Abstract
The transfer of lipid-linked oligosaccharide to asparagine residues of polypeptide chains is catalyzed by oligosaccharyltransferase (OTase). In most eukaryotes, OTase is a hetero-oligomeric complex composed of eight different proteins, in which the STT3 component is believed to be the catalytic subunit. In the parasitic protozoa Leishmania major, four STT3 paralogues, but no homologues to the other OTase components seem to be encoded in the genome. We expressed each of the four L. major STT3 proteins individually in Saccharomyces cerevisiae and found that three of them, LmSTT3A, LmSTT3B, and LmSTT3D, were able to complement a deletion of the yeast STT3 locus. Furthermore, LmSTT3D expression suppressed the lethal phenotype of single and double deletions in genes encoding other essential OTase subunits. LmSTT3 proteins did not incorporate into the yeast OTase complex but formed a homodimeric enzyme, capable of replacing the endogenous, multimeric enzyme of the yeast cell. Therefore, these protozoan OTases resemble the prokaryotic enzymes with respect to their architecture, but they used substrates typical for eukaryotic cells: N-X-S/T sequons in proteins and dolicholpyrophosphate-linked high mannose oligosaccharides.
INTRODUCTION
Asparagine (N)-linked glycosylation is a highly conserved protein modification in eukaryotic cells. It is initiated at the membrane of the endoplasmic reticulum (ER), in which the oligosaccharide Glc3Man9GlcNAc2 is assembled on the lipid carrier dolichol pyrophosphate and then transferred to asparagine side chains in certain glycosylation sequons (N-X-S/T; X≠P) in substrate proteins (Tanner and Lehle, 1987; Gavel and von Heijne, 1990; Cummings, 1992; Herscovics and Orlean, 1993; Burda and Aebi, 1999). The transfer of the oligosaccharide is catalyzed by the oligosaccharyltransferase (OTase) complex in the lumen of the ER. OTase of animals, plants, and fungi is a hetero-oligomeric protein complex. In the well-studied model organism S. cerevisiae, it consists of at least eight different subunits: Ost1p, Ost2p, Wbp1, Stt3p, Swp1p, Ost4p, Ost5p, and Ost3p/Ost6p (Silberstein and Gilmore, 1996; Knauer and Lehle, 1999; Dempski and Imperiali, 2002; Yan and Lennarz, 2005; Kelleher and Gilmore, 2006; Weerapana and Imperiali, 2006). Recent findings provided evidence that STT3 protein is the catalytic subunit of this enzyme (Yan and Lennarz, 2002; Kelleher et al., 2003; Nilsson et al., 2003). The most direct support for this hypothesis is the observation that a prokaryotic homologue of yeast Stt3p is an active oligosaccharyltransferase in the absence of any other accessory proteins (Wacker et al., 2002; Kowarik et al., 2006a). Proteins homologous to yeast Stt3p are encoded in almost all eukaryotic genomes (Kelleher and Gilmore, 2006), but based on comparative genome analysis, it has been suggested that OTase composition became increasing complex during the evolutionary divergence of eukaryotes. Single subunit OTases seem to be present in Giardia and kinetoplastids, whereas four subunit OTases consisting of STT3, OST1, OST2, and WBP1 homologues are found in diplomonads, entamoebas, and apicomplexan species. Additionally, multiple forms of the putative STT3 proteins can be encoded in trypanosomatid genomes: three STT3 homologues are found in Trypanosoma brucei and four in Leishmania major (McConville et al., 2002; Berriman et al., 2005; Ivens et al., 2005; Samuelson et al., 2005; Kelleher and Gilmore, 2006). Based on this phylogenetic analysis, it can be proposed that the simplest eukaryotic OTase is a single subunit STT3 protein, similar to the situation found in bacterial N-glycosylation systems. In this report, we addressed this hypothesis and studied the function of L. major OTase in yeast.
In trypanosomatid parasites, N-linked glycosylation principally follows the pathway described for fungal or animal cells, but with different oligosaccharide structures transferred to protein (Parodi, 1993; McConville et al., 2002). It has been shown that, depending on the species, either Man6GlcNAc2 or Man7GlcNAc2 is the largest glycan transferred to protein in the genus Leishmania (Parodi, 1993). Indeed, L. major lacks the ER lumenal mannosyltransferase gene ALG12 and all ER lumenal glucosyltransferase genes: ALG6, ALG8, and ALG10 (Samuelson et al., 2005). From this genetic information, it may be inferred that Man7GlcNAc2 is transferred in L. major. Unlike the yeast and mammalian OTase that preferably use Glc3Man9GlcNAc2, the trypanosome OTase is not selective and transfers different lipid-linked oligosaccharides (LLO) at the same rate (Bosch et al., 1988).
Heterologous expression of single subunit OTase has been used to address the function of this enzyme in vivo. When expressed in Escherichia coli in combination with the biosynthetic pathway of oligosaccharide assembly, PglB, the Campylobacter jejuni OTase, mediates N-glycosylation of defined proteins in the heterologous host (Wacker et al., 2002). Similarly, expression of the Toxoplasma gondii or Trypanosoma cruzi STT3 protein in the yeast Saccharomyces cerevisiae can complement the lethal phenotype of an stt3 deletion (Shams-Eldin et al., 2005; Castro et al., 2006). Evidence was presented indicating that the T. cruzi STT3 protein integrates into the yeast OTase complex. These results support the functional equivalence of the protozoan and yeast STT3 proteins.
In the present study, we have expressed the Leishmania major (Lm) STT3 paralogues LmSTT3A, LmSTT3B, LmSTT3C, or LmSTT3D individually in S. cerevisiae and found that three complemented the yeast stt3 deletion. In addition, LmSTT3D expression suppressed deletions of other essential OTase subunits (Wbp1p, Ost1p, Ost2p, and Swp1p). We showed that the L. major STT3 paralogues were active enzymes that did not incorporate into the yeast OTase complex, but instead formed dimers. These enzymes have different specificities with respect to the peptides and the LLO substrate.
MATERIALS AND METHODS
Plasmid Constructs
The open reading frames encoded in the L. major genome LmgF35.1130 (LmSTT3A), LmgF35.1140 (LmSTT3B), LmgF35.1150 (LmSTT3C), and LmgF35.1160 (LmSTT3D) were cloned in a pRS425_GPD yeast expression vector with a C-terminal fusion of a triple hemagglutinin (HA)-tag (Knop et al., 1999) followed by 12 additional vector-derived amino acids (KLIDTVDLESCN). The resulting plasmids were called pLmSTT3A, pLmSTT3B, pLmSTT3C, and pLmSTT3D.
Cloning was performed by two overlapping polymerase chain reaction (PCR) fragments and homologous recombination in yeast (Oldenburg et al., 1997). The first PCR fragment was obtained using the genomic DNA of L. major as a template, and primers LmSTT3_for and LmSTT3_rev (Supplemental Table 1). The second PCR fragment was obtained using plasmid pYM2 as a template (which contains a triple HA-tag) and primers LmSTT3_HA and CYC1_HA (Supplemental Table 1). Both PCR fragments contained 50 base pairs of overlap to the vector sequence. Integration of the PCR fragments into the HindIII-PstI linearized expression vector was achieved by homologous recombination in W303 yeast strain during transformation. Transformants were selected on synthetic minimal medium (SD; Guthrie and Fink, 1991; Sherman, 2002) lacking leucine (SD-Leu), and recombined plasmids were isolated from the yeast cells.
For the plasmid shuffle technique, pSTT3, a YEp352-derived high copy number plasmid containing the yeast STT3 locus (Zufferey et al., 1995), was used.
Yeast Strains and Media
Media.
Standard yeast media and genetic techniques were used (Guthrie and Fink, 1991). For selection of ura− cells, media containing 100 μg/ml 5-fluoroorotic acid (5-FOA; Boeke et al., 1987) and 1 M sorbitol were used.
Strains.
The strains used in this study are detailed in Supplemental Table 2.
YBS10 and YBS11 strains were generated by introducing the pSTT3 plasmid in the heterozygous STT3 diploid strain Y24390 (Euroscarf, Frankfurt, Germany; Supplemental Table 2). Sporulation of these cells was induced, and after tetrad dissection, haploid strains harboring the complementing plasmid and the stt3 deletion were identified. Strains YBS10 and YBS11 were chosen for further analysis. These strains were individually transformed with all four pLmSTT3 plasmids, and transformants were selected on synthetic minimal medium lacking leucine and uracil (SD-Leu-Ura) plates. Individual transformants were then plated on 5-FOA–containing media selecting for cells that had lost the STT3-containing URA3 plasmid.
To generate haploid strains with genomic deletions of other essential OTase subunits, diploid strains heterozygous for genes encoding essential subunits were purchased (Euroscarf): Y20242 (WBP1), Y26770 (OST1), Y22359 (OST2), and Y20730 (SWP1). These strains were transformed with pLmSTT3D; sporulation was induced and tetrads were dissected on YPD plates containing 1 M sorbitol. Haploid strains harboring the plasmid and the deletion in the OTase loci were identified based on the resistance toward G418. Haploid double mutant strains were generated by crossing individual single mutant strains, after inducing sporulation in these cells spore tetrads were dissected on YPD/1 M sorbitol plates. Haploid spores harboring the plasmid and the two deletions were identified on G418-containing media in nonparental ditype tetrads. For both single and double mutant strains, the absence of the specific S. cerevisiae genes was confirmed by PCR and the absence of the corresponding protein was verified by immunoblot analysis by using specific antisera.
Whole Cell Protein Extract and Immunological Methods
Cells were grown in synthetic minimal medium lacking leucine (SD-Leu) at permissive temperature to mid-log phase, corresponding to an OD600 nm of 1. Cells from 5-ml cultures were harvested by centrifugation, and cell pellets were resuspended in 0.2 ml of reducing sample buffer (0.0625 M Tris-HCl, pH 6.8, 2% SDS [vol/wt], 5% β-mercaptoethanol [vol/vol], 10% glycerol [vol/vol], and 0.01% bromphenol blue [wt/vol]), supplemented with 1 mM phenylmethylsulfonyl fluoride [PMSF] and 1× protease inhibitor complete cocktail used as was recommended by the supplier (PIC; Complete EDTA-free protease inhibitor cocktail; Roche Diagnostics, Mannheim, Germany). Incubation was done for 5 min at 95°C. For the analysis of membrane proteins, cells from 5-ml cultures were harvested and resuspended in 0.2 ml of reducing sample buffer supplemented with 7 M urea, 1 mM PMSF, and PIC and incubated for 20 min at 40°C.
Ten microliters of protein extracts was used for SDS-polyacrylamide gel electrophoresis (PAGE) (Lämmli, 1970), electroblotted to nitrocellulose membranes, and probed with anti-carboxypeptidase Y (CPY) (Burda et al., 1996b) and anti-HA antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:1000, and goat anti-rabbit immunoglobulin G-horseradish peroxidase (Santa Cruz Biotechnology) was used as a secondary antibody at a dilution of 1:3000. Visualization was done by chemiluminescence detection (ECL; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Preparation of Membrane Extracts
Yeast strains were grown in synthetic minimal medium lacking leucine (SD-Leu) to mid-log phase at 30°C to an OD600 nm of 1. Cells from 800 ml were harvested and washed once in 50 ml of 50 mM Tris-HCl, pH 7.5, 1 mM MgCl2, and 1 mM MnCl2. The cells were resuspended in the same buffer supplemented with 1 mM dithiothreitol (DTT), 1 mM PMSF, and 1× PIC, and they were lysed with acid washed glass beads by 2 × 4-min pulse of the bead beater with 1 min of pause between the pulses at 4°C. Intact cells, cell debris, and nuclei were removed by centrifugation for 5 min at 1000 × g. To obtain the membrane pellet, the supernatant was then centrifuged twice for 30 min at 50,000 × g at 4°C. The supernatant was removed, and the pellet was resuspended and homogenized in 800 μl of storage buffer (50 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM MnCl2, 35% glycerol, 1 mM DTT, 1 mM PMSF, and PIC). Extracts were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Solubilization of Membrane Proteins
To 100 μl of membrane suspension, we added 300 μl of 50 mM Tris-HCl, pH 7.4, 0.2 M mannitol, 0.1 M NaCl, 1 mM MgCl2, 1 mM CaCl2, 1 mM MnCl2, 1 mM DTT, 1 mM PMSF, and PIC. DNA was digested with 0.2 mg/ml DNAse I (3000 U/mg; Fluka, Buchs, Switzerland) for 45 min at 25°C. Membrane proteins were solubilized with 1.5% digitonin in the presence of 750 mM 6-aminocaproic acid for 45 min at 4°C with agitation. Insoluble material was removed by centrifugation for 30 min at 100,000 × g at 4°C. Samples were frozen in liquid nitrogen and stored at −80°C.
Blue Native PAGE
Blue native electrophoresis was carried out in the Protean II cell from Bio-Rad (Hercules, CA), with gel dimensions 20 × 15 × 0.1 cm. The gels consisted of a separating gel with a 5–9% acrylamide gradient and a stacking gel (4% acrylamide). Buffers and gel compositions were as described previously (Spirig et al., 2005). Protein concentration of the solubilized membrane protein samples was adjusted to 1 μg/μl with TM buffer (50 mM Tris-HCl, pH 7.4, 0.2 M mannitol, 0.1 M NaCl, 1 mM MgCl2, 1 mM CaCl2, and 1 mM MnCl2), supplemented with 10% glycerol, 750 mM 6-aminocaproic acid, 1.5% digitonin, 1 mM DTT, 1 mM PMSF, and PIC. Sample buffer consisting of 100 mM Tris-HCl, pH 7.5, 500 mM 6-aminocaproic acid, and 5% Serva blue G was added to 15% of the final volume, gently mixed, and the samples were loaded on the gel. For protein size marker, a mixture of 50 μl of albumin (1 mg/ml), 5 μl of ferritin (25 mg/ml), and 5 μl of thyroglobulin (35 mg/ml) was used. The electrophoresis was carried out at 4°C with the current limited to 10 mA for 24 h. After six running hours, the cathode buffer (50 mM Tricine, 15 mM Tris-HCl, and 0.02% Serva blue G; pH 7.5; 4°C) was removed, and the electrophoresis was continued with a cathode buffer without Serva blue G (50 mM Tricine and 15 mM Tris-HCl; pH 7.5; 4°C). After electrophoresis, the gels were soaked in transfer buffer (25 mM Tris-base, 200 mM glycine, 0.1% SDS, and 20% methanol), and proteins were transferred onto polyvinylidene difluoride (PVDF) membrane by using a semidry blotter. Removal of Coomassie dye from the PVDF membrane was achieved by soaking the blots for 3 × 15 min in 50% methanol, 10% acetic acid and subsequent washing in 90% methanol and finally in phosphate-buffered saline (136 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, and 0.1% Tween 20 [vol/vol]). Blots were dried and incubated with appropriate antisera against Wbp1p (te Heesen et al., 1991, 1992), Swp1p (te Heesen et al., 1993), Stt3p (Spirig et al., 2005), and Sec61p (provided by T. Rapoport, Harvard Medical School).
Cell Wall Protein Sample Preparation and Mass Spectrometry
Proteins covalently linked to the cell wall polysaccharide matrix were prepared from yeast cells, detected with liquid chromatography-electrospray ionization-tandem mass spectrometry, and the relative glycosylation occupancy of their N-glycosylation sites were determined as described previously (Schulz and Aebi, unpublished data). Peptides containing N-glycosylation sequons identified by mass spectrometry found in this study are listed in Supplemental Table 3.
RESULTS
Heterologous Expression of L. major STT3 Proteins Complements stt3 Mutations in S. cerevisiae
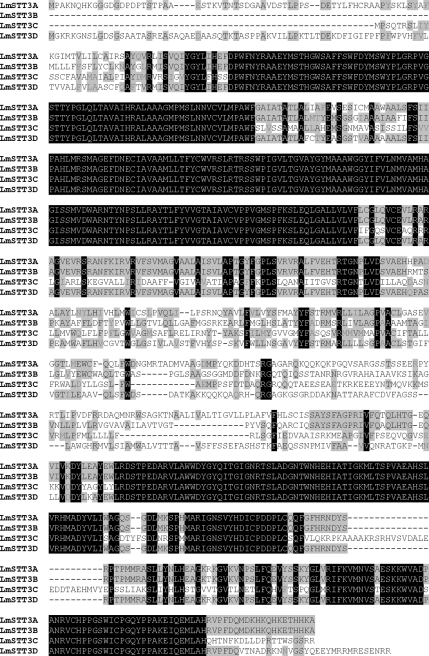
In the genome of L. major, four STT3 paralogues are encoded by adjacent loci: LmSTT3A, LmSTT3B, LmSTT3C, and LmSTT3D. The four STT3 paralogues of L. major have a high sequence identity (Figure 1).
Figure 1.
Sequence alignment of the four L. major STT3 paralogues LmSTT3A, LmSTT3B, LmSTT3C, and LmSTT3D. Identical amino acid residues conserved in all four proteins are shown in white letters on dark background, identical amino acid residues conserved in two or three proteins are shown in black letters on gray background, and similar residues are shown in dark gray on gray background. Alignment was calculated with Multalin (Corpet, 1988) by using Blosum62-12-2 alignment parameters.
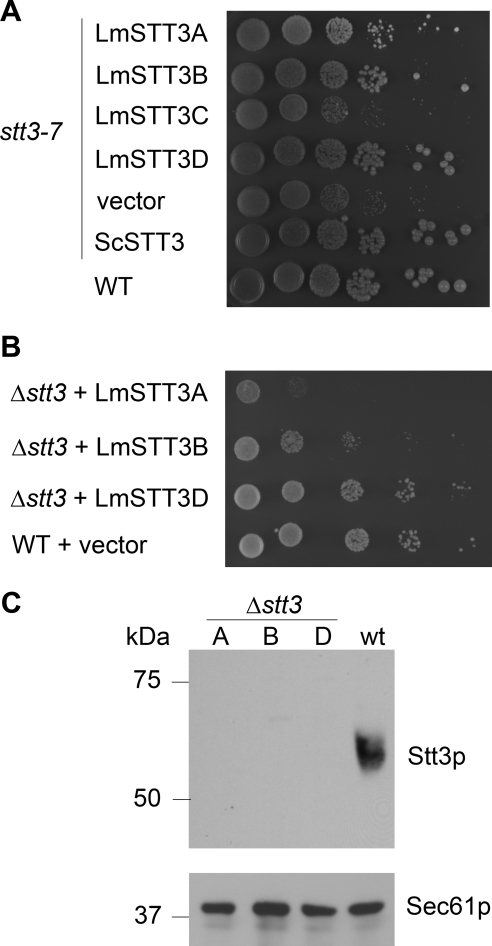
In this study, we expressed the four LmSTT3 proteins including C-terminal fusion of a triple HA-tag followed by 12 additional vector-derived amino acids from a high copy number yeast plasmid, by using the yeast GPD promoter to drive heterologous gene expression. All four heterologous proteins were expressed at comparable levels as judged by immunoblot analysis of yeast cell extracts by using HA-specific antiserum (data not shown; but see below). We tested the ability of the four L. major proteins to suppress the temperature-sensitive phenotype of the stt3-7 allele in strain YG543 (Spirig et al., 1997). Indeed, expression of LmSTT3A, LmSTT3B, and LmSTT3D but not of LmSTT3C suppressed the temperature-sensitive phenotype (Figure 2A). We concluded that the LmSTT3 proteins conferred different OTase activity.
Figure 2.
Complementation of mutations in OTase loci by expression of LmSTT3 proteins. Serial dilutions of the haploid yeast strains carrying the stt3-7 allele (A) or a deletion of the STT3 locus (B) and expressing LmSTT3 proteins (indicated at the left) were spotted. Plates were incubated at 30°C. Strains: YG543 (stt3-7), YG2052 (Δstt3 LmSTT3A), YG2054 (Δstt3 LmSTT3B), and YG2058 (Δstt3 LmSTT3D). (C) Analysis of yeast Stt3p expression in Δstt3 strains expressing LmSTT3A (A), LmSTT3B (B), and LmSTT3D (D); strains used were YG2052 (Δstt3 LmSTT3A), YG2054 (Δstt3 LmSTT3B), and YG2058 (Δstt3 LmSTT3D). Wild-type (wt; BY4741) served as a control. Protein extracts of the strains indicated were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with anti-Stt3p antiserum (top). The membrane was stripped and reprobed with anti-Sec61p antiserum to control protein loading (bottom).
We then asked whether expressing LmSTT3 proteins rescued the lethal phenotype of Δstt3 mutant cells as was reported for the T. cruzi STT3 protein. Strains YBS10 and YBS11 that contain a deletion of the STT3locus but harbor a plasmid carrying both the URA3 and the STT3 locus (relevant genotype ura3-52 Δstt3::kanMX4 pURA3/STT3; see Materials and Methods) were transformed with a plasmid expressing one of the four LmSTT3 proteins. The plasmid shuffle technique was used by selecting for cells growing on 5-FOA–containing plates and therefore lacking the pURA3/STT3 plasmid. 5-FOA–resistant cells were recovered from strains expressing LmSTT3A, LmSTT3B, or LmSTT3D, but not from cells expressing LmSTT3C or the empty vector controls. The absence of the endogenous STT3 locus in these cells was confirmed by polymerase chain reaction (PCR) analysis using primers specific for the yeast STT3 locus and immunoblot analysis using anti-Stt3p antiserum (Figure 2). Although the expression level of the LmSTT3 proteins was similar, their complementation efficiencies as judged by growth rate were different, with LmSTT3D being the highest, LmSTT3B intermediate, and LmSTT3A the lowest (Figure 2B).
We concluded that three of the four LmSTT3 proteins were able to partially replace the function of endogenous yeast Stt3p.
Deletions of the Essential, Non-STT3 OTase Subunits Are Suppressed by Expressing LmSTT3D
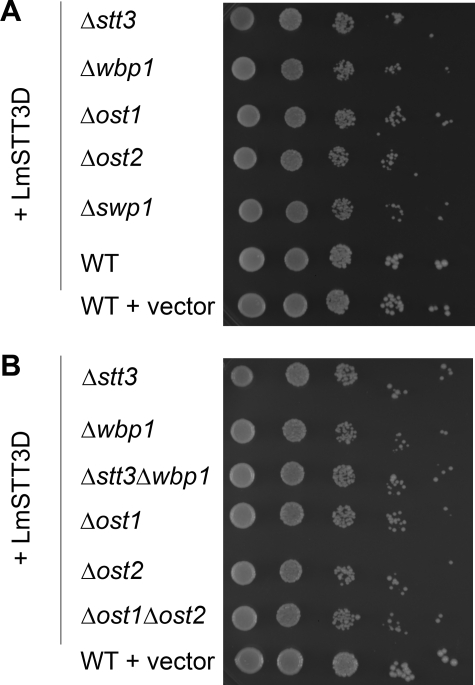
The functional replacement of the yeast Stt3p by LmSTT3 proteins was explained by either an integration of the LmSTT3 proteins into the yeast OTase complex, as was reported for the T. cruzi STT3 (Castro et al., 2006), or by a complete OTase functionality of the LmSTT3 proteins not requiring other yeast OTase components. The latter hypothesis was addressed directly by testing the ability of the LmSTT3D protein to suppress the lethal phenotype of deletions in loci encoding other essential subunits of the OTase.
Plasmid encoded LmSTT3D was expressed in diploid yeast strains heterozygous for a deletion in one of the essential loci WBP1, OST1, OST2, or SWP1. Sporulation was induced, spore tetrads were dissected and the resulting strains were analyzed. Haploid progeny carrying a deletion in the essential OTase subunit loci were recovered and characterized genetically. PCR and immunoblot experiments using sera directed against the different OTase subunits confirmed the absence of the essential OTase loci in these strains (data not shown; see below).
Interestingly, LmSTT3D expression rescued individual deletions of all essential OTase subunits (Stt3p, Ost1p, Wbp1p, Swp1p, and Ost2p). The growth rate of the yeast strains bearing LmSTT3D with a deletion in one of the essential OTase subunits was independent of the missing subunit: all strains grew at a similar rate slightly slower than wild-type cells (Figure 3A). To extend this analysis, we generated haploid yeast strains harboring deletions of two essential OTase loci (Δstt3Δwbp1 and Δost1Δost2) but containing the LmSTT3D expression plasmid. The growth rate of these double mutant strains was again similar to that of the single mutants, and no temperature-sensitive phenotype was observed (Figure 3B).
Figure 3.
Suppression of deletions in essential OTase loci by expression of LmSTT3 proteins. Serial dilutions of the haploid yeast strains carrying deletions in different OTase subunits encoding loci (given at the left) containing the LmSTT3D-expressing plasmid were spotted, and plates were incubated. Wild-type cells (BY4741) carrying LmSTT3D-expressing plasmid or the empty vector served as controls. Strains spotted: Δstt3 (YG2057), Dwbp1 (YG2072), Δost1 (YG2073), Δost2 (YG2075), Δswp1 (YG2080), Δstt3Δwbp1 (YG2084), and Δost1Δost2 (YG2099).
This genetic analysis suggested that the LmSTT3D protein was able to act in yeast as an OTase that did not require endogenous components of the yeast OTase for activity.
L. major STT3 Proteins Were Not Incorporated into the Yeast OTase Complex
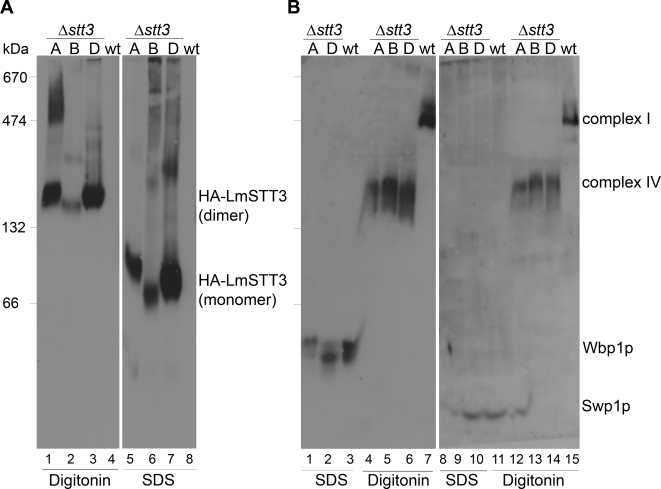
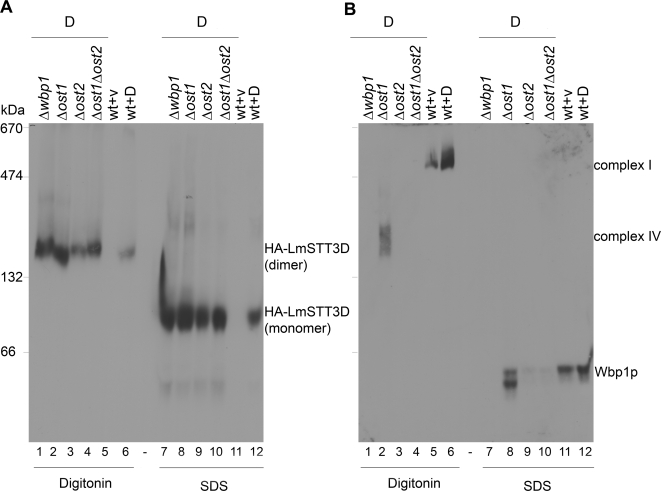
We addressed OTase composition in LmSTT3-expressing cells by blue native gel electrophoresis. This is a powerful technique to resolve membrane protein complexes by gel electrophoresis (Schägger and von Jagow, 1991), and we have previously used it to analyze the OTase complex in mutant yeast cells (Spirig et al., 2005).
We first analyzed wild type and Δstt3 strains expressing HA-tagged LmSTT3A, LmSTT3B, and LmSTT3D. Membrane proteins were solubilized with either 1.5% digitonin or 1.5% SDS, and separated by blue native gel electrophoresis using a 5–9% polyacrylamide gradient gel. Expression of HA-tagged LmSTT3 proteins was visualized by anti-HA antiserum. In nondenaturing conditions (1.5% digitonin), LmSTT3 proteins migrated at a position corresponding to a molecular mass of ∼160 kDa, compatible with the mass of a dimer. For LmSTT3A, slower moving protein resulting in a smear at higher molecular weight was also detected (Figure 4A, lane 1). The potential LmSTT3 complexes were disrupted in samples solubilized with SDS.
Figure 4.
Complex formations of LmSTT3 proteins, Wbp1p, and Swp1p in Δstt3 cells. Δstt3 mutant strains expressing HA-tagged LmSTT3A, LmSTT3B, or LmSTT3D (lanes 1, 2, and 3, respectively) and wild-type strains harboring empty vector (lane 4) were grown, membrane fractions were prepared and solubilized with 1.5% digitonin or 1.5% SDS. Solubilized proteins were separated by blue native gel electrophoresis using a 5–9% polyacrylamide gradient gel and transferred to PVDF membranes. Anti-HA antiserum (A), anti-Wbp1 (left), and Anti-Swp1 (right) antisera (B) were used for protein detection. OTase complex I and IV (see text), respectively, in wild-type and Δstt3 strains expressing LmSTT3A/B/D were detected. Strains: wild-type strain (BY4742) with empty vector, Δstt3 strains expressing LmSTT3A (YG2052), LmSTT3B (YG2054), or LmSTT3D (YG2058). Molecular mass of size marker is indicated at the left.
We analyzed the same extracts for the presence of yeast OTase complex (Figure 4B). In wild-type cells, sera directed against Wbp1p and Swp1p, respectively, detected the complete OTase complex migrating at a position equivalent to 550 kDa (Figure 4B, lanes 7 and 15; Spirig et al., 2005). Importantly, complete OTase complex was absent in Δstt3 cells, instead, we detected a subcomplex, previously termed complex IV (Spirig, 1999) that contains Wbp1p, Swp1p, and Ost2p (Figure 4B, lanes 4–6 and 12–14).
The mobility of the LmSTT3 proteins in blue native gel electrophoresis was in the same range as observed for Wbp1p and Swp1p in mutant cells, raising the possibility that the LmSTT3 proteins formed a complex with these endogenous OTase components. To address this issue, we analyzed OTase complex in yeast strains lacking these essential OTase subunits but expressing LmSTT3D. The mobility of the LmSTT3D complex was not influenced by the absence of different OTase components. In all cases tested we observed the same complex (Figure 5A, lanes 1–4). However, complex formation of the endogenous components was drastically affected by the absence of the different OTase subunits, as exemplified by the analysis of Wbp1p containing complexes (Figure 5B). Normal OTase complex was not observed in these deletion strains. Complex IV was detected in cells lacking Ost1p (Figure 5B, lane 8), but not in cells lacking Ost2p (Figure 5B, lanes 9 and 10). In the latter case, Wbp1p seemed to be degraded, because it was absent in SDS treated membranes (Figure 5B, lanes 9 and 10). As expected, Wbp1p was absent in Δwbp1 cells (Figure 5B, lane7).
Figure 5.
Complex formations of LmSTT3D protein and Wbp1p in yeast strains with different deletions in OTase-subunit encoding loci. Δwbp1, Δost1, Δost2, and Δost1Δost2 mutant and wild-type strains expressing HA-tagged LmSTT3D (lanes 1, 2, 3, 4, and 6, respectively), wild-type strains harboring empty vector (lane 5) were grown, and membrane fractions were prepared and solubilized with 1.5% SDS or 1.5% digitonin. Solubilized membrane proteins were separated by blue native gel electrophoresis by using a 5–9% polyacrylamide gradient gel and transferred to PVDF membrane. Anti-HA antiserum (A) and anti-Wbp1 antiserum (B) were used for protein detection. Strains used: wild-type strain (BY4742) with empty vector or LmSTT3D-expressing plasmid, and strains expressing LmSTT3D including Dwbp1 (YG2072), Δost1 (YG2073), Δost2 (YG2075), and Δost1Δost2 (YG2099). Molecular mass of size marker is indicated at the left.
These results confirmed our genetic data indicating that LmSTT3 proteins did not incorporate into the yeast OTase complex or subcomplexes. Most likely, the LmSTT3A, LmSTT3B, and LmSTT3D proteins formed active OTase in the heterologous host without the need of yeast OTase components.
Glycosylation Efficiency in Δstt3 Yeast Strains Expressing L. major STT3 Proteins
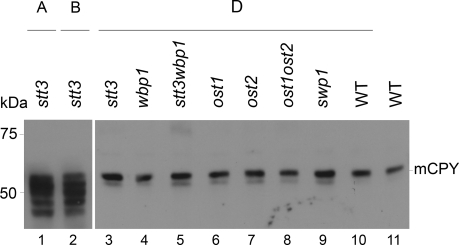
Our novel experimental system allowed us to study the function of the protozoan OTase in a genetically tractable, well defined heterologous system. We first tested the N-glycosylation of the model protein CPY, a vacuolar protease that contains four N-linked glycans. Hypoglycosylation of CPY can be monitored by the analysis of the mobility in SDS-PAGE. Glycoforms lacking one to four N-linked glycans migrate faster and are visualized as distinct bands after immunological detection of the protein (te Heesen et al., 1992). CPY showed hypoglycosylation in Δstt3 mutant strain complemented by LmSTT3A and LmSTT3B (Figure 6, lanes 1 and 2), whereas LmSTT3D restored CPY glycosylation to almost wild-type levels (Figure 6, lanes 3, 10, and 11). Similarly, expressing LmSTT3D rescued CPY glycosylation in strains lacking the other essential OTase subunits Wbp1p, Ost1p, Ost2p, and Swp1p, as well as in Δstt3Δwbp1 and Δost1Δost2 double mutant strains (Figure 6, lanes 4–9).
Figure 6.
Glycosylation of CPY in yeast strains lacking essential OTase subunits. Yeast strains with deletions in the OTase-subunit encoding loci indicated and expressing LmSTT3A (lane1), LmSTT3B (lane 2), LmSTT3D (lanes 3–10), or containing the vector DNA (lane 11) were grown to mid-log phase, total protein extracts were prepared, and proteins were separated by SDS-PAGE. After transferring to nitrocellulose membranes CPY protein was detected by immunoblotting using anti-CPY antiserum. Molecular mass of size marker is indicated at the left. Strains used: wild-type strain (BY4742) with empty vector or LmSTT3D-expressing plasmid, Δstt3 strains expressing LmSTT3A (YG2052), LmSTT3B (YG2054), and mutant strains expressing LmSTT3D were Δstt3 (YG2057), Dwbp1 (YG2072), Δstt3Δwbp1 (YG2084), Δost1 (YG2073), Δost2 (YG2075), Δost1Δost2 (YG2099), and Δswp1 (YG2080).
To obtain quantitative data for the glycosylation efficiency of LmSTT3 proteins in yeast and to detect potential alterations in protein substrate recognition, we applied a quantitative glycoproteomic approach. Proteins covalently linked to the yeast cell wall matrix were isolated and digested with Endoglycosidase H (EndoH), leaving a single N-acetylglucosamine (GlcNAc) residue covalently linked to the modified N-glycosylation sites. After digestion with specific proteases and mass spectrometric analysis, site occupancy at different sites was determined by comparing the levels of modified (GlcNAc-containing) and unmodified peptides (Schulz and Aebi, unpublished data).
As reported previously, wild-type cells glycosylate a majority of potential acceptor sites that can be analyzed (Table 1), and the efficiency of glycosylation at these sites was not altered by the introduction of the additional functionality, the heterologous L. major OTase (LmOTase) in wild-type cells (Table 1). However, we observed partial hypoglycosylation at specific sites in strains lacking the yeast Stt3p. The site specific level of glycosylation was dependent on the specific LmSTT3 isoform expressed (Table 1). In accordance with the growth phenotypes (Figure 2B), the most severe hypoglycosylation was observed in the LmSTT3A-expressing strain, whereas almost wild-type level of glycosylation was measured in LmSTT3D cells. We determined the glycosylation efficiency of strains carrying deletions of different OTase loci (Supplemental Table 4). Our quantitative data revealed that N-glycosylation by LmSTT3D was independent of the presence of yeast OTase components, because the same sites were incompletely glycosylated in these various strains.
Table 1.
Relative glycosylation site occupancy in different yeast strains
| Peptide | WT + vec | Δstt3 + A | Δstt3 + B | Δstt3 + D |
|---|---|---|---|---|
| CRH2_28 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.57 ± 0.03 | 1.00 ± 0.00 |
| CRH2_96 | 1.00 ± 0.00 | 0.82 ± 0.03 | 0.94 ± 0.06 | 0.99 ± 0.02 |
| CRH2_233 | 1.00 ± 0.00 | 0.58 ± 0.08 | 0.80 ± 0.20 | 0.96 ± 0.04 |
| CRH2_310 | 1.00 ± 0.00 | 0.28 ± 0.04 | 0.10 ± 0.02 | 1.00 ± 0.00 |
| ECM33_57 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.97 ± 0.03 | 1.00 ± 0.00 |
| ECM33_83 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| ECM33_197 | 1.00 ± 0.00 | 0.96 ± 0.04 | 0.86 ± 0.14 | 0.70 ± 0.01 |
| ECM33_210 | 1.00 ± 0.00 | 0.67 ± 0.03 | 0.42 ± 0.03 | 0.96 ± 0.02 |
| ECM33_268 | 1.00 ± 0.00 | 0.80 ± 0.02 | 0.75 ± 0.06 | 1.00 ± 0.00 |
| ECM33_329 | 1.00 ± 0.00 | 0.27 ± 0.04 | 0.05 ± 0.01 | 0.67 ± 0.06 |
| GAS1_40 | 1.00 ± 0.00 | 0.93 ± 0.07 | 0.77 ± 0.39 | 0.98 ± 0.02 |
| GAS1_57 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.97 ± 0.03 |
| GAS1_95 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.97 ± 0.03 | 1.00 ± 0.00 |
| GAS1_149 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.98 ± 0.02 | 1.00 ± 0.00 |
| GAS1_253 | 0.99 ± 0.01 | 0.99 ± 0.02 | 0.98 ± 0.02 | 1.00 ± 0.00 |
| GAS3_269 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.68 ± 0.05 | 1.00 ± 0.00 |
| GAS3_350 | 1.00 ± 0.00 | 0.73 ± 0.05 | 0.98 ± 0.03 | 1.00 ± 0.00 |
| GAS5_24 | 1.00 ± 0.00 | 0.79 ± 0.03 | 0.25 ± 0.01 | 1.00 ± 0.00 |
| GAS5_60 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| GAS5_344 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| PLB2_47 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| PLB2_193 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| CRH1_177 | 1.00 ± 0.00 | 0.95 ± 0.04 | 0.83 ± 0.09 | 1.00 ± 0.00 |
| EXG2_50 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| SAG1_79 | 0.72 ± 0.04 | nd | nd | 1.00 ± 0.00 |
| YJR1_99 | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.28 ± 0.03 | 1.00 ± 0.00 |
| CCW14_87 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| CWP1_45 | 0.00 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.00 ± 0.00 |
| TOS1_417 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
The relative occupancy of a site given at the left is shown for wild-type strain (BY4742) with empty vector, Δstt3 strains expressing LmSTT3A (YG2052), LmSTT3B (YG2054), or LmSTT3D (YG2057). nd, not determined.
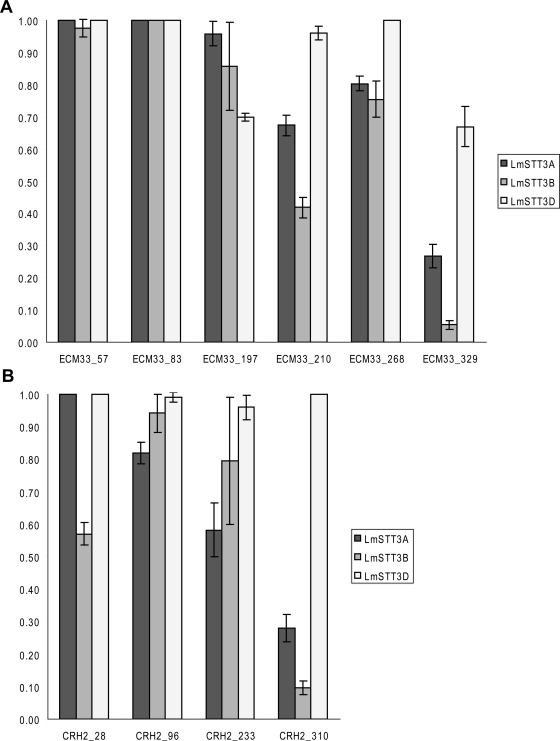
Our analytical method allowed us to compare the glycosylation efficiency of different sites located in the same protein. We observed that it was the individual site rather than the glycoprotein that determined the affinity toward the individual oligosaccharyltransferase (Figure 7).
Figure 7.
Site occupancy of glycosylation sites in two selected cell wall proteins. The relative occupancy of N-glycosylation sites was determined in Δstt3 strains expressing LmSTT3A (YG2052), LmSTT3B (YG2054), or LmSTT3D (YG2057). Values are given for the sites indicated of the proteins Ecm33p (A) and Crh2p (B) and represent the mean of three independent measurements. Standard deviations are indicated by bars. In wild-type yeast cells, the relative occupancy of all sites is 1.
Additionally, our analysis showed that LmSTT3D expression not only rescued OTase deficiencies but also increased glycosylation of the SAG1_79 site that was partially glycosylated by yeast OTase (Table 1 and Supplemental Table 4).
Although there was a strong overexpression of the LmSTT3 proteins, we can conclude that the affinity of three LmOTase toward different acceptor sites differed from the endogenous yeast OTase. This differential site-specific glycosylation was independent of the remaining OTase subunits of the host cell but was affected by the particular LmSTT3 isoform expressed. This suggested that the various LmOTases have different protein substrate affinities when expressed in yeast.
Glycan Specificity of the L. major OTase
The N-glycosylation pathways in kinetoplastids differ from those in higher eukaryotes with respect to the glycan structures that are transferred to protein substrates. Genetic information suggests that in L. major the lipid-linked Man7GlcNAc2 oligosaccharide is the mature substrate for OTase (Parodi, 1993; McConville et al., 2002; Ivens et al., 2005; Samuelson et al., 2005). Our experimental system offered the opportunity to modify the LLO assembly pathway to evaluate the substrate preferences of LmOTase in vivo.
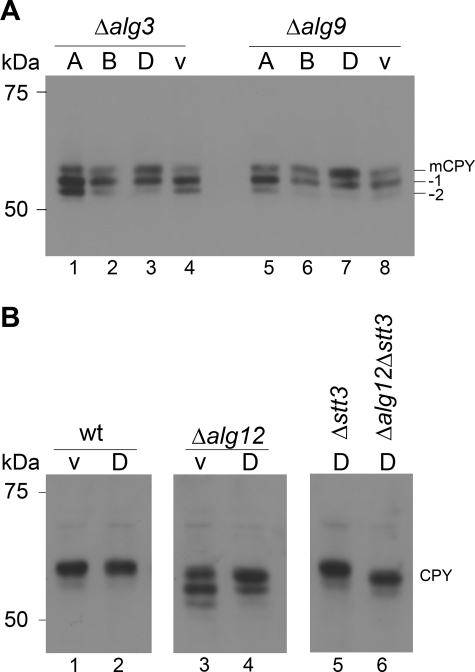
We tested whether the expression of LmOTases in various alg mutant strains improved the glycosylation efficiency of the model protein CPY. Due to the preference of the yeast OTase for the mature Glc3Man9GlcNAc2 lipid-linked glycan, hypoglycosylation was observed in strains deficient in specific steps of LLO biosynthesis. Δalg3, Δalg9, and Δalg12 mutations result in the biosynthesis of defined, incomplete LLO substrates (Man5GlcNAc2 in Δalg3, Man6GlcNAc2 in Δalg9, and Man7GlcNAc2 in Δalg12 cells) and subsequent hypoglycosylation (Huffaker and Robbins, 1983; Burda et al., 1996a; Burda et al., 1999). Expression of LmSTT3A and LmSTT3B did not improve the glycosylation of CPY in strains with alg3 or alg9 deficiency, whereas LmSTT3D expression significantly increased the steady state-level of CPY glycosylation (Figure 8A, lanes 3 and lane 7). Because these experiments were performed in strains with fully active yeast OTase, the increased glycosylation observed with expression of LmSTT3D was a dominant gain-of-function phenotype and suggested that this OTase isoform had different or relaxed glycan substrate specificity.
Figure 8.
Glycosylation of CPY in alg mutant strains expressing LmSTT3 proteins. (A) Δalg3 cells (Y03108; left) and Δalg9 cells (Y01993; right) expressing LmSTT3A, LmSTT3B, LmSTT3D or carrying empty vector (v) were grown, protein extracts were separated by SDS-PAGE, transferred to PVDF membrane, and anti-CPY antiserum was used for protein detection. (B) Wild-type (BY4741), Δalg12 (Y05405), Δstt3 (YG2058), and Δalg12Δstt3 (YG2082) strains carrying empty vector (v) or plasmid expressing LmSTT3D were grown, protein extracts were separated by SDS-PAGE, transferred to nitrocellulose membrane, and anti-CPY antiserum was used for protein detection. The position of fully glycosylated CPY (mCPY) and CPY molecules lacking 1 (−1) or 2 (−2) oligosaccharide chains are indicated. Molecular mass of size marker are given at the left.
We evaluated the substrate specificity of LmSTT3D in more detail in Δalg12 mutant cells assembling lipid-linked Man7GlcNAc2 oligosaccharide (Burda et al., 1999), the predicted product of LLO biosynthesis in L. major cells. As was the case for Δalg3 and Δalg9 cells, expression of LmSTT3D improved the glycosylation of CPY, independently of the presence or absence of the endogenous OTase (Figure 8B, lanes 3, 4, and 6). In addition, LmSTT3D-dependent CPY glycosylation was not affected by the structure of LLO substrates: the glycosylation efficiency was very similar in Δalg12 and ALG12 cells expressing LmSTT3D (Figure 8B, lanes 5 and 6). However, in wild-type cells, complete oligosaccharide was predominantly transferred, as visualized by the ALG12-dependent mobility shift of glycosylated CPY in SDS-PAGE (Figure 8B, lanes 5 and 6).
Together, these results suggested that biosynthetic intermediates as well as the complete Glc3Man9GlcNAc2 oligosaccharide were transferred efficiently to protein by the LmSTT3D OTase isoform without any apparent substrate specificity.
DISCUSSION
LmSTT3D Acts Independently of Endogenous OTase Subunits in Yeast
Heterologous expression of STT3 proteins from protozoan organisms in yeast revealed the functional equivalence of these proteins to the endogenous Stt3p. Although the molecular basis for the complementation of stt3 mutation by the expression of the T. gondii STT3 protein was not analyzed in detail (Shams-Eldin et al., 2005), Castro et al. (2006) showed that the T. cruzi STT3 protein most likely replaced the yeast Stt3p in the complex. Our analysis revealed that such integration into the complex was not observed for the LmSTT3 proteins when expressed in yeast.
Three of the four LmSTT3 proteins were able to suppress the lethal phenotype of a Δstt3 mutation, but they were not a part of the yeast OTase, as revealed by blue native gel electrophoresis and by the ability of LmSTT3D to suppress the lethal phenotype of missing essential OTase subunits such as Wbp1p, Ost1p, Swp1p, and Ost2p.
Our data were consistent with the model that the LmOTase is composed of a single subunit, active as a dimeric complex. We did not analyze possible heterodimer formation in L. major or in yeast, but the high sequence identity in different regions of the LmSTT3 paralogues makes it possible that these proteins form heterodimers.
To the best of our knowledge, our data provided the first direct experimental evidence for an active eukaryotic OTase consisting of a single subunit. Phylogenetic analysis suggests that this is also the case in other trypanosomatid protozoa as no homologues of the additional yeast or mammalian OTase subunits are encoded in their genomes. The bacterial OTase from C. jejuni is also a single subunit enzyme (Wacker et al., 2002; Kowarik et al., 2006b), but it differs from the kinetoplastid LmSTT3D OTase with respect to LLO, and, more importantly, protein substrate specificity. The prokaryotic enzyme requires the “extended” acceptor sequence D/E-X1-N-X2-S/T (Kowarik et al., 2006b), whereas the L. major enzymes glycosylated the “minimal” consensus N-X-S/T in yeast and used LLO substrates typical for eukaryotic cells.
The various LmOTase isoforms displayed different protein substrate specificities at the level of individual glycosylation sequons. These data confirmed that besides the essential N-X-S/T sequon motif recognized by STT3 protein, additional structural features of protein substrates may influence the acceptor properties of a given sequon. It is likely that these additional features are different for each of the LmOTase isoforms. More detailed quantitative analyses will define the substrate specificity of the LmOTase isoforms.
Our genetic analysis revealed a relaxed specificity of the LmSTT3D OTase with respect to LLO substrate. As reported for the native T. cruzi enzyme, glucosylation of the LLO did not seem to affect glycosylation efficiency (Bosch et al., 1988; Kelleher et al., 2007). Within the limits of our experimental in vivo system, we also did not detect a reduced transfer efficiency of glycans lacking α1,2-linked mannose on the B (and C) branch as reported for the T. cruzi enzyme (Kelleher et al., 2007).
Besides the identification and preliminary characterization of the protozoan OTase, our experiments revealed novel aspects of yeast OTase activity. Very surprisingly, the single subunit protozoan enzyme could functionally replace the yeast OTase complex. This raised questions regarding the functional significance of the additional subunits in the yeast enzyme. As elegantly shown by Kelleher et al. (2007), one prominent functionality of the complex eukaryotic enzyme is the LLO substrate specificity that can be associated with the presence of the Swp1p–Wbp1p–Ost2p subcomplex, the complex IV we observed in Stt3p-deficient cells (Figure 4B, lanes 4–6 and 12–14). This subcomplex possibly contains a regulatory binding site for completely assembled LLO and acts as a modulator of the catalytic OTase (Stt3p) subunit (Kelleher et al., 2007). However, absence of this regulatory function does not explain the lethal phenotype of swp1, wbp1, or ost2 deletions because the absence of completely assembled oligosaccharide in alg mutant cells is not lethal for the cells.
It is possible that yeast Stt3p is not catalytically active on its own but requires the presence of other proteins in the OTase complex for activity. This view was supported by the experimental finding that overexpression of the yeast STT3 subunit did not improve glycosylation in a Δalg12 strain nor did it rescue the lethal phenotype of a wbp1 mutation (data not shown).
Alternatively, the lethal phenotype of nonSTT3 OTase subunits could be due to degradation of the remaining “orphan” OTase subunits (Figure 5B) and therefore a loss of OTase activity. The lethal phenotype of missing noncatalytic subunits would therefore be the consequence of complex instability and subsequent degradation of subunits. This hypothesis is supported by the finding that deficiencies in the ER-associated protein degradation pathway can suppress stt3-7 temperature-sensitive phenotype (Jakob et al., 2001), although deletion of essential, non-STT3 subunits are not suppressed by these mutations.
The results reported here provide the basis for a novel experimental system that allows a more direct analysis of the N-glycosylation process in eukaryotic cells. Similar to the functional transfer of the bacterial N-glycosylation machinery from C. jejuni to E. coli, this heterologous system makes it possible to investigate the basic features and mechanisms of various eukaryotic OTases. Structure–function analysis of the single subunit eukaryotic OTase isoforms is now possible, independent of complications arising from possible interactions with the endogenous yeast OTase complex. Vice versa, the functional expression of the LmOTase in yeast provides novel experimental tools to study the yeast OTase complex in greater detail. Because the lethal phenotype of subunit mutations can be suppressed by expression of the protozoan OTase, experimental analyses of complex formation and the function of the essential, noncatalytic OTase subunits will now be possible.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Functional Genomics Center Zurich for the excellent support and the members of the Aebi lab for fruitful discussions. This work was supported by the Swiss National Science Foundation grant 3100A0-105541 (to M. A.) and the ETH Zurich. F. G. acknowledges support by ISCIII-Red de Investigación Cooperativa en Enfermedades Tropicales grant RD06/0021/0002.
Abbreviations used:
- CPY
carboxypeptidase Y
- LLO
lipid-linked oligosaccharide
- LmOTase
L. major oligosaccharyltransferase
- OTase
oligosaccharyltransferase
- PIC
protease inhibitor cocktail.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0467) on July 2, 2008.
REFERENCES
- Berriman M., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bosch M., Trombetta S., Engstrom U., Parodi A. J. Characterization of dolichol diphosphate oligosaccharide: protein oligosaccharyltransferase and glycoprotein-processing glucosidases occurring in trypanosomatid protozoa. J. Biol. Chem. 1988;263:17360–17365. [PubMed] [Google Scholar]
- Burda P., Aebi M. The dolichol pathway of N-glycosylation. Biochim. Biophys. Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Burda P., Jakob C. A., Beinhauer J., Hegemann J. H., Aebi M. Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology. 1999;9:617–625. doi: 10.1093/glycob/9.6.617. [DOI] [PubMed] [Google Scholar]
- Burda P., te Heesen S., Aebi M. Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: identification of the ALG9 gene encoding a putative mannosyl transferase. Proc. Natl. Acad. Sci. USA. 1996a;93:7160–7165. doi: 10.1073/pnas.93.14.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P., te Heesen S., Brachat A., Wach A., Dusterhoft A., Aebi M. Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: identification of the ALG9 gene encoding a putative mannosyl transferase. Proc. Natl. Acad. Sci. USA. 1996b;93:7160–7165. doi: 10.1073/pnas.93.14.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro O., Movsichoff F., Parodi A. J. Preferential transfer of the complete glycan is determined by the oligosaccharyltransferase complex and not by the catalytic subunit. Proc. Natl. Acad. Sci. USA. 2006;103:14756–14760. doi: 10.1073/pnas.0607086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R. D. Synthesis of asparagine-linked oligosaccharides: Pathway, genetics, and metabolic regulation. In: Allen H. J., Kisailus E. C., editors. Glycoconjugates. New York: Marcel Dekker; 1992. pp. 333–360. [Google Scholar]
- Dempski R. E., Jr., Imperiali B. Oligosaccharyl transferase: gatekeeper to the secretory pathway. Curr. Opin. Chem. Biol. 2002;6:844–850. doi: 10.1016/s1367-5931(02)00390-3. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Methods in Enzymology. Volume 194. Cold Spring, NY: Cold Spring Harbor Laboratory Press; 1991. Guide to Yeast Genetics and Molecular Biology. [Google Scholar]
- Herscovics A., Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Huffaker T. C., Robbins P. W. Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. USA. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens A. C., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001;2:423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher D. J., Banerjee S., Cura A. J., Samuelson J., Gilmore R. Dolichol-linked oligosaccharide selection by the oligosaccharyltransferase in protist and fungal organisms. J. Cell Biol. 2007;177:29–37. doi: 10.1083/jcb.200611079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher D. J., Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Karaoglu D., Mandon E. C., Gilmore R. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell. 2003;12:101–111. doi: 10.1016/s1097-2765(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Knauer R., Lehle L. The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta. 1999;1426:259–273. doi: 10.1016/s0304-4165(98)00128-7. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kowarik M., Numao S., Feldman M. F., Schulz B. L., Callewaert N., Kiermaier E., Catrein I., Aebi M. N-Linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 2006a;314:1148–1150. doi: 10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., Aebi M. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006b;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McConville M. J., Mullin K. A., Ilgoutz S. C., Teasdale R. D. Secretory pathway of trypanosomatid parasites. Microbiol. Mol. Biol. Rev. 2002;66:122–154. doi: 10.1128/MMBR.66.1.122-154.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I., Kelleher D. J., Miao Y., Shao Y., Kreibich G., Gilmore R., von Heijne G., Johnson A. E. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J. Cell Biol. 2003;161:715–725. doi: 10.1083/jcb.200301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg K. R., Vo K. T., Michaelis S., Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi A. J. N-Glycosylation in trypanosomatid protozoa. Glycobiology. 1993;3:193–199. doi: 10.1093/glycob/3.3.193. [DOI] [PubMed] [Google Scholar]
- Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Shams-Eldin H., Blaschke T., Anhlan D., Niehus S., Muller J., Azzouz N., Schwarz R. T. High-level expression of the Toxoplasma gondii STT3 gene is required for suppression of the yeast STT3 gene mutation. Mol. Biochem. Parasitol. 2005;143:6–11. doi: 10.1016/j.molbiopara.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Silberstein S., Gilmore R. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 1996;10:849–858. [PubMed] [Google Scholar]
- Spirig U. ETH Zurich. Zurich, Switzerland: 1999. The oligosaccharyltransferase complex of Saccharomyces cerevisiae. [Google Scholar]
- Spirig U., Bodmer D., Wacker M., Burda P., Aebi M. The 3.4-kDa Ost4 protein is required for the assembly of two distinct oligosaccharyltransferase complexes in yeast. Glycobiology. 2005;15:1396–1406. doi: 10.1093/glycob/cwj025. [DOI] [PubMed] [Google Scholar]
- Spirig U., Glavas M., Bodmer D., Reiss G., Burda P., Lippuner V., te Heesen S., Aebi M. The STT3 protein is a component of the yeast oligosaccharyltransferase complex. Mol. Gen. Genet. 1997;256:628–637. doi: 10.1007/s004380050611. [DOI] [PubMed] [Google Scholar]
- Tanner W., Lehle L. Protein glycosylation in yeast. Biochim. Biophys. Acta. 1987;906:81–99. doi: 10.1016/0304-4157(87)90006-2. [DOI] [PubMed] [Google Scholar]
- te Heesen S., Janetzky B., Lehle L., Aebi M. The yeast WBP1 is essential for oligosaccharyltransferase activity in vivo and in vitro. EMBO J. 1992;11:2071–2075. doi: 10.1002/j.1460-2075.1992.tb05265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S., Knauer R., Lehle L., Aebi M. Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyltransferase activity. EMBO J. 1993;12:279–284. doi: 10.1002/j.1460-2075.1993.tb05654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S., Rauhut R., Aebersold R., Abelson J., Aebi M., Clark M. W. An essential 45kDa yeast transmembrane protein reacts with anti-nuclear pore antibodies: purification of the protein, immunolocalisation and cloning of the gene. Eur. J. Cell Biol. 1991;56:8–18. [PubMed] [Google Scholar]
- Wacker M., et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- Weerapana E., Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Yan A., Lennarz W. J. Unraveling the mechanism of protein N-glycosylation. J. Biol. Chem. 2005;280:3121–3124. doi: 10.1074/jbc.R400036200. [DOI] [PubMed] [Google Scholar]
- Yan Q., Lennarz W. J. Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 2002;277:47692–47700. doi: 10.1074/jbc.M208136200. [DOI] [PubMed] [Google Scholar]
- Zufferey R., Knauer R., Burda P., Stagljar I., te Heesen S., Lehle L., Aebi M. STT3, a highly conserved protein required for yeast oligosaccharyltransferase activity in vitro. EMBO J. 1995;14:4949–4960. doi: 10.1002/j.1460-2075.1995.tb00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.