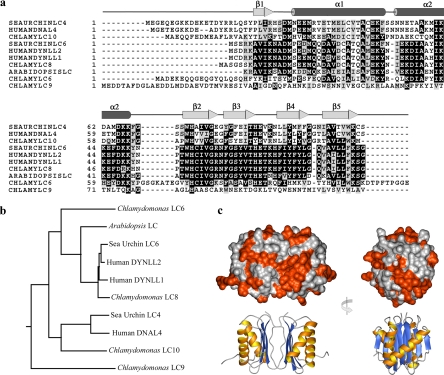
Figure 2.
Phylogenetic and structural analysis of LC10. (a) CLUSTALW alignment of the three Chlamydomonas members of the LC8 family and homologues from humans, sea urchin, and Arabidopsis. Protein sequences used for the alignment are Chlamydomonas outer arm LC6 (Q39579), LC8 (Q39580) and LC10 (EU448319), sea urchin (Anthocidaris crassispina) outer arm LC4 (BAA24152) and LC6 (AB004830), Arabidopsis LC (NP_193328), and human DYNLL1 (NM_003746), DYNLL2 (NM_080677), and DNAL4 (AAP88849). The Chlamydomonas Tctex1-related outer arm dynein protein LC9 was used as the out-group. The LC8/DYNLL1 secondary structure is shown above the alignment. (b) Neighbor-joining phylogenetic tree of the LC6/LC8/LC10 homologues aligned in panel a. (c) Two views of the molecular surface of rat LC8/DYNLL1 (PDB accession 1F3C) colored to indicate those residues (red-orange) that are not conserved between LC8 and LC10. Orientation of the LC8 dimer is indicated by the ribbon diagrams at bottom.
The residues that differ between LC8 and LC10 were mapped onto the rat LC8/DYNLL1 molecular surface (PDB accession 1F3C; Figure 2c). This revealed considerable variation between the two proteins spread over all external faces including the intermonomer groove involved in binding dynein ICs and the α-helical face that contains most of the sequence differences between mammalian DYNLL1 and DYNLL2; note that for a particular isoform this region is highly conserved between species.
LC10 Is a Component of the Outer Dynein Arm
To define the flagellar location of LC10, we raised a rabbit polyclonal antiserum (CT247) against the full-length protein expressed as a C-terminal fusion with MBP and blot-purified the antibody against recombinant LC10. Immunoblot analysis revealed that this antibody preparation specifically recognized LC10 and importantly did not react with recombinant LC6 or LC8 (Figure 3a). The CT247 antibody recognized a single protein band of Mr ∼12,000 in whole flagella samples (Figure 3b). After detergent treatment to remove the flagellar membrane, essentially all of this protein was associated with the axoneme and was solubilized by 0.6 M NaCl (Figure 3c).
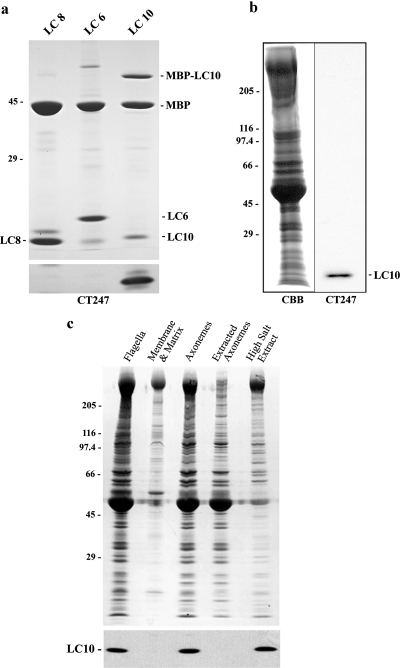
Figure 3.
LC10 is an axonemal component. (a) MBP-LC6, MBP-LC8, and MBP-LC10 fusion proteins were incubated with Factor Xa, electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue (top panel) or blotted to nitrocellulose and probed with the CT247 antibody (bottom panel). This antibody detected LC10 and specifically did not recognize either LC6 or LC8. (b) Wild-type Chlamydomonas flagella were electrophoresed in a 5–15% acrylamide gradient gel and either stained with Coomassie blue (left panel) or blotted to nitrocellulose and probed with the CT247 antibody (right panel). Only a single band at Mr ∼12,000 corresponding to LC10 was detected. (c) Wild-type Chlamydomonas flagella were demembranated with 1% IGEPAL CA-630, and the resulting axonemes extracted with 0.6 M NaCl. Samples were electrophoresed in a 5–15% acrylamide gradient gel and either stained with Coomassie blue (top panel) or blotted to nitrocellulose and probed with antibody CT247 to detect LC10 (bottom panel). This protein is exclusively present in the axonemal fraction and is essentially completely extracted by the high salt treatment.
Analysis of mutant axonemes lacking various substructures revealed that LC10 was specifically missing only in those strains that lack the outer dynein arm (oda1, oda2, oda6, oda9, and pf28 pf30 ssh1) or are null for LC10 (oda12-1; Figure 4). This protein was present in mutants defective for inner arms (ida1 and ida4) and radial spokes (pf14). Additional outer arm mutants defective for the α HC (oda11), the β HC motor domain (oda4-s7), and the oda4-s7 oda11 double mutant all contained LC10 (Figure 4). Previously, we found that oda6-r88, which contains an altered segment of 23-residues within the N-terminal region of IC2 (Mitchell and Kang, 1993), does not incorporate LC2, LC6, and LC9 into the outer arm, whereas a second IC2 mutant (oda6-r75) altered in a neighboring region did (DiBella et al., 2005). Immunoblot analysis revealed that both oda6-r75 (not shown) and oda6-r88 (Figure 4) retain LC10, although the amount present in oda6-r88 appears significantly reduced compared with wild type (Figure 4).
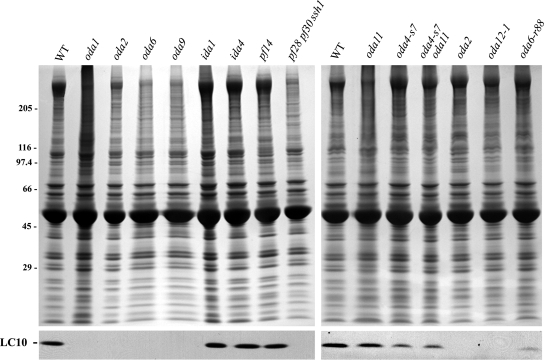
Figure 4.
LC10 is missing in mutant axonemes lacking outer dynein arms. Axonemes were prepared from wild-type Chlamydomonas and from mutant strains lacking various axonemal components. After electrophoresis, samples were stained with Coomassie blue (top panels) or blotted and probed with CT247 (bottom panels). LC10 is completely missing in strains lacking outer dynein arms (oda1, oda2, oda6, oda9, pf28pf30ssh1) and in the oda12-1 mutant that is null for both LC2 and LC10. However, this protein is present in strains lacking other substructures such as the inner arms (ida1 and ida4) and radial spokes (pf14) or only segments of the outer arm (oda4-s7 and oda11).
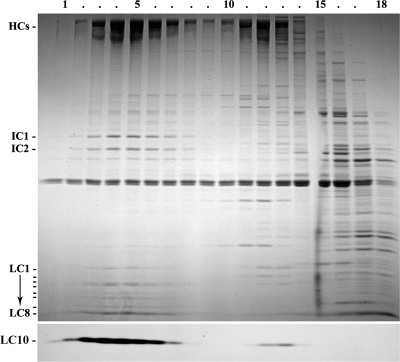
To confirm that LC10 is indeed an outer arm component, we purified this motor from the 0.6 M NaCl axonemal extract by sucrose density gradient centrifugation in the presence of Mg2+ and at low hydrostatic pressure; conditions designed to retain association of the entire outer dynein arm and docking complex (Takada et al., 1992). Most LC10 comigrated with known outer dynein arm components in fractions 3–7 at ∼23 S (Figure 5). A minor amount of LC10 was also present higher in the sucrose gradient and sedimented at ∼8 S (fractions 12 and 13); this likely represents a small pool of LC that dissociated from the main complex, as we have observed similar levels of dissociation products with other dynein LCs (e.g., LC7b; DiBella et al., 2004).
Figure 5.
LC10 copurifies with outer arm dynein in sucrose density gradients. An axonemal high-salt extract was sedimented in a 5–20% sucrose density gradient. Fractions were electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue (top panel) or blotted to nitrocellulose and probed with CT247 to detect LC10 (bottom panel). This LC comigrates with components of the outer dynein arm at ∼23 S; a minor fraction was also observed at ∼8 S and likely represents a partial dissociation product.
Lack of LC10 Leads to Motility Defects
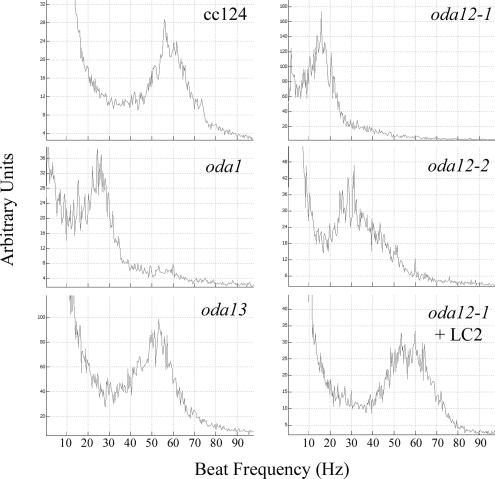
Previously, the swimming velocity of the oda12-1 strain rescued with a large genomic clone containing both the LC2 and LC10 genes was restored to wild-type levels (Pazour et al., 1999). To further test whether the lack of LC10 altered dynein function, we examined the flagellar beat frequency of wild type, oda1, oda12-1, oda12-2, and oda12-1 rescued for LC2 (but not LC10) with pBD14 and oda13 using the population-based fast Fourier transform method (Figure 6); to ensure that the power spectra were directly comparable all strains were grown and analyzed under identical conditions. The oda1 strain, which lacks the entire outer arm and docking complex, had a beat frequency of ∼25 Hz, whereas that of oda12-1 was consistently lower (∼15–20 Hz); oda12-2 was less impaired and showed a single peak in the power spectrum centered at ∼30 Hz, with a broad shoulder extending toward 50 Hz. The oda13 mutant that lacks only LC6 had a peak frequency slightly less than wild type as reported previously (DiBella et al., 2005). The oda12-1 strain that had been rescued for LC2 with the pBD14 plasmid exhibited a broad peak from ∼50–60 Hz that was subtly distinct from wild type. Thus, the lack of LC10 alone has only a minor effect but in combination with an LC2 defect results in a beat frequency that is less than that of strains that lack the entire outer arm. To further confirm this result, we compared the beat frequency of oda12-1 to that of oda1-oda9 when all these strains were grown under identical conditions and assayed at the same time of day. We obtained a mean beat frequency of 16 Hz for oda12-1 compared with 20 Hz (oda1), 23 Hz (oda2), 19 Hz (oda3), 21 Hz (oda4), 20 Hz (oda5), 23 Hz (oda6), 19 Hz (oda7), 20 Hz (oda8), and 19 Hz (oda9).
Figure 6.
Flagellar beat frequency effects due to loss of LC10. Beat frequency of wild-type Chlamydomonas (strain cc124) and the oda12-2 (lacks LC2), oda12-1 (lacks LC2 and LC10), oda12-1 rescued for LC2 (lacks LC10), oda13 (lacks LC6), and oda1 (lacks the entire outer arm and docking complex) mutants were assessed using the population-based fast Fourier transform method. The lack of both LC2 and LC10 in oda12-1 is more detrimental than the lack of the entire outer arm and docking complex in oda1.
Role of LC6, 8, and 10 in the Cytoplasmic Stability of Axonemal Dynein Complexes
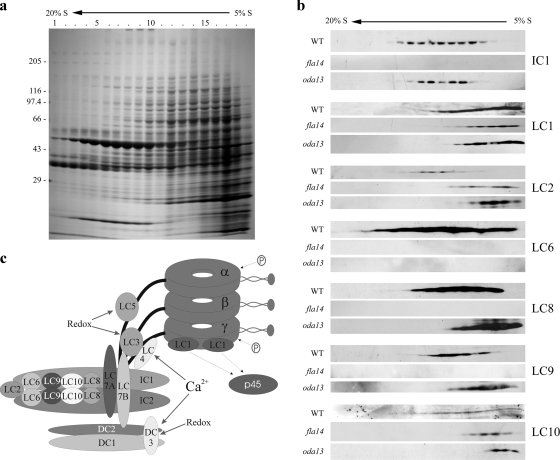
To assess whether these LCs are involved in stabilizing dynein complexes during assembly, we fractionated cytoplasm from wild-type Chlamydomonas and from strains specifically lacking LC6 or LC8 (Figure 7a). Immunoblot analysis (Figure 7b) revealed that the lack of LC8 in fla14 had a significant effect on the stability of some dynein components (Figure 7c) such that IC1, LC6, and LC9 were undetectable, and LC2 and LC10 migrated near the top of the gradient; the γ HC-associated LC1 protein was unaffected. In contrast, lack of LC6 in oda13 had essentially no effect on the migration of IC1 but did result in dissociation of LC2/LC10 from the IC complex under these conditions. Thus lack of LC8 leads to instability of some dynein components and the failure of outer arm assembly, whereas lack of LC6 results in complex dissociation in sucrose gradients but does not inhibit axonemal incorporation.
Figure 7.
Dynein stability in cytoplasm. (a) Cytoplasmic extract from wild-type Chlamydomonas was sedimented in a 5–20% sucrose density gradient. Equal volumes of each fraction were electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue. (b) Immunoblot analysis of cytoplasmic extracts from wild-type, fla14, and oda13 strains. Samples were probed with antibodies 1878A, R5932, R5391, R4928, R4058, CT231, and CT247 to detect IC1, LC1, LC2, LC6, LC8, LC9, and LC10, respectively. (c) Model of the Chlamydomonas outer dynein arm and docking complex illustrating the approximate locations and associations of the various components. LC2, LC6, LC8, LC9, and LC10 all interact with the N-terminal regions of the ICs, whereas LC7a and LC7b are thought to associate near the C-terminal WD-repeat domains. Modified from King and Kamiya (2008).
Mammalian DNAL4 (DNALC4) is a Ciliary/Flagellar Component
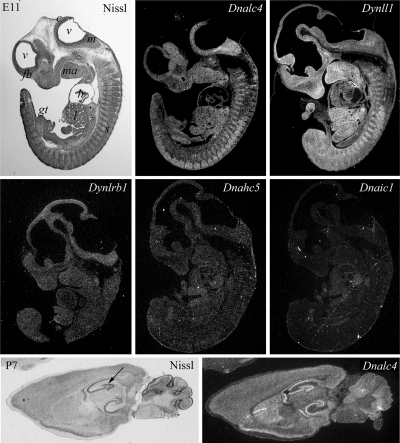
The DNAL4 protein expressed in mammals shares 50% identity (65% similarity) with Chlamydomonas LC10 (Figure 2a). To assess whether DNAL4 is also present in cilia and flagella, we generated a polyclonal antibody (CT254) against a MBP-DNAL4 fusion. After blot purification, this antibody was found to exhibit weak cross-reactivity with LC10 but did not recognize either LC6 or LC8 (Figure 8a). DNAL4 could be detected in a variety of murine tissue homogenates including testis, ovary, and brain (Figure 8b), and histological analysis revealed that it is highly expressed in the ciliated epithelial layer lining the bronchi of the lungs and in sperm flagella and spermatids within testis (Figure 8, c and d). These observations suggest that DNAL4 is associated with motile cilia and flagella. However, it is possible that DNAL4 plays additional roles in mammals as in situ hybridization studies indicate that this protein is highly expressed throughout the developing mouse embryo as is the ubiquitous DYNLL1 (aka LC8) protein (Figure 9). In contrast, other exclusively axonemal dynein components such as DNAHC5 and DNAIC1 exhibit much lower expression. Furthermore, DNAL4 is also expressed in adult mouse brain and is especially prominent in the hippocampus (Figure 9).
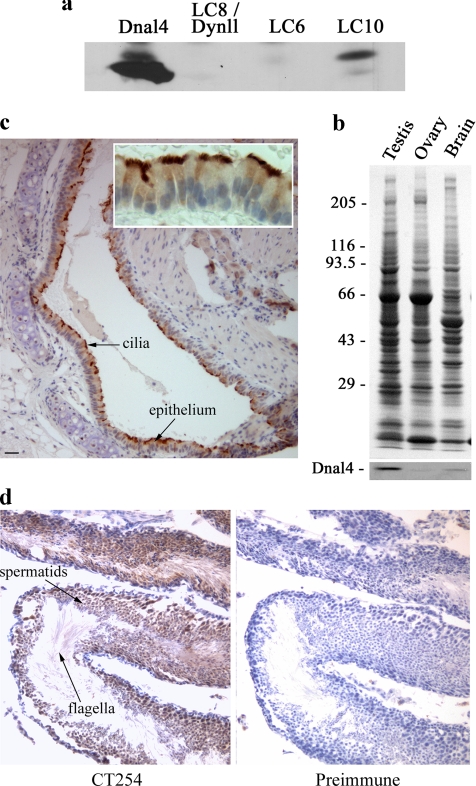
Figure 8.
DNAL4 is present in motile mammalian cilia and flagella. (a) Immunoblot analysis of recombinant murine DNAL4 and Chlamydomonas LC6, LC8, and LC10. The CT254 antibody recognizes DNAL4 and also exhibits weak cross-reactivity with LC10; in contrast, neither LC6 nor LC8 were detected. (b) Tissue homogenates from murine testis, ovary, and brain were electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue (top panel) or blotted to nitrocellulose and probed with the CT254 antibody (bottom panel). DNAL4 is especially prominent in testis but was also readily detected in the other tissues. (c) Histological analysis of a section through a bronchus from murine lung stained with hematoxylin to detect nuclei and for DNAL4 reactivity (brown). This protein is localized to the cilia at the apical face of the epithelium lining the lung. Bar, 50 μm. The inset shows an enlargement of the ciliated epithelium. (d) Histological analysis of murine testis sections stained either with CT254 (left panel) or with the preimmune serum (right panel) and counterstained with hematoxylin. DNAL4 is very prominent in sperm flagella and in spermatids.
Figure 9.
Expression of DNAL4 in mouse embryos and brain. In situ hybridization analysis of E11 mouse embryos (top panels) probed to reveal DNAL4, DYNLL1, DYNLRB1, DNAHC5, and DNAIC1 expression. A Nissl-stained section is shown for orientation: c, cerebellum; fb, forebrain; gt, genital tubercule; h, heart; l, liver; m, medulla; ma, mandible; s, somites; v, ventricles. Components specifically found only in cells bearing motile cilia/flagella have very low signals, whereas both DYNLL1 and DNAL4 exhibit a ubiquitous and very high expression. Furthermore, DNAL4 is readily detected throughout P7 murine brain (bottom panels) and is highly expressed in the hippocampal region (arrow). These images are from the publicly available BGEM database (Magdaleno et al., 2006; http://www.stjudebgem.org).
DISCUSSION
Here we report the identification of a third member of the LC8/DYNLL family in Chlamydomonas. LC10 orthologues are missing in nematodes and higher plants, suggesting that this protein is present only in those organisms that have motile cilia/flagella. LC10 copurifies with the outer dynein arm and is completely absent from axonemes of Chlamydomonas strains lacking only this motor, indicating that it is specifically associated with outer arm dynein and is not also present in the inner arm system as are LC7a, LC7b, and LC8. A Chlamydomonas LC10-null mutant exhibits subtle defects in motility indicating that this protein is indeed necessary for wild-type motor function and the combined LC2/LC10 deficiency is highly deleterious. However, LC10 does not appear necessary for dynein arm assembly per se. In contrast, lack of LC8 results in the complete failure of outer arm assembly, whereas the loss of LC6 has only a very minor effect on motility but does alter cytoplasmic stability of dynein subcomplexes. These results indicate that the three members of the LC8/DYNLL family in Chlamydomonas are functionally distinct.
The Light Chain Complement of Outer Arm Dynein
Analysis of 35S-labeled αβ HC dimer from the Chlamydomonas outer arm dynein using 2D nonequilibrium pH gradient electrophoresis revealed nine LC spots in addition to the two LCs associated with the γ HC (Pfister et al., 1982). With the identification of LC10 as an integral component of the IC/LC complex, we can now account for all the LC components that copurify with this dynein (Table 1 and see Figure 7C). The IC/LC complex consists of two WD-repeat ICs (IC1 and IC2) associated with dimers of the three LC8/DYNLL-related proteins (LC6, LC8, and LC10), two members of the LC7/Roadblock/DYNLRB family (LC7a and LC7b), the dimeric Tctex1/DYNLT1 homologue (LC9), and the apparently monomeric LC2 protein that is related to murine Tctex2. The other LCs that have distinct regulatory functions interact directly with the N-terminal regions of the HCs (LC3, LC4, and LC5) or with the AAA+ domains of the γ HC motor unit (LC1).
Table 1.
Light chains of Chlamydomonas outer arm dynein
| Light chaina | Mass (Da) | Predicted pI | Mutant | Interactions within outer arm dynein | Properties | Outer arm specific? |
|---|---|---|---|---|---|---|
| LC1 | 22,150 | 5.54 | −−− | γ HC, p45b | Leucine-rich repeat protein; motor domain-associated | Yes |
| LC2 | 15,825 | 6.01 | oda12-1 | LC6, IC1, IC2 | Tctex2; required for outer arm function; oda12-1 also is null for LC10; absent in oda6-r88 | Yes |
| oda12-2 | ||||||
| LC3 | 17,365 | 7.76 | −−− | β HC, γ HC, LC7b | Thioredoxin | Yes |
| LC4 | 17,788 | 4.22 | −−− | γ HC | Ca2+ binding | Yes |
| LC5 | 14,179 | 8.34 | −−− | α HC | Thioredoxin | Yes |
| LC6 | 13,856 | 6.80 | oda13 | LC2, IC2 | LC8 homologue; not required for arm assembly; absent in oda6-r88 | Yes |
| LC7a | 11,928 | 7.85 | oda15 | LC7b, IC1 and/or IC2 | Roadblock homologue; present in outer arms and inner arm I1 | No |
| LC7b | 11,147 | 5.56 | −−− | LC3, LC7a, DC2 | Roadblock homologue; present in outer arms and inner arm I1 | No |
| LC8 | 10,321 | 7.51 | fla14 | IC1, IC2 | Highly conserved, not dynein-specific | No |
| LC9 | 12,981 | 4.37 | −−− | IC1, IC2 | Tctex1; not required for arm assembly; absent in oda6-r88 | Yes |
| LC10 | 12,086 | 5.28 | oda12-1 | nk | LC8 homologue; oda12-1 also is null for LC2; reduced in oda6-r88 | Yes |
a The light chain nomenclature used here is specific for the Chlamydomonas outer dynein arm.
b The LC1-interacting protein p45 (Benashski et al., 1999) has now been identified as tubulin (R. S. Patel-King and S. M. King, unpublished data).
Structural Features of the LC8/DYNLL Family
Structural analysis has revealed that LC8/DYNLL proteins contain two α helices and five β strands and form symmetric dimers. The interface consists of strand-switched β sheets and leads to formation of two identical grooves into which short segments of the target protein bind (Liang et al., 1999; Fan et al., 2001). The α helical face of each monomer protrudes away from this interaction site and no associations via this region have yet been reported, even though these surfaces have been very highly conserved. The monomer-dimer transition is modulated by pH in vitro (Liang et al., 1999; Nyarko et al., 2005) and has been reported to be sensitive to phosphorylation (Song et al., 2008). The LC10 sequence aligns readily with that of LC8, suggesting that it has a similar overall architecture. Although both LC6 and LC10 have N-terminal extensions of 11–13 residues, the LC6 protein more closely resembles LC8 with the exception of an enlarged loop region between α2 and β2 elements and an extended C-terminal segment. In each LC8 monomer, the β strands interact to form an antiparallel β sheet that mediates dimerization. However, there are significant differences between LC8 and LC10 in the β3 strand, which is switched out to form the strand-switched dimer interface, suggesting that these two proteins are unlikely to form heterodimers. Furthermore, the properties of the molecular surface of LC8 and LC10 are quite distinct and are thus predicted to mediate distinct interactions within the dynein complex.
The Role of LC10 in Dynein Function
Previously, we described the comparative analysis of oda12-1 and oda6-r88 strains both of which lack LC2 (DiBella et al., 2005). We found that oda12-1 has a beat frequency of <∼20 Hz, whereas that of oda6-r88 was 30–35 Hz. This observation was surprising in that although both strains lack LC2, oda6-r88 is in addition missing LC6 and LC9. Our current finding that oda12-1 is also a complete null for LC10, whereas this protein is present in oda6-r88, suggests that the combined lack of LC2 and LC10 is more detrimental to outer arm function than is the LC2, LC6, and LC9 combination. Furthermore, we have now found that when assayed under identical conditions, the oda12-1 strain exhibits a consistently lower beat frequency than do the oda1-oda9 strains that lack the docking complex and/or the entire outer arm. Even though the amount of γ HC is reduced from wild type, all the motor and other regulatory components are present in oda12-1. Thus, the combined lack of LC2/LC10 has a further negative effect on flagellar function. This might be caused by dysfunction of the remaining outer arm motor units providing in essence a braking effect or potentially by negative feedback onto the inner arm system. Intriguingly, although dynein function is abrogated in oda12-1, outer arm assembly can still occur although not to wild-type levels. Only in the oda6-r88 oda12-1 double mutant that is missing LCs 2, 6, 9, and 10 did we observe a complete lack of dynein arm incorporation into the flagellum (DiBella et al., 2005).
The lack of LC6 alone or in combination with LC2 and LC9 results in flagellar beat frequencies that are either little altered from wild type or remain greater than that of outer arm-less mutants. In contrast, LC8 null cells are completely unable to assemble outer dynein arms into the short flagella stubs that form in this mutant. Thus, there appears to be a very distinct difference between the roles played by the three members of this dynein LC family within the same dynein holoenzyme.
Organization of the Intermediate Chain/Light Chain Complex
Structural studies of the LC8(DYNLL1) and Tctex1(DYNLT1) dimers in complex with a small region of the cytoplasmic dynein N-terminal domain revealed an extended conformation with the LCs packed close together between the two IC strands (Williams et al., 2007). In the Chlamydomonas outer arm, alteration of residues 31–54 within IC2 leads to the failure of LC2, LC6, and LC9 assembly and a marked reduction in LC10 levels. This IC region is predicted to form an extended β strand. We found previously using chemical cross-linking that LC2 and LC6 are within 9.2 Å of each other (DiBella et al., 2001) and that the LC9 dimer directly associates with both IC1 and IC2. Thus it is likely that these LCs bind in a sequential manner similar to that observed in cytoplasmic dynein. Alteration of the adjoining residues 54–64 of IC2 had no effect on LC incorporation (DiBella et al., 2005), further suggesting that these four LCs all bind IC2 within the N-terminal 54 residues.
The location at which LC8 associates with the outer arm ICs is uncertain. However, the sea urchin orthologue of IC1 contains a perfect copy of the canonical LC8 binding motif K/RxTQT and sequence alignment consequently allows for the tentative identification of the motif 184RETFT188 as the likely site in Chlamydomonas IC1. However, neither of these motifs is present in Chlamydomonas IC2 or its sea urchin equivalent. Similarly, the sites at which the LC7a/b proteins bind the ICs also remain to be defined, although based on analysis of cytoplasmic dynein (Susalka et al., 2002) and inner arm I1/f (Hendrickson et al., 2004), the interaction site is likely close to the C-terminal WD-repeat β-propellers.
DNAL4 is the Mammalian Orthologue of LC10
The human DNAL4 gene maps to chromosome 22q13.1 and encodes a protein that is 50% identical to Chlamydomonas LC10. Immunostaining demonstrated that DNAL4 is highly concentrated in the apical cilia of epithelial cells lining the bronchi of the lungs and in sperm flagella and spermatids. These observations support the conclusion that DNAL4 is likely associated with axonemal dyneins. Surprisingly, however, examination of the BGEM in situ hybridization database (Magdaleno et al., 2006) revealed that in murine embryos DNAL4 is ubiquitously expressed at a very high level (as is DYNLL1) in contrast to known axonemal dynein components such as the DNAHC5 dynein HC. Furthermore, DNAL4 is also highly expressed in brain and especially the hippocampus. Thus, it is likely that DNAL4 plays additional roles in mammals beyond acting as an outer arm dynein LC.
In conclusion, we have demonstrated that the Chlamydomonas outer dynein arm contains three members of the LC8/DYNLL family that have distinct roles in motor assembly and activity. These data also provide strong evidence for the essential role of the IC/LC complex in controlling dynein motor function. Furthermore, we have found that DNAL4 represents the mammalian orthologue of Chlamydomonas LC10 and that its expression patterns suggest an additional role in processes other than ciliary/flagellar motility.
ACKNOWLEDGMENTS
We thank Dr. Nada Zecevic (UCHC) for discussion of embryonic anatomy. This study was supported by Grants GM51293 (to S.M.K.) and GM060992 (to G.J.P.) from the National Institutes of Health.
Abbreviations used:
- HC
heavy chain
- IC
intermediate chain
- LC
light chain
- MBP
maltose-binding protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0362) on June 25, 2008.
REFERENCES
- Beckwith S. M., Roghi C. H., Liu B., Morris N. R. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J. Cell Biol. 1998;143:1239–1247. doi: 10.1083/jcb.143.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benashski S. E., Patel-King R. S., King S. M. Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the γ heavy chain. Biochemistry. 1999;38:7253–7264. doi: 10.1021/bi990466y. [DOI] [PubMed] [Google Scholar]
- Day A. A transposon-like sequence with short terminal inverted repeats in the nuclear genome of Chlamydomonas reinhardtii. Plant Mol. Biol. 1995;28:437–442. doi: 10.1007/BF00020392. [DOI] [PubMed] [Google Scholar]
- DiBella L. M., Benashski S. E., Tedford H. W., Harrison A., Patel-King R. S., King S. M. The Tctex1/Tctex2 class of dynein light chains. Dimerization, differential expression, and interaction with the LC8 protein family. J. Biol. Chem. 2001;276:14366–14373. doi: 10.1074/jbc.M011456200. [DOI] [PubMed] [Google Scholar]
- DiBella L. M., Gorbatyuk O., Sakato M., Wakabayashi K., Patel-King R. S., Pazour G. J., Witman G. B., King S. M. Differential light chain assembly influences outer arm dynein motor function. Mol. Biol. Cell. 2005;16:5661–5674. doi: 10.1091/mbc.E05-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella L. M., Sakato M., Patel-King R. S., Pazour G. J., King S. M. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol. Biol. Cell. 2004;15:4633–4646. doi: 10.1091/mbc.E04-06-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick T., Ray K., Salz H. K., Chia W. Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol. Cell. Biol. 1996a;16:1966–1977. doi: 10.1128/mcb.16.5.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick T., Surana U., Chia W. Molecular and genetic characterization of SLC1, a putative Saccharomyces cerevisiae homolog of the metazoan cytoplasmic dynein light chain 1. Mol. Gen. Genet. 1996b;251:38–43. doi: 10.1007/BF02174342. [DOI] [PubMed] [Google Scholar]
- Espindola F. S., Suter D. M., Partata L. B., Cao T., Wolenski J. S., Cheney R. E., King S. M., Mooseker M. S. The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil. Cytoskelet. 2000;47:269–281. doi: 10.1002/1097-0169(200012)47:4<269::AID-CM2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Fan J. -S., Zhang Q., Tochio H., Li M., Zhang M. Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain. J. Mol. Biol. 2001;306:97–108. doi: 10.1006/jmbi.2000.4374. [DOI] [PubMed] [Google Scholar]
- Havecker E. R., Gao X., Voytas D. F. The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain 8. Plant Physiol. 2005;139:857–868. doi: 10.1104/pp.105.065680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson T. W., Perrone C. A., Griffin P., Wuichet K., Mueller J., Yang P., Porter M. E., Sale W. S. IC138 Is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell. 2004;15:5431–5442. doi: 10.1091/mbc.E04-08-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey S. R., Snyder S. H. PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science. 1996;274:774–777. doi: 10.1126/science.274.5288.774. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods. 2000;22:383–387. doi: 10.1006/meth.2000.1090. [DOI] [PubMed] [Google Scholar]
- King S. M. Large-scale isolation of Chlamydomonas flagella. Methods Cell Biol. 1995;47:9–12. doi: 10.1016/s0091-679x(08)60783-9. [DOI] [PubMed] [Google Scholar]
- King S. M., Barbarese E., Dillman J. F., III, Patel-King R. S., Carson J. H., Pfister K. K. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J. Biol. Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- King S. M., Kamiya R. Axonemal dyneins: assembly, structure and force generation. In: Witman G. B., editor. Cell Motility and Behavior. The Chlamydomonas Source Book. vol. III. San Diego: Elsevier; 2008. (in press) [Google Scholar]
- King S. M., Patel-King R. S. The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 1995;270:11445–11452. doi: 10.1074/jbc.270.19.11445. [DOI] [PubMed] [Google Scholar]
- King S. M., Wilkerson C. G., Witman G. B. The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alpha-tubulin in situ. J. Biol. Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Liang J., Jaffrey S. R., Guo W., Snyder S. H., Clardy J. Structure of the PIN/LC8 dimer with a bound peptide. Nat. Struct. Biol. 1999;6:735–740. doi: 10.1038/11501. [DOI] [PubMed] [Google Scholar]
- Magdaleno S., et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Okazaki K., Shimanuki M., Yamamoto A., Hiraoka Y., Niwa O. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell. 2002;13:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. R., Kang Y. Reversion analysis of dynein intermediate chain function. J. Cell Sci. 1993;105:1069–1078. doi: 10.1242/jcs.105.4.1069. [DOI] [PubMed] [Google Scholar]
- Nyarko A., Cochrun L., Norwood S., Pursifull N., Voth A., Barbar E. Ionization of His 55 at the dimer interface of dynein light-chain LC8 is coupled to dimer dissociation. Biochemistry. 2005;44:14248–14255. doi: 10.1021/bi0512694. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Koutoulis A., Benashski S. E., Dickert B. L., Sheng H., Patel-King R. S., King S. M., Witman G. B. LC2, the Chlamydomonas homologue of the t complex-encoded protein Tctex2, is essential for outer dynein arm assembly. Mol. Biol. Cell. 1999;10:3507–3520. doi: 10.1091/mbc.10.10.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Wilkerson C. G., Witman G. B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J. Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Witman G. B. Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods. 2000;22:285–298. doi: 10.1006/meth.2000.1081. [DOI] [PubMed] [Google Scholar]
- Pfister K. K., Fay R. B., Witman G. B. Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. 1982;2:525–547. doi: 10.1002/cm.970020604. [DOI] [PubMed] [Google Scholar]
- Pfister K. K., et al. Cytoplasmic dynein nomenclature. J. Cell Biol. 2005;171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillis R., Statton D., Caruccio P., Murphey R. K. Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development. 1996;122:2955–2963. doi: 10.1242/dev.122.10.2955. [DOI] [PubMed] [Google Scholar]
- Piperno G., Luck D. J. Axonemal adenosine triphosphatases from flagella of Chlamydomonas reinhardtii. Purification of two dyneins. J. Biol. Chem. 1979;254:3084–3090. [PubMed] [Google Scholar]
- Poisson N., Real E., Gaudin Y., Vaney M. C., King S., Jacob Y., Tordo N., Blondel D. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8, dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 2001;82:2691–2696. doi: 10.1099/0022-1317-82-11-2691. [DOI] [PubMed] [Google Scholar]
- Puthalakath H., Huang D. C., O'Reilly L. A., King S. M., Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- Raux H., Flamand A., Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P., Pedersen L., Patel-King R. S., King S. M. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J. Cell Sci. 2007;120:3653–3665. doi: 10.1242/jcs.012773. [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Bohmann K., Nusslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- Song C., Wen W., Rayala S., Chen M., Ma J., Zhang M., Kumar R. Serine 88 phosphorylation of the 8 kDa dynein light chain is a molecular switch for its dimerization status and functions. J. Biol. Chem. 2008;283:4004–4013. doi: 10.1074/jbc.M704512200. [DOI] [PubMed] [Google Scholar]
- Susalka S. J., Nikulina K., Salata M. W., Vaughan P. S., King S. M., Vaughan K. T., Pfister K. K. The roadblock light chain binds a novel region of the cytoplasmic dynein intermediate chain. J. Biol. Chem. 2002;277:32939–32946. doi: 10.1074/jbc.M205510200. [DOI] [PubMed] [Google Scholar]
- Takada S., Sakakibara H., Kamiya R. Three-headed outer arm dynein from Chlamydomonas that can functionally combine with outer-arm-missing axonemes. J. Biochem. (Tokyo) 1992;111:758–762. doi: 10.1093/oxfordjournals.jbchem.a123832. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., King S. M. Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J. Cell Biol. 2006;173:743–754. doi: 10.1083/jcb.200603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson C. G., King S. M., Koutoulis A., Pazour G. J., Witman G. B. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. C., Roulhac P. L., Roy A. G., Vallee R. B., Fitzgerald M. C., Hendrickson W. A. Structural and thermodynamic characterization of a cytoplasmic dynein light chain intermediate chain complex. Proc. Natl. Acad. Sci. USA. 2007;104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G. B. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Wu H., Maciejewski M. W., Takebe S., King S. M. Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure. 2005;13:213–223. doi: 10.1016/j.str.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Yang P., Diener D. R., Rosenbaum J. L., Sale W. S. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 2001;153:1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]