Abstract
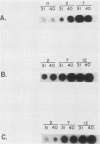
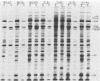
To facilitate the determination of the genomic location of the vaccinia virus gene(s) encoding alpha-amanitin resistance (alpha r) (Villarreal et al., J. Virol. 51:359-366, 1984), a collection of alpha r, temperature-sensitive (ts) mutants were isolated. The premise of these experiments was that mutants might be found whose dual phenotypes were the result of a single or two closely linked mutations. Genetic analyses of the alpha rts mutant library revealed two mutants, alpha rts7 and alpha rts12, that apparently fit this criterion; in alpha rts7 the two lesions were indistinguishable, whereas in alpha rts12 the two mutations were closely linked but separable. Cloned vaccinia virus HindIII DNA fragments were used to marker rescue the temperature-sensitive phenotype of these two dual mutants. The temperature-sensitive lesion of alpha rts7 was rescued by the HindIII N fragment (1.5 kilobases), whereas alpha rts12 was rescued by the neighboring HindIII M fragment (2.0 kilobases). The progeny virions of the alpha rts7 HindIII-N rescue reverted to an alpha-amanitin-sensitive phenotype, whereas the alpha rts12 HindIII-M progeny were still resistant to the drug. Taken together, these data indicate that the gene encoding alpha-amanitin resistance maps to the HindIII N fragment and provides evidence for the existence of essential vaccinia virus genes in a region of the genome previously believed to be nonessential for replication in tissue culture. Biochemical analyses revealed that both mutants were capable of synthesizing DNA as well as early and late viral proteins at the permissive and nonpermissive temperatures. At the nonpermissive temperature alpha rts12 and alpha rts7 were unable to process the major core precursors P94 and P65 into VP62 and VP60.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Condit R. C., Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981 Aug;113(1):224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Drillien R., Spehner D., Kirn A. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology. 1982 Jun;119(2):372–381. doi: 10.1016/0042-6822(82)90096-4. [DOI] [PubMed] [Google Scholar]
- Ensinger M. J. Isolation and genetic characterization of temperature-sensitive mutants of vaccinia virus WR. J Virol. 1982 Sep;43(3):778–790. doi: 10.1128/jvi.43.3.778-790.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard S., Spehner D., Drillien R. Mapping of a vaccinia host range sequence by insertion into the viral thymidine kinase gene. J Virol. 1985 Jan;53(1):316–318. doi: 10.1128/jvi.53.1.316-318.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973 Aug;54(2):536–539. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- Hruby D. E., Guarino L. A., Kates J. R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979 Feb;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruby D. E., Lynn D. L., Kates J. R. Vaccinia virus replication requires active participation of the host cell transcriptional apparatus. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1887–1890. doi: 10.1073/pnas.76.4.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isle H. B., Venkatesan S., Moss B. Cell-free translation of early and late mRNAs selected by hybridization to cloned DNA fragments derived from the left 14 million to 72 million daltons of the vaccinia virus genome. Virology. 1981 Jul 15;112(1):306–317. doi: 10.1016/0042-6822(81)90636-x. [DOI] [PubMed] [Google Scholar]
- Morgan J. R., Roberts B. E. Organization of RNA transcripts from a vaccinia virus early gene cluster. J Virol. 1984 Aug;51(2):283–297. doi: 10.1128/jvi.51.2.283-297.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicali D., Davis S. W., Mercer S. R., Paoletti E. Two major DNA variants present in serially propagated stocks of the WR strain of vaccinia virus. J Virol. 1981 Mar;37(3):1000–1010. doi: 10.1128/jvi.37.3.1000-1010.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington T. H., Follett E. A. Vaccinia virus replication in enucleate BSC-1 cells: particle production and synthesis of viral DNA and proteins. J Virol. 1974 Feb;13(2):488–493. doi: 10.1128/jvi.13.2.488-493.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M., Dales S. Evidence against involvement of host transcription in the replication of vaccinia and herpes simplex viruses. Virology. 1982 Apr 15;118(1):214–218. doi: 10.1016/0042-6822(82)90334-8. [DOI] [PubMed] [Google Scholar]
- Silver M., McFadden G., Wilton S., Dales S. Biogenesis of poxviruses: role for the DNA-dependent RNA polymerase II of the host during expression of late functions. Proc Natl Acad Sci U S A. 1979 Aug;76(8):4122–4125. doi: 10.1073/pnas.76.8.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. W., Hirst G. K. Temperature-sensitive mutants of influenza A virus: isolation of mutants and preliminary observations on genetic recombination and complementation. Virology. 1968 May;35(1):41–49. doi: 10.1016/0042-6822(68)90303-6. [DOI] [PubMed] [Google Scholar]
- Sridhar P., Condit R. C. Selection for temperature-sensitive mutations in specific vaccinia virus genes: isolation and characterization of a virus mutant which encodes a phosphonoacetic acid-resistant, temperature-sensitive DNA polymerase. Virology. 1983 Jul 30;128(2):444–457. doi: 10.1016/0042-6822(83)90269-6. [DOI] [PubMed] [Google Scholar]
- Villarreal E. C., Roseman N. A., Hruby D. E. Isolation of vaccinia virus mutants capable of replicating independently of the host cell nucleus. J Virol. 1984 Aug;51(2):359–366. doi: 10.1128/jvi.51.2.359-366.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]