Abstract
Background
One potential site of convergence of the nicotine and alcohol actions is the family of the neuronal nicotinic acetylcholine receptors. Our study examines the genetic association between variations in the genomic region containing the CHRNA5, A3 and B4 gene cluster (A5A3B4) and several phenotypes of alcohol and tobacco use in an ethnically diverse young adult sample. Significant results were then replicated in a separate adult population-representative sample.
Methods
In a selected sample, nine single nucleotide polymorphisms (SNPs) were tested for association with various nicotine and alcohol phenotypes, including age of initiation and measures of frequency, quantity and subjective responses to the substances. Analysis was conducted using the statistical genetics program WHAP in the full sample (1075 subjects) including ethnicities as covariates and within each ethnic group sub-sample. Replication of the significant results in a separate population-based sample was carried out using the PBAT statistical genetics program.
Results
Two linked SNPs (rs8023462 and rs1948) located in a conserved region of the A5A3B4 gene cluster, significantly predicted early age of initiation for tobacco with a hazard ratio (HR) of 1.35 (95%CI;1.08–1.70) for the TT genotype of rs8023462 and a HR of 1.29 (95%CI;1.01–1.63) for the CC genotype of rs1948. These findings were then replicated in a separate population-representative sample, showing rs1948 and rs8023462 to be associated with age of initiation for both tobacco and alcohol use (p < 0.01 and p < 0.001).
Conclusion
Variations in A5A3B4 genes may influence behaviors that promote early age of experimentation with drugs.
Introduction
Alcohol and tobacco are the most commonly used drugs in the world and a substantial proportion of those who use these substances go on to develop dependence on them (1). The fact that genetic factors contribute to tobacco and alcohol problem use has been well established through adoption, twin, and family studies (2–10). Additionally, behavioral genetics studies strongly suggest that problem use of alcohol and tobacco may be due in part to genetic factors common to the etiology of use of both substances (11–18). However, our understanding of the specific genetic factors and underlying molecular mechanisms remains limited.
The neuronal nicotinic acetylcholine receptors (nAChRs ) belong to the large superfamily of ligand-gated ion channels that bind the neurotransmitter acetylcholine and the alkaloid nicotine. Different combinations of subunits generate subtypes of nAChRs with diverse functional and pharmacological properties, which in vivo may have selective roles in specific brain pathways. The phylogenetically conserved cluster of nAChRs subunit genes, the α5 α3 and β4 gene-cluster (A5A3B4), encodes heteromeric channels important in fast cholinergic synaptic transmission. The three subunits are co-expressed in autonomic ganglia and several structures of the brain (19).
In this study, we first used a young adult Colorado based sample to test individual single nucleotide polymorphisms (SNPs) for association with various nicotine and alcohol phenotypes, including age of initiation, DSM-IV dependence symptoms, quantity, frequency, and measures of response to the substances in the period shortly after initiation. Significant results with early age of drug initiation were subsequently replicated in a separate sample representative of the US population, underscoring the significance of this association.
Materials and Methods
Center for the Genetics of Antisocial Drug Dependence
Participants
We evaluated 1075 unrelated individuals, all participants in the Center for the Genetics of Antisocial Drug Dependence (CADD), an ongoing multicomponent, collaborative study at the University of Colorado (20,21). The pool of potential subjects encompassed over 5000 youth; we selected for inclusion in this study those assessed between ages 17 and 21 (mean age 18 ± 1.50). A more detailed description of this sample has been published elsewhere (22). A description of the study was presented to all subjects, who signed written informed assent (minors) or consent (adults) to participate. Table 1 shows the characteristics of the CADD sample used for this study.
Table 1.
Characteristics of the Colorado CADD sampler.
| Sample | Male (%) | Female (%) | Control (%) | Clinical (%) |
|---|---|---|---|---|
| Pooled (1075) | 625 (58) | 450 (42) | 792(74) | 283 (26) |
| Caucasian (775) | 452 (58) | 329 (42) | 624 (79) | 151 (21) |
| Hispanic (169) | 115 (68) | 54 (32) | 80 (47) | 89 (53) |
| African-American (43) | 27 (63) | 16 (37) | 19 (44) | 24 (56) |
Assessments
Substance use patterns (e.g., onset and frequency) were assessed using the Composite International Diagnostic Interview - Substance Abuse Module (CIDI-SAM), a structured, face-to-face diagnostic assessment designed to be administered by trained, lay interviewers (23). This assessment procedure has been shown to be valid for adolescent subjects (24). Subjects were asked also questions related to their subjective responses to each drug (25) that were subjected to principal-components factor analysis. Three factors were generated for each substance (26), as indicated in the Supplementary Table 1 online, with a summary of all phenotypes.
National Youth Survey – Family Study
Participants
Significant SNP associations in the CADD sample were subsequently examined in a genetic supplemental sample participating in the National Youth Survey Family Study (NYS-FS) (27,28). The NYS is a nationally representative probability-sample of subjects aged 11–17 in 1976 and living in the United States in 1977. In 2002, a follow-up interview was conducted (35–44 years) during which behavioral data and DNA samples were collected on a voluntary basis by buccal swabs. A total of 1071 individuals both agreed to follow-up interviews and provided DNA samples; 990 of these had tried alcohol, and 856 had tried cigarettes. The sample consists of 227 families with sibships ranging from 2–5 offspring per family (592) and 479 individuals without siblings..
Assessments
Alcohol and tobacco use behaviors were assessed during a face-to-face structured interview including an adaptation of the CIDI-SAM (23). Two age of onset questions were used. The first was asked in the initial phase of the interview: “How old were you when you first tried tobacco/alcohol?”, referred to here as “Age first tobacco (or alcohol)”. The second was asked in the section of the interview devoted to the particular substance, and was “How old were you when you began smoking?” and “How old were you when you first had any wine/beer/other alcohol at least once a month (for 6 months or more)?”, referred to here as “Age of initiation for smoking (or drinking)”. There were very few individuals who reported ages of “first” or “initiation” for tobacco who did not also report a similar age for smoking.
Genotyping
Candidate polymorphisms for the CHRNA5/A3/B4 genes were identified using the SNPbrowser Software version 3.5 from Applied Biosystems (http://www.appliedbiosystems.com) and the public database, dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). The CHRNA5 and CHRNA3 genes partially overlap in a tail-to-tail configuration, sharing their 3′ ends. These two genes are transcribed in opposite directions and are clustered on chromosome 15q25.1 with the CHRNB4 gene (29,30). The structures of the CHRNA5/A3/B4 genes, and the SNPs selected, are shown in Figure 1. Genomic DNA was preamplified using the method of Zheng et al. (31,32). TaqMan®assays for allelic discrimination (Applied Biosystems) were used to determine SNP genotypes.
Figure 1.
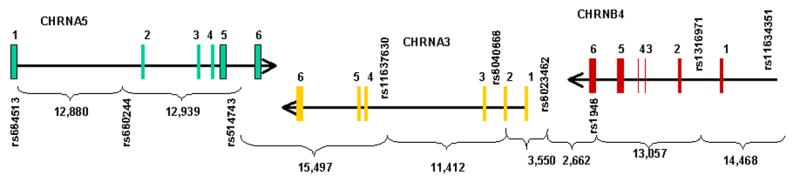
Schematic representation of the CHRNA5/A3B4 gene cluster structure. Boxes represent exons separated by intronic regions. Nine SNPs were genotyped in the cluster, with their reference sequence numbers and gene locations indicated. The number of nucleotide base pairs (bp) between each SNP is also indicated.
Analytic Methods
Single marker and haplotype analyses of the CADD sample were performed using the statistical genetics program WHAP (http://pngu.mgh.harvard.edu/~purcell/whap/) (33–35). All analyses were first conducted on the entire sample using the ethnic group information as a covariate. A secondary analysis was conducted separately for each group.. All phenotypic measures were age- and sex-corrected based on the distribution of the community sample data (i.e., z-scores of clinical subjects were expressed as deviations from the means in the community samples).
All reported p-values are empirical values obtained from completing 500 permutations except in the supplementary online table (supplementary Table 2). Significance levels were set at 0.0085 for the pooled, Caucasian and Hispanic samples and at 0.0073 for the African-American sample. These levels were estimated using the SNP spectral decomposition (SNPSpD) approach (36), which maintains the Type I error rate at 5% in the context of multiple correlated markers. Supplementary Table 3 online indicates the intercorrelation matrix for the alcohol and tobacco phenotypes of the CADD sample.
For the NYS-FS sample, single marker analyses were performed using PBAT time-to-onset analysis (37,38) to take full advantage of the sibling structure and family information. All analyses were first conducted on the entire sample using the self-reported ethnic group information as covariates, followed by separate analyses for the Caucasians and each sex. All reported p-values are asymptotic values, not adjusted for multiple testing. Only the four phenotypes reported here were tested in the NYS-FS (for nine SNPs).
Pairwise linkage disequilibrium (r2) for both samples (CADD and NYS-FS) was calculated using Haploview (39). Haplotype analysis in the CADD sample was carried out with the WHAP program, which assigns weighted haplotypes to each individual. In order to reduce the number of tests performed, we focused the haplotype analysis on the two phenotypes (“age at first use”) that provided the most significant results from the single marker analysis.
Time-to-Onset Analysis
Time-to-onset analyses focused on the most significant SNPs (rs8023462, rs1948) from the single-SNP analysis of the CADD sample (above). Censored subjects were defined as those who did not start to drink or smoke at the time of their study interview, and were assigned the age at the time of the interview. Based on the evidence from the single-SNP analyses in WHAP, genotypes were coded as recessive for both rs8023462 and rs1948 markers. A visual inspection of the data using Kaplan-Meier survival curves were generated using PROC LIFETEST of SAS version 9.1. Estimates of hazard ratios (HRs) were carried out using PROC PHREG of SAS version 9.1. In the NYS-FS sample, time-to-onset analysis was carried out with the program PBAT (38) as described above.
Results
Individual SNP frequencies in different ethnic groups
The allele frequencies and their relative positions for the nine polymorphisms studied in the gene cluster are listed in Table 3 (CADD) and Table 4 (NYS-FS replication). All markers were in Hardy-Weinberg equilibrium. However, frequency calculations in the CADD sample revealed a significant difference in allele frequency for the SNP markers between the major ethnic groups: Caucasians, Hispanics and African-Americans (last two columns of Table 3). In view of these allelic frequency differences, we analyzed the data with WHAP using the pooled sample (1075 subjects with ethnicities included as covariates), the Caucasian sample (775), the African-American sample (43) and the Hispanic sample (168) separately. The allele frequencies in the NYS were similar to those found in CADD and reported in the literature (Table 4). Only rs11634351 showed significantly different allele frequencies between African-Americans and Caucasians (tested by χ2), so the NYS-FS sample was not divided into groups by self reported ethnicity, though ethnicity was included as a covariate in the association analysis. The representation of other ethnic groups, such as Hispanics, was too small to obtain accurate allele frequencies and test for frequency differences.
Table 3.
Markers and allele frequencies in the whole sample and ethnic groups.
| Reference sequence | Description | Gene | Relative SNP position | MAF whole sample | MAF Caucasian | MAF AA | MAF Hispanic | p-value of MAFa Hisp. vs Cauca. | p-value of MAFb AA vs. Cauca. |
|---|---|---|---|---|---|---|---|---|---|
| rs684513 | intron 1 | CHRNA5 | 0 | 0.251 | 0.228 | 0.112 | 0.385 | 0.0003 | 0.006 |
| rs680244 | intron 1 | CHRNA5 | 12880 | 0.401 | 0.43 | 0.475 | 0.286 | 0.007 | 0.336 |
| rs514743 | intron 5 | CHRNA5 | 25819 | 0.34 | 0.36 | 0.276 | 0.277 | 0.11 | 0.110 |
| rs11637630 | intron 3 | CHRNA3 | 41316 | 0.294 | 0.242 | 0.232 | 0.482 | <0.0001 | 0.789 |
| rs8040868 | Exon 2 | CHRNA3 | 52728 | 0.368 | 0.386 | 0.402 | 0.256 | 0.013 | 0.784 |
| rs8023462 | Intergenic | CHRNA3/B4 | 56278 | 0.33 | 0.364 | 0.176 | 0.253 | 0.023 | 0.0002 |
| rs1948 | UTR-3′ | CHRNB4 | 58940 | 0.321 | 0.35 | 0.22 | 0.248 | 0.044 | 0.0088 |
| rs1316971 | intron 1 | CHRNB4 | 71997 | 0.277 | 0.211 | 0.475 | 0.476 | <0.0001 | <0.0001 |
| rs11634351 | Upstream | CHRNB4 | 86465 | 0.362 | 0.399 | 0.154 | 0.256 | 0.006 | <0.0001 |
MAF – minor allele frequency.
p-value of the Chi-square statistic that evaluates the differences in allele frequencies between the Hispanic and Caucasian populations.
p-value of the Chi-square statistic that evaluates the differences in allele frequencies between the African-American and Caucasian populations.
Table 4.
Allele frequencies and ethnic differences in the NYS-FS
| Reference sequence (rs) | Location | Gene | MAF sample | MAF Caucasian | MAF AA | P-value Eth. diff |
|---|---|---|---|---|---|---|
| rs684513 | intron 1 | CHRNA5 | 0.203 | 0.214 | 0.182 | 0.57 |
| rs680244 | intron 1 | CHRNA5 | 0.467 | 0.459 | 0.479 | 0.78 |
| rs514743 | intron 5 | CHRNA5 | 0.367 | 0.388 | 0.267 | 0.02 |
| rs11637630 | intron 3 | CHRNA3 | 0.240 | 0.238 | 0.268 | 0.626 |
| rs8040868 | Exon 2 | CHRNA3 | 0.355 | 0.386 | 0.296 | 0.279 |
| rs8023462 | Intergenic | CHRNA3/B4 | 0.366 | 0.38 | 0.236 | 0.006 |
| rs1948 | UTR-3′ | CHRNB4 | 0.358 | 0.357 | 0.250 | 0.100 |
| rs1316971 | intron 1 | CHRNB4 | 0.266 | 0.207 | 0.401 | <0.0001 |
| rs11634351 | Upstream | CHRNB4 | 0.356 | 0.418 | 0.077 | <0.0001 |
Single Marker Analyses in the CADD sample
Single marker analyses performed using WHAP were χ2 (1 degree of freedom) tests with 500 permutation-derived p-values. Results for association tests with all phenotypes examined in the CADD and the nine individual SNPs are presented in the Supplementary Table 2 online. Table 5 presents the significant and noteworthy findings of the single SNP analysis with the age of first use of tobacco and alcohol phenotypes in the CADD sample. Table 6 shows the results with the age of first use and age of initiation phenotypes in the NYS-FS sample. Tests performed in the pooled CADD sample (1075 subjects) included ethnicities as covariates and the bold italicized p-values indicate statistical significance after correction for multiple SNP testing. The adjusted p-value for the 9 SNPs is 0.0085 for the pooled and ethnic sample, according to the SNP spectral decomposition method (36) mentioned above.
Table 6.
Significant genetic associations in the NYS-FS from PBAT*
| Phenotype | Gene | SNP | p-value additive | p-value recessive | Risk allele |
|---|---|---|---|---|---|
| Age first alcohol | CHRNB4 | rs1948 | 0.0278 | 0.017 | C |
| Age first alcohol | CHRNA3/B4 | rs8023462 | 0.007 | 0.001 | T |
| Age init. “drinking” | CHRNB4 | rs1948 | 0.028 | 0.0248 | C |
| Age init. “drinking” | CHRNA3/B4 | rs8023462 | 0.0034 | 0.0008 | T |
| Age init. “drinking” | CHRNA5 | rs514743 | 0.028 | 0.03 | T |
| Age first tobacco | CHRNB4 | rs1948 | 0.024 | 0.0081 | C |
| Age first tobacco | CHRNA3/B4 | rs8023462 | 0.017 | 0.0015 | T |
| Age init. “smoking” | CHRNB4 | rs1948 | 0.0242 | 0.3 | C |
| Age init. “smoking” | CHRNA3/B4 | rs8023462 | 0.014 | 0.0016 | T |
| Age init. “smoking” | CHRNA5 | rs514743 | 0.024 | 0.015 | T |
PBAT incorporates censoring with time-to-onset in the genetic model.
A first examination (see online supplementary Table 2) of the single SNP analysis results points to the synonymous SNP (rs8040868) of the CHRNA3 gene as a variation that may influence both alcohol and tobacco phenotypes. Additionally, the CHRNB4 gene promoter SNP (rs11634351) is the only one showing an association trend with the positive and negative emotions related to alcohol consumption. However, the most interesting results were observed for the age of first use variables.
In Table 5, results of the analysis of the “alcohol age first use” variable with the cluster markers in the pooled sample revealed a trend of association (p ≤ 0.02, not significant after multiple testing adjustment) for rs514743 (CHRNA5), rs8023462 (intergenic) and rs1948 (CHRNB4), and significant association (p<0.0085) for the rs11634351 (CHRNB4) marker. More interestingly, results obtained with the “tobacco age first use” phenotype and the CHRNA5/A3/B4 gene cluster in the CADD pooled sample indicate that rs680244 (CHRNA5), rs514743 (CHRNA5), rs8040868 (CHRNA3), rs8023462 (intergenic) and rs1948 (CHRNB4) are significantly associated with the age of tobacco initiation (p ≤ 0.022), although rs514743 and rs8040868 are not statistically significant after correction for multiple testing. Therefore, these results indicate that three SNPs of the cluster (rs514743, rs8023462 and rs1948) overlap in their putative association with the age of tobacco and alcohol initiation phenotypes.
Table 5.
Single Marker Results for A5A3B4 cluster in pooled and ethnic samples from CADD.
| Sample (N) | Phenotype | SNP (gene location) | LRT df=1 | Beta-estimate | p-value |
|---|---|---|---|---|---|
| Pooled* (1075) | Alcohol age first use | rs514743 (Intron 5) | 5.21 | −0.120 | 0.022 |
| rs8023462 (inter) | 5.35 | −0.123 | 0.020 | ||
| rs1948 (UTR-3′) | 5.78 | −0.129 | 0.016 | ||
| rs11634351 (Upstr) | 7.20 | 0.145 | 0.007 | ||
| Tobacco age first use | rs680244 (Intron 1) | 9.64 | −0.185 | 0.002 | |
| rs514743 (Intron 5) | 6.61 | −0.150 | 0.010 | ||
| rs8040868 (Exon 2) | 5.24 | 0.136 | 0.022 | ||
| rs8023462 (inter) | 12.98 | −0.211 | <0.001 | ||
| rs1948 (UTR-3′) | 10.95 | −0.198 | <0.001 | ||
| Caucasian (775) | Alcohol age first use | rs514743 (Intron 5) | 5.11 | −0.145 | 0.023 |
| rs8023462(inter) | 5.95 | −0.154 | 0.014 | ||
| rs1948 (UTR-3′) | 7.17 | −0.174 | 0.007 | ||
| rs11634351 (Upst) | 6.88 | 0.173 | 0.007 | ||
| Tobacco age first use | rs680244 (Intron 1) | 9.02 | −0.214 | 0.002 | |
| rs514743 (Intron 5) | 5.32 | −0.162 | 0.02 | ||
| rs8040868 (Exon 2) | 3.54 | 0.136 | 0.059 | ||
| rs8023462 (inter) | 12.22 | −0.242 | <0.001 | ||
| rs1948 (UTR-3′) | 10.30 | −0.231 | <0.001 | ||
| Hispanic (168) | Alcohol age first use | rs514743 (Intron 5) | 1.02 | −0.133 | 0.313 |
| rs8023462 (inter) | 1.00 | −0.139 | 0.317 | ||
| rs1948 (UTR-3′) | 0.00 | −0.156 | 1.00 | ||
| rs11634351 (Upst) | 2.77 | 0.222 | 0.096 | ||
| Tobacco age first use | rs680244 (Intron 1) | 4.17 | −0.310 | 0.041 | |
| rs514743 (Intron 5) | 5.94 | −0.365 | 0.014 | ||
| rs8040868 (Exon 2) | 4.07 | 0.297 | 0.043 | ||
| rs8023462 (inter) | 3.27 | −0.275 | 0.070 | ||
| rs1948 (UTR-3′) | 4.09 | −0.300 | 0.043 | ||
| African-American (43) | Alcohol age first use | rs514743 (Intron 5) | 2.058 | 0.355 | 0.151 |
| rs8023462 (inter) | 3.161 | 0.549 | 0.075 | ||
| rs1948 (UTR-3′) | 3.21 | 0.437 | 0.073 | ||
| rs11634351 (Upst) | 7.74 | −0.824 | 0.005 | ||
| Tobacco age first use | rs680244 (Intron 1) | 0.727 | 0.168 | 0.394 | |
| rs514743 (Intron 5) | 0.239 | 0.100 | 0.625 | ||
| rs8040868 (Exon 2) | 0.476 | −0.142 | 0.490 | ||
| rs8023462 (inter) | 1.019 | 0.256 | 0.313 | ||
| rs1948 (UTR-3′) | 0.919 | 0.217 | 0.338 |
Pooled sample (1075) analyzed using ethnicities as covariates in Whap. Adjusted nominal p-value for the pooled, Caucasian and Hispanic samples is 0.0085 and for the African-American sample is 0.0073, according to SNPSpD approach (36). Significant p-values are italicized and bold.
Results of the single marker analysis for the three separate ethnic groups of the study are presented also in Table 5. Empirical permutation p-values were adjusted according to the SNPSpD approach (36). The association trend of the rs8040868 (CHRNA3), rs8023462 (intergenic) and rs1948 (CHRNB4) markers with the age at first time use of alcohol and/or tobacco variables appears to be consistent across the ethnic samples.
Single Marker Analyses in the NYS-FS replication sample
Results from analyses with age of initiation variables for the NYS-FS sample are shown in Table 6. SNPs were initially tested with an additive genetic model, followed by a secondary analysis assuming a recessive model. Under an additive model, markers rs1948 and rs8023462 were associated with age of first alcohol (p=0.028 and p=0.007) and first tobacco use (p=0.024 and p=0.017). The strongest association was between rs8023462 and age of initiation for “drinking” with the T allele modeled as recessive (p=0.0008). Strong evidence for association was also found between rs1948 and rs8023462 and age of first use for alcohol (p=0.017, p=0.001) and tobacco (p=0.0081 and p=0.0015) under a recessive model. Marker rs514743 is associated with age of initiation for “drinking” (p=0.028) and “smoking” (p=0.024) assuming additive allelic effects. The significance of this association was roughly equivalent when modeled as recessive (p=0.015 and p=0.03). The strength of these associations did not depend strongly on whether “age first” or “age of initiation” is used, with the exception of rs514743, which showed no association with age first alcohol nor age first tobacco. The minority groups within the NYS-FS sample were too small for individual analyses, but results did not differ in significance within the Caucasian sample, nor within each sex. Neither the marker rs8040868 located in exon 2 of the CHRNA3 gene nor the marker rs680244 located in intron 1 of CHRNA5 was not found to be significantly associated with age of onset in the NYS-FS sample.
Haplotype structure
Pairwise linkage disequilibrium (LD) estimates, r2, for the gene markers were obtained from Haploview (39) and are shown in Supplementary online Figures 1.1 – 1.5. Significant LD was found for markers rs8023462 (intergenic) and rs1948 (CHRNB4) in the pooled, Caucasian and Hispanic samples. Two additional blocks were found only in the CADD Hispanic sample for markers rs684513 (CHRNA5) and rs680244 (CHRNA5) and for markers rs1316971 (CHRNB4) and rs11634351 (CHRNB4). These results are in agreement with the HapMap (www.hapmap.org) LD estimates for the CHRNA5/A3/B4 locus. Block structure and SNP correlations in the NYS-FS were similar to those shown in the CADD and in the HapMap website (Suplementary Figure 1.5). All SNPs were in HWE.
Haplotype-based tests in the CADD sample
In order to study the possible allele combinations of the CHRNA5/A3/B4 loci that may be associated with age at onset of tobacco/alcohol use, haplotype tests were performed using the WHAP analysis program and a haplotype-frequency cut off of 5%. Haplotype analysis in WHAP is performed using two tests; the primary test is a regression-based analysis of association between haplotype and trait, with one regression coefficient per haplotype. Therefore, for H haplotypes, a primary single omnibus test is performed to test jointly for any difference in haplotype effect, which is a single H-1 degrees of freedom test. As indicated above, the default omnibus test assesses the overall haplotype frequency profile differences in the sample (i.e.; using all haplotypes above 5%) for the phenotypic scores at hand. Using the haplotype block information from the LD analysis (Supplementary Figure 1 online) we tested the “age of first use” variables for tobacco and alcohol with the rs8023462-rs1948 block (markers 6–7, Supplementary Figure 1 online) of the CHRNA5/A3/B4 locus. Significant omnibus results for “age of first use of tobacco” were observed in the pooled (LRT = 10.17, df =1, p= 0.001) and Caucasian sample (LRT = 11.12, df =1, p= 0.0008), but not in the Hispanic sample (LRT = 1.1, df =1, p= 0.3). Regarding the “age of first use of alcohol” variable, omnibus results were modestly significant for the pooled (LRT = 3.64, df =1, p= 0.056) and Caucasian samples (LRT = 4.84, df =1, p= 0.027) but not significant for the Hispanic sample (LRT = 0.49, df =1, p= 0.5).
An alternative haplotype-based hypothesis test focuses on the differences of individual haplotype frequencies. This haplotype-specific test (option -hs in WHAP) performs all possible 1 degree of freedom haplotype-specific tests and can be used to test the effect of each haplotype individually against all others, (i.e. constraining all other haplotypes to have equal β weights). As shown in Supplementary Table 4, there is evidence that two main haplotypes (out of four possible combinations: CC, CT, TC, TT) are significantly associated with the age of first use variables for alcohol and tobacco. In the pooled sample, the common (66.6 %) haplotype CT appears to confer protection (positive β weights, older age) for the early initiation of tobacco use (LRT= 11.221, β= 0.197, p= 0.0008) whereas the slightly less common (32 %) TC haplotype may confer risk for a younger age at drug initiation (β=−0.200, p = 0.0008 and β= −0.113, p= 0.036 for tobacco and alcohol respectively). Haplotype analysis in the ethnic groups indicated that the CT and TC haplotypes were associated with the age for initiation phenotype in Caucasians only (Supplementary Table 4).
Time-to-Onset Results
The effect of the genotypic variants rs8023462 and rs1948 (that form the haplotypes CT and TC) on the relative risk of early onset of drinking and smoking in our study subjects was evaluated with a Cox proportional hazard regression analysis with censoring. Regarding the “age at first drink” variable, 96% of the participants (n=1038) were included in the analysis. Of these, 21.77 % (n=226) participants were censored because at the time of the interview they did not report they had initiated drinking. Mean age at the time of first drink was 14.39 years (SD=2.64). A total of 1044 subjects (97%) were included in the analysis for the variable “age at first tobacco use” survival analysis. Of these, 36.3% were censored (n=379) and the mean age at the time of first use of tobacco was 13.43 years (SD=2.68). Results of the Cox proportional hazard regression model are reported in Table 7. The homozygous TT genotype of the rs8023462 marker emerged as the most potent predictor of early initiation of tobacco use in our sample of young adults aged 17 to 21years (Figure 2.1). The CC genotype of the rs1948 marker located in the 3′UTR of CHRNB4 was also a significant predictor of the age at first tobacco use (Figure 2.2). These genotypes, however, were not significant predictors for the early initiation of alcohol drinking in our sample (Figures of plots shown in Figure 2 of supplementary information online). For the NYS-FS replication sample, the time to onset analyses were performed using PBAT time-to-onset analysis tools and the results are presented in Table 6. The mean age of onset (± SD) for each phenotype of the NYS-FS sample were the following:’ Age first tobacco’, 14.7 (± 4.05); ‘Age first alcohol’ 17.5(± 6.8); ‘Age initiation of smoking’, 17.4(± 5.28) and ‘Age initiation of drinking’, 21.5(± 6.0). Differences in the ages-of-onset observed between samples are likely due to both the ascertained nature of the CADD sample and generational differences between individuals who were teenagers in the late 1970s (NYS-FS) versus in the 1990s (CADD).
Table 7.
Cox proportional hazards model for “tobacco age first use” and “alcohol age first use” in the pooled sample.
| Tobacco age first use | ||
|---|---|---|
| Genotype | Hazard ratio (95% CI) | p-value of ChiSq |
| rs8023462 (TT vs. CT + CC) | 1.355 (1.078, 1.704) | 0.009 |
| rs1948 (CC vs. TC + TT) | 1.287 (1.015, 1.632) | 0.037 |
| Alcohol age first use | ||
| rs8023462 (TT vs. CT + CC) | 1.125 (0.908, 1.394) | 0.3 |
| rs1948 (CC vs. TC + TT) | 1.116 (0.895, 1.394) | 0.3 |
Proportional hazards assumptions were not violated since the proportionality of the predictors was maintained, as indicated by the parallelism of the curves in the survival distribution function plots.
Figure 2.
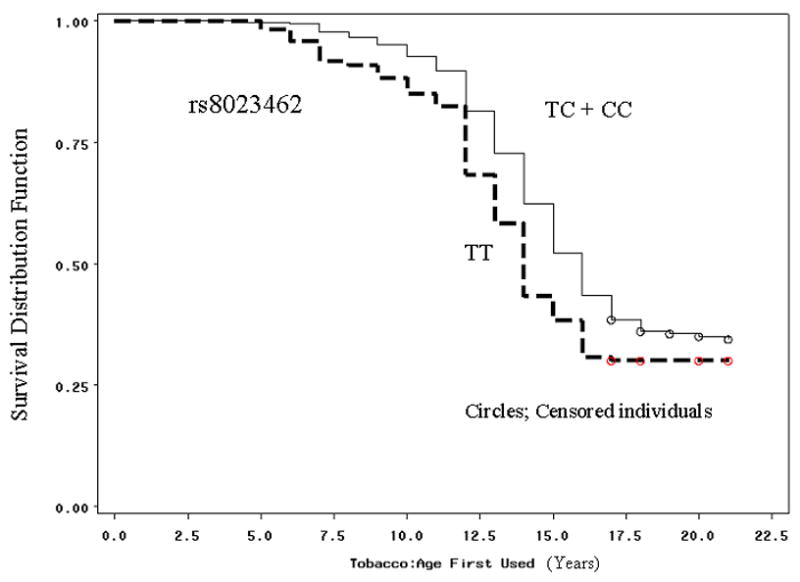
Survival density function plots for all subjects of the study by age of tobacco initiation and a recessive genetic model of markers rs8023462 (2.1) and rs1948 (2.2). The dashed lines represent the early age of initiation genotypes for rs8023462 (TT) and rs1948 (CC). Hazard ratios, confidence intervals and p-values are shown in Table 7. Survival plots for the age of alcohol initiation are shown in the supplementary information (Supplementary Figures 2.1 and 2.2) online at the Journal’s web site.
Discussion
Co-morbidity of tobacco and alcohol use has been recognized for many years (40,41), but the possible underlying common biological mechanisms for tobacco and alcohol use are not well understood. In this report, we present evidence of an association of age of initiation of alcohol and tobacco use phenotypes with the CHRNA5/A3/B4 locus polymorphisms in two separate samples: a selected sample of young adults and a population-representative adult sample.
In an exploratory fashion using the CADD sample, we examined nine SNPs for a possible association with alcohol and tobacco phenotypes, in addition to three factor scores (25). Our results with the single marker analysis using the pooled sample indicated that three SNPs are associated with the age at initiation of both tobacco use and alcohol drinking. These markers are rs514743 (CHRNA5), rs8023462 (intergenic) and rs1948 (CHRNB4), where rs8023462 and rs1948 are in high linkage disequilibrium in the pooled, Caucasian and Hispanic samples. The similar trend of results in both larger ethnic subgroups (Caucasians and Hispanics) underscores the potential importance of the CHRNA5/A3/B4 locus in the initiation of both alcohol and tobacco use in young adults. To provide validation of these findings, we have replicated our results in a separate population-based sample, the NYS-FS sample.
Other genetic association studies, including the recent study by Saccone et al. (42), also identified the rs514743 (CHRNA5) variation as one of the “top association markers with nicotine dependence”. Since molecular studies have shown that the sequence around this SNP is involved in antisense formation between the CHRNA5 and CHRNA3 mRNAs (30), one might speculate that the rs514743 variation in this regulatory sense-antisense mRNA interaction could be involved in protein translation. Another SNP significantly associated with the age at initiation of tobacco use in our present study is rs680244, which has also been associated with nicotine dependence in young adults (43)
The intergenic SNP (rs8023462) is located in the promoter region of CHRNA3 and the downstream region of the CHRNB4 gene, potentially affecting regulatory elements of both loci. This marker is in linkage disequilibrium with the rs1948 SNP of the CHRNB4 untranslated region (3′UTR), located 2,662 bp downstream of rs8023462, (figures 1 and 2) and 80 bp beyond the stop codon of the CHRNB4 transcript. The importance of this tightly linked region in the potential regulation of the CHRNA5/A3/B4 gene cluster stems from transcriptional analysis of nicotinic receptors in rodents, where McDonough and Deneris (44) discovered a novel enhancer positioned in the 3′-untranslated exon of the CHRNB4 gene. The location of this enhancer within the CHRNB4 gene may be under selective pressure for maintaining tight linkage of the clustered neuronal CHRNA5/A3/B4 genes (19,44). To date, there is no functional evidence for a human CHRNB4 3′UTR-enhancer, but this region appears to be conserved between rodents and humans (Dr. Deneris, personal communication). We can only speculate that these sequence variations could be responsible for increased or decreased expression of the α3 subunit in cells of the central nervous system where the α3 protein is known to be found, like the ventral tegmental area (VTA) and the medial habenula, which are components of dopaminergic pathways associated with drug reinforcing actions (45). This possibility needs to be investigated with functional studies.
The potential relevance of the markers rs8023462 and rs1948 in the early age at first use of tobacco is also highlighted in our proportional hazard ratios (Table 7). These results indicate that the TT genotype of rs8023462 and the CC genotype of the rs1948 SNP are significant predictors of the early age at smoking initiation. In fact, the combination of these markers generates a T-C haplotype that our haplotype analysis predicted as a significant risk haplotype for early initiation of tobacco and alcohol use in our pooled and Caucasian samples (supplementary Table 4). Examination of the genotypes and risk alleles in the NYS-FS sample supports the same model as the one observed in the CADD sample.
Elucidation of the genes that contribute to smoking (46) and alcohol drinking (47–49) initiation is critical for disentangling the full etiology of development of these disorders. Furthermore, it is possible that initiation of smoking and drinking may be part of a broader spectrum of phenotypes that includes a vulnerability to developing behavioral problems (50,51). Future studies should be aimed at exploring this possibility, and whether or not these nicotinic receptor variations might contribute to a generalized behavioral disinhibition phenotype.
Supplementary Material
Note: Supplementary information is available at the Biological Psychiatry website.
Table 2.
Characteristics of the NYS-FS sample.
| Sample | Male (%) | Female (%) | Ever Tobacco (%) | Ever Alcohol (%) |
|---|---|---|---|---|
| Pooled (1051) | 506 (49) | 545 (51) | 856 (81) | 990 (94) |
| Caucasian (860) | 410 (48) | 450 (52) | 727 (85) | 824 (96) |
| Hispanic (28) | 11 (39) | 17 (61) | 23 (82) | 27 (96) |
| African-American (132) | 67 (51) | 65 (49) | 82 (62) | 109 (83) |
Acknowledgments
The authors gratefully acknowledge the comments and suggestions made by Dr. Thomas J. Crowley (TJC), Dr. Scott Menard (SM) and Dr. David Huizinga during the preparation of the manuscript.
This work was supported by Colorado Tobacco Research Program IDEA grant 2I-034 and Supplement 4S-003 (MAE), NIH grants DA011015 (TJC), DA012845 (TJC), HD010333, EY012562 (JKH), DA03194 (ACC), MH001865 (SEY), DA13956 (SHR), AA007464 (NRH), DA017637 (NRH), MH15442 (BH), NIH AA11949-03 (NYS-FS, SM) and DA015522 (CJH).
Footnotes
FINANCIAL DISCLOSURES AND POTENTIAL CONFLICTS OF INTEREST. Isabel R. Schlaepfer and, Dr. A. Collins, Dr. Robin P. Corley, Dr. John K. Hewitt, Dr. Christian J. Hopfer, Dr. Jeffrey Lessem, Dr. Mathew B. McQueen Dr. Soo Hyun Rhee, Dr. Nicole R. Hoft and Dr. Marissa A. Ehringer do not have any potential conflicts of interest, financial or otherwise, relevant to the subject matter of this work.
References
- 1.Anthony JC, Echeagaray-Wagner F. Epidemiologic analysis of alcohol and tobacco use. Alcohol Res Health. 2000;24:201–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9:64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- 3.Gynther LM, Hewitt JK, Heath AC, Eaves LJ. Phenotypic and genetic factors in motives for smoking. Behav Genet. 1999;29:291–302. doi: 10.1023/a:1021601715308. [DOI] [PubMed] [Google Scholar]
- 4.Heath AC, Bucholz KK, Madden PA, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–96. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 5.Madden PA, Bucholz KK, Martin NG, Heath AC. Smoking and the genetic contribution to alcohol-dependence risk. Alcohol Res Health. 2000;24:209–14. [PMC free article] [PubMed] [Google Scholar]
- 6.Madden PA, Heath AC. Shared genetic vulnerability in alcohol and cigarette use and dependence. Alcohol Clin Exp Res. 2002;26:1919–21. doi: 10.1097/01.ALC.0000040960.15151.30. [DOI] [PubMed] [Google Scholar]
- 7.Madden PA, Heath AC, Martin NG. Smoking and intoxication after alcohol challenge in women and men: genetic influences. Alcohol Clin Exp Res. 1997;21:1732–41. [PubMed] [Google Scholar]
- 8.True WR, Heath AC, Scherrer JF, et al. Genetic and environmental contributions to smoking. Addiction. 1997;92:1277–87. [PubMed] [Google Scholar]
- 9.True WR, Xian H, Scherrer JF, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–61. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- 10.Kendler KS, Thornton LM, Pedersen NL. Tobacco consumption in Swedish twins reared apart and reared together. Arch Gen Psychiatry. 2000;57:886–92. doi: 10.1001/archpsyc.57.9.886. [DOI] [PubMed] [Google Scholar]
- 11.Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 12.Hopfer CJ, Stallings MC, Hewitt JK. Common genetic and environmental vulnerability for alcohol and tobacco use in a volunteer sample of older female twins. J Stud Alcohol. 2001;62:717–23. doi: 10.15288/jsa.2001.62.717. [DOI] [PubMed] [Google Scholar]
- 13.Madden PA, Heath AC, Martin NG. Smoking and intoxication after alcohol challenge in women and men: genetic influences. Alcohol Clin Exp Res. 1997;21:1732–41. [PubMed] [Google Scholar]
- 14.Swan GE, Carmelli D, Cardon LR. The consumption of tobacco, alcohol, and coffee in Caucasian male twins: a multivariate genetic analysis. J Subst Abuse. 1996;8:19–31. doi: 10.1016/s0899-3289(96)90055-3. [DOI] [PubMed] [Google Scholar]
- 15.Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–90. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- 16.Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60:1256–64. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- 17.Young SE, Rhee SH, Stallings MC, Corley RP, Hewitt JK. Genetic and environmental vulnerabilities underlying adolescent substance use and problem use: general or specific? Behav Genet. 2006;36:603–15. doi: 10.1007/s10519-006-9066-7. [DOI] [PubMed] [Google Scholar]
- 18.Elkins IJ, King SM, McGue M, Iacono WG. Personality traits and the development of nicotine, alcohol, and illicit drug disorders: prospective links from adolescence to young adulthood. J Abnorm Psychol. 2006;115:26–39. doi: 10.1037/0021-843X.115.1.26. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Scott MM, Deneris ES. Shared long-range regulatory elements coordinate expression of a gene cluster encoding nicotinic receptor heteromeric subtypes. Mol Cell Biol. 2006;26:5636–49. doi: 10.1128/MCB.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stallings MC, Corley RP, Dennehey B, et al. A genome-wide search for quantitative trait Loci that influence antisocial drug dependence in adolescence. Arch Gen Psychiatry. 2005;62:1042–51. doi: 10.1001/archpsyc.62.9.1042. [DOI] [PubMed] [Google Scholar]
- 21.Stallings MC, Corley RP, Hewitt JK, et al. A genome-wide search for quantitative trait loci influencing substance dependence vulnerability in adolescence. Drug Alcohol Depend. 2003;70:295–307. doi: 10.1016/s0376-8716(03)00031-0. [DOI] [PubMed] [Google Scholar]
- 22.Schlaepfer IR, Clegg HV, Corley RP, et al. The human protein kinase C gamma gene (PRKCG) as a susceptibility locus for behavioral disinhibition. Addict Biol. 2007;12:200–9. doi: 10.1111/j.1369-1600.2007.00063.x. [DOI] [PubMed] [Google Scholar]
- 23.Cottler LB, Keating SK. Operationalization of alcohol and drug dependence criteria by means of a structured interview. Recent Dev Alcohol. 1990;8:69–83. [PubMed] [Google Scholar]
- 24.Crowley TJ, Mikulich SK, Ehlers KM, Whitmore EA, MacDonald MJ. Validity of structured clinical evaluations in adolescents with conduct and substance problems. J Am Acad Child Adolesc Psychiatry. 2001;40:265–73. doi: 10.1097/00004583-200103000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Lyons MJ, Toomey R, Meyer JM, et al. How do genes influence marijuana use? The role of subjective effects. Addiction. 1997;92:409–17. [PubMed] [Google Scholar]
- 26.Ehringer MA, Rhee SH, Young S, Corley R, Hewitt JK. Genetic and Environmental Contributions to Common Psychopathologies of Childhood and Adolescence: A Study of Twins and Their Siblings. J Abnorm Child Psychol. 2006:1–17. doi: 10.1007/s10802-005-9000-0. [DOI] [PubMed] [Google Scholar]
- 27.Elliott DS, Huizinga D, Menard SW. Multiple problem youth: delinquency, substance use, and mental health problems. New York: Springer-Verlag; 1989. [Google Scholar]
- 28.Huizinga D, Haberstick BC, Smolen A, et al. Childhood maltreatment, subsequent antisocial behavior, and the role of monoamine oxidase A genotype. Biol Psychiatry. 2006;60:677–83. doi: 10.1016/j.biopsych.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Duga S, Solda G, Asselta R, et al. Characterization of the genomic structure of the human neuronal nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster and identification of novel intragenic polymorphisms. J Hum Genet. 2001;46:640–8. doi: 10.1007/s100380170015. [DOI] [PubMed] [Google Scholar]
- 30.Solda G, Boi S, Duga S, et al. In vivo RNA-RNA duplexes from human alpha3 and alpha5 nicotinic receptor subunit mRNAs. Gene. 2005;345:155–64. doi: 10.1016/j.gene.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Zheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev. 2001;10:697–700. [PubMed] [Google Scholar]
- 32.Anchordoquy HC, McGeary C, Liu L, Krauter KS, Smolen A. Genotyping of Three Candidate Genes After Whole-Genome Preamplification of DNA Collected from Buccal Cells. Behav Genet. 2003;33:73–78. doi: 10.1023/a:1021007701808. [DOI] [PubMed] [Google Scholar]
- 33.Curran S, Purcell S, Craig I, Asherson P, Sham P. The serotonin transporter gene as a QTL for ADHD. Am J Med Genet B Neuropsychiatr Genet. 2005;134:42–7. doi: 10.1002/ajmg.b.30118. [DOI] [PubMed] [Google Scholar]
- 34.Sham PC, Rijsdijk FV, Knight J, Makoff A, North B, Curtis D. Haplotype association analysis of discrete and continuous traits using mixture of regression models. Behav Genet. 2004;34:207–14. doi: 10.1023/B:BEGE.0000013734.39266.a3. [DOI] [PubMed] [Google Scholar]
- 35.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2006 doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 36.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–94. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 38.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–9. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Istvan J, Matarazzo JD. Tobacco, alcohol, and caffeine use: a review of their interrelationships. Psychol Bull. 1984;95:301–26. [PubMed] [Google Scholar]
- 41.Battjes RJ. Smoking as an issue in alcohol and drug abuse treatment. Addict Behav. 1988;13:225–30. doi: 10.1016/0306-4603(88)90049-4. [DOI] [PubMed] [Google Scholar]
- 42.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenbaum L, Kanyas K, Karni O, et al. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11:312–22. 223. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- 44.McDonough J, Deneris E. beta43′: An enhancer displaying neural-restricted activity is located in the 3′-untranslated exon of the rat nicotinic acetylcholine receptor beta4 gene. J Neurosci. 1997;17:2273–83. doi: 10.1523/JNEUROSCI.17-07-02273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsson A, Engel JA. Neurochemical and behavioral studies on ethanol and nicotine interactions. Neurosci Biobehav Rev. 2004;27:713–20. doi: 10.1016/j.neubiorev.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 46.Khuder SA, Dayal HH, Mutgi AB. Age at smoking onset and its effect on smoking cessation. Addict Behav. 1999;24:673–7. doi: 10.1016/s0306-4603(98)00113-0. [DOI] [PubMed] [Google Scholar]
- 47.Hingson RW, Heeren T, Winter MR. Age at drinking onset and alcohol dependence: age at onset, duration, and severity. Arch Pediatr Adolesc Med. 2006;160:739–46. doi: 10.1001/archpedi.160.7.739. [DOI] [PubMed] [Google Scholar]
- 48.Schmid B, Hohm E, Blomeyer D, et al. Concurrent alcohol and tobacco use during early adolescence characterizes a group at risk. Alcohol Alcohol. 2007;42:219–25. doi: 10.1093/alcalc/agm024. [DOI] [PubMed] [Google Scholar]
- 49.Sartor CE, Lynskey MT, Heath AC, Jacob T, True W. The role of childhood risk factors in initiation of alcohol use and progression to alcohol dependence. Addiction. 2007;102:216–25. doi: 10.1111/j.1360-0443.2006.01661.x. [DOI] [PubMed] [Google Scholar]
- 50.McGue M, Iacono WG, Legrand LN, Malone S, Elkins I. Origins and consequences of age at first drink. I. Associations with substance-use disorders, disinhibitory behavior and psychopathology, and P3 amplitude. Alcohol Clin Exp Res. 2001;25:1156–65. [PubMed] [Google Scholar]
- 51.Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96:684–95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available at the Biological Psychiatry website.


