Abstract
The spindle assembly checkpoint monitors the state of spindle–kinetochore interaction to prevent premature onset of anaphase. Although checkpoint proteins, such as Mad2, are localized on kinetochores that do not interact properly with the spindle, it remains unknown how the checkpoint proteins recognize abnormalities in spindle–kinetochore interaction. Here, we report that Mad2 localization on kinetochores in fission yeast is regulated by two partially overlapping but distinct pathways: the Dam1/DASH and the Bub1 pathways. We show that Mad2 is localized on “unattached” as well as “tensionless” kinetochores. Our observations suggest that Bub1 is required for Mad2 to detect tensionless kinetochores, whereas Dam1/DASH is crucial for Mad2 to detect unattached kinetochores. In cells lacking both Bub1 and Dam1/DASH, Mad2 localization on kinetochores is diminished, and mitotic progression appears to be accelerated despite the frequent occurrence of abnormal chromosome segregation. Furthermore, we found that Dam1/DASH is required for promotion of spindle association with unattached kinetochores. In contrast, there is accumulating evidence that Bub1 is involved in resolution of erroneous spindle attachment on tensionless kinetochores. These pathways may act as molecular sensors determining the state of spindle association on each kinetochore, enabling proper regulation of the checkpoint activation as well as promotion/resolution of spindle attachment.
INTRODUCTION
The kinetochore, a multiprotein complex assembled on centromeric DNA, is essential for accurate chromosome segregation, as it mediates the interaction between chromosomes and spindle microtubules during mitosis (Cleveland et al., 2003). At the very beginning of prometaphase, chromosomes stand unattached to microtubules. Subsequently, a kinetochore is captured by microtubules emanating from a spindle pole. The bioriented interaction between the chromosome and the spindle is established when its sister kinetochore is captured by microtubules from the opposite pole (Rieder and Salmon, 1994; Zhou et al., 2002).
The kinetochore is not just an interface connecting the chromosome and the spindle, but also plays a central role in the spindle assembly checkpoint (SAC), a cell cycle control mechanism crucial for the accuracy of chromosome segregation (Musacchio and Hardwick, 2002; May and Hardwick, 2006). The SAC is evolutionarily conserved among eukaryotes from yeasts to mammals. For sister chromosomes to be divided equally into daughter cells, each kinetochore must interact with the spindle in a bioriented manner before the onset of anaphase. The SAC prevents metaphase–anaphase transition until all the chromosomes establish biorientation. As kinetochore–spindle attachment is a stochastic process, most chromosomes are transiently mono-oriented before the establishment of biorientation. Mono-oriented attachments include syntelic and monotelic attachment; in a syntelic kinetochore, both of the sisters are connected to the same spindle pole, whereas, in a monotelic kinetochore, only one of the sisters is captured by the spindle (Pinsky and Biggins, 2005; Salmon et al., 2005). During early stages of mitosis, many of the SAC components, such as Mad2 and Bub1, accumulate on kinetochores (Chen et al., 1996; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998; Meraldi et al., 2004). The accumulation of the components on kinetochores appears to be the initial step of SAC activation (Yu, 2002). Activated SAC prevents transition from metaphase to anaphase by inhibiting Anaphase-promoting complex/Cyclosome (APC/C). Although there is accumulating evidence regarding how the SAC components inhibit the APC/C (Hwang et al., 1998; Kim et al., 1998; Yu, 2002; Tang et al., 2004), the mechanism controlling the activation of SAC remains unclear. To date, two evolutionarily conserved kinetochore complexes, the Mis6 complex and the Nuf2-Ndc80 complex, have been reported to be required for the accumulation of Mad2 on the kinetochore (Martin-Lluesma et al., 2002; DeLuca et al., 2003; Hori et al., 2003; Liu et al., 2003; Saitoh et al., 2005). The fission yeast Aurora kinase, Ark1, has been also shown to be important for accumulation of Mad2 on unattached kinetochores (Petersen and Hagan, 2003). Gaining an understanding of the roles of molecules essential for the kinetochore accumulation of the SAC components appears crucial to determine the mechanism controlling SAC activation.
A model has been proposed in which two physical properties of the kinetochore trigger SAC activation: the absence of tension and the lack of microtubule attachment (Musacchio and Hardwick, 2002). This model predicts the existence of two SAC signaling pathways: one pathway that detects the absence of tension, which should be generated across bioriented sister kinetochores by spindle pulling forces toward opposing poles, and another that detects the absence of microtubule attachment. Due to the interdependence between tension and microtubule attachment, it is still unclear whether these pathways are separable (Pinsky and Biggins, 2005). It has been argued that the absence of microtubule attachment may be the primary signal for the SAC activation in the budding yeast Saccharomyces cerevisiae; tensionless kinetochores may be converted into unattached kinetochores by Aurora B kinase that promote detachment of a microtubule from the tensionless kinetochore (Pinsky et al., 2006). In contrast, it has been reported that Mad2 specifically recognizes unattached kinetochores, whereas Bub1 and BubR1 recognize tensionless kinetochores in mammalian cells (Waters et al., 1998; Skoufias et al., 2001), implicating the presence of two distinct signaling pathways in higher eukaryotes.
Errors in the spindle–kinetochore interaction causing SAC activation must be corrected while the metaphase–anaphase transition is blocked. As mentioned above, Aurora B kinase has been suggested to be involved in the resolution of tensionless spindle–kinetochore interaction in budding yeast (Tanaka et al., 2002). In vertebrates, kinesin-13 family proteins, which have microtubule-depolymerizing activity regulated by Aurora B, are thought to function in the resolution of deleterious interactions between the spindle and the kinetochore (Desai et al., 1999; Andrews et al., 2004; Lan et al., 2004). Previously, we showed that ectopic expression of Hos2 protein, an integral component of the Dam1/DASH complex, partially rescues various types of kinetochore-defective mis mutants in the fission yeast Schizosaccharomyces pombe (Kobayashi et al., 2007). This observation suggests that the Dam1/DASH complex may also be involved in the correction of abnormalities in mis mutant kinetochores.
The Dam1/DASH complex was originally identified in budding yeast (Cheeseman et al., 2001; Janke et al., 2002; Li et al., 2002). This complex consists of 10 subunits and forms a ring encircling a microtubule in vitro (Miranda et al., 2005; Westermann et al., 2005). In fission yeast, the complex binds specifically to the spindle and the central core domain of the centromere in mitosis (Liu et al., 2005; Sanchez-Perez et al., 2005; Kobayashi et al., 2007). Unlike the budding yeast homolog, the complex is not essential for viability in fission yeast, but deletion of the Dam1/DASH complex caused cold-sensitive cell growth (Kobayashi et al., 2007). It has been reported that the fission yeast Dam1/DASH complex is essential for chromosome biorientation and that depletion of the complex in cells lacking Klp5 or Klp6 proteins, members of the kinesin-8 family, caused synthetic lethality (Sanchez-Perez et al., 2005; Griffiths et al., 2008). Klp5 and Klp6 have been suggested to possess microtubule-depolymerizing activity and to function in generating a force that pulls the chromosome toward the spindle pole (Garcia et al., 2002b). Very recently, it has been proposed that the Dam1/DASH complex couples the force generated by microtubule depolymerization to direct chromosome movement in fission yeast (Franco et al., 2007). Thus, the Dam1/DASH complex may also be involved in generation of the pulling force or in its transmission to the kinetochore. The budding yeast Dam1/DASH has been proposed to promote “end-on” pulling of kinetochores by the spindle (Tanaka et al., 2007).
In an attempt to understand the roles of kinetochore proteins in fission yeast, we found that two proteins, Bub1 and Hos2, are crucial for the accumulation of Mad2 on kinetochores. Here, we report that fission yeast Mad2 is recruited to mono-oriented as well as unattached kinetochores. Our observations suggest that Bub1 and the Dam1/DASH complex may play partially overlapping but distinct roles in the control of Mad2 localization. Furthermore, we found that the Dam1/DASH complex is required for efficient capture of detached kinetochores by the spindle, but not for motion of the captured kinetochores, during prometaphase. On the basis of these results, we propose that Bub1 and the Dam1/DASH complex may independently promote the accumulation of Mad2 on kinetochores in response to different types of kinetochore–spindle abnormality, mono-oriented and unattached, and coordinate Mad2-dependent cell cycle control with error-correcting machinery that facilitates the establishment of chromosome biorientation.
MATERIALS AND METHODS
General Techniques, Strains, and Plasmids
Fission yeast methods and media were described previously (Moreno et al., 1991). YES or YPD was used as rich medium, and EMM2 with appropriate supplements was used as minimal medium. For C-terminal gene tagging and gene disruption, the PCR-mediated method described previously (Krawchuk and Wahls, 1999) was used. The strains used in this study are listed in Table 1. In SP3548, the native bub1 gene was disrupted, and a plasmid containing the bub1-Δ28-160 allele, which was generated by PCR-mediated mutagenesis, was integrated at the aur1+ locus. To construct the strains expressing fluorescent tubulin (SP3259, SP3260, SP3261, and SP3262), a single copy of a DNA fragment containing the ctr4+ promoter-driven CFP-atb2 (α-tubulin) fusion gene and ura4+ selection marker was integrated into the genome. A modified version of CFP named cerulean was used (Rizzo et al., 2004). To minimize the effect of ectopic expression of cyan fluorescent protein (CFP)-tubulin, 10 μM CuSO4, which reduces the expression from the ctr4-promoter without affecting cell growth (Zhou and Thiele, 2001), was added to cultures of these strains unless otherwise noted. The pmCherry vector haboring Cherry red fluorescent protein gene (Shaner et al., 2004) was purchased from Clontech (Mountain View, CA). For cell cycle synchronization in S phase, cells were incubated in YES medium containing 12 mM hydroxyurea (HU) for 4 h and then washed three times in fresh YES. For treatment with CBZ (Sigma-Aldrich, St. Louis, MO), a freshly prepared 5 mg/ml solution of carbendazim (CBZ) in DMSO was diluted in medium to 50 μg/ml unless otherwise noted.
Table 1.
Strains used in this study
| Name | Genotype |
|---|---|
| SP3259 | h+leu1 ura4 his2 nda3-KM311 nuf2-YFP::NAT atb2+::ctr4prom-CFP-atb2::ura4+ |
| SP3260 | h+leu1 ura4 his2 nda3-KM311 Δhos2::kanR nuf2-YFP::NAT atb2+::ctr4prom-CFP-atb2::ura4+ |
| SP3261 | h+leu1 ura4 his2 nda3-KM311 mad2-YFP::NAT atb2+::ctr4prom-CFP-atb2::ura4+ |
| SP3539 | h+leu1 ura4 his2 nda3-KM311 hos2-YFP::NAT atb2+::ctr4prom-CFP-atb2::ura4+mis12-cherry::kanR |
| SP3540 | h+leu1 ura4 his2 nda3-KM311 mad2-YFP::NAT atb2+::ctr4prom-CFP-atb2::ura4+mis12-cherry::kanR |
| SP3580 | h−leu1 ura4 cut9-665 mad2-GFP:: kanR atb2+::ctr4prom-CFP-atb2::ura4+ |
| SP3262 | h−leu1 ura4 cut9-665 Δbub1::hph mad2-GFP:: kanR atb2+::ctr4prom-CFP-atb2::ura4+ |
| SP3263 | h−leu1 ura4 mis12-GFP::LEU2 pcp1-CFP::NAT |
| SP3264 | h−leu1 ura4 Δhos2::hph mis12-GFP::LEU2 pcp1-CFP::NAT |
| SP3265 | h−leu1 ura4 Δbub1:: kanR mis12-GFP::LEU2 pcp1-CFP::NAT |
| SP3266 | h−leu1 ura4 Δbub1:: kanR Δhos2::hph mis12-GFP::LEU2 pcp1-CFP::NAT |
| SP3557 | h−leu1 ura4 Δmad2:: hph mis12-GFP::LEU2 pcp1-CFP::NAT |
| SP3560 | h−leu1 ura4 Δmad2:: hph Δhos2::hph mis12-GFP::LEU2 pcp1-CFP::NAT |
| SP1028 | h−leu1 cut9-665 mad2-GFP:: kanR |
| SP2094 | h−leu1 cut9-665 mis4-242 mad2-GFP:: kanR |
| SP2095 | h−leu1 cut9-665 mis4-242 Δbub1::hph mad2-GFP:: kanR |
| SP634 | h−leu1 mad2-GFP:: kanR |
| SP1730 | h−leu1 Δhos2::hph mad2-GFP:: kanR |
| SP2058 | h−leu1 ura4 Δbub1::ura4+mad2-GFP:: kanR |
| SP2354 | h−leu1 ura4 Δbub1::ura4+ Δhos2::hph mad2-GFP:: kanR |
| SP3581 | h−leu1 ura4 bub1-K762R Δhos2::hph mad2-GFP:: kanR |
| SP3548 | h−leu1 ura4 Δbub1::ura4+aur1+::bub1-Δ28-160 Δhos2::hph mad2-GFP:: kanR |
| SP2187 | h−leu1 Δmad3::hph mad2-GFP:: kanR |
| SP2231 | h−leu1 Δhos2::hph Dmad3::hph mad2-GFP:: kanR |
Strains listed were created as described in Material and Methods and by crossing.
Fluorescence Microscopy
For immunostaining and DAPI staining methods, see references in Saitoh et al. (1997). The methanol fixation method was used to preserve Mad2-green fluorescent protein (GFP) fluorescence. For observation, an Olympus IX81 microscope (Olympus, Tokyo, Japan) equipped with a 100× objective lens (NA 1.35) and RETIGA EXi FAST cooled CCD camera (Roper Scientific, Tucson, AZ) was used. Three-dimensional stacks of Z-sectioned images (0.3-μm intervals) were collected, deconvolved, and flattened using Metamorph software (Molecular Devices, Sunnyvale, CA).
Live Cell Analysis
Living S. pombe cells were cultured in EMM2 and observed using a Leica ASMDW live cell imaging system (Leica Microsystems, Wetzlar, Germany) equipped with a 100× objective lens (NA 1.4) and a temperature control unit. Three-dimensional stacks of Z-sections (0.4-μm interval) were collected every 20 s unless otherwise noted. For deconvolution, the nonblind algorithm of ASMDW software was applied. Nonprocessed image data were used to measure fluorescence intensity.
For the nda3 block and release experiments, nda3 mutant cells were incubated in YPD medium at 18°C for 14–16 h, washed once in ice-cold EMM2, and then resuspended in a small volume of ice-cold EMM2 to which CBZ was added at 10 μg/ml to prevent reformation of the spindle. Aliquots of 20 μl of the cell suspension were mounted on concanavalin A–coated glass-bottomed dishes and set on a microscope placed in an incubation chamber prewarmed at 33°C. For release of the cells from mitotic arrest, CBZ was diluted out by addition of 1 ml of prewarmed EMM2 medium. Time-lapse recording started immediately when a cell with the metaphase spindle (∼2.5 μm in length) and a detached kinetochore was found. It took 10–15 min to find such a cell. The cut9 mutant cells were incubated in EMM2 medium at 36°C for 3 h and then for 1 h after addition of 50 μg/ml CBZ. The cells were then treated as described above.
RESULTS
Dissection of the Process for Establishment of Kinetochore–Spindle Biorientation
When cells enter mitosis, kinetochores are captured by spindle microtubules in a mono-oriented manner and subsequently establish bioriented interaction. In wild-type fission yeast cells, kinetochores of all three chromosomes are packed in a short linear path of the spindle (∼2.5 μm) during the early stages of mitosis, and thus it is difficult to determine how each kinetochore interacts with the spindle because of the resolution limit of optical microscopes. To circumvent such limitations and dissect the processes involved in the establishment of biorientation of kinetochores, we used the method described by Grishchuk and McIntosh (2006). Briefly, we examined how a detached kinetochore reestablishes the interaction with the spindle upon recovery from microtubule depolymerization caused by cold-sensitive nda3 (β-tubulin) mutation (Hiraoka et al., 1984). The search for a cell harboring the metaphase spindle and a clearly detached kinetochore was initiated immediately after release from nda3 block, and time-lapse recording was started at the moment of finding such a cell, which was designated as time = 0. Time-lapse images of a representative cell are shown in Supplemental Figure S1, which shows kinetochores and spindle microtubules labeled with Nuf2-YFP and CFP-Atb2 (α-tubulin), respectively. No microtubules appeared to associate with the lost kinetochore at the beginning of recording (time = 0–220 s), because the movement of the kinetochore was not restricted. The kinetochore then suddenly showed unidirectional movement toward one of the spindle pole bodies (SPBs; time = 240–280 s). This rapid movement was called “p-movement,” or poleward movement, indicating that the kinetochore was captured by a spindle microtubule fiber (Grishchuk and McIntosh, 2006), and the rate of p-movement was ∼1 μm/min. The captured kinetochore reached the SPB and remained there for a few minutes (time = 280–520 s, indicated by black arrowheads); the interaction between the kinetochore and the microtubule at this point was likely to be mono-oriented. Subsequently, the kinetochore moved to the middle region of the spindle, which indicated that the kinetochore established the bioriented interaction with the spindle and was pulled toward the other pole. This behavior of the lost kinetochore was reproducibly observed in multiple samples (n = 7) and was consistent with that reported previously (Grishchuk and McIntosh, 2006). Lost kinetochores were captured stochastically by the spindle, and the probability of such an event was estimated as 0.1 event/min in nda3 mutant cells at the permissive temperature (Table 2).
Table 2.
The Dam1/DASH complex is required for efficient capture of kinetochores
| Strain | No. of examples of the rescuea | Average time point of the rescue (min)b | Probability of the rescue (events/min)c |
|---|---|---|---|
| nda3 (SP3259) | 7/7 | 8.9 ± 4.7 | 0.11 |
| nda3 Δhos2 (SP3260) | 4/11 | >38.6 | <0.026 |
a The number of examples in which the detached kinetochore was captured within the recording period (45 min) is shown. Seven (nda3) or 11 (nda3 Δhos2) samples were examined in total.
b The average time point at which the detached kinetochore was captured is shown.
c The probability of the kinetochore-rescue event is calculated, given that the time points of the rescue followed a geometric distribution.
Mono-oriented Capture Is Not Sufficient for the Release of Mad2 from a Kinetochore
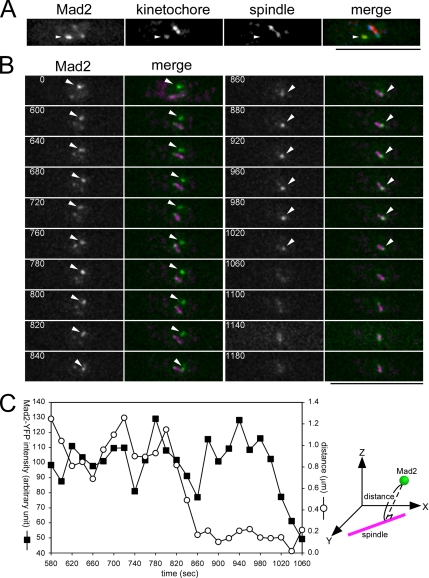
In fission yeast, the spindle assembly checkpoint protein Mad2 is recruited to unattached kinetochores at the beginning of mitosis and released before the onset of anaphase (Ikui et al., 2002; Saitoh et al., 2005; Tange and Niwa, 2007). Although, in many higher eukaryotes, it has been suggested that Mad2 is released from the kinetochore upon spindle attachment regardless of whether such attachment generates tension (Waters et al., 1998; Logarinho et al., 2004), the precise timing of Mad2 release is obscure in fission yeast, and therefore we attempted to determine the timing using the above method. Triple labeling of Mad2, kinetochores, and the spindle shown in Figure 1A clearly indicated that Mad2 specifically accumulated on a detached kinetochore and along the spindle, but not on kinetochores associated with the spindle upon recovery from nda3 block. A representative example of time-lapse analysis is shown in Figure 1B, where the Mad2 and the spindle were visualized by Mad2-YFP and CFP-Atb2, respectively. At time 0, a bright Mad2-YFP signal was observed, along with faint signals on the nuclear periphery and along the spindle. This bright spot was presumed to be concomitant with a kinetochore detached from the spindle. Subsequently, the Mad2 spot moved quickly toward the SPB (time = 820–860 s), and this movement appeared to correspond with the p-movement of the captured kinetochore. The intensity of the Mad2 spot after reaching the spindle pole was comparable to that before capture by the spindle (time = 880–980 s, Figure 1, B and C), which indicated that Mad2 localizes on not only detached kinetochores but also mono-oriented kinetochores. Mad2 localization on mono-oriented kinetochores was reproducibly observed in multiple samples (n = 8, Supplemental Figure S2). Therefore, the mono-oriented attachment of the spindle microtubule was apparently insufficient for release of Mad2 from the kinetochore. Thereafter, the spot faded gradually (time = 980–1180 s). It was difficult to determine precisely what event(s) triggered the disappearance of the Mad2 spot. Judging from the spindle dynamics, anaphase chromosome separation took place soon after this disappearance. Thus, the release of Mad2 from the kinetochore was presumed to be coincident with the establishment of kinetochore biorientation. We concluded that kinetochore biorientation, but not the mono-oriented attachment, triggers the release of Mad2 from the kinetochore.
Figure 1.
Mono-oriented capture is not sufficient for release of Mad2 from the kinetochore. (A) Snapshot image of an nda3 mutant cell expressing Mad2-YFP, Mis12-Cherry (a kinetochore component), and CFP-Atb2 (α-tubulin) after recovery from mitotic arrest. Nine serial Z-sections with an interval of 0.3 μm were flattened after deconvolution. The position of the kinetochore detached from the spindle is indicated by an arrowhead. In the merged panel, Mad2-YFP, Mis12-Cherry, and CFP-Atb2 are pseudocolored green, red, and blue, respectively. Bar, 10 μm. (B) Time-lapse images of an nda3 cell expressing Mad2-YFP and CFP-Atb2 after recovery from mitotic arrest. Panels show the YFP channel alone (Mad2), and YFP merged with CFP (merge: YFP and CFP are pseudocolored green and magenta, respectively). Eleven serial Z-sections with an interval of 0.4 μm were collected every 20 s, deconvolved, and flattened using a Leica ASMDW microscopy system. The bright Mad2 spot coincident with the detached/mono-oriented kinetochore is indicated by an arrowhead. The numbers indicate the duration in seconds. Bar, 10 μm. (C) The fluorescence intensity of the Mad2-YFP spot was measured at each time point using three-dimensional images of the cell shown in B and is plotted in arbitrary units (left axis). Vertical distance between the Mad2 spot and the spindle was also calculated (right axis).
Mad2 Was Able to Localize on Unattached Kinetochores But Not on Mono-oriented Kinetochores When the bub1 Gene Was Deleted
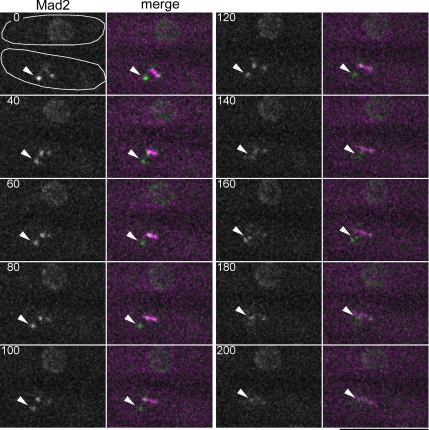
The results shown in Figure 1 indicated that Mad2 localized on not only unattached but also mono-oriented kinetochores. Although it has been reported that kinetochore localization of Mad2 requires Bub1, a protein kinase crucial for the SAC, in higher eukaryotes (Sharp-Baker and Chen, 2001; Johnson et al., 2004), the relationship between Mad2 and Bub1 is not clear in fission yeast; fission yeast Bub1 is also essential for the SAC, but it was reported to be dispensable for localization of Mad2 and Mad1, which is a binding partner of Mad2, on to unattached kientochores (Yamaguchi et al., 2003; Kadura et al., 2005). Tension across sister kinetochores, which is generated by spindle pulling forces toward opposing poles, is not exerted on mono-oriented kinetochores, and Bub1 has been suggested to be involved in a pathway that senses tension (Skoufias et al., 2001; Garcia et al., 2002a). Thus, we examined whether Bub1 was required for the retention of Mad2 on mono-oriented kinetochores. As nda3 mutant cells lacking Bub1 failed to prevent premature activation of APC/C due to a defect in the SAC, we utilized cut9 mutation, which inactivates APC/C, in combination with treatment with a microtubule depolymerizing drug, Carbendazim (CBZ), to mimic nda3 mutation. cut9 Δbub1 mutant cells in which Mad2 and the spindle were labeled with fluorescent proteins were incubated at the restrictive temperature (36°C) for 3 h, and for an additional 1 h in the presence of 50 μg/ml CBZ. After CBZ treatment, the cells were mounted on a glass-bottomed dish and set on a microscope prewarmed at 33°C. Spindle reformation was initiated by diluting out CBZ. Before CBZ was diluted out, most of the mitotic cells possessed bright Mad2-GFP spots that appeared concomitant with unattached kinetochores and/or the SPBs. Although Mad2 spots adjacent to the spindle pole disappeared immediately upon reformation of the spindle (data not shown), a bright Mad2 spot remained in a small percentage of mitotic cells. A representative time-lapse image of a cell with the Mad2 spot is shown in Figure 2. This remaining spot was localized neither along a spindle nor on the SPBs, suggesting that the spot colocalized with a kinetochore detached from the spindle. An interphase cell is also shown as a control indicating the degree of photobleaching during observation. The Mad2 spot was observed clearly at time 0 and then diminished quickly during p-movement (time = 180–200 s). This diminishment was not caused by photobleaching of Mad2-GFP, because the intensity of nuclear Mad2-GFP fluorescence did not change significantly in the control cell. In 11 of 16 cut9 Δbub1 cells examined, the Mad2 spot on a detached kinetochore disappeared before completion of p-movement; in the remaining five examples, the Mad2 spot persisted but did not show p-movement during observation, suggesting that the kinetochore was not captured in these cells. As the Mad2 spot persisted during p-movement and for a few minutes after completion of p-movement in cut9 single mutant cells (Supplemental Figure S3, white arrows), cut9 mutation was unlikely to be responsible for the premature diminishment of the Mad2 spot. Taken together, these observations indicated that the localization of Mad2 on mono-oriented kinetochores requires Bub1. In contrast, deletion of bub1+ did not appear to impair the localization of Mad2 on unattached kinetochores because Mad2 was observed to accumulate on a detached kinetochore in cut9 Δbub1 cells (time = 0 s). Interestingly, in a few cut9 Δbub1 cells escaping from mitotic arrest, the Mad2 spot, which was not associated with the spindle or SPBs, persisted even after the onset of anaphase spindle extension (Supplemental Figure S4). Thus, Mad2 may be able to localize on unattached kinetochores in Δbub1 cells even during anaphase.
Figure 2.
Bub1 is required for retention of Mad2 on mono-oriented kinetochores. Time-lapse images of cut9 Δbub1 cells expressing Mad2-GFP and CFP-Atb2 after removal of CBZ. Panels show the GFP channel (Mad2) and GFP merged with CFP (merge: GFP and CFP are colored green and magenta, respectively). Stacks of Z-sections with an interval of 0.4 μm were taken every 20 s. Three-dimensional images were flattened after deconvolution. Two cells are shown in each panel and the outlines of the cells are drawn in the panel of time = 0; the upper cell is in interphase, and the other is in mitosis. The Mad2 spot presumably coincident with a detached kinetochore is indicated by an arrowhead. The numbers indicate the duration in seconds. Bar, 10 μm.
Bub1 Is Required for Localization of Mad2 on Cohesionless Kinetochores
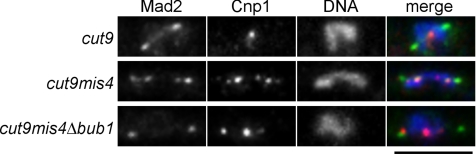
The results described above indicated that the localization of Mad2 on mono-oriented kinetochores is dependent on Bub1, which is proposed to be involved in the tension-sensing pathway. It has been reported that Mad2 accumulates on kinetochores in mis4 mutant cells (Toyoda et al., 2002), which have a defect in the establishment of sister chromatid cohesion (Furuya et al., 1998). Cohesion between sister chromatids is a prerequisite for chromosome biorientation, and tension is not expected to be exerted across the cohesionless sister kinetochores. Thus, we next examined whether Mad2 localization on cohesion-defective kinetochores also requires Bub1. The localization of Mad2-GFP in mis4-deficient cells is shown in Figure 3. To determine the precise positions of kinetochores, Cnp1, the fission yeast CENP-A homolog (Takahashi et al., 2000), was immunostained with anti-Cnp1 polyclonal antibody. The cut9 mutation inactivating APC/C was used to prevent premature entry into anaphase. In cut9 single mutant cells, Mad2-GFP was localized strongly on the SPBs and weakly along the spindle. In these cells, Mad2 did not accumulate on kinetochores, which were clustered in the middle of the spindle. In contrast to cut9 single mutant cells, Mad2 accumulated on cohesion-defective kinetochores as well as the SPBs in cut9 mis4 double mutant cells. Mad2 localization on the kinetochores in mis4 mutant cells required the bub1+ gene, because Mad2 no longer accumulated on kinetochores in cut9 mis4 Δbub1 triple mutant cells, in which Mad2 was still able to localize on the SPBs. cut9 mis4 Δbub1 triple mutation did not impair the targeting of Mad2 to unattached kinetochores because Mad2 localized on kinetochores in the triple mutant cells treated with CBZ (data not shown). Therefore, Bub1 appears to be essential for localization of Mad2 on cohesionless as well as mono-oriented kinetochores. As both cohesionless and mono-oriented kinetochores lack tension, Mad2 is likely to localize on tensionless kinetochores in a Bub1-dependent manner.
Figure 3.
Bub1 is indispensable for localization of Mad2 on cohesion-defective kinetochores. The cut9 single mutant, cut9 mis4 double mutant, and cut9 mis4 Δbub1 triple mutant cells were incubated at the restrictive temperature for two generations (5, 6, and 8 h, respectively), and fixed for immunostaining of Cnp1 (s.p. CENP-A). Mad2 was visualized by GFP fusion in these cells. Stacks of Z-sections were taken and flattened after deconvolution. In merge panels, Mad2, Cnp1, and DNA are colored green, red, and blue, respectively. Bar, 5 μm.
Mad2 Accumulation on Unattached Kinetochores Was Diminished when hos2+ and bub1+ Genes Were Deleted Simultaneously
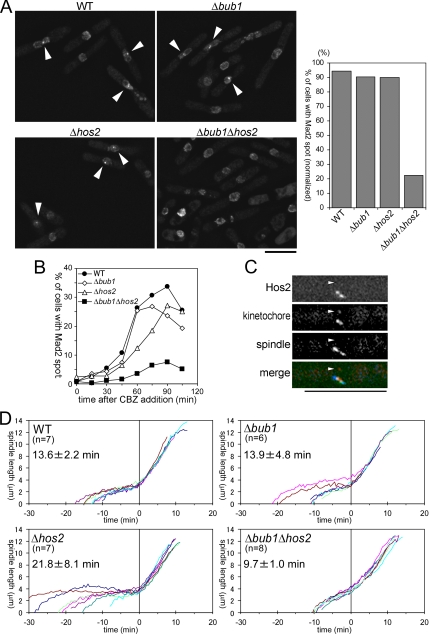
Fission yeast Mad2 localized on tensionless kinetochores as well as unattached kinetochores, and the Mad2 localization on tensionless kinetochores required Bub1. These observations raise the question of what is required for the localization of Mad2 on unattached kinetochores. The unattached kinetochore is inevitably tensionless, and therefore depletion of molecules responsible for Mad2 localization on unattached kinetochores is predicted to impair Mad2 localization only when the tension-sensing pathway is disabled simultaneously. In the course of our studies of Hos2 function, we noticed that cells with bright Mad2 spots, which appeared to be localized on kinetochores, were not found in an exponentially growing population of a Δhos2 Δbub1 double mutant (Supplemental Figure S5A). Similarly, Mad2 spots on kinetochores were not observed in Δask1 Δbub1 or Δduo1 Δbub1 double mutant cells (data not shown); Hos2, Duo1, and Ask1 are integral components of the Dam1/DASH complex. Cells with bright Mad2 spots were detected in populations of either Δhos2 or Δbub1 single mutants, and thus double deletion of hos2+ and bub1+ genes was thought to synergistically impair accumulation of Mad2 on kinetochores. To test this hypothesis, we examined whether Mad2 was localized to kinetochores in CBZ-treated wild-type (WT), Δbub1, Δhos2, and Δbub1 Δhos2 cells. For precise analyses, synchronized cell populations were used as described by Vanoosthuyse et al. (2004). Briefly, the cells were synchronized in S phase by treatment with HU, which inhibits deoxyribonucleotide synthesis, and cultures were then split into two upon release from HU block. One of these cultures was incubated without addition of CBZ and stained with DAPI to monitor the cell cycle progression after release, whereas CBZ was added to the other culture that was prepared for measuring the frequency of cells with bright Mad2 spots. Hos2 deletion appeared to slightly slow down cell cycle progression; 55.6% of WT cells and 35.1% of Δbub1 cells entered/completed mitosis at 60 min after release from HU block, whereas 42.1% of Δhos2 and 34.0% of Δhos2 Δbub1 did so at 90 min after release. At these time points, the frequencies of cells harboring bright Mad2 spots were 53.0% in WT, 31.8% in Δbub1, 37.8% in Δhos2, and 7.6% in Δbub1 Δhos2 cells (n > 400).
To compensate for the differences in cell cycle progression, the frequency of cells with Mad2 spots was divided by that of cells that had entered/completed mitosis, and these normalized frequencies are shown in Figure 4A (right panel). The results indicated that simultaneous deletion of bub1+ and hos2+ genes severely impaired the accumulation of Mad2 on unattached kinetochores. We also performed the time course analysis of the frequency of cells with Mad2 spots (Figure 4B) and confirmed the deficiency of Δbub1 Δhos2 cells in Mad2 accumulation on kinetochores. As mentioned above, Δask1 Δbub1 or Δduo1 Δbub1 double deletion also impaired the accumulation of Mad2, and therefore the Dam1/DASH complex is thought to be important for the regulation of Mad2 localization. As single deletion of either the bub1 or hos2 gene did not affect Mad2 accumulation on unattached kinetochores, the Dam1/DASH complex may be involved in a pathway controlling Mad2 localization in parallel with Bub1. We next attempted to examine the localization of the Dam1/DASH complex upon recovery from nda3 block. Because of severe photobleaching, however, we failed to perform time-lapse imaging of triply labeled cells in which Hos2, kinetochores, and the spindle were fused with different fluorescent proteins. A three-dimensional snapshot of the triply labeled cell was taken upon recovery from nda3 block (Figure 4C). Little or no Hos2-YFP signal was detected on the unattached kinetochore (arrowhead), whereas strong YFP signals were detected in the vicinity of the kinetochores that associated with the spindle. This result suggests that the Dam1/DASH complex may accumulate preferentially on attached kinetochores, and thus the complex may somehow distinguish attached from unattached kinetochores. Taking these observations together, we presume that the Dam1/DASH pathway monitors the state of spindle–kinetochore attachment and controls Mad2 accumulation on unattached kinetochores.
Figure 4.
Δbub1 Δhos2 double gene deletion impairs accumulation of Mad2 on unattached kinetochores. (A) Cells with the indicated genotype were treated with CBZ after S phase synchronization. Details are described in the text. Images of Mad2-GFP in these cells are shown, and bright Mad2 spots are indicated by arrowheads. Bar, 10 μm. Calculated percentages of cells with the Mad2 spot in the mitotic population are presented in the right panel. (B) Percentages of cells with Mad2 spot in total cells were counted every 15 min after addition of CBZ. More than 250 cells were examined for each sample. Cells were presynchronized in S phase as described in the text. (C) Snapshot of an nda3 mutant cell expressing Hos2-YFP, Mis12-Cherry (a kinetochore component) and CFP-Atb2 (α-tubulin) after recovery from mitotic arrest. Nine serial Z-sections with an interval of 0.3 μm were flattened after deconvolution. The position of the kinetochore detached from the spindle is indicated by an arrowhead. In the merged panel, Hos2-YFP, Mis12-Cherry, and CFP-Atb2 are pseudocolored green, red, and blue, respectively. Bar, 10 μm. (D) Spindle lengths from multiple movies of exponentially growing cells with the indicated genotype were analyzed. Cells were cultured in minimal medium at 26°C, and Pcp1-CFP (SPB marker) was used to determine the length. The time point at which phase 3 spindle extension began was defined as 0. The number of samples examined (n) and average time required for phase 1 + 2 (average ± SD) in each strain are shown.
It has been suggested that Bub1 consists of two parts: the C-terminal kinase domain essential for accurate chromosome segregation and the N-terminal domain essential for the SAC (Vanoosthuyse et al., 2004). We found that bub1-Δ28-160 mutation, which specifically impairs N-terminal function of Bub1 (Vanoosthuyse et al., 2004), abolished the localization of Mad2 on kinetochores in cells lacking Hos2, whereas the kinase-defective bub1-K762R mutation did not (Yamaguchi et al., 2003; Supplemental Figure S5A). Thus, N-terminal SAC function of Bub1 appears to be important for the kinetochore localization of Mad2. Fission yeast Mad3 is a putative homolog of BubR1 (Millband and Hardwick, 2002), which is also thought to function in a tension-sensing pathway in higher eukaryotes (Skoufias et al., 2001). In contrast to Δbub1 Δhos2 double deletion, simultaneous deletion of mad3+ and hos2+ genes did not impair the Mad2 localization on kinetochores (Supplemental Figure S5B), suggesting that Mad3 is not involved in the regulation of Mad2 localization.
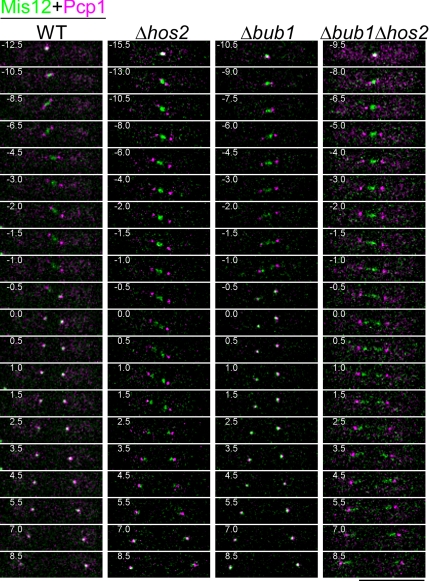
Δbub1 Δhos2 Double Deletion Accelerates the Onset of Phase 3 Spindle Extension
As described above, Δbub1 Δhos2 double deletion disabled localization of Mad2 on kinetochores. Accumulation on kinetochores was thought to be the initial step by which Mad2 blocks metaphase–anaphase transition (Yu, 2002), and thus deletion of bub1+ and hos2+ genes may influence the normal mitotic progression. To test this possibility, the extension of the spindle, which is correlated with mitotic progression (Nabeshima et al., 1998), was monitored in growing WT, Δbub1, Δhos2, and Δbub1 Δhos2 cells (Figure 4D). These cells, in which the kinetochores and the SPBs were marked by Mis12-YFP and Pcp1-CFP, respectively (Goshima et al., 1999; Flory et al., 2002), were incubated under a fluorescence microscope at 26°C, and the behavior of kinetochores and the spindle during mitosis was recorded. The length of the mitotic spindle was determined as the linear distance between the SPBs and is plotted against time. Representative time-lapse images of each strain are shown in Figure 5. As reported previously (Nabeshima et al., 1998), the spindle extension occurred in three phases, phases 1–3. In the case of WT cells, the spindle elongated rapidly to ∼2.5 μm in phase 1, had a relatively constant length during phase 2, and began to extend again reaching a length of ∼13 μm in phase 3. Rapid sister chromatid separation in anaphase A occurred just before the onset of phase 3 (Nabeshima et al., 1998), the moment of which was defined as time = 0. The times required for phase 1 + 2 were 13.6 ± 2.2, 21.8 ± 8.1, 13.9 ± 4.8, and 9.7 ± 1.0 min (average ± SD) in WT, Δhos2, Δbub1, and Δbub1 Δhos2 cells, respectively. The differences were largely due to the differences in length of prometaphase, in which the kinetochores moved along the spindle. The length of phase 1 + 2 was shortest and the cell-to-cell variation was smallest in Δbub1 Δhos2 cells, indicating that the metaphase–anaphase transition did not slow down in these cells despite abnormalities in chromosome segregation (Figure 5). Considering the impairment of Mad2 accumulation on unattached kinetochores, this result suggests the total dysfunction of the mitotic checkpoint control in Δbub1 Δhos2 cells. Interestingly, although deletion of the mad2+ gene also greatly reduced the length of phase 1 + 2 in most of cells lacking Hos2, two of the nine Δmad2 Δhos2 cells showed a slight delay in phase 2 (Supplemental Figure S6B). Such delay was also observed in some of Δmad2 cells, and thus it may be caused by Mad2-independent SAC pathways, which have been reported to require Bub1 and to respond to incorrect spindle orientation and/or defects caused by the deletion of EB1 protein, Mal3 (Rajagopalan et al., 2004; Tournier et al., 2004; Asakawa et al., 2005).
Figure 5.
Δhos2 cells are defective in poleward movement of kinetochores during anaphase A. Time-lapse images of mitosis in exponentially growing cells with the indicated genotype are presented. Kinetochores and the SPB were visualized by Mis12-GFP (green) and Pcp1-CFP (magenta), respectively. Cells were cultured in minimal medium at 26°C, and stacks of Z-sections with an interval of 0.4 μm were taken every 0.5 min. The numbers indicate the duration in minutes. The time point at which phase 3 spindle extension began was defined as 0. Bar, 10 μm.
Surprisingly, in these observations, we noticed that poleward movement of kinetochores during anaphase A was severely impaired in cells lacking hos2+ (Figure 5 and Supplemental Figure S6A). Separated sister kinetochores concentrated in the vicinity of the spindle poles, but failed to reach the SPB before the onset of phase 3 (time = 0) in Δhos2 cells, whereas they instantly reached the SPB at the end of phase 2 in WT and Δbub1 cells. Such abnormalities in kinetochore separation during anaphase A were exaggerated in Δhos2 Δbub1 and Δmad2 Δhos2 double deletion cells; although the separated kinetochores moved apart as the spindle extended during phase 3 in the double deletion strain, they did not reach the poles and were scatted along the spindle in proximity to the SPB. Therefore, the Dam1/DASH complex is required for effective poleward movement of kinetochores during anaphase A.
Hos2 Is Required for the Efficient Capture of Kinetochores
As described above, the Dam1/DASH complex may be involved in regulation of the SAC. It was reported previously that the introduction of a multicopy plasmid harboring the hos2+ gene rescued a broad range of kinetochore-defective mutants, implying that the Dam1/DASH complex may also correct some kinetochore defects in the mutants (Kobayashi et al., 2007). These results suggested that the Dam1/DASH complex coupled SAC activation with correction of certain types of error in spindle–kinetochore interaction. As shown in Figure 4C, the Dam1/DASH complex appears to distinguish unattached from attached kinetochores. Thus, to examine the possibility that the Dam1/DASH complex is somehow involved in rescue of unattached kinetochores, we examined how the detached kinetochores were captured by the spindle in nda3 (control) and nda3 Δhos2 mutant cells upon recovery from spindle disassembly. The results are summarized in Table 2. In all hos2+ control cells examined (n = 7), the detached kinetochore was rescued efficiently within 20 min after the beginning of time-lapse recording. The average time point at which the kinetochore was recaptured was 8.9 ± 4.7 min in hos2+ cells, and the probability of the rescue event was estimated as 0.11 events/min, given that the time points of the rescue followed a geometric distribution with the probability constant during the experiments. On the other hand, a detached kinetochore failed to be rescued in 7 of 11 Δhos2 cells during the recording period (45 min). The average timing of recapture was later than 38.6 min in Δhos2 cells, and the estimated probability of the rescue event was <0.026 event/min. Thus, deletion of the hos2+ gene severely reduced the efficiency of rescuing a detached kinetochore. It is noteworthy that a detached kinetochore was eventually captured and delivered to the SPB in 4 of 11 Δhos2 cells examined, although the rate of p-movement appeared slightly slower than that in hos2+ cells (Supplemental Figure S7). Thus, in contrast to poleward movement of kinetochores during anaphase A, the Dam1/DASH complex was dispensable for p-movement of kinetochores during prometaphase, consistent with a previous report in budding yeast (Tanaka et al., 2005). Controversially, it has been reported that detached kinetochores are not delivered to the SPB in cells lacking the Dam1/DASH complex, whereas it binds to the lateral face of a microtubule (Franco et al., 2007; Gachet et al., 2008). We cannot yet explain this discrepancy at this moment. Differences in strains used in the analyses might cause this discrepancy. Other possibility is that the length of time-lapse recording in the previous reports might not be sufficient to detect p-movement of the recaptured kinetochores in Dam1/DASH deficient cells. On the basis of our observations, we suppose that the Dam1/DASH complex functions rather in increasing the probability of the reassociation of detached kinetochores with the spindle microtubules.
DISCUSSION
In this study, we showed that Bub1 and the Dam1/DASH complex play important roles in regulating the accumulation of Mad2 on kinetochores; double deletion of bub1+ and hos2+ genes severely impaired the localization of Mad2 on unattached kinetochores in CBZ-treated cells. We have also shown that the Dam1/DASH complex is required for the efficient capture of unattached kinetochores by the spindle, whereas Bub1 has been reported to be essential for kinetochore targeting of Sgo2, which ensures kinetochore biorientation (Kitajima et al., 2004). Bub1 and the Dam1/DASH complex, therefore, appear to be involved in not only regulation of Mad2 localization but also in the promotion of chromosome biorientation. Thus, we propose that Bub1 and the Dam1/DASH complex couples the Mad2 checkpoint pathway with the error-correcting mechanisms that facilitate proper spindle–kinetochore interaction.
We found that Mad2 localized on mono-oriented kinetochores, on which the localization of Mad2 required Bub1. In many organisms, including mammals, Mad2 has been reported to be released from kinetochores upon spindle attachment, regardless of whether such attachment generates tension (Waters et al., 1998; Skoufias et al., 2001). Although we could not determine how the microtubule(s) interacted with the mono-oriented kinetochore, it is possible that the interaction was monotelic; only one of the sister kinetochores was captured by a single or a few microtubules and the other remained unattached. In this case, Mad2 could be absent on the captured sister kinetochore and retained only on the unattached kinetochore. This appears unlikely, however, because the amount of Mad2 on the kinetochore was not reduced to half upon mono-oriented attachment of the spindle microtubule; that is, Mad2 is supposed to remain on both sister kinetochores even after mono-oriented attachment. It is noteworthy that maize Mad2 monitors tension in meiosis, whereas it monitors attachment in mitosis (Yu et al., 1999). Thus, abnormalities in spindle–kinetochore interactions that trigger Mad2 accumulation may differ between mitosis and meiosis, and among species. Another explanation is that mono-oriented kinetochores may associate with the spindle microtubules “immaturely,” and such immature attachment may not be sufficient for the release of Mad2, although it is capable of supporting p-movement of the kinetochore. Two types of kinetochore-microtubule interaction have been described: “lateral” and “end-on” attachment. Lateral attachment, in which a kinetochore binds to the lateral surface of microtubules, matures into an end-on attachment, in which the plus ends of microtubules are embedded in the kinetochore (Maiato et al., 2004a). Bub1 may recognize a kinetochore that does not interact with microtubules in end-on manner. Alternatively, occupancy of microtubules on mono-oriented kinetochores may not be sufficient for the release of Mad2. Unlike the case in budding yeast, fission yeast kinetochores are occupied by several microtubules in early mitosis (Ding et al., 1993). Fission yeast Mad2 may be retained on kinetochores until the number of associated microtubules reaches a certain threshold. In this scenario, Bub1 may monitor the occupancy or determine the threshold of Mad2 removal. It has been reported that application of tension on kinetochores can enhance both the stability of individual microtubule attachments and the overall occupancy of kinetochores (Nicklas and Ward, 1994; King and Nicklas, 2000), and thus the occupancy is closely correlated with tension across sister kinetochores.
In cells lacking both Bub1 and Hos2, Mad2 did not accumulate on kinetochores even when the cells were treated with the spindle-depolymerizing drug, CBZ. Δbub1 single deletion did not abolish Mad2 accumulation on unattached kinetochores in CBZ-treated cells, whereas it abolished the Mad2 accumulation on a mono-oriented and a cohesionless kinetochore. Thus, although both Bub1 and the Dam1/DASH complex appear to be involved in triggering Mad2 accumulation on kinetochores, they may monitor different types of abnormality in spindle–kinetochore interaction. We presume that Bub1 recognizes the absence of tension, whereas the Dam1/DASH complex detects the lack of spindle attachment (Figure 6). Dis1, a microtubule-associated XMAP215/TOG family protein, localizes on kinetochores and along the spindle during mitosis (Nabeshima et al., 1995; Nakaseko et al., 2001; Aoki et al., 2006), and its kinetochore localization requires the Dam1/DASH complex (Kobayashi et al., 2007). This suggests the physical interaction of the Dam1/DASH complex with Dis1 on the kinetochore, and this interaction may be important for the complex to recognize spindle–kinetochore attachment. We found that the Dam1/DASH complex appeared to accumulate preferentially on spindle-associated kinetochores upon recovery from nda3 block. In arrested nda3 mutant cells, Liu et al. (2005) reported that the Dam1/DASH complex binds unstably on kinetochores detached from the SPBs and strongly on kinetochores associating with the SPBs. Thus, the Dam1/DASH complex appears capable of binding to kinetochores that do not associate with microtubules or SPBs, whereas strong or stable binding may require association of the kinetochores with the spindle. These observations suggest that the Dam1/DASH complex is involved in the molecular mechanism distinguishing unattached from attached kinetochores. Likewise, the state of Bub1 binding may distinguish mono- or maloriented from bioriented kinetochores. Although the molecular mechanism controlling Bub1 localization on kinetochores is still unclear, the physical tension across sister kinetochores appears to be a key element regulating the localization of Bub1 (Skoufias et al., 2001). Bub1 may promote not only the accumulation of Mad2 but also the resolution of tensionless microtubule attachment, as discussed below.
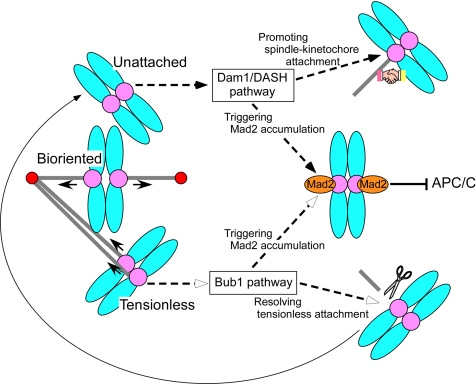
Figure 6.
Model: Bub1 and Dam1/DASH may coordinate the checkpoint control with error-correcting machineries. Mad2 accumulation on kinetochores are regulated by two distinct pathways: the Dam1/DASH pathway and the Bub1 pathway. Unattached kinetochores are thought to be detected by the Dam1/DASH pathway, which may trigger Mad2 accumulation on the kinetochores and also facilitate spindle–kinetochore attachment. On the other hand, tensionless kinetochores are detected by the Bub1 pathway, which may promote both Mad2 accumulation on the kinetochore and the resolution of tensionless spindle attachment. This resolution of the attachment results in conversion of the tensionless kinetochore into the unattached kinetochore.
The regulation of kinetochore targeting of Mad2 by the Dam1/DASH complex and Bub1 appeared to play a pivotal role in the control of cell cycle progression. Live cell analysis revealed that one-third of Δbub1 cells, which lacked the Bub1-dependent checkpoint pathway, showed a delay in metaphase–anaphase transition. In contrast, the onset of anaphase spindle extension in cells lacking both Bub1 and the Dam1/DASH complex was not delayed, despite abnormalities in chromosome segregation, such as lagging chromosomes (Figure 5). Therefore, mitotic delay in Δbub1 cells appears to require the Dam1/DASH complex. Bub1 has been reported to have both noncheckpoint and checkpoint functions at the kinetochore (Vanoosthuyse et al., 2004; Yu and Tang, 2005). Therefore, depletion of Bub1 may cause a kinetochore defect that activates the remaining Mad2 checkpoint in a Dam1/DASH-dependent manner and consequently result in transient mitotic delay.
Ectopic expression of Hos2 rescued a variety of kinetochore-defective mutants, suggesting that Dam1/DASH somehow corrects abnormalities on the kinetochore (Kobayashi et al., 2007). Hos2 depletion reduced the frequency of kinetochore association with the spindle in prometaphase. Thus, the Dam1/DASH complex appears to be involved in a molecular mechanism that facilitates spindle–kinetochore association. We have shown previously that the deletion of hos2+ in mis mutant strains defective in inner-kinetochore resulted in the dis phenotype, in which the SAC was active (Kobayashi et al., 2007). This dis phenotype may be caused by the accumulation of unattached kinetochores that should have been repaired by the Dam1/DASH-dependent error-correcting mechanism. How the Dam1/DASH facilitates spindle–kinetochore association remains to be clarified. In higher eukaryotes, it has been shown that short microtubule fibers form in the vicinity of kinetochores (Maiato et al., 2004b), and presumably the fibers facilitate interaction between kinetochores and the spindle. If the kinetochore-driven fibers are formed in fission yeast, the Dam1/DASH may be involved in their formation or may mediate lateral interaction between the kinetochore-derived fibers and the microtubules from the SPB. It is also possible that the Dam1/DASH complex promotes the emanation of microtubules toward kinetochores from the SPB. It is noteworthy that normal prometaphase p-movement of the captured kinetochore was observed in Δhos2 cells. Our observations do not deny the possibility that the Dam1/DASH complex is involved in generation of kinetochore-pulling force for the p-movement, but, if so, other molecules, such as Klp5 and Klp6 (Sanchez-Perez et al., 2005), could replace the Dam1/DASH complex. In contrast, the poleward motion of separated kinetochores in anaphase A was severely impaired in Δhos2 strains. These observations indicate that the molecular mechanisms for kinetochore movement in anaphase A may differ from those for p-movement during prometaphase and that the Dam1/DASH complex may play an irreplaceable role in the former. Interestingly, in budding yeast, it has been suggested that dephosphorylation of Ask1, a component of the Dam1/DASH complex, contributes to the modulation of spindle microtubule dynamics and successful anaphase A (Higuchi and Uhlmann, 2005). The Dam1/DASH complex might regulate the dynamics of kinetochore microtubules in anaphase A.
Bub1 may also be involved in the correction of errors in spindle–kinetochore interaction. In fission yeast and mammalian cells, Bub1 was reported to be required for the kinetochore localization of Sgo2 (Kitajima et al., 2004; Huang et al., 2007). Mammalian Sgo2 recruits MCAK, a microtubule depolymerase, to the inner centromere (Huang et al., 2007). Fission yeast Sgo2 is required for the maintenance of the SAC responding tensionless spindle–kinetochore attachment and for kinetochore localization of chromosome passenger proteins, including Aurora B kinase (Kawashima et al., 2007; Vanoosthuyse et al., 2007). Fission yeast Aurora kinase (Ark1) has been reported to be important for the localization of Mad2 on kinetochores and for the resolution of syntelic spindle–kinetochore interaction (Petersen and Hagan, 2003; Hauf et al., 2007). Thus, Bub1 appears to regulate MCAK and/or Aurora B directly or indirectly, and consequently to disassemble syntelically attached microtubules.
For the effective establishment of chromosome biorientation, a suitable error-correcting mechanism must be chosen depending on the state of the kinetochore-spindle interaction. That is, a mechanism that destabilizes kinetochore-microtubule attachment is required to resolve syntelic interaction, whereas a mechanism that increases the probability of a kinetochore interacting with spindle microtubules should be activated on unattached kinetochores and monotelic kinetochores. Although further studies are required to determine the molecular mechanisms specifying the status of kinetochore-spindle interaction (i.e., unattached, monotelic, syntelic, merotelic, or bioriented), we speculate that Bub1 and the Dam1/DASH complex are integral parts of such a mechanism. These molecules are also involved in regulation of the Mad2 checkpoint. Thus, we suppose that these molecules couple Mad2 checkpoint control with the error-correcting mechanisms. The most suitable mechanism can be selected on each kinetochore. For example, on unattached kinetochores, the Dam1/DASH complex may activate the mechanism that facilitates kinetochore-spindle association and also trigger the Mad2 checkpoint pathway to block cell cycle progression until the kinetochore is captured by the spindle. Similarly, Bub1 may couple the cell cycle block with the resolution of erroneous spindle–kinetochore interaction on syntelic kinetochores. Such coordination of the SAC with error-correcting machinery enables the efficient establishment of chromosome biorientation, while avoiding unnecessary cell cycle delay.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. A. M. Carr (University of Sussex, Brighton, United Kingdom), P. Hentges (University of Sussex), P. Nurse (Rockefeller University, New York, NY), and M. Yanagida (Kyoto University, Kyoto, Japan) for providing the materials used in this study. This work was supported by a Grant-in-Aid for “Time's Arrow and Biosignaling” in Precursory Research for Embryonic Science and Technology Agency (K.T.), a Grant-in-Aid for Scientific Research on Priority Areas “Nuclear Dynamics” (K.T.), “Bio-nanosystems” and “Chromosome Cycle” (S.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and a Grant-in-Aid for Young Scientists (B) (S.S.) from the Japan Society for the Promotion of Science.
Abbreviations used:
- CBZ
carbendazim
- HU
hydroxyurea
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0298) on July 16, 2008.
REFERENCES
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Aoki K., Nakaseko Y., Kinoshita K., Goshima G., Yanagida M. CDC2 phosphorylation of the fission yeast dis1 ensures accurate chromosome segregation. Curr. Biol. 2006;16:1627–1635. doi: 10.1016/j.cub.2006.06.065. [DOI] [PubMed] [Google Scholar]
- Asakawa K., Toya M., Sato M., Kanai M., Kume K., Goshima T., Garcia M. A., Hirata D., Toda T. Mal3, the fission yeast EB1 homologue, cooperates with Bub1 spindle checkpoint to prevent monopolar attachment. EMBO Rep. 2005;6:1194–1200. doi: 10.1038/sj.embor.7400540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Brew C., Wolyniak M., Desai A., Anderson S., Muster N., Yates J. R., Huffaker T. C., Drubin D. G., Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Waters J. C., Salmon E. D., Murray A. W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Howell B. J., Canman J. C., Hickey J. M., Fang G., Salmon E. D. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T. J., Walczak C. E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Ding R., McDonald K. L., McIntosh J. R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory M. R., Morphew M., Joseph J. D., Means A. R., Davis T. N. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 2002;13:47–58. [PubMed] [Google Scholar]
- Franco A., Meadows J. C., Millar J. B. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J. Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- Furuya K., Takahashi K., Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Reyes C., Courtheoux T., Goldstone S., Gay G., Serrurier C., Tournier S. Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring dam1 and klp2. Mol. Biol. Cell. 2008;19:1646–1662. doi: 10.1091/mbc.E07-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002a;21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 2002b;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths K., Masuda H., Dhut S., Toda T. Fission yeast dam1–A8 mutant is resistant to and rescued by an anti-microtubule agent. Biochem. Biophys. Res. Commun. 2008;368:670–676. doi: 10.1016/j.bbrc.2008.01.156. [DOI] [PubMed] [Google Scholar]
- Grishchuk E. L., McIntosh J. R. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Biswas A., Langegger M., Kawashima S. A., Tsukahara T., Watanabe Y. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 2007;26:4475–4486. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Toda T., Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Huang H., Feng J., Famulski J., Rattner J. B., Liu S. T., Kao G. D., Muschel R., Chan G. K., Yen T. J. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J. Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L. H., Lau L. F., Smith D. L., Mistrot C. A., Hardwick K. G., Hwang E. S., Amon A., Murray A. W. Budding yeast Cdc20, a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Ikui A. E., Furuya K., Yanagida M., Matsumoto T. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 2002;115:1603–1610. doi: 10.1242/jcs.115.8.1603. [DOI] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Tanaka T. U., Lechner J., Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V. L., Scott M. I., Holt S. V., Hussein D., Taylor S. S. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Kadura S., He X., Vanoosthuyse V., Hardwick K. G., Sazer S. The A78V mutation in the Mad3-like domain of Schizosaccharomyces pombe Bub1p perturbs nuclear accumulation and kinetochore targeting of Bub1p, Bub3p, and Mad3p and spindle assembly checkpoint function. Mol. Biol. Cell. 2005;16:385–395. doi: 10.1091/mbc.E04-07-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S. A., Tsukahara T., Langegger M., Hauf S., Kitajima T. S., Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Lin D. P., Matsumoto S., Kitazono A., Matsumoto T. Fission yeast Slp1, an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King J. M., Nicklas R. B. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 2000;113(Pt 21):3815–3823. doi: 10.1242/jcs.113.21.3815. [DOI] [PubMed] [Google Scholar]
- Kitajima T. S., Kawashima S. A., Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Saitoh S., Ogiyama Y., Soejima S., Takahashi K. The fission yeast DASH complex is essential for satisfying the spindle assembly checkpoint induced by defects in the inner-kinetochore proteins. Genes Cells. 2007;12:311–328. doi: 10.1111/j.1365-2443.2007.01053.x. [DOI] [PubMed] [Google Scholar]
- Krawchuk M. D., Wahls W. P. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Li Y., Bachant J., Alcasabas A. A., Wang Y., Qin J., Elledge S. J. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Liu S. T., Hittle J. C., Jablonski S. A., Campbell M. S., Yoda K., Yen T. J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003;5:341–345. doi: 10.1038/ncb953. [DOI] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J. R., 3rd, He X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J. M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C. E. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- Maiato H., DeLuca J., Salmon E. D., Earnshaw W. C. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004a;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- Maiato H., Rieder C. L., Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004b;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- May K. M., Hardwick K. G. The spindle checkpoint. J. Cell Sci. 2006;119:4139–4142. doi: 10.1242/jcs.03165. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Draviam V. M., Sorger P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Millband D. N., Hardwick K. G. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka H., Takeuchi M., Kinoshita K., Nakaseko Y., Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A. F., Murray A., Chikashige Y., Yamashita Y. M., Hiraoka Y., Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Goshima G., Morishita J., Yanagida M. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Ward S. C. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J., Hagan I. M. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 2003;13:590–597. doi: 10.1016/s0960-9822(03)00205-7. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S., Bimbo A., Balasubramanian M. K., Oliferenko S. A potential tension-sensing mechanism that ensures timely anaphase onset upon metaphase spindle orientation. Curr. Biol. 2004;14:69–74. doi: 10.1016/j.cub.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Salmon E. D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M. A., Springer G. H., Granada B., Piston D. W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Ishii K., Kobayashi Y., Takahashi K. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol. Biol. Cell. 2005;16:3666–3677. doi: 10.1091/mbc.E05-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Takahashi K., Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Salmon E. D., Cimini D., Cameron L. A., DeLuca J. G. Merotelic kinetochores in mammalian tissue cells. Philos. Trans. R Soc. Lond. B Biol. Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., Meadows J. C., Franco-Sanchez A., Fleig U., Toda T., Millar J. B. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R. H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D. A., Andreassen P. R., Lacroix F. B., Wilson L., Margolis R. L. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E. S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T. U. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C., Tanaka T. U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Tange Y., Niwa O. Novel mad2 alleles isolated in a Schizosaccharomyces pombe gamma-tubulin mutant are defective in metaphase arrest activity, but remain functional for chromosome stability in unperturbed mitosis. Genetics. 2007;175:1571–1584. doi: 10.1534/genetics.106.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Tournier S., Gachet Y., Buck V., Hyams J. S., Millar J. B. Disruption of astral microtubule contact with the cell cortex activates a Bub1, Bub3, and Mad3-dependent checkpoint in fission yeast. Mol. Biol. Cell. 2004;15:3345–3356. doi: 10.1091/mbc.E04-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda Y., Furuya K., Goshima G., Nagao K., Takahashi K., Yanagida M. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 2002;12:347–358. doi: 10.1016/s0960-9822(02)00692-9. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., Prykhozhij S., Hardwick K. G. Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Mol. Biol. Cell. 2007;18:1657–1669. doi: 10.1091/mbc.E06-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Valsdottir R., Javerzat J. P., Hardwick K. G. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol. Cell. Biol. 2004;24:9786–9801. doi: 10.1128/MCB.24.22.9786-9801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., Drubin D. G., Nogales E., Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Decottignies A., Nurse P. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J. 2003;22:1075–1087. doi: 10.1093/emboj/cdg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- Yu H., Tang Z. Bub1 multitasking in mitosis. Cell Cycle. 2005;4:262–265. [PubMed] [Google Scholar]
- Yu H. G., Muszynski M. G., Kelly Dawe R. The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 1999;145:425–435. doi: 10.1083/jcb.145.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Thiele D. J. Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 2001;276:20529–20535. doi: 10.1074/jbc.M102004200. [DOI] [PubMed] [Google Scholar]
- Zhou J., Yao J., Joshi H. C. Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 2002;115:3547–3555. doi: 10.1242/jcs.00029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.