Abstract
Nuclear export of mRNA is tightly linked to transcription, nuclear mRNA processing, and subsequent maturation in the cytoplasm. Tip-associated protein (TAP) is the major nuclear mRNA export receptor, and it acts coordinately with various factors involved in mRNA expression. We screened for protein factors that associate with TAP and identified several candidates, including RNA helicase DDX3. We demonstrate that DDX3 directly interacts with TAP and that its association with TAP as well as mRNA ribonucleoprotein complexes may occur in the nucleus. Depletion of TAP resulted in nuclear accumulation of DDX3, suggesting that DDX3 is, at least in part, exported along with messenger ribonucleoproteins to the cytoplasm via the TAP-mediated pathway. Moreover, the observation that DDX3 localizes transiently in cytoplasmic stress granules under cell stress conditions suggests a role for DDX3 in translational control. Indeed, DDX3 associates with translation initiation complexes. However, DDX3 is probably not critical for general mRNA translation but may instead promote efficient translation of mRNAs containing a long or structured 5′ untranslated region. Given that the DDX3 RNA helicase activity is essential for its involvement in translation, we suggest that DDX3 facilitates translation by resolving secondary structures of the 5′-untranslated region in mRNAs during ribosome scanning.
INTRODUCTION
Eukaryotic mRNAs are transcribed as precursors that undergo extensive nuclear processing to become mature and subsequently are exported to the cytoplasm for translation. Some mRNAs may be regulated by means of their turnover rate, subcellular localization, or translation efficiency in the cytoplasm. Much progress has been made recently toward understanding the molecular basis for mRNA export, which is coordinately coupled to several other events of mRNA metabolism (Maniatis and Reed, 2002; Reed, 2003).
Tip-associated protein (TAP) is considered as the major receptor for bulk mRNA export (Katahira et al., 1999; Stutz and Izaurralde, 2003). TAP is highly conserved from yeast to humans. All known TAP homologues contain an RNA-binding domain and a leucine-rich region in the N-terminal part, and a nuclear transport factor 2-like domain and a ubiquitin-associated domain in the C-terminal–most region (Stutz and Izaurralde, 2003). Through the two C-terminal domains, TAP forms a heterodimer with p15 and docks to the nucleoporins of the nuclear pore complex (NPC) (Wiegand et al., 2002). Although TAP contains an RNA binding domain, its association with messenger ribonucleoproteins (mRNPs) largely involves various adaptor proteins (Strasser and Hurt, 2000; Rodrigues et al., 2001; Stutz and Izaurralde, 2003; Huang and Steitz, 2005). The exon junction complex (EJC), which is deposited onto spliced mRNA during precursor mRNA (pre-mRNA) splicing, can recruit TAP before mRNA export (Dreyfuss et al., 2002). In addition, several RNA-binding proteins, including nucleocytoplasmic shuttling serine/arginine dipeptide-rich proteins (SR proteins), can also serve as adaptors of TAP (Huang et al., 2003; Huang and Steitz, 2005). Nevertheless, TAP can directly bind to the constitutive transport element that resides within the Mason-Pfizer monkey virus RNA and also within its own intron 10-retained mRNA via the leucine-rich repeat domain to facilitate the nuclear export of these RNAs (Gruter et al., 1998; Li et al., 2006).
Nuclear export of mRNA is closely linked to transcription and mRNA processing. In vertebrate cells, pre-mRNA splicing may greatly enhance the export of the resultant mature mRNA (Luo and Reed, 1999). Splicing factor UAP56 associates with the primary transcripts to facilitate spliceosome assembly and subsequently recruits the EJC component REF/Aly (Luo et al., 2001). Several EJC factors, such as RNPS1 and Pinin, can function in pre-mRNA splicing, perhaps before EJC formation (Mayeda et al., 1999; Li et al., 2003). After splicing, REF/Aly or the EJC recruits export receptor TAP/p15 to the spliced mRNPs for export (Dreyfuss et al., 2002; Reed, 2003); however, such activity may be redundant with activities of other export adaptors, i.e., shuttling RNA-binding proteins. REF/Aly facilitates nuclear export of intronless RNAs through its direct interaction with TAP, suggesting that splicing is not the only route to promote mRNA export (Rodrigues et al., 2001). Indeed, UAP56 and REF/Aly can associate with the nascent transcripts cotranscriptionally through their interaction with the transcription/export complex, TREX, a factor involved in transcription elongation (Erkmann and Kutay, 2004; Reed and Cheng, 2005). Such an interaction suggests that transcription and mRNA export are coupled. In addition, mutations of several mRNA 3′-end processing factors result in defective mRNA export (Hammell et al., 2002). Therefore, the 3′-end maturation process of mRNA may also influence its subsequent nuclear export (Hammell et al., 2002). Furthermore, mRNA export can impact downstream steps of mRNA biogenesis in the cytoplasm. For example, several EJC components participate in nonsense-mediated decay, an mRNA surveillance pathway that targets premature termination codon-bearing mRNA for degradation (Hentze and Kulozik, 1999). Furthermore, the export receptor TAP can selectively enhance translation of mRNAs that contain its binding target, the constitutive transport element (Jin et al., 2003). Therefore, the molecular basis of how these mRNA biosynthesis steps—including mRNA export—are coupled still remains to be studied.
To better understand mRNA export pathways, in this study we isolated and identified TAP-associated proteins. The RNA helicase DDX3 was one of the candidates that associated with TAP. RNA helicases participate in various aspects of mRNA metabolism, including transcription, pre-mRNA splicing, mRNA export and decay, and translation (Rocak and Linder, 2004; Cordin et al., 2006). They are presumed to consume energy from ATP hydrolysis to unwind RNA-duplex structures or to remodel RNA–protein complexes (Rocak and Linder, 2004; Cordin et al., 2006). In recent years, several RNA helicases have been reported to play a role in mRNA export. Among them, Dbp5 has been the most extensively characterized. Dbp5 facilitates mRNA export likely by remodeling mRNPs at the cytoplasmic filaments of the NPC (Cole and Scarcelli, 2006). Dbp5 also functions in translation by promoting stop codon recognition for termination (Gross et al., 2007). Another RNA helicase, UAP56, plays a role in both pre-mRNA splicing and mRNA export, and perhaps links transcription, splicing, and mRNA export (Reed, 2003; Erkmann and Kutay, 2004; Reed and Cheng, 2005).
DDX3 belongs to the DEAD-box family of RNA helicases. This family of RNA helicases contains a highly conserved catalytic core domain that mediates the ATPase and helicase activities (Rocak and Linder, 2004; Cordin et al., 2006). The yeast DDX3 homologue, Ded1, has been identified as a genetic suppressor of a pre-mRNA splicing factor prp8 mutant (Jamieson et al., 1991). A Ded1 mutation resulted in altered splicing profiles, suggesting its role in pre-mRNA splicing (Burckin et al., 2005). Moreover, Ded1 is likely involved in global translation (Chuang et al., 1997). Like yeast Ded1, mammalian DDX3 may participate in several cellular activities, including transcription, mRNA export, and translation (Fukumura et al., 2003; Chao et al., 2006; Shih et al., 2008). DDX3 facilitates nuclear export of human immunodeficiency virus RNAs via a specific pathway involving the importin-β family receptor CRM1 (Yedavalli et al., 2004). More recently, DDX3 was shown to suppress cap-dependent translation but activate the internal ribosome entry site-mediated translation of hepatitis C virus RNA (Shih et al., 2008). Nevertheless, the molecular mechanisms underlying DDX3 involvement in gene expression remain to be fully determined.
MATERIALS AND METHODS
Plasmid Construction
The FLAG-tagged TAP expression plasmid was a generous gift of J. A. Steitz (Yale University, New Haven, CT). A sequence encoding the C-terminally truncated TAP (TAPΔC), which lacks the C-terminal 36 amino acids, was inserted into pcDNA3.1 (Invitrogen, Carlsbad, CA) with the appropriate restriction enzymes. Full-length TAP and TAPΔC were subcloned individually into pCEP4 (Invitrogen) to generate constructs encoding hemagglutinin (HA)-tagged proteins. The plasmids expressing FLAG-tagged DDX3, GST-DDX3, and a series of truncated DDX3 fragments have been described previously (Chao et al., 2006, Shih et al., 2008). Full-length DDX3 was subcloned into the pEGFP-C1 (Clontech, Mountain View, CA) to create a construct expressing DDX3 fused with green fluorescent protein (GFP-DDX3). The plasmid expressing HA-tagged TIA-1 was kindly provided by N. Kedersha (Brigham and Women's Hospital, Boston, MA). The open reading frames of HuR, PABP1, and eukaryotic initiation factor (eIF)4A were obtained by polymerase chain reaction (PCR) by using a human fetal brain cDNA library (Clontech), and each was inserted into pCEP4 (Invitrogen) to create constructs encoding HA-tagged proteins. The plasmid encoding myc-tagged eIF2α was kindly provided by W. Chang (Academia Sinica, Taipei, Taiwan). For in vivo translation assay, the reporter pFL-SV40 was generated by replacing the coding sequence of the Renilla luciferase in pRL-SV40 (Promega, Madison, WI) with the firefly luciferase derived from pGL3-Basic (Promega). The pFL-SV40 reporter containing a hairpin was created by insertion of a self-annealed double-stranded oligonucleotide (5′-CTAGCGGATCCGGCCGGATCCGGCCGGATCCG-3′) into the 5′-proximal Hind III site in the 5′-untranslated region (UTR). The DNA fragments encoding human transforming growth factor (TGF) β1 and rat ornithine decarboxylase (ODC) 5′-UTRs were obtained by PCR by using human embryonic kidney (HEK) 293 cells and PC12 genomic DNA as template, respectively; each 5′-UTR DNA was inserted into pFL-SV40. The pSilencer 1.0-U6 (Ambion, Austin, TX) vector was used to express short hairpin RNAs (shRNAs) targeting human DDX3 (shRNA1, open reading frame nucleotides 1699-1717; shRNA2, nucleotides 131-149) or human TNPO3 (nucleotides 240-258). The plasmids expressing mutant (S382L) and shRNA-resistant DDX3 were generated using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA). The shRNA-resistant DDX3 was created by introducing silent mutations (as underlined) into the target sequence of DDX3 shRNA1 (nucleotides 1699-GGCCAAGCTGGCTGGAAA-1717). The small interfering RNA (siRNA) duplex for targeting TAP was purchased from Invitrogen, corresponding to nucleotides 950–974 of the TAP coding sequence.
Cell Culture and Transfection
HeLa and HEK293 cells were grown at 37°C in DMEM containing 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Transfection was performed using the Lipofectamine 2000 (Invitrogen).
Preparation of Recombinant Proteins and Antibodies against DDX3
Glutathione transferase (GST) and GST-DDX3 fusion proteins were overproduced in Escherichia coli strain BL21 and purified over glutathione-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). His-tagged DDX3 protein was overexpressed in E. coli strain BL21 and purified on HisBind resin (Novagen, Madison, WI) according to the procedure recommended by the manufacturer. A polyclonal antiserum against DDX3 was generated by immunizing rabbits with purified His-tagged DDX3. To purify anti-DDX3, rabbit antiserum was incubated with nitrocellulose membranes containing immobilized GST-DDX3. Absorbed antibodies were eluted with 0.2 M glycine-HCl, pH 2.3, as described previously (Lai et al., 2000).
Indirect Immunofluorescence and Fluorescence In Situ Hybridization
Immunostaining of HeLa cells was performed essentially as described previously (Lai et al., 2000). To examine the nuclear export pathway of DDX3, HeLa cells were treated with 40 nM leptomycin B for 3 h. The primary antibodies used were polyclonal anti-DDX3 (0.5 μg/ml; purified from anti-serum), eIF4AI (2.5 μg/ml; Abcam, Cambridge, United Kingdom), and CRM1 (0.4 μg/ml; Abcam), and monoclonal anti-HA (1:200 dilution; gift from S.-C. Cheng, Academia Sinica, Taipei, Taiwan), PABP (2 μg/ml; Sigma), eIF4E (2.5 μg/ml; BD Transduction Laboratories, Lexington, KY), and cyclin B1 (0.4 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibodies used (Cappel Laboratories, Durham, NC) included fluorescein-conjugated anti-mouse immunoglobulin G (IgG) (20 μg/ml), fluorescein-conjugated anti-rabbit IgG (24 μg/ml), and rhodamine-conjugated anti-mouse IgG (20 μg/ml). The images of double-stained cells were obtained using a laser confocal microscope (MRC1000; Bio-Rad, Hercules, CA).
To detect the distribution of poly(A)+ RNA, fluorescence in situ hybridization was performed as described previously (Li et al., 2003). Digoxigenin (DIG)-labeled oligo(dT) (50-mer) was used as probe.
Immunoprecipitation and Immunoblotting
FLAG-tagged proteins were expressed alone or coexpressed with either HA-tagged or myc-tagged protein in HEK293 cells for 2 d. Immunoprecipitation was performed essentially according to Lykke-Andersen et al. (2001). In brief, 0.5 ml of cell lysates prepared from ∼5 × 106 cells (i.e., from a >90% confluent 60-mm culture dish) were incubated with 10 μl of anti-FLAG M2-agarose (Sigma Chemical, Poole, Dorset, United Kingdom). Immunoprecipitates were treated with 1 mg/ml RNase A at 37°C for 30 min. For immunoblotting, the primary antibodies used were against the following proteins or epitope tags: DDX3 (purified from anti-serum), TAP, nucleolin, lamin A/C, eIF2α (the above four from Santa Cruz Biotechnology), α-tubulin (NeoMarkers, Fremont, CA), eIF4G, eIF4E (the above two from BD Transduction Laboratories), eIF5 (Abnova, Taipei City, Taiwan), FLAG (Sigma Chemical), HA (Covance Research Products, Princeton, NJ), and myc (Millipore, Billerica, MA).
In Vitro Pull-down Assay
The TAP protein was in vitro translated and 35S-labeled in the TNT-coupled reticulocyte lysate systems (Promega) and then incubated with 2 μg of GST-DDX3 in a 60-μl mixture at 30°C for 30 min for the pull-down analysis as described previously (Lai et al., 2000).
Subcellular Fractionation
HEK293 cells were washed twice with cold phosphate-buffered saline (PBS) and then harvested. To obtain the cytoplasmic and nuclear fractions, cells were resuspended in PBS containing digitonin (40 μg/ml; Calbiochem, San Diego, CA) and protease inhibitors (Complete; Roche Diagnostics, Mannheim, Germany). After incubation on ice for 5 min, extracts were centrifugated at 2000 × g for 10 min at 4°C. The supernatants were saved as the cytoplasmic fractions. The pellets were resuspended in PBS containing 0.2% NP-40 and protease inhibitors (Complete; Roche Diagnostics) and then disrupted by sonication to yield the nuclear fraction.
Protein-RNA UV Cross-Linking and mRNA Affinity Chromatography
The in vivo UV cross-linking procedure was essentially as described in Pinol-Roma et al. (1989). Irradiation was performed using a UV cross-linker (Stratagene). Total cell lysate or subcellular fractions were subsequently prepared as described above. The fractions were heated at 65°C for 10 min in the presence of 0.5% SDS and 1% β-mercaptoethanol. After chilling on ice, LiCl was added to the fractions at a final concentration of 0.5 M. Poly(U)-Sepharose beads (GE Healthcare) were previously swelled and equilibrated in buffer A (10 mM Tris-HCl, pH 7.5, 0.5% SDS, 1 mM EDTA, and 0.5 M LiCl). The fractions were incubated with beads at room temperature for 30 min. After extensive washing with buffer A, bound RNP complexes were eluted with buffer A lacking LiCl. Subsequently, RNP complexes were precipitated with 0.2 M of LiCl and 3 volumes of ethanol at −20°C overnight. After digestion with 50 μg/ml RNase A at 37°C for 1 h, RNA-binding proteins were precipitated with 10% trichloroacetic acid (TCA) and analyzed by immunoblotting.
Sucrose Gradient Centrifugation and Polysome Profile Analysis
HEK293 cells were harvested in PBS containing 100 μg/ml cycloheximide at 4°C, and then resuspended in RSB-150 (10 mM Tris-HCl, pH 7.4, 3 mM MgCl2, and 150 mM NaCl) containing 100 μg/ml cycloheximide, 40 μg/ml digitonin (Calbiochem), 20 U/ml RNasin (Promega), and protease inhibitors (Complete; Roche Diagnsotics). After incubation on ice for 5 min, the cells were disrupted by passage five times through a 26-gauge needle. The lysates were centrifuged at 3000 × g for 2 min at 4°C. The cytoplasmic fractions were further clarified by 15-min centrifugation at 11,000 × g and then loaded onto a linear gradient of 15–40% (wt/wt) sucrose in RSB-150 and centrifuged at 38,000 rpm for 3 h at 4°C in a Beckman SW41 rotor. After centrifugation, RNAs and proteins were recovered from the gradient fractions for analysis. For polysome profile analysis, the gradients were monitored at 254 nm using a fractionation system (ISCO, Lincoln, NE).
Pulse-labeling Assay
HeLa cells were transfected with shRNA expression vectors for 48 h and then were seeded in a six-well plate (3 × 105 cells/well) for an overnight incubation at 37°C. For pulse labeling, cells were washed and preincubated with methionine- and cysteine-free DMEM (Invitrogen) for 30 min, and then labeled with 100 μCi/ml [35S]methionine/cysteine (35S-ProMix; GE Healthcare) for 30 min. Subsequently, cells were lysed in passive lysis buffer (Promega), and total proteins were resolved on 10% SDS-polyacrylamide gel electrophoresis and subjected to autoradiography. Moreover, radiolabeled proteins were precipitated by 10% TCA and quantified by scintillation counting.
In Vivo Translation Assay
HeLa cells were cotransfected with a pFL-SV40 derived reporter (0.2 μg), the control vector pRL-SV40 (0.2 μg; Promega), and an shRNA expression vector (1 μg). For Figure 7C, 0.6 μg of shRNA-resistant DDX3 expression vector was used in transfection. At 48 h after transfection, cells were lysed in passive lysis buffer (Promega). The activities of firefly luciferase and Renilla luciferase were measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
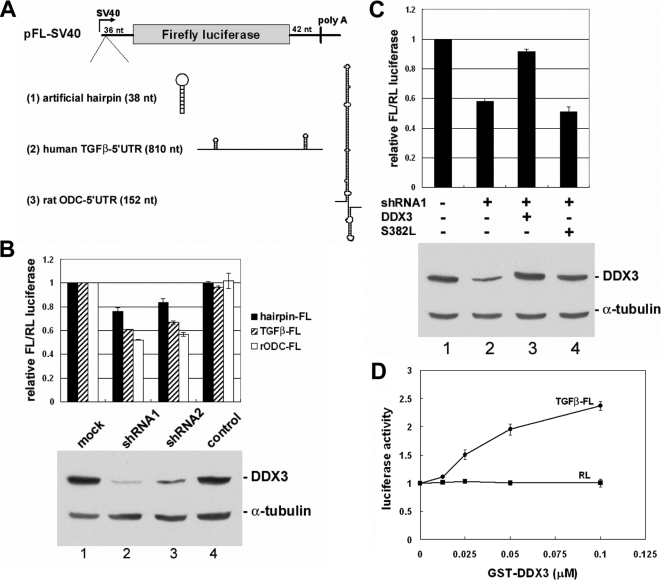
Figure 7.
DDX3 is required for efficient translation initiation of mRNAs harboring a long or structured 5′-UTR. (A) Schematics illustrate the luciferase reporters and the possible secondary structures for the 5′-UTR in the engineered pFL-SV40 reporters. (B) Transient transfection was performed essentially similar to Figure 6A, except that an FL reporter (hairpin, TGFβ, or ODC) and pRL-SV40 were cotransfected. The averaged FL activity was normalized to that of the RL activity in individual transfectants. The bar graph shows the average FL/RL value of each transfection relative to that of the mock transfectant. The data were obtained from three independent experiments. A representative immunoblot is shown at the bottom. (C) The ODC-containing FL reporter was used. Lane 1 is similar to lane 1 of B. Lanes 2–4, transfection was performed similarly to lane 2 of B, except that each transfection contained one additional vector (lane 2, empty pcDNA3.1 vector; lane 3, vector encoding shRNA-resistant wild-type DDX3; lane 4, vector encoding a DDX3 helicase mutant, S382L). The bar graph and immunoblot are presented similarly to B. (D) The in vitro translation was performed in the reticulocyte lysate using in vitro transcribed RL control mRNA from pRL-SV40 (Figure 6A) and TGFβ1 5′-UTR–containing FL mRNA as reporter in the presence of recombinant GST-DDX3. Luciferase activity of each reaction was normalized to the control reaction without DDX3.
In Vitro Translation Assay
The in vitro translation system was used to examine the effect of recombinant DDX3 on translation. Each 10 μl-translation reaction contained 5 μl of rabbit reticulocyte lysate (Promega), 50 ng of in vitro transcribed luciferase reporter mRNA with the TGFβ 5′-UTR, and recombinant GST-DDX3. The reaction was performed according to the manufacturer's recommendation. Luciferase activity was measured as described above.
RESULTS
Identification of TAP-interacting Proteins
TAP and its associated complexes may act to coordinate different steps of gene expression. To gain a more comprehensive understanding of the function(s) of TAP and its associated factors, we used immunoprecipitation to identify TAP-interacting proteins. The vector encoding FLAG-tagged TAP was transiently transfected into HEK293 cells, and the cell lysates were then prepared and subjected to immunoprecipitation with anti-FLAG. Mass spectrometric analysis identified a number of nucleoporins, including Nup358/RanBP2, Nup214, Nup153, Nup133, Nup88, p62, and RanGAP1 (Table 1). This observation was consistent with the subcellular localization of TAP in HeLa cells (Figure 1A), indicating that TAP stably associates with the NPC in the nuclear envelope.
Table 1.
Identification of TAP-interacting proteins by mass spectrometry
| TAP | TAPΔC |
|---|---|
| NPC proteins | RNA-binding proteins |
| Nup358/RanBP2 | hnRNP U |
| Nup214/CAN | ASF/SF2 |
| Nup153 | TRAP150 |
| Nup133 | Caprin-1 |
| Nup88 | Nucleolin |
| Nup62/p62 | RNA helicases |
| RanGAP1 | RHA/DHX9 |
| DDX3 | |
| Ribosomal proteins | |
| rpS3 | |
| rpS6 | |
| rpL3 | |
| rpL4 | |
| rpL6 | |
| rpL7a | |
| rpL8 |
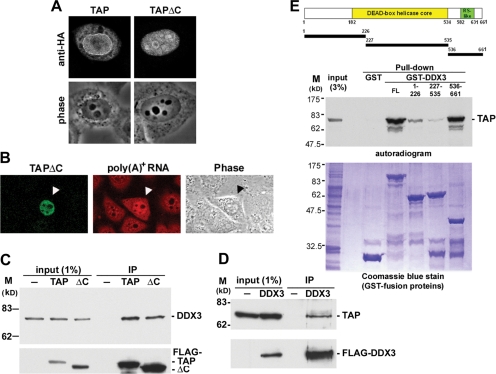
Figure 1.
DDX3 interacts with TAP in vivo and in vitro. (A) HeLa cells that transiently expressed HA-tagged TAP and TAPΔC proteins for 24 h were subjected to immunostaining with anti-HA. (B) In situ hybridization of poly(A)+ RNA was performed with HeLa cells that transiently expressed HA-tagged TAPΔC protein (arrowhead). (C) HEK293 cells were transiently transfected with empty vector (−) or the vector encoding FLAG-tagged TAP or TAPΔC (ΔC). Immunoprecipitation was performed at 24 h after transfection by using anti-FLAG. Immunoblotting was performed using polyclonal anti-DDX3 (top) and anti-FLAG (bottom). (D) Mock-transfected HEK293 cell lysate (−) or lysate containing transiently expressed FLAG-tagged DDX3 were subjected to immunoprecipitation as in C. Endogenous TAP was detected by anti-TAP. (E) Schematic diagram shows DDX3 fragments that were used for the pull-down assay. The assay using recombinant GST, GST-DDX3, or GST-DDX3 fragments as bait to pull-down 35S-labeled TAP protein (autoradiogram). Coomassie Blue staining of the GST-fusion proteins is shown at bottom.
To avoid the tight interaction of TAP with the NPC, we exploited a truncated TAP mutant (hereafter termed TAPΔC) that lacks the C-terminal 36 amino-acid residues and exhibits a dominant-negative effect on export of constitutive transport element-containing RNAs (Kang and Cullen, 1999). TAPΔC was essentially localized in the nucleus and accumulated in the nucleoli as well as splicing factor-containing speckles (Figure 1A). The subcellular localization pattern of TAPΔC was similar to that of a truncated TAP lacking the entire ubiquitin-associated domain (Bachi et al., 2000). Restricted localization of TAPΔC in the nucleus may reflect its inefficient nuclear export and may enable its stable binding to nuclear adaptor proteins. Accordingly, we observed by fluorescence in situ hybridization that poly(A)+ RNAs accumulated in the nucleus of TAPΔC-expressing HeLa cells (Figure 1B). Therefore, global mRNA export was probably impaired when TAPΔC was overexpressed. Nevertheless, immunoprecipitation of FLAG-tagged TAPΔC from HEK293 cell lysates showed a protein profile that differed from that observed with full-length TAP (data not shown). Mass spectrometric identification revealed ASF, hnRNP U, TRAP150, caprin-1, nucleolin, RHA, and DDX3 as potential TAPΔC-interacting proteins (Table 1). Interestingly, most of these proteins have been implicated in mRNA metabolism. In addition, identification of certain ribosomal proteins may reflect the role of TAP in ribosome subunit export and translation (Jin et al., 2003, Yao et al., 2007).
DDX3 Interacts with TAP
Among the identified TAPΔC-interacting proteins, we primarily focused on the DEAD-box RNA helicase DDX3. The association of TAP with DDX3 was examined by immunoprecipitation of transiently expressed FLAG-tagged TAP or TAPΔC followed by immunoblotting with anti-DDX3. Endogenous DDX3 coprecipitated with anti-FLAG even in the presence of RNase A, indicating that DDX3 directly interacted with full-length as well as truncated TAP (Figure 1C). A reciprocal experiment showing that endogenous TAP associated with FLAG-DDX3 further confirmed the TAP–DDX3 interaction (Figure 1D).
DDX3 contains a highly conserved helicase core domain in the central region and, especially, an RS dipeptide-rich segment in the C-terminal domain (Figure 1E). We next investigated, which DDX3 domains are involved in TAP interaction using an in vitro pull-down assay. GST was fused to full-length DDX3 as well as to each of the DDX3 fragments, including the N-terminal region (residues 1-226), central helicase core (227-535), and C-terminal region (536-661) (Figure 1E). The GST-fusion proteins were overproduced in E. coli and purified over glutathione resins. Each of these GST fusions was incubated with in vitro translated 35S-labeled TAP followed by affinity selection. TAP bound to full-length and the C-terminal region of DDX3. We concluded that the RS-like domain-containing C-terminal region of DDX3 is responsible for interaction with TAP.
DDX3 Associates with TAP and mRNPs
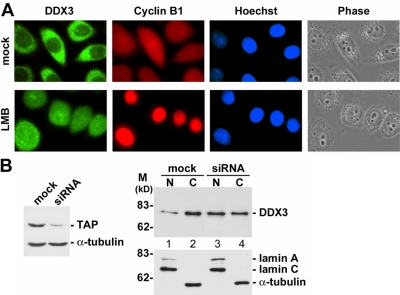
Using affinity-purified anti-DDX3 antibodies, we observed that endogenous DDX3 primarily distributed in the cytoplasm of HeLa cells (Figure 2A). TAP is located in both the nucleus and the cytoplasm and is particularly enriched in the nuclear envelope (Figure 1A; Katahira et al., 1999; Bachi et al., 2000). Nevertheless, both DDX3 and TAP shuttle between the nucleus and the cytoplasm (Kang and Cullen, 1999; Katahira et al., 1999; Sekiguchi et al., 2004; Yedavalli et al., 2004); therefore, we attempted to examine the cellular compartments where TAP interacts with DDX3. HEK293 cells were transiently transfected with the vector encoding FLAG-tagged TAP. Transfected cells were sequentially extracted with digitonin and the nonionic detergent NP-40 to yield the cytoplasmic and the nuclear fractions, respectively. Immunoblotting of subcellular fractions revealed that nucleolin and lamin A/C were largely in the nucleus and α-tubulin was in the cytoplasm (Figure 2B). Nevertheless, DDX3 was more abundant in the cytoplasm than in the nucleus (Figure 2B, lanes 1–4), consistent with the observation of indirect immunofluorescence (Figure 2A). Immunoprecipitation with anti-FLAG showed that DDX3 bound to TAP primarily in the nucleus (Figure 2B, lane 5), which can explain the interaction of DDX3 with nuclear localized TAPΔC (Table 1). However, the DDX3-TAP interaction might also occur in the cytoplasm (Figure 2B, lane 6), in line with their colocalization in cytoplasmic SGs (see below; Figure 4).
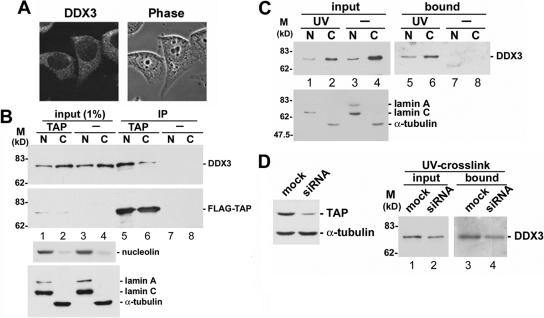
Figure 2.
DDX3 associates with TAP and mRNPs in both the nucleus and the cytoplasm. (A) Immunostaining of HeLa cells was performed using affinity-purified anti-DDX3. (B) HEK293 cells were transiently transfected with the vector expressing FLAG-tagged TAP or with empty vector (−). Immunoprecipitation of the nuclear (N) and cytoplasmic (C) fractions of the cell lysate was performed using monoclonal anti-FLAG, followed by immunoblotting to detect endogenous DDX3 and FLAG-TAP (top). The subcellular fractions were also subjected to immunoblotting using antibodies against nucleolin, lamin A/C, and α-tubulin (bottom). (C) Cell fractionation was performed in UV-irradiated (UV) or mock-treated (−) HEK293 cells as in B. The lysate fractions were then subjected to chromatography over poly(U)-Sepharose. Proteins cross-linked to poly(A)+ RNAs were detected by anti-DDX3 (top). The efficiency of cell fractionation was also determined as in B (bottom). (D) The TAP-targeting siRNA was transiently transfected into HEK293 cells. Immunoblotting of the cell lysate was performed using anti-TAP (left). UV-cross-linking and poly(U)-Sepharose-affinity selection were performed as described in C, except that total cell lysate was used.
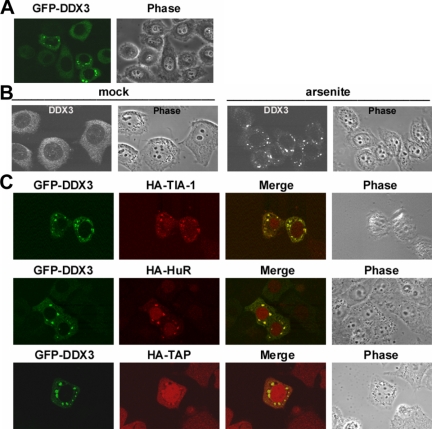
Figure 4.
DDX3 is a component of stress granules. (A) HeLa cells were transiently transfected with a vector encoding GFP-DDX3 for 24 h. Subcellular localization of GFP-DDX3 was detected by fluorescence microscopy. (B) HeLa cells were treated with 0.5 mM sodium arsenite (arsenite) or mock-treated (mock) for 1 h. Cells were subsequently subjected to immunostaining using anti-DDX3. (C) HA-tagged TIA-1, HuR, and TAP were each transiently coexpressed with GFP-DDX3 in HeLa cells. Immunostaining was carried out using monoclonal anti- HA to detect HA-tagged proteins. B and C were obtained using a laser confocal microscope.
TAP is recruited to mRNPs in the nucleus and participates in their export (Katahira et al., 1999; Stutz and Izaurralde, 2003), and thus we next examined whether DDX3 binds directly to mRNA in cells. After UV cross-linking in vivo, mRNPs were selected followed by ribonuclease treatment to release cross-linked proteins. On exposure to UV light, DDX3 copurified with poly(A)+ RNAs (Figure 2C, lanes 5 and 6), indicating that DDX3 indeed bound to mRNAs in both the nucleus and the cytoplasm and thus behaved similarly to its Chironomus tentans homologue hrp84 (Nashchekin et al., 2006). Next, we examined whether DDX3 interaction with mRNAs depends on TAP. When TAP expression was substantially diminished by siRNA, DDX3 was still cross-linked to mRNAs (Figure 2D). These results suggested that DDX3 might associate with mRNAs as well as TAP in the nucleus and subsequently accompany mRNPs to the cytoplasm.
Nuclear Export of DDX3 May in Part Undergo the TAP-mediated Pathway
Previous reports have indicated that the importin-β protein CRM1 participates in nuclear export of DDX3 (Yedavalli et al., 2004). When HeLa cells were treated with the CRM1 inhibitor leptomycin B, nuclear retention of DDX3 was indeed detected, but its cytoplasmic signal was not completely diminished (Figure 3A). In contrast, cyclin B1, another CRM1 export cargo, was exclusively retained in the nucleus (Figure 3A). Our observation was consistent with previous reports (Sekiguchi et al., 2004; Yedavalli et al., 2004), suggesting that another pathway exists for nuclear export of DDX3. We therefore investigated whether nuclear export of DDX3 involves TAP. As shown in Figure 3B, when TAP expression was reduced by siRNA, DDX3 protein level was increased in the nuclear fraction and, accordingly, reduced in the cytoplasm (Figure 3B, lanes 3 and 4). Therefore, TAP may be, at least partially, involved in nuclear export of DDX3.
Figure 3.
DDX3 may be in part exported to the cytoplasm via the TAP-mediated pathway. (A) HeLa cells were treated with 40 nM leptomycin B (LMB) or mock-treated (mock) for 3 h and subsequently subjected to immunostaining by using anti-DDX3 and anti-cyclin B1. DNA staining with Hoechst 33258 indicates the cell nucleus. (B) Knockdown of TAP expression and cell fractionation were performed in HEK293 cells as described in Figure 2, D and B, respectively. Immunoblotting was carried out using antibodies against DDX3, lamin A/C, and α-tubulin.
Moreover, the association of DDX3 with TAP and mRNPs led a possibility that DDX3 may participate in nuclear export of mRNAs. We therefore used fluorescence in situ hybridization to detect cellular distribution of poly(A)+ RNAs in DDX3-depleted HeLa cells. However, no significant change was observed with poly(A)+ RNA distribution when DDX3 protein level was reduced to ∼30% by transiently expressed shRNA (Supplemental Figure S1). In addition, we microinjected recombinant DDX3 together with radiolabeled RNAs into Xenopus oocyte nuclei. Regardless of whether injected RNA contained introns, its nuclear export efficiency was not significantly changed (data not shown). Perhaps DDX3 is not critical for general mRNA export, but the possibility that DDX3 is involved in export of specific mRNAs cannot be excluded. Together, DDX3 might associate with mRNPs to export to the cytoplasm via the TAP-mediated pathway, but it does not play a critical role for mRNA export.
DDX3 Is Targeted to Cytoplasmic Stress Granules
We next took advantage of GFP fusion to monitor subcellular localization of DDX3. We found that overexpressed GFP-DDX3 distributed diffusely in the cytoplasm and, in addition, it could be concentrated in discrete cytoplasmic foci in ∼10% of the transfected HeLa cells (Figure 4A). These foci were reminiscent of stress granules (SGs) that could be induced upon cell exposure to environmental stress (Anderson and Kedersha, 2006). Indeed, when cells were treated with sodium arsenite (Figure 4B) or exposed to heat shock (data not shown), endogenous DDX3 formed cytoplasmic foci almost in all cell samples. We therefore examined whether DDX3 is colocalized with SG proteins. Indirect immunofluorescence showed that GFP-DDX3 was well colocalized with coexpressed HA-tagged TIA-1 and HuR (Figure 4C), two SG markers in cytoplasmic foci, suggesting that DDX3 is a component of SGs, whereas overexpressed or under cell stress conditions. Moreover, we observed that HA-tagged TAP relocated to SGs when GFP-DDX3 was coexpressed (Figure 4C). This result further suggested the TAP-DDX3 interaction in cells. Although DDX3 may not be essential for nuclear mRNA export in general, the observation of colocalization of DDX3 and TAP in SGs provided a hint to their potential function in the cytoplasm.
DDX3 Associates with Translation Initiation Complexes
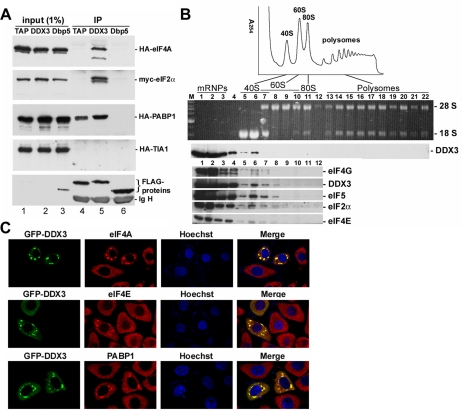
SGs contain several translation initiation factors and 40S ribosomal subunits, representing stalled translation preinitiation complexes, as well as a number of RNA binding proteins (Anderson and Kedersha, 2006). Therefore, we examined whether DDX3 could interact with those SG factors. We transiently coexpressed FLAG-tagged DDX3 with each of the HA- or myc-tagged eIF4A, eIF2α, and PABP1 and TIA-1 in HEK293 cells. Immunoprecipitation with anti-FLAG showed that DDX3 interacted with eIF4A, eIF2α, and PABP1 but not with TIA-1 (Figure 5A, lane 5). FLAG-DDX3 also coprecipitated endogenous PABP1 (Supplemental Figure S2). These coprecipitations were resistant to RNase A, suggesting direct interactions with DDX3. We also examined TAP and Dbp5, an RNA helicase required for mRNA export (Tseng et al., 1998). TAP interacted only with PABP1, whereas no interaction was detected between Dbp5 and any of the factors examined (Figure 5A, lanes 4 and 6). Furthermore, sucrose gradient sedimentation analysis of HEK293 cytoplasmic extracts showed that DDX3 distributed together with several translation initiation factors in fractions enriched with the 40S ribosome subunit (Figure 5B). However, DDX3 was not detectable in polysomes (Figure 5B). Therefore, DDX3 might only be associated with translation initiation complexes and not participate in the elongation or subsequent steps.
Figure 5.
Association of DDX3 with translation initiation complexes. (A) FLAG-tagged TAP, DDX3 or Dbp5 was transiently coexpressed with HA- or myc-tagged eIF4A, eIF2α, PABP1, and TIA-1 in HEK293 cells. Immunoprecipitation was performed using anti-FLAG, followed by immunoblotting using antibodies against the HA, myc, or FLAG epitope. Ig H represents the immunoglobulin heavy chain. The input of FLAG-tagged TAP and DDX3 was visible upon longer exposure (lanes 1 and 2). (B) HEK293 cytoplasmic extract was centrifuged through a continuous 15–40% sucrose gradient. The polysome profile was plotted by A254 values (top). RNAs and proteins were recovered from 22 fractions for analysis (middle). RNAs were resolved on a 1% formaldehyde/agarose gel and rRNAs were visualized by ethidium bromide staining. Immunoblotting was performed using anti-DDX3. Immunoblotting of the first 12 fractions is shown at the bottom using antibodies against eIF4G, DDX3, eIF5, eIF2α, and eIF4E. (C) GFP-DDX3 was transiently expressed in HeLa cells. Immunostaining was carried out using antibodies against eIF4A, eIF4E, or PABP1.
We also examined whether DDX3 colocalizes with some of the translation initiation factors in SGs. Immunofluorescence showed that endogenous eIF4A, eIF4E, and PABP1 indeed translocated to GFP-DDX3-induced cytoplasmic granules in HeLa cells (Figure 5C). However, knockdown of DDX3 did not impair arsenite-induced SG localization of PABP1 (Supplemental Figure S3). Therefore, DDX3 is likely not a nucleator for SG assembly during cell stress. Nevertheless, the observation that overexpression of DDX3 induced SGs formation may be in line with the role of DDX3 in general translation repression (Shih et al., 2008).
Depletion of DDX3 Does Not Significantly Affect General Translation
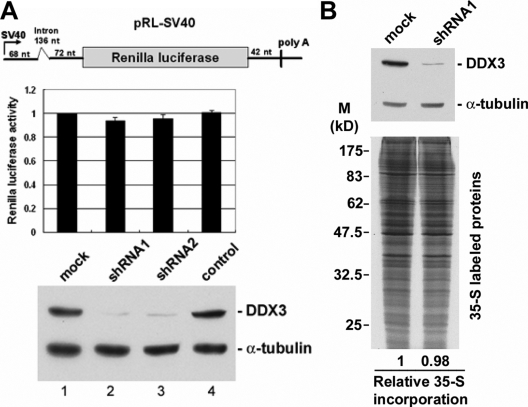
To further understand the role of DDX3 in translation, we depleted DDX3 from HeLa cells using the shRNA strategy. DDX3 protein level was significantly reduced by >90% in HeLa cells when either of the DDX3-targeting shRNAs was transiently expressed (Figure 6A, immunoblotting). However, under these conditions, the expression level of a Renilla luciferase (RL) reporter had no significant change (Figure 6A, bar graph).
Figure 6.
DDX3 is dispensable for general translation. (A) HeLa cells were cotransfected with the Renilla luciferase vector (the scheme) and the empty pSilencer vector (mock) or the vector expressing DDX3 shRNA1 or shRNA2 or TNPO3 shRNA (a control shRNA, lane 4) for 48 h. The bar graph shows the average Renilla luciferase activity in each transfection (lanes 2–4) relative to that of the mock transfectant (lane 1). Average values with SD were obtained from three independent experiments. Immunoblotting was performed using anti-DDX3 and anti-α-tubulin. (B) HeLa cells were transfected with pSilencer (mock) or the vector expressing DDX3-targeting shRNA1. DDX3 and α-tubulin were detected by corresponding antibodies (top). The transfectants were subjected to the pulse-labeling assay. Autoradiography shows radiolabeled proteins of the cell lysates (bottom).
Next, a pulse-labeling experiment was performed to examine whether DDX3 knockdown would alter the pattern of newly synthesized proteins. However, no significant change could be detected in the global translation pattern and efficiency at a diminished level of DDX3 (Figure 6B). We also observed that depletion of DDX3 did not change the polysome profile of cytoplasmic extracts (data not shown). Therefore, mammalian DDX3 seemed to be dispensable for general translation. However, our analysis may not have detected changes in the expression of low-abundance proteins, and thus the possibility that DDX3 is involved in specific mRNA translation still remained.
DDX3 Promotes Translation of mRNAs Containing a Long or Structured 5′-UTR
Yeast Ded1 and its close homologue Dbp1 have been shown to affect ribosome scanning in translation initiation (Berthelot et al., 2004). Therefore, we attempted to investigate whether DDX3 could specifically facilitate translation of mRNAs containing a long or highly structured 5′-UTR by using a reporter assay. The reporters of the firefly luciferase (FL) contained an artificial hairpin in the 5′-UTR or harbored the 5′-UTR derived from human TGFβ1 and rat ODC (Figure 7A). The hairpin contained a palindrome sequence that presumably formed a stem-loop structure with a free energy value of −29 Kcal/mol. The 5′-UTR of human TGFβ1 mRNA is unusually long (∼810 nucleotides) and contains two stem-loops (Kim et al., 1992); this 5′-UTR has been shown to inhibit translation (Kim et al., 1992). The rat ODC mRNA has an ∼300-nucleotide-long and GC-rich 5′-UTR, of which the 5′ half probably forms a stable secondary structure that interferes with ribosome scanning (Van Steeg et al., 1991). Therefore, this 5′ half region (∼150 nucleotides) was inserted into the FL reporter. Meanwhile, the RL reporter, which encodes the RL mRNA without any specific structure in the 5′-UTR, was cotransfected with the FL reporters as a control.
As predicted, when structured 5′-UTR was included, FL reporter expression was somewhat impaired (data not shown). Although normalized to the RL control, we observed that DDX3 depletion reduced the expression of the structured 5′-UTR reporters by 20–50% (Figure 7B, lanes 2 and 3) without changing the level of the reporter mRNAs in the cytoplasm (Supplemental Figure S4). Therefore, DDX3 might function in the translation process rather than RNA export or stability control. Because a more significant reduction in expression efficiency was observed with the TGFβ1 and ODC 5′-UTRs, a longer and highly structured 5′-UTR may increase the dependency of mRNA translation on DDX3. A similar result was obtained from reciprocal experiments in which RL mRNAs harbored a structured 5′-UTR, whereas the FL mRNA with a nonmodified 5′-UTR served as a control (data not shown). Introduction of shRNA-resistant DDX3 largely relieved the suppression of ODC 5′-UTR–containing FL expression in DDX3-depleted HeLa cells (Figure 7C, lane 3), indicating the specificity of DDX3 in this translation control.
We also examined whether DDX3 could facilitate translation of the TGFβ1 5′-UTR-containing FL mRNA by using the in vitro translation assay. Addition of recombinant GST-DDX3 in the reticulocyte lysate showed that FL reporter translation was activated in a dose-dependent manner (Figure 7D). This result further suggested that DDX3 is required for translation of mRNAs containing a long or structured 5′-UTR.
The RNA Helicase Motif of DDX3 Is Required for Its Activity in Promoting Translation of mRNAs Containing a Structured 5′-UTR
From the above-mentioned result, we presumed that DDX3 exerts an RNA helicase activity to resolve RNA structures, thereby facilitating ribosome scanning. To test this hypothesis, we examined whether a DDX3 mutant (S382L) that has been shown to lack helicase activity (Yedavalli et al., 2004) is able to activate the translation of structured 5′-UTR-containing mRNAs. In contrast to wild-type DDX3, the shRNA-resistant S382L mutant failed to rescue the effects of endogenous DDX3 depletion on translation of the FL reporter containing the ODC 5′-UTR (Figure 7C, lane 4). Therefore, in line with our hypothesis, the RNA helicase activity of DDX3 may be required for efficient expression of the mRNAs containing secondary structures in the 5′-UTR.
DISCUSSION
Nuclear Export of DDX3
Previous reports have indicated that DDX3 is a component of the spliceosome and prefers to bind mRNAs that have undergone splicing (Stevens et al., 2002; Zhou et al., 2002; Merz et al., 2007). Here, we demonstrate that DDX3 directly interacts with TAP and associates with mRNPs (Figures 1 and 2). Therefore, DDX3 may be recruited to spliced mRNAs to become a component of TAP-containing mRNPs and subsequently accompany mRNPs to be exported to the cytoplasm. The interaction between the EJC component REF/Aly and DDX3 also supports the idea that DDX3 is a cofactor of export-competent mRNPs (Masuda et al., 2005). However, unlike yeast Dbp5 that actively promotes mRNA export (Tseng et al., 1998), DDX3 may not be critical for general mRNA export (Supplemental Figure S1), but whether DDX3 is involved in export of specific cellular mRNAs still remains to be studied.
DDX3 promotes the nuclear export of incompletely spliced human immunodeficiency virus RNAs through a CRM1-dependent pathway (Yedavalli et al., 2004). CRM1 is not involved in bulk mRNA export, but it has been implicated in export of certain AU-rich element-containing mRNAs such as c-fos and cyclooxygenase-2 (Brennan et al., 2000; Jang et al., 2003). Therefore, DDX3 may participate in export of these mRNAs and/or act merely as an export cargo of CRM1. Indeed, nuclear export of the Xenopus DDX3, An3, can be mediated by CRM1 (Askjaer et al., 1999). Consistent with this, treatment of cultured mammalian cells with leptomycin B results in nuclear accumulation of DDX3 (Sekiguchi et al., 2004; Yedavalli et al., 2004). However, residual DDX3 in the cytoplasm after leptomycin B treatment (Figure 3; Sekiguchi et al., 2004; Yedavalli et al., 2004) implied alternative pathways exist for nuclear export of DDX3. Our observation that nuclear export of DDX3 was partially impaired by depletion of TAP (Figure 3) indicated that DDX3 may export in part through the TAP-mediated pathway. Similarly, yeast Dbp5 acts as an essential factor in Mex67/TAP-mediated mRNA export but can be exported by Xpo1/CRM1 as well (Hodge et al., 1999). Perhaps export cofactors may use distinct export pathways for specific mRNAs or for specific functions.
DDX3 Is a Component of Stress Granules
DDX3 has been identified as a component of RNA-transporting granules in murine neurons (Kanai et al., 2004). Therefore, DDX3 may participate in cytoplasmic mRNA localization and translational control. In this study, we demonstrated that endogenous DDX3 is recruited to SGs under cell stress conditions and that overexpression of DDX3 induces relocalization of its interacting partners TAP and several translation factors to SGs (Figures 4 and 5). Therefore, DDX3 may translocate together with mRNPs and stalled translation complexes to SG particularly when cells encounter stress. In contrast, Dbp5 neither interacted with the translation factors we examined (Figure 5) nor localized to SGs (data not shown). Therefore, different export mRNP-associated RNA helicases likely have distinct functions in mRNA metabolism. Moreover, previous work has shown that DDX3 interacts and colocalizes with the hepatitis C virus core protein in the endoplasmic reticulum (Mamiya and Worman, 1999). Notably, unfolded protein accumulation in the endoplasmic reticulum can induce eIF2α phosphorylation and thereby attenuates protein synthesis (Holcik and Sonenberg, 2005). The detection of the interaction between DDX3 and eIF2α (Figure 5) provides a possibility that they may colocalize in the endoplasmic reticulum in the presence of the hepatitis C virus core protein. Perhaps DDX3 localizes to various cytoplasmic compartments that contain translation initiation factors for translational control.
DDX3 Functions in Translational Control
Yeast Ded1 is essential for general translation and likely functions in the initiation step (Chuang et al., 1997). Interestingly, human DDX3 and its murine homologue PL10 can rescue the lethality of the ded1 null yeast (Mamiya and Worman, 1999), suggesting functional conservation between the DDX3 homologues. However, in vivo knockdown of DDX3 did not substantially affect ribosome biogenesis as revealed by sedimentation (data not shown) or general translation as judged by reporter assays as well as by new protein synthesis (Figure 6). Consistent with our result, a previous study has shown that total protein synthesis is not impeded in a BHK21-derived cell line that carries a temperature-sensitive mutant of DDX3 (Fukumura et al., 2003). Perhaps mammalian DDX3 is not critical for general translation, but this point still needs further investigation.
In contrast, previous work also has shown that the replication of brome mosaic virus is inhibited in a ded1 mutant of yeast due to its defective expression of the polymerase-like 2a protein encoded by RNA2 (Noueiry et al., 2000). The 5′-noncoding region of brome mosaic virus RNA2 accounted for this translational repression, suggesting that Ded1 functions in the noncoding region. In the fission yeast Schizosaccharomyces pombe, the translation of B-type cyclins, Cig2 and Cdc13, is particularly sensitive to the inactivation of Ded1 (Grallert et al., 2000). Notably, the Cig2 mRNA harbors an unusually long 5′-UTR (∼1 kb) and the Cdc13 mRNA is predicted to contain a complex structure in the 5′-UTR (Daga and Jimenez, 1999). Thus, these observations suggest a role of Ded1 in selective translation of specific mRNAs. We here provide evidence that human DDX3 may be required for efficient translation of mRNAs that contain a long or structured 5′-UTR (Figure 7). Therefore, mammalian DDX3 and its yeast homologues likely have a conserved function in translational regulation by remodeling the 5′-UTR of mRNAs during translation initiation.
Eukaryotic translation initiation factor eIF4A is an RNA helicase that facilitates ribosome scanning by unwinding mRNA secondary structures in the 5′-UTR. Recent evidence suggests that Ded1 and its highly related protein Dbp1 are more effective than eIF4A at unwinding an RNA stem-loop and thereby can facilitate translation of mRNAs containing a long 5′-UTR (Berthelot et al., 2004; Marsden et al., 2006). Analogous to DDX3, the DExH-box RNA helicase RHA can also promote translation of selected mRNAs by binding to a structured post-transcriptional control element in the 5′-UTR (Hartman et al., 2006). Therefore, translation may involve multiple RNA helicases that exert their specific activities. For example, eIF4A may unwind 5′-proximal secondary structures in general mRNAs for 40S ribosome subunit recruitment, whereas scanning of a long 5′-UTR containing multiple structured regions requires additional RNA helicases such as DDX3 and RHA. However, the possibility that these RNA helicases have redundant activities still remains.
A recent report has shown that overexpression of DDX3 inhibits cap-dependent translation by its direct interaction with eIF4E, which disrupts the integrity of the cap-binding complex (Shih et al., 2008). Consistently, we showed that overexpressed DDX3 recruited translation initiation factors to SGs where translation is attenuated. An overdose of recombinant DDX3 indeed inhibited translation of capped mRNAs in vitro (data not shown). Nevertheless, DDX3 could facilitate translation of mRNA comprising a long or structured 5′-UTR (this study), and also activates internal ribosome entry site-mediated translation (Shih et al., 2008). Moreover, DDX3 interacts with initiation factors eIF4A (this study) and eIF4E (Shih et al., 2008), consistent with the observation of yeast Ded1, which genetically interacts with Tif1 (eIF4A) and Cdc33 (eIF4E) (De La Cruz et al., 1997). Therefore, DDX3 may have phylogenetically conserved function in various types of translation control through interactions with specific translation initiation factors.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Chang, S.-C. Cheng, N. Kedersha, and J. A. Steitz for plasmids or antibodies. We thank Kuo-Ming Lee for technical assistance and Dr. Tim C. Taylor for editing the manuscript. This work was supported by Grant NSC 95-2311-B001-019 from the National Science Council of Taiwan.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1264) on July 2, 2008.
REFERENCES
- Anderson P., Kedersha N. RNA granules. J. Cell. Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askjaer P., et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A., et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J.E.G. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 2004;51:987–1001. doi: 10.1046/j.1365-2958.2003.03898.x. [DOI] [PubMed] [Google Scholar]
- Brennan C. M., Gallouzi I. E., Steitz J. A. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell. Biol. 2000;151:1–13. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T., Nagel R., Mandel-Gutfreund Y., Shiue L., Clark T. A., Chong J. L., Chang T. H., Squazzo S., Hartzog G., Ares M., Jr. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Chao C. H., Chen C. M., Cheng P. L., Shih J. W., Tsou A. P., Wu Lee Y. H. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer. Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Scarcelli J. J. Unravelling mRNA export. Nat. Cell. Biol. 2006;8:645–647. doi: 10.1038/ncb0706-645. [DOI] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N. K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Daga R. R., Jimenez J. Translational control of the Cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J. Cell Sci. 1999;112:3137–3146. doi: 10.1242/jcs.112.18.3137. [DOI] [PubMed] [Google Scholar]
- De La Cruz J., Iost I., Kressler D., Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V. N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Erkmann J. A., Kutay U. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp. Cell. Res. 2004;296:12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Fukumura J., Noguchi E., Sekiguchi T., Nishimoto T. A temperature-sensitive mutant of the mammalian RNA helicase, DEAD-box X isoform, DBX, defective in the transition from G1 to S phase. J. Biochem. 2003;134:71–82. doi: 10.1093/jb/mvg126. [DOI] [PubMed] [Google Scholar]
- Grallert B., Kearsey S. E., Lenhard M., Carlson C. R., Nurse P., Boye E., Labib K. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J. Cell. Sci. 2000;113:1447–1458. doi: 10.1242/jcs.113.8.1447. [DOI] [PubMed] [Google Scholar]
- Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J. J., Seedorf M., Cole C. N., Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- Gruter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B. K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Hammell C. M., Gross S., Zenklusen D., Heath C. V., Stutz F., Moore C., Cole C. N. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T. R., Qian S., Bolinger C., Fernandez S., Schoenberg D. R., Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Kulozik A. E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Hodge C. A., Colot H. V., Stafford P., Cole C. N. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell. Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Huang Y., Steitz J. A. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huang Y., Gattoni R., Stevenin J., Steitz J. A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., Rahe B., Pringle J., Beggs J. D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Jang B. C., Munoz-Najar U., Paik J. H., Claffey K., Yoshida M., Hla T. Leptomycin B, an inhibitor of the nuclear export receptor CRM1, inhibits COX-2 expression. J. Biol. Chem. 2003;278:2773–2776. doi: 10.1074/jbc.C200620200. [DOI] [PubMed] [Google Scholar]
- Jin L., Guzik B. W., Bor Y. C., Rekosh D., Hammarskjold M. L. Tap and NXT promote translation of unspliced mRNA. Genes. Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kang Y., Cullen B. R. The human TAP protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes. Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser K., Podtelejnikov A., Mann, Jung M., J. U., Hurt E. The Mex67p-mediated nuclear amRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Park K., Koeller D., Kim K. Y., Wakefield L. M., Sporn M. B., Roberts A. B. Post-transcriptional regulation of the human transforming growth factor-β1 gene. J. Biol. Chem. 1992;267:13702–13707. [PubMed] [Google Scholar]
- Lai M. C., Lin R. I., Huang S. Y., Tsai C. W., Tarn W. Y. A human importin-β family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Li C., Lin R. I., Lai M. C., Ouyang P., Tarn W. Y. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol. Cell. Biol. 2003;23:7363–7376. doi: 10.1128/MCB.23.20.7363-7376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bor Y. C., Misawa Y., Xue Y., Rekosh D., Hammarskjold M. L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- Luo M. J., Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. J., Zhou Z., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M. D., Steitz J. A. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Mamiya N., Worman H. J. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Marsden S., Nardelli M., Linder P., McCarthy J.E.G. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J. Mol. Biol. 2006;361:327–335. doi: 10.1016/j.jmb.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes. Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Badolato J., Kobayashi R., Zhang M. Q., Gardiner E. M., Krainer A. R. Purification and characterization of human RNPS 1, a general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz C., Urlaub H., Will C. L., Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashchekin D., Zhao J., Visa N., Daneholt B. A novel Ded1-like RNA helicase interacts with the Y-box protein ctYB-1 in nuclear mRNP particles and in polysomes. J. Biol. Chem. 2006;281:14263–14272. doi: 10.1074/jbc.M600262200. [DOI] [PubMed] [Google Scholar]
- Noueiry A. O., Chen J., Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S., Adam S. A., Choi Y. D., Dreyfuss G. Ultraviolet-induced cross-linking of RNA to protein in vivo. Methods Enzymol. 1989;180:410–418. doi: 10.1016/0076-6879(89)80114-4. [DOI] [PubMed] [Google Scholar]
- Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell. Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Reed R., Cheng H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- Rodrigues J. P., Rode M., Gatfield D., Blencowe B. J., Carmo-Fonseca M., Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T., Iida H., Fukumura J., Nishimoto T. Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp. Cell. Res. 2004;300:213–222. doi: 10.1016/j.yexcr.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Shih J. W., Tsai T. Y., Chao C. H., Wu Lee Y. H. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- Stevens S. W., Ryan D. E., Ge H. Y., Moore R. E., Young M. K., Lee T. D., Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Strasser K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- Tseng S. S., Weaver P. L., Liu Y., Hitomi M., Tartakoff A. M., Chang T. H. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steeg H., Van Oostrom C.T.M., Hodemaekers H. M., Peters L., Thomas A.A.M. The translation in vitro of rat ornithine decarboxylase mRNA is blocked by its 5′ untranslated region in a polyamine-independent way. Biochem. J. 1991;274:521–526. doi: 10.1042/bj2740521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H. L., Coburn G. A., Zeng Y., Kang Y., Bogerd H. P., Cullen B. R. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., Roser D., Kohler A., Bradatsch B., Bassler J., Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider L. J., Gygi S. P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.