Abstract
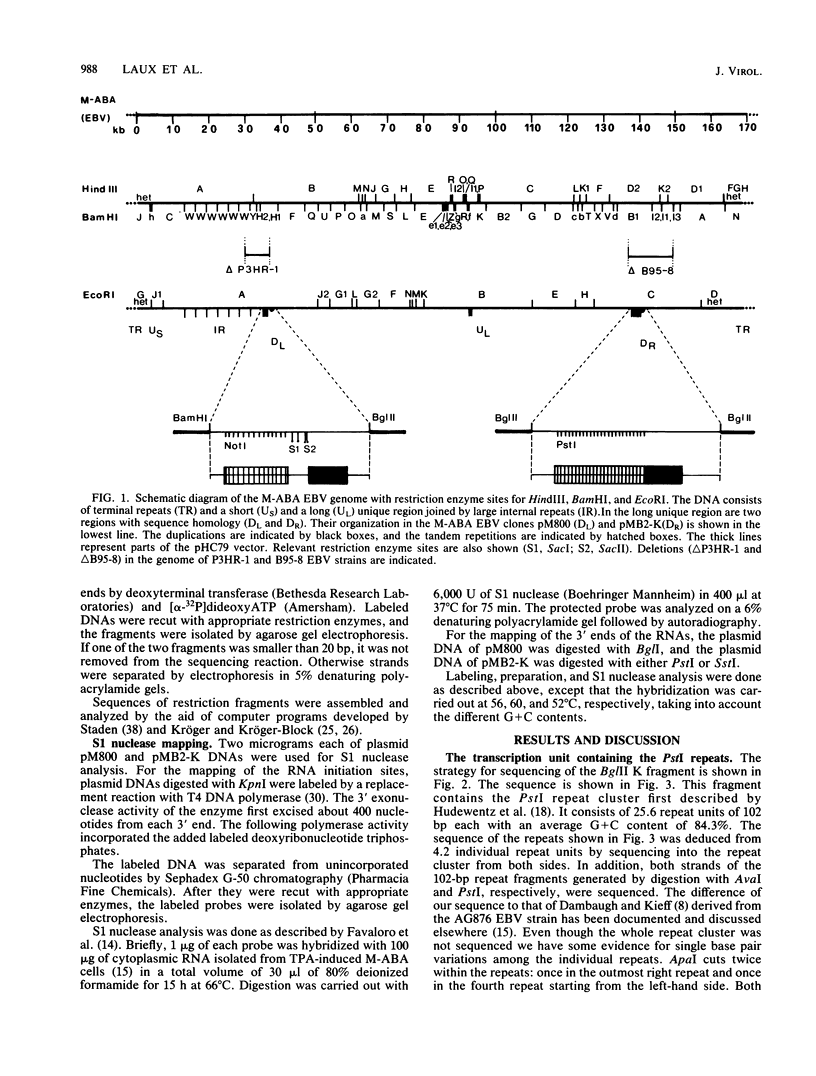
Two regions of the Epstein-Barr virus (EBV) genome carrying partially homologous clusters of short tandem repeats (NotI and PstI repeats) flanked by 1044 and 1045 base pairs with almost complete homology (DL and DR, left and right duplication, respectively) were most abundantly transcribed into poly(A)+ mRNA after induction with the tumor promoter 12-O-tetradecanoyl-phorbol-13-acetate. The nucleotide sequence of both repeat clusters and the conserved upstream regulatory sequences from the M-ABA EBV strain are presented. Nearly the whole part of the sequences coding for the RNAs is covered by the NotI and PstI repeats, respectively. The regulatory sequences for these genes are located in the homologous regions of 1044 and 1045 base pairs (DL and DR, respectively). A CAAT box, a TATA box, and other herpes simplex virus-like elements were identified for both transcription units. The initiation points and the 3' ends of both inducible RNAs were mapped by S1 nuclease analysis. Both genes have open reading frames and may potentially code for proteins with repetitive amino acid compositions. The structure of these two inducible EBV genes is discussed, and an evolutionary model is proposed for the generation of gene duplication in the M-ABA strain of EBV.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adldinger H. K., Delius H., Freese U. K., Clarke J., Bornkamm G. W. A putative transforming gene of Jijoye virus differs from that of Epstein-Barr virus prototypes. Virology. 1985 Mar;141(2):221–234. doi: 10.1016/0042-6822(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Zimber U., Hudewentz J., Epstein M. A. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980 Sep;35(3):603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Draper K. G., Banks L., Powell K. L., Cohen G., Eisenberg R., Wagner E. K. High-resolution characterization of herpes simplex virus type 1 transcripts encoding alkaline exonuclease and a 50,000-dalton protein tentatively identified as a capsid protein. J Virol. 1983 Dec;48(3):591–603. doi: 10.1128/jvi.48.3.591-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford D. H., Epstein M. A., Bornkamm G. W., Achong B. G., Finerty S., Thompson J. L. Biological and biochemical observations on isolates of EB virus from the malignant epithelial cells of two nasopharyngeal carcinomas. Int J Cancer. 1979 Sep 15;24(3):294–302. doi: 10.1002/ijc.2910240305. [DOI] [PubMed] [Google Scholar]
- Dambaugh T. R., Kieff E. Identification and nucleotide sequences of two similar tandem direct repeats in Epstein-Barr virus DNA. J Virol. 1982 Dec;44(3):823–833. doi: 10.1128/jvi.44.3.823-833.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Farrell P. J., Bankier A., Séguin C., Deininger P., Barrell B. G. Latent and lytic cycle promoters of Epstein-Barr virus. EMBO J. 1983;2(8):1331–1338. doi: 10.1002/j.1460-2075.1983.tb01588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P. J., Deininger P. L., Bankier A., Barrell B. Homologous upstream sequences near Epstein-Barr virus promoters. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1565–1569. doi: 10.1073/pnas.80.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Freese U. K., Laux G., Hudewentz J., Schwarz E., Bornkamm G. W. Two distant clusters of partially homologous small repeats of Epstein-Barr virus are transcribed upon induction of an abortive or lytic cycle of the virus. J Virol. 1983 Dec;48(3):731–743. doi: 10.1128/jvi.48.3.731-743.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty D., Peifer M. A., Rubenstein I., Messing J. The primary structure of a plant storage protein: zein. Nucleic Acids Res. 1981 Oct 10;9(19):5163–5174. doi: 10.1093/nar/9.19.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. D., Lazarowitz S. G., Hayward G. S. Organization of the Epstein-Barr virus DNA molecule. II. Fine mapping of the boundaries of the internal repeat cluster of B95-8 and identification of additional small tandem repeats adjacent to the HR-1 deletion. J Virol. 1982 Jul;43(1):201–212. doi: 10.1128/jvi.43.1.201-212.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudewentz J., Delius H., Freese U. K., Zimber U., Bornkamm G. W. Two distant regions of the Epstein-Barr virus genome with sequence homologies have the same orientation and involve small tandem repeats. EMBO J. 1982;1(1):21–26. doi: 10.1002/j.1460-2075.1982.tb01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M., Kieff E. Mapping of polypeptides encoded by the Epstein-Barr virus genome in productive infection. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5698–5702. doi: 10.1073/pnas.79.18.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Hayward S. D. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J Virol. 1983 Oct;48(1):135–148. doi: 10.1128/jvi.48.1.135-148.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. D., Griffin B. E. Clustered repeat sequences in the genome of Epstein Barr virus. Nucleic Acids Res. 1983 Jun 25;11(12):3919–3937. doi: 10.1093/nar/11.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., Cox G. N., Hirsh D. Comparisons of the complete sequences of two collagen genes from Caenorhabditis elegans. Cell. 1982 Sep;30(2):599–606. doi: 10.1016/0092-8674(82)90256-2. [DOI] [PubMed] [Google Scholar]
- Kröger M., Kröger-Block A. A flexible new computer program for handling DNA sequence data. Nucleic Acids Res. 1982 Jan 11;10(1):229–236. doi: 10.1093/nar/10.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger M., Kröger-Block A. Simplified computer programs for search of homology within nucleotide sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):193–201. doi: 10.1093/nar/12.1part1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C., Simonsen C. C., Levinson A. D. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984 May 3;309(5963):82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- Lizardi P. M. Genetic polymorphism of silk fibroin studied by two-dimensional translation pause fingerprints. Cell. 1979 Oct;18(2):581–589. doi: 10.1016/0092-8674(79)90074-6. [DOI] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Structural features of the herpes simplex virus alpha gene 4, 0, and 27 promoter-regulatory sequences which confer alpha regulation on chimeric thymidine kinase genes. J Virol. 1982 Dec;44(3):939–949. doi: 10.1128/jvi.44.3.939-949.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Clements J. B. DNA sequence homology between two co-linear loci on the HSV genome which have different transforming abilities. EMBO J. 1983;2(11):1953–1961. doi: 10.1002/j.1460-2075.1983.tb01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki L. S., Svec P., Nussenzweig R. S., Nussenzweig V., Godson G. N. Structure of the plasmodium knowlesi gene coding for the circumsporozoite protein. Cell. 1983 Oct;34(3):815–822. doi: 10.1016/0092-8674(83)90538-x. [DOI] [PubMed] [Google Scholar]
- Polack A., Hartl G., Zimber U., Freese U. K., Laux G., Takaki K., Hohn B., Gissmann L., Bornkamm G. W. A complete set of overlapping cosmid clones of M-ABA virus derived from nasopharyngeal carcinoma and its similarity to other Epstein-Barr virus isolates. Gene. 1984 Mar;27(3):279–288. doi: 10.1016/0378-1119(84)90072-6. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. The end of the message and beyond. Nature. 1984 Feb 2;307(5950):412–413. doi: 10.1038/307412a0. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N., Dambaugh T., Kieff E. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell. 1980 Nov;22(1 Pt 1):257–267. doi: 10.1016/0092-8674(80)90173-7. [DOI] [PubMed] [Google Scholar]
- Seibl R., Wolf H. Mapping of Epstein-Barr virus proteins on the genome by translation of hybrid-selected RNA from induced P3HR1 cells and induced Raji cells. Virology. 1985 Feb;141(1):1–13. doi: 10.1016/0042-6822(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Roth M. B., Manning R. F., Gage L. P. Alleles of the fibroin gene coding for proteins of different lengths. Cell. 1979 Jun;17(2):407–413. doi: 10.1016/0092-8674(79)90167-3. [DOI] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sümegi J., Wieslander L., Daneholt B. A hierarchic arrangement of the repetitive sequences in the Balbiani ring 2 gene of Chironomus tentans. Cell. 1982 Sep;30(2):579–587. doi: 10.1016/0092-8674(82)90254-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Whitton J. L., Clements J. B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984 Feb 24;12(4):2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Avvedimento V. E., Mudryj M., Ohkubo H., Vogeli G., Irani M., Pastan I., de Crombrugghe B. The collagen gene: evidence for its evolutinary assembly by amplification of a DNA segment containing an exon of 54 bp. Cell. 1980 Dec;22(3):887–892. doi: 10.1016/0092-8674(80)90565-6. [DOI] [PubMed] [Google Scholar]
- Ycas M. Origin of periodic proteins. Fed Proc. 1976 Aug;35(10):2139–2140. [PubMed] [Google Scholar]