Abstract
Background
Type 1 fimbriae are the most commonly found fimbrial appendages on the outer membrane of Salmonella enterica serotype Typhimurium. Previous investigations indicate that static broth culture favours S. Typhimurium to produce type 1 fimbriae, while non-fimbriate bacteria are obtained by growth on solid agar media. The phenotypic expression of type 1 fimbriae in S. Typhimurium is the result of the interaction and cooperation of several genes in the fim gene cluster. Other gene products that may also participate in the regulation of type 1 fimbrial expression remain uncharacterized.
Results
In the present study, transposon insertion mutagenesis was performed on S. Typhimurium to generate a library to screen for those mutants that would exhibit different type 1 fimbrial phenotypes than the parental strain. Eight-two mutants were obtained from 7,239 clones screened using the yeast agglutination test. Forty-four mutants produced type 1 fimbriae on both solid agar and static broth media, while none of the other 38 mutants formed type 1 fimbriae in either culture condition. The flanking sequences of the transposons from 54 mutants were cloned and sequenced. These mutants can be classified according to the functions or putative functions of the open reading frames disrupted by the transposon. Our current results indicate that the genetic determinants such as those involved in the fimbrial biogenesis and regulation, global regulators, transporter proteins, prophage-derived proteins, and enzymes of different functions, to name a few, may play a role in the regulation of type 1 fimbrial expression in response to solid agar and static broth culture conditions. A complementation test revealed that transforming a recombinant plasmid possessing the coding sequence of a NAD(P)H-flavin reductase gene ubiB restored an ubiB mutant to exhibit the type 1 fimbrial phenotype as its parental strain.
Conclusion
Genetic determinants other than the fim genes may involve in the regulation of type 1 fimbrial expression in S. Typhimurium. How each gene product may influence type 1 fimbrial expression is an interesting research topic which warrants further investigation.
Background
Salmonellosis is one of the important causes of food-borne diseases throughout the world [1]. Salmonella enterica contains more than 2,300 serotypes among which Typhimurium is an important causative agent of gastroenteritis. Adhesion of bacteria to the host epithelial cells is a prerequisite step in establishing infection. Specific adhesion requires the interaction of specialized complementary molecules in a ligand-receptor interaction between bacterial surfaces and host tissues [2]. Proteinaceous hair-like structures called fimbriae on the surface of bacteria have been implicated in such an event [3]. Many members of the family Enterobacteriaceae including Salmonella produce type 1 fimbriae, the most commonly found type of fimbrial appendages [4]. Type 1 fimbriae comprise a family of rod-shaped organelles which are 7 nm in diameter and 0.2–2.0 μm long [5]. Type 1 fimbriae adhere to different cell types including erythrocytes, leukocytes, intestinal cells, respiratory cells, protozoa, yeast, fungal hyphae, and plant root hairs [6]. Several studies indicated that type 1 fimbriae also contribute to virulence [7-9]. For example, type 1 fimbriae expressing S. Typhimurium caused persistent infection in swine [7]. Type 1 fimbriae may also modulate the bacterial tropism to the gut of the host [8], and type 1-fimbriated Salmonella were more virulent than the fim-minus ones [9]. The fact that more than 80% of Salmonella isolates produce this fimbrial type suggests that type 1 fimbriae play an important role at some stage in the life cycle of bacteria [10].
Phenotypic variation of the expression of type 1 fimbriae in S. Typhimurium was first characterized by Old et al. [11,12]. They described a biphasic growth pattern associated with the outgrowth of fimbriate bacteria incubated in static, liquid broth culture. Briefly, strongly type 1 fimbriate phase S. Typhimurium cells were isolated following serial passage every 48 h in static broth culture. Non-fimbriate phase bacteria were obtained by growth on solid media.
Current data indicate that the phenotypic expression of type 1 fimbriae in S. Typhimurium is the result of the interaction and cooperation of several genes in the fim gene cluster [13-18]. The FimZ transcriptional factor activates the fimA expression by binding to the fimA promoter, and in addition, FimZ also positively regulates its own transcription [13]. FimZ requires another co-activator FimY to activate fimA expression, however, no evidence has revealed any interaction of FimY and the fimA promoter [19]. Interestingly, fimY gene possesses five rarely used arginine codon which is recognized by an arginine tRNA gene fimU. A S. Typhimurium fimU mutant did not produce type 1 fimbriae and the fimY translation was inhibited [16,17]. fimU modulates the fimY expression in the translational level. A fimW deleted strain, on the contrary, overproduced type 1 fimbriae as compared to the parental strain. This negative regulation was proposed to be mediated through an inhibitory effect on the FimZ protein by a FimW-FimZ protein-protein interaction [15]. Other than gene products within the fim gene cluster, McFarland et al. recently reported that a knockout mutation in lrp, encoding the leucine-responsive regulatory protein (LRP), was non-fimbriated [20]. They demonstrated that LRP positively regulated type 1 fimbrial expression by binding to the promoter region of fimZ [20]. Other gene products that may play a role in type 1 fimbrial expression were seldom characterized. Our long-term goal in the laboratory is focused on finding any genetic determinants, other than those previously described within the fim gene cluster, which may also play a role in the phenotypic expression of type 1 fimbriae in S. Typhimurium. Initially we were interested in characterizing the genetic elements that may be associated with the ability of S. Typhimurium to switch type 1 fimbrial expressions in response to solid agar and static broth culture conditions. We used transposon mutagenesis to generate a library for S. Typhimurium and screened mutants that exhibited different type 1 fimbrial phenotypes than the parental strain between solid agar and static broth culture conditions. The flanking DNA fragments of the transposon insertion sites in the mutants of interest were cloned and sequenced. Herein, we report on the genetic determinants in S. Typhimurium that may play a role in the regulation of type 1 fimbrial expression between solid agar and static broth culture conditions.
Results
Construction of a transposon mutant library of S. Typhimurium
In order to identify any genetic determinant that is responsible for S. Typhimurium to regulate type 1 fimbrial expression between solid agar and static broth cultures, we used a transposon mutagenesis technique to construct a transposon library. We obtained a library with approximately 8,000 clones.
Screening of S. Typhimurium transposon mutants that exhibited different type 1 fimbrial phenotypes than the parental strain
In total, 7,239 transposon mutants were screened for those which exhibited different type 1 fimbrial phenotypes as compared to the parental strain S. Typhimurium LB5010. The expression of type 1 fimbriae was determined by yeast agglutination test. S. Typhimurium LB5010, when grown in static broth culture for 36–48 h, agglutinated Candida albicans on a glass slide. These bacterial cells produced type 1 fimbriae on the outer membrane when negatively stained with phosphotungstic acid and observed under electron microscopy (Figure 1, Panel A). On the contrary, S. Typhimurium LB5010 did not agglutinate C. albicans on a glass slide when bacterial cells were prepared from solid agar medium grown for 16–18 h. The absence of agglutination correlated with the fact that no type 1 fimbriae were observed by electron microscopy (Figure 1, Panel B). Fifty-four mutants verified by Southern blot to have a single unique transposon insertion were selected for further investigation (Figure 2). For those strains with multiple transposon insertions (Figure 2, lane 6 for example) were currently excluded from the present study. This could be due to partial digestion of the genomic DNA or multiple transposons inserted into one bacterial strain. These 54 mutants no longer exhibited the same type 1 fimbrial phenotype as the parental strain, among which 31 mutants produced type 1 fimbriae in both culture conditions, while another 23 mutants produced type 1 fimbriae in neither culture condition. The genes inactivated in the mutants were identified by cloning and sequencing the DNA fragments adjacent to the transposon insertion sites. The DNA sequences were used to search against the genomic sequence of S. Typhimurium. The position of the transposon insertion site was determined along with whether or not a transposon insertion site was located in or near an open reading frame (ORF), as predicted by the annotated genomic sequences. In the present study, we only focused on those mutants that demonstrated disruptions in ORFs; these types of mutation are expected to disrupt the functions of the ORFs involved. Table 1 lists those mutants. Transposons which disrupted between ORFs were excluded in the present study.
Figure 1.
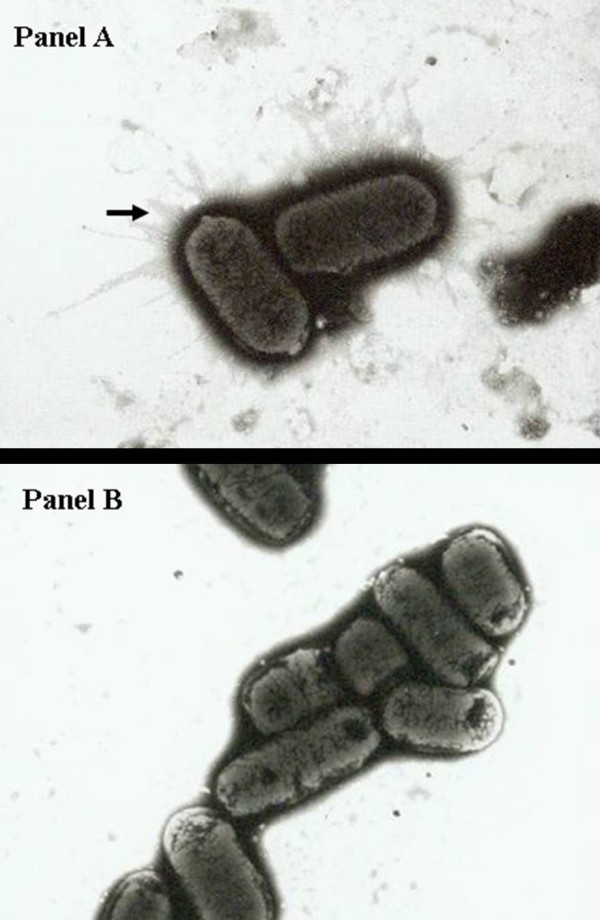
Observation of type 1 fimbriae in S. Typhimurium LB5010 by electron microscopy. Panel A. S. Typhimurium LB5010 cells produced type 1 fimbrial appendages on the outer membrane (arrow) when cultured in LB static broth at 37°C for 48 hr (20,000 ×). Panel B. S. Typhimurium LB5010 cells did not produce type 1 fimbrial appendages on the outer membrane when cultured on LB agar at 37°C for 18 hr (20,000 ×). Bacterial cells were negatively stained with 2% of phosphotungstic acid.
Figure 2.
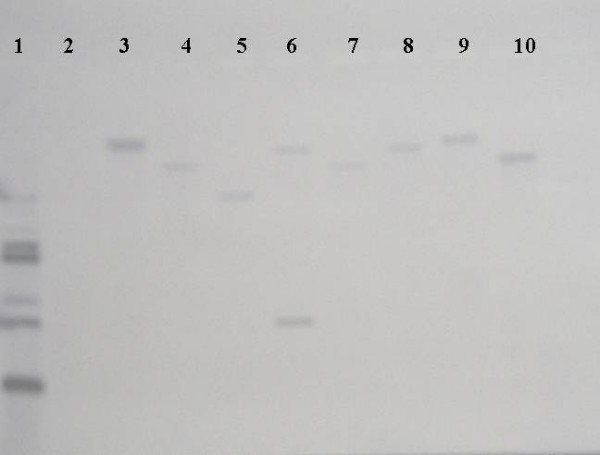
A representative Southern blot demonstrating hybridization of the genomic DNA cleaved with EcoRI with a DIG-labeled 1.3-kb kanamycin-resistance cassette DNA probe. Lane 1: pUC4K plasmid DNA (positive control); lane 2: S. Typhimurium LB5010 (negative control); lanes 3 to 10: K1 to K8 strains, respectively.
Table 1.
S. Typhimurium mutants that exhibited different type 1 fimbrial phenotypes than the parental strain
| Category | Mutant | Phenotype | Genea | Function |
| (agar, broth) | ||||
| Fimbrial biosynthesis and regulation | K2 | -, - | fimZ | Positive regulator of type 1 fimbriae major subunit gene fimA |
| K14 | -, - | fimY | Positive regulator of type 1 fimbriae major subunit gene fimA | |
| K40 | -, - | fimA | Major subunit of type 1 fimbriae | |
| K66 | -, - | fimD | Fimbrial usher protein of FimA | |
| K75 | -, - | fimC | Fimbrial chaperone protein of FimA | |
| K44 | -, - | fimH | Adhesin of type 1 fimbriae | |
| K70 | +, + | lpfB | Fimbrial chaperone protein of long polar fimbriae | |
| K61 | +, + | pefI | Regulator of plasmid-encoded fimbriae | |
| K23 | +, + | stbC | Fimbrial usher protein of Stb fimbriae | |
| Putative cytoplasmic protein | K1 | +, + | STM1078 | Putative cytoplasmic protein |
| K28 | +, + | STM4529 | Putative cytoplasmic protein | |
| Global regulator | K46 | -, - | lrp | Leucine-responsive regulatory protein |
| K15 | -, - | hupA | Histone-like DNA binding protein | |
| Membrane associated protein | K17, | +, + | yhjV | Putative transporter protein |
| K39 | -, - | STM1128 | Putative sodium/glucose cotransporter | |
| K45 | -, - | yihO | Putative glycoside-pentoside-hexuronide (GPH) family transport protein | |
| K50 | +, + | STM2532 | Putative inner membrane lipoprotein | |
| K52 | +, + | STM2486 | Putative inner membrane protein | |
| K54 | -, - | yjiH | Putative inner membrane protein | |
| K72 | +, + | mgtB | Mg2+ transporter protein | |
| K30 | +, + | narK | Nitrate/nitrite transporter protein | |
| Cell envelope associated protein | K60 | +, + | ytfB | Putative cell envelope opacity-associated protein A |
| Periplasmic protein | K69 | +, + | ydbH | Putative periplasmic protein |
| Ribosome modulation factor | K56 | -, - | rmf | Ribosome modulation factor |
| Type III secretion system | K55 | -, - | ssaB | Effector protein of type III secretion system |
| K59 | +, + | invB | Chaperone protein for SopE/SopE2 | |
| K63 | -, - | ssaV | Apparatus protein for intracellular trafficking and secretion | |
| Prophage-derived protein | K11 | +, + | STM4200 | Putative phage tail protein H |
| K33 | +, + | sseI | Gifsy-2 prophage putative type III secreted effector protein | |
| K19 | -, - | STM1041 | Gifsy-2 prophage probable minor tail protein | |
| Sensor protein | K51 | +, + | ssrA | Sensor component of type III secretion system |
| K58 | +, + | yehU | Sensor component | |
| Transcription termination factor | K73 | -, - | nusA | Transcription termination factor |
| Enzymes | K3 | -, - | cafA | Endoribonuclease |
| K5 | +, + | res | Subunit of type III restriction-modification enzyme | |
| K8 | +, + | STM4467 | Putative arginine deiminase | |
| K16 | +, + | STM1627 | Alcohol dehydrogenase class III | |
| K31 | +, + | STM2446 | Putative iron-dependent peroxidase | |
| K41 | -, - | nuoE | NADH dehydrogenase I chain E | |
| K37 | +, + | yafA | Putative hydrolase | |
| K43 | -, - | miaA | Isopentenylpyrophosphate transferase | |
| K38 | -, - | prC | C-terminal protease for penicillin binding protein | |
| K47 | -, - | atpH | Membrane-bound ATP synthase | |
| K48 | +, + | ubiB | NAD(P)H-flavin reductase | |
| K49 | -, - | ydiF | Putative acetyl-CoA: acetoacetyl-CoA transferase | |
| K53 | +, + | tdK | Thymidine kinase | |
| K65 | +, + | pps | Phosphoenolpyruvate synthase | |
| K57 | +, + | ybjX | Putative enzyme with unknown function | |
| K67 | +, + | pncA | Nicotinamidase/pyrazinamidase | |
| K68 | +, + | rfaB | Lipopolysaccharide 1,6-galactosyltransferase | |
| K71 | +, + | xapB | Xanthosine permease | |
| K64 | +, + | STM1940 | Putative cell wall-associated hydrolase | |
| K74 | +, + | cbiA | Cobyrinic acid a, c-diamide synthase | |
| K42 | -, - | aroA | 3-enolpyruvylshikimate-5-phosphate synthetase |
a: The gene that was disrupted by the transposon insertion is listed. The annotation of S. Typhimurium LT2 genome is used if the transposon insertion was present in a gene which is uncharacterized and unnamed.
Classification of the mutants according to the functions or putative functions of genes disrupted by the transposon insertions
The 54 mutants listed in Table 1 were classified according to the functions or putative functions of the ORFs disrupted by the transposons. We classified the mutants into the 12 categories: fimbrial biosynthesis and regulation, putative cytoplasmic protein, global regulator protein, cell membrane associated protein, cell envelope associated protein, periplasmic protein, ribosome modulation factor, type III secretion system protein, prophage-derived protein, sensor protein, transcription termination factor, and enzymes. Nine mutants that had defects in fimA, fimC, fimD, fimH, fimY, fimZ, lpfB, pefI, and stbC were placed in the fimbrial biosynthesis and regulation category. FimA is the major subunit of the type 1 fimbrial shaft, while the adhesion protein, FimH, confers the binding specificity of type 1 fimbriae [18,21]. FimC and FimD serve as the chaperone and molecular usher for type 1 fimbriae [18]. FimZ and FimY are positive regulators of fimA [14,19]. Since all these genes are involved in type 1 fimbrial biosynthesis and regulation, disruption of these genes caused the mutants to produce no fimbriae at all in either culture condition. Three mutants produced type 1 fimbriae in both culture conditions and had transposons inserted in other fimbrial related genes. K70 had a defect in lpfB, the chaperone protein for long polar fimbriae [22]. K61 had a defect in pefI, which involves the regulation of plasmid-encoded fimbriae [23]. K23 had a transposon inserted in stbC that encodes an outer membrane usher protein for Stb fimbriae in S. Typhimurium. The usher protein facilitates anchoring the developing fimbrial structure to the outer membrane [24].
The K1 and K28 mutants exhibited type 1 fimbriae in both static broth and on solid agar media. The gene products involved are putative cytoplasmic proteins, and their functions remain to be characterized. The K15 mutant had a transposon inserted in the hupA gene whose product is the histone-like DNA-binding protein, HU, a small, basic, thermostable protein involving DNA replication [25]. The K15 mutant and another mutant K46 that has disruption in lrp encoding leucine-responsive regulatory protein (LRP) did not exhibit type 1 fimbriae in either culture condition. Membrane associated proteins possess transmembrane domains. K17 had interruption of yhjV encoding protein similar to the tryptophan/tyrosine permease family in Escherichia coli [26]. The putative protein affected in K39 is a sodium/glucose cotransporter protein which belongs to a superfamily of membrane proteins responsible for the uphill transport of substrates coupled to the downhill transport of Na+ [27]. The mgtB and narK genes, defective in K72 and K30 respectively, are involved in Mg2+ and nitrate/nitrite transportation, respectively [28]. Disruption of the rmf gene impaired the 70S ribosome dimerization and resulted in inhibition of S. Typhimurium to produce type 1 fimbriae as seen in K56 [29]. The ssaB, ssaV, and invB belong to members of the type III secretion system, which is important in the pathogenesis of S. Typhimurium. The ssaB and ssaV mutants did not produce type 1 fimbriae in either culture condition, whereas the invB mutant produced type 1 fimbriae in both culture conditions. Two mutants, K11 and K19, possessed insertions in genes involved in synthesis of putative phage tail proteins [30]. K33 had an insertion in the gene encoding for a type III translocated protein, SseI, carried by the prophage Gifsy-2 [31]. Two mutants, K51 and K58, had transposons inserted into genes which encode the sensor part of the two-component regulatory system. SsrA is associated with the type III secretion system for the pathogenicity island II [32], while YehU in S. Typhimurium remains to be characterized. NusA binds to the RNA polymerase core enzyme and functions as a transcription termination factor [33]. The K73 carrying transposon in nusA did not produce type 1 fimbriae in either culture condition.
Among the mutants, 21 of 54 mutants (almost 39%) were in the category of enzymes comprising the largest group of the mutants. The enzymes detected in these mutants could be further divided into endoribonuclease (K3), restriction-modification enzyme (K5), protease (K38), tRNA modification enzyme (K43), the enzyme for lipopolysaccharide synthesis (K68), and primarily the enzymes required for general metabolism.
Complementation test
Table 2 summarizes the results of the complementation test for K3 cafA, K5 res, K43 miaA, K48 ubiB and K73 nusA mutant strains. The K48 ubiB mutant strain produced type 1 fimbriae constitutively regardless grown in static broth or on solid agar culture condition. Transforming the plasmid pUbiB that contained the coding sequence of ubiB conferred the K48ubiB mutant to exhibit the type 1 fimbrial phenotype as seen in S. Typhimurium LB5010. Transforming a plasmid possessing the coding sequence of res to K5 res mutant did repress the type 1 fimbriae to express on solid agar as the parental strain. However, the complemented strain did not produce type 1 fimbriae in static broth culture. The complemented strains K3 cafA (pCafA), K43 miaA (pMiaA) and K73 nusA (pNusA) remained unchanged as their respective mutant strains, which did not produce type 1 fimbriae in either culture condition.
Table 2.
Phenotypic expression of type 1 fimbriae by the selected mutant and the complemented strains
| Strain | Plasmid transformed | Phenotypic expression of type 1 fimbriae by bacteria grown on or ina | |
| agar | broth | ||
| LB5010 | none | - | + |
| K3 cafA | none | - | - |
| K3 cafA | pCafA | - | - |
| K5 res | none | + | + |
| K5 res | pRes | - | - |
| K43 miaA | none | - | - |
| K43 miaA | pMiaA | - | - |
| K48 ubiB | none | + | + |
| K48 ubiB | pUbiB | - | + |
| K73 nusA | none | - | - |
| K73 nusA | pNusA | - | - |
a: phenotypic expression of type 1 fimbriae was determined by mannose-sensitive yeast agglutination test.
Reverse transcription polymerase chain reaction (RT-PCR) analysis
Total RNA from the parental strain S. Typhimurium LB5010, K48 ubiB mutant, and K48 ubiB (pUbiB) were prepared and analyzed for fimA mRNA and 16S rRNA expression by RT-PCR. Figure 3 showed the RT-PCR analysis results. As for the parental strain LB5010, the fimA expression obtained from the static broth was approximately 4.5 fold (1: 0.22) of that obtained from the agar culture condition. For the K48 ubiB mutant, expression of fimA (static broth) was about 60% (1: 1.74) of the fimA (agar). When K48 ubiB strain harboured the plasmid pUbiB, fimA expression (broth) was about 2.6 fold (1: 0.39) of fimA (agar). As a control, 16S rRNA was constantly expressed in all the strains tested.
Figure 3.
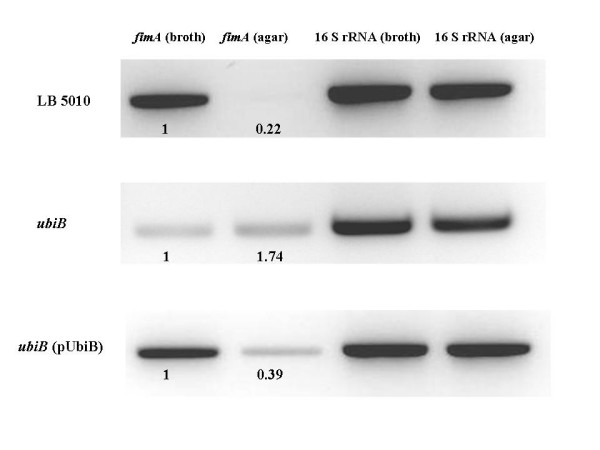
RT-PCR for fimA transcription. RT-PCR assays were used to monitor fimA and 16S rRNA transcription in the parental strain LB5010, ubiB mutant, and ubiB (pUbiB) strain. The intensities of the bands for each strain were determined by densitometry and expressed (Arabic numbers) relative to the value for fimA transcription obtained from the static LB broth culture condition. The intensities of 16S rRNA shown indicates that equivalent amounts of total RNA were used in the experiment.
Discussion
It is not surprising that many bacterial fimbriae have the ability to switch expression from a fimbriate to a non-fimbriate phenotype. Since bacterial population may face unpredictable milieu during their life cycle, having cells in both fimbriate and non-fimbriate phases offers the bacterial population an advantage of surviving in the diverse situations it will encounter [34]. The mechanisms that control the fimbrial regulation have been described in detail for only few fimbrial types. For example, the regulation in E. coli type 1 fimbriae is primarily controlled by inversion of a 314-bp segment of DNA (fim switch region) upstream of the fimA gene coding for the major fimbrial subunit [35]. Accessory proteins including the integration host factor (IHF), LRP and the histone-like nucleoid-structuring (H-NS) protein, also bind to the fim switch region to re-orientate the DNA into a configuration favourable for inversion [36-38]. Interestingly, the E. coli K-12 strain also possesses the sfm gene cluster (sfmACDHFZU), which is the orthologue of S. Typhimurium fim gene cluster [39]. Nevertheless not much characterization has been performed on sfm. The control system for type 1 fimbriae in S. Typhimurium differs from that of E. coli. No DNA inversion event has been detected and several fim gene products have been implicated as regulators of type 1 fimbrial expression [13-18,40]. Previous studies did not reveal that global regulator like IHF or H-NS play a role in S. Typhimurium type 1 fimbrial expression. In order to explore other elements outside the fim gene cluster that might also participate in type 1 fimbrial expression, we constructed a transposon library of S. Typhimurium. From the mutants that no longer exhibited the same type 1 fimbrial phenotypes as the parental strain whether grown on solid agar and in static broth cultures, 12 groups were classified according to the putative functions of the genes that were interrupted by the transposons. Not surprisingly, several mutants that had defects in the fim genes did not produce type1 fimbriae in either condition. The mutants possessing transposons in fimA, fimC, fimD, fimH, fimZ, and fimY all exhibited the predicted phenotypes. These fim genes are required for type 1 fimbrial biogenesis or regulation, and disruption of these genes causes the bacteria to be non-fimbriate in either condition. Interestingly, the present study identified 3 mutants that had defective genes for other fimbrial systems, and they produced type 1 fimbriae in both culture conditions. K23 had a transposon inserted in stbC that encodes the fimbrial usher protein for Stb fimbriae [41]. K70 had insertion in fimbrial chaperone protein for long polar fimbriae (Lpf). Those two gene products are involved in fimbrial biogenesis. How disruption of the outer membrane usher gene, stbC, of the Stb fimbrial system affects another fimbrial system poses an interesting research topic. We did try to complement the stbC mutant with a recombinant plasmid possessing the stbC coding sequence. However, the stbC mutant exhibited resistance to a battery of antibiotics, including ampicillin and chloramphenicol those frequently used in molecular cloning. This unexpected finding impeded us to transforming available plasmids into this strain for the time being. PefI identified in K61 is the homolog of PapI in E. coli. PefI negatively regulates the production of PefA, the major fimbrial subunit of plasmid-encoded fimbriae (Pef) [42]. Cross-talk between different fimbrial systems within the same microorganism has been demonstrated previously. For example, minor fimbrin of type 1 and F1C fimbriae of E. coli can be reciprocally exchanged to generate hybrid fimbriae with changed receptor specificities [43]. The regulator PapB of pyelonephritis-associated pili (Pap) prevents inversion of the fim switch controlling the expression of type 1 fimbriae in the same E. coli strain [44]. Recently Nuccio et al. also demonstrated that the presence of type 1 fimbrial biosynthesis genes interfered with the expression of plasmid-encoded fimbriae in S. Typhimurium [45]. Although additional research is required to elucidate the interaction of different fimbrial systems, our results did stress the observation that different types of fimbriae may coordinate for their own benefit.
A global regulator, LRP, has been demonstrated to play a role in the regulation of both type 1 fimbriae and P fimbriae of E. coli and Pef of S. Typhimurium. Recently McFarland et al. demonstrated that an lrp knockout S. Typhimurium strain did not produce type 1 fimbriae and Lrp is a positive regulator for fim gene expression by interacting with the fimZ promoter region [20]. FimZ is a positive regulator for fimA [14]. Their data suggested that a positive contribution of fim gene expression by Lrp/fimZ promoter interaction may be due to the displacement of H-NS, a repressor for fimZ [20,46]. The K46 mutant with the transposon inserted into lrp did not produce type 1 fimbriae in either culture condition. Our result concurred with their finding. The role that lrp plays in terms of type 1 fimbrial regulation in S. Typhimurium becomes apparent.
Several S. Typhimurium strains had transposons inserted in the genes that encode enzymes. The K3 strain had a defect in cafA that encodes a ribonuclease named RNase G in E. coli. Umitsuki et al. reported that RNase G was involved in the in vivo degradation of adhE mRNA which encodes a fermentative alcohol dehydrogenase, AdhE [47]. A mutation at cafA resulted in accumulation of adhE mRNA and prolonged the half-life of its mRNA. Interestingly, the K16 strain had a mutation in the gene that encodes alcohol dehydrogenase. The K16 strain constitutively expressed type 1 fimbriae. Overexpression of AdhE inhibited type 1 fimbrial expression, while the absence of AdhE caused type 1 fimbrial expression on solid agar or in static broth. How an enzyme involved in metabolism influences fimbrial expression is not clear, but previous studies demonstrated that type 1 fimbrial expression was linked to the nutrient status of the bacterial encounter. Broth culture supplemented with glucose favours Shigella flexneri remaining in a non-fimbriate state [6]. Morgenroth and Duguid described a S. Typhimurium biotype called the FIRN strain; it is non-type 1 fimbriate, non-inositol-fermenting, and non-rhamnose-fermenting [48]. The genes controlling fimbrial expression may be linked to the genes involved in carbohydrate metabolism. Interestingly, similar to the K16 strain, a variety of mutants in the present study having transposons inserted in the genes encoding "house-keeping" enzymes all constitutively exhibited type 1 fimbriae. Barak et al. used transposon mutagenesis to identify genetic determinants required for S. Newport to attach to plant tissue [49]. They also found that alcohol dehydrogenase, hydrolase, bacteriophage protein, and several membrane associated proteins, including transporter protein, similar to our findings, were the essential components for this Salmonella serotype to adhere to plant tissue [49]. The ability of Salmonella to colonize plant tissue and modulate type 1 fimbrial expression may share a common regulatory network.
A complementation test allowed us to examine if providing the coding sequence of the gene carried in a plasmid back to the mutant stain defective in such a gene would switch the phenotype of type 1 fimbrial expression to that originally observed. A comprehensive complementation test is not within the scope of the present study; however, we did test some strains. The complemented K3 cafA (pCafA), K43 miaA (pMiaA), and K73 nusA (pNusA) strains did not exhibit the type 1 fimbrial phenotype as the parental strain. Polar effect caused by the transposon could be one of the reasons. Another possibility could be due to the gene dosage effect conferred by the cloning vector. The K5 res (pRes) strain did repress the type 1 fimbrial production on solid agar as the parental strain LB5010. However, in static broth culture, the complemented strain did not exhibit type 1 fimbriae. The res gene, encoding for the restriction subunit of type III restriction-modification (R-M) system of S. Typhimurium, locates upstream of the mod gene which encodes the methyltransferase subunit of R-M system. Since Res subunit requires Mod to form a complex to recognize and cleave DNA [50], failure of mod expression caused by polar effect could account for the partially complemented phenotype of the K5res (pRes) strain. NusA is a transcription termination factor and under some circumstance it may also function as a transcription elongation factor [51]. The ability of NusA to enhance RNA polymerase pausing would result in tight coupling of transcription and translation, thus interferes with the Rho-mRNA interactions and consequently blocks termination of transcription [52]. The transposon inserted in nusA in K73 strain could cause transcription/translation coupling defect in a variety of genes including those that may affect fim expression. The pNusA transformed in K73 strain may not produce precise stoichiometric ration of NusA, leading K73 nusA (pNusA) strain remained unchanged as the K73 nusA strain. MiaA is an isopentenylpyrophosphate transferase that is required for tRNA modification. Ericson and Björk demonstrated that a miaA mutant in S. Typhimurium LT2 which was deficient in 2-methylthio-N6-(4-hydroxyisopentenyl) adenosine (ms2io6A37) in its tRNA induced pleiotropic effects on bacterial physiology such as reduced growth rate and polypeptide chain elongation rate [53]. Non-fimbriate phenotype exhibited by K43 miaA strain in both culture conditions could be the result of such pleiotropic effects. The K48 ubiB mutant transformed with the pUbiB plasmid restored the type 1 fimbrial phenotype as the parental strain LB5010. The K48 ubiB (pUbiB) cultured on agar did not agglutinate yeast cells. Although signals of both LB5010 and K48 ubiB (pUbiB) from agar could be detected by densitometry in RT-PCR assay, we argued that such fimA amount would not be enough for scoring positive in yeast agglutination test. Our complementation assay did prove that the ubiB mutant could exhibit the same phenotype as LB5010 when it harboured a plasmid containing the coding sequence of ubiB. The ubiB in S. Typhimurium could encode a NAD(P)H-flavin-reductase, whose homologue in E. coli has been proved to be required for the first monooxygenase step in ubiquinone biosynthesis [54]. How a gene involving in biosynthesis of ubiquinone, an essential component of electron transport chain influences type 1 fimbrial expression is currently unknown. However, Leonard et al. reported that UbiB protein contains motifs found in eukaryotic-type protein kinase [55]. It was speculated that phosphorylation may be required for ubiquinone biosynthesis-activating proteins [55]. Possibly UbiB would also phosphorylate another substrate protein that affects fimA expression. The function of ubiB in S. Typhimurium and the relationship of ubiB and fim expression demands more careful characterization. Our data clearly showed that an ubiB mutant produced fimA at the condition that either favours or inhibits type 1 fimbrial expression and complementing this mutant with a plasmid possessing ubiB coding sequence restored it to the phenotype similar to the parental strain.
The present study revealed that the genetic determinants other than the fim genes could have the potential to affect type 1 fimbrial expression in response to different standard laboratory culture conditions. However, a battery of genes identified by transposon mutagenesis still requires detail characterization to validate its linkage to fimbrial expression. The phenotype caused by polar effect is one of the drawbacks in this kind of study. Validation of the genes of interest that are associated with type 1 fimbrial expression in S. Typhimurium is presently under investigation in our laboratory.
Conclusion
The ability to switch between fimbrial expressions in bacteria may be beneficial for survival. Fimbriate bacteria may facilitate colonizing host cells at certain stages of infection, while non-fimbriate bacteria may also have their own role to play, for example, as in avoiding recognition by phagocytes, or allowing the bacteria to shed and colonize another site on the host. Compared to E. coli, Salmonella has a higher survival rate in the external environment, which promotes transmission to a new host [56]. Persistent infection of Salmonella in the environment is enhanced by adhesion and biofilm formation, both of which involve fimbrial appendages [21]. How S. Typhimurium fluctuates between type 1 fimbriate phase and non-fimbriate phase in vivo remains to be uncovered. Nonetheless the present study reveals that several genetic determinants may be associated with the ability of S. Typhimurium to modulate type 1 fimbrial expression between solid agar and static broth culture conditions. These two laboratory culture conditions may to some extent mimic the environmental milieu that Salmonella would encounter. We report here that other gene products besides those in the fim gene cluster are also required for type 1 fimbrial expression. How each gene influences type 1 fimbrial expression is an interesting research topic which warrants further investigation.
Methods
Bacterial strains, media, culture conditions, primers and plasmids
The S. Typhimurium strain used in the present study is S. Typhimurium LB5010, a LT2 strain derivative [57]. This strain produces type 1 fimbriae and is fimbrial phase variable [57]. E.coli JM109 strain was used for molecular cloning [58]. The primers and plasmids used in the present study are listed in Table 3 and Table 4, respectively. Bacteria were grown in Luria-Bertani (LB) broth (Difco/Becton Dickinson, Franklin Lakes, NJ) or plated on LB agar [59]. Media were supplemented with antibiotics when required at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml. The above antibiotics were obtained from Sigma (St. Louis, MO).
Table 3.
Oligonucleotide primers used in the present study
| Primer | Sequence (5'-3') |
| kan-5 | TAACATCATTGGCAACGCTACCT |
| kan-6 | GCATCGGGCTTCCCATACAATCG |
| kan-7 | GTCGCACCTGATTGCCCGACATT |
| cafA-F | ATACGACTCACAACCTTGCTTTGCCGGACG |
| cafA-R | TTTCTGCGCAGGATATTAGTGGCTATGTCG |
| res-F | CATTGTCATTTACGGCTACTCT |
| res-R | ACGATAACCTTCAAGTCAAC |
| miaA-F | CTGCTGGCGGATGTTGAGCGGCTATGT |
| miaA-R | CGCAATGCGTTCAGGAACGGATCTTGT |
| nusA-F | CCGTCCTATGTTCACTGCCGAT |
| nusA-R | CTGCTGTACCAGGCGATCCACGGAAAC |
| ubiB-F | GGTCGTCCTATTGTTAAAGATCCTGA |
| ubiB-R | CGCAATACAAAGCCTGTAGATATTCA |
| 16S-F | TTCCTCCAGATCTCTCTACGCA |
| 16S-R | GTGGCTAATACCGCATAACG |
| fimA-F | ACTATTGCGAGTCTGATGTTTG |
| fimA-R | CGTATTTCATGATAAAGGTGGC |
Table 4.
The plasmids used in the present study
| Plasmid | Genotype or relevant featuresa | Reference or source |
| pUC4K | 3.9 kb vector, Kanr Amr | Amersham Biosciences |
| yT&A | 2.7 kb cloning vector; Amr | Yeasten Biotech |
| pCafA | 1.3 kb DNA fragment containing cafA cloned into yT&A; Amr | This study |
| pRes | 3.7 kb DNA fragment containing res cloned into yT&A; Amr | This study |
| pMiaA | 1.2 kb DNA fragment containing miaA cloned into yT&A; Amr | This study |
| pNusA | 1.8 kb DNA fragment containing nusA cloned into yT&A; Amr | This study |
| pUbiB | 988 bp DNA fragment containing ubiB cloned into yT&A; Amr | This study |
a: Kanr, kanamycin resistant; Amr, ampicillin resistant.
Construction of a transposon mutant library of S. Typhimurium
A transposon mutant library of S. Typhimurium was constructed using the EZ::TN <KAN-2> Tnp Transposome system (Epicentre, Madison, WI). This system contains the transposon Tn903 which possesses a kanamycin resistance cassette. Briefly, 1 μl of the transposome mixture was placed in 40 μl of S. Typhimurium-competent cells and electroporated into the cells for transposition to occur. The electroporation conditions were set as follows using ECM 630 (BTX, Pittsburgh, PA): voltage of 1.70 kV, resistance of 125 ohms, and capacitance of 50 μF. Immediately after electroporation, 1 ml of S.O.C medium (Invitrogen, Carlsbad, CA) was added to the cuvette and the medium/cell mixture was transferred to a tube and incubated at 37°C with constant shaking for 1 h. Four milliliters of LB broth was added to dilute the bacterial cells, and a-100 μl of aliquot was evenly spread on the kanamycin-containing LB agar.
Screening of S. Typhimurium transposon mutants that exhibited different type 1 fimbrial phenotypes than the parental strain
The transformants grown on LB agar supplemented with kanamycin were randomly selected. Each colony was streaked on LB agar and then inoculated into 10 ml of LB broth. The LB agar plates were incubated at 37°C for 18 h while the broth preparations were incubated statically at 37°C for 48 h. Bacterial cells from the solid agar were collected by a sterile loop and resuspended in 100 μl of 1 × phosphate-buffered saline (PBS). Cells in the broth medium were collected by centrifugation, and the pellet was resuspended in 100 μl of 1 × PBS. Subsequently, 30 μl of a 3% (vol/vol) suspension of C.albicans in PBS and an equal amount of bacterial cells to be tested were mixed together on a glass slide [11]. Visible agglutination after gentle agitation indicated a positive reaction for the presence of type 1 fimbriae. Any bacterial suspension that produced type 1 fimbriae was further mixed with C. albicans along with 3% (wt/vol) of a D-mannose solution (Sigma). The mannose-sensitive agglutination conferred by type 1 fimbriae was inhibited in the presence of mannose.
Southern hybridization
Southern hybridization analysis was used to detect the presence of the kanamycin resistance cassette on the S. Typhimurium genomic DNA. Genomic DNAs from S. Typhimurium LB5010 and the mutant strains that were no longer type 1 fimbrial phase variable were isolated using a MasterPure DNA Purification Kit (Epicentre). The genomic DNA preparations were cleaved by EcoRI, and the DNA fragments were separated by electrophoresis in a 0.7% agarose gel. EcoRI was chosen to cut the genomic DNA since there was no EcoRI site within the kanamycin resistance cassette of Tn903. A DNA probe derived from the kanamycin resistance cassette sequence was hybridized to one band in the Southern blot if the transposition had occurred only once. The DNA was soaked in a denaturing solution (0.5 M NaOH and 1 M NaCl) twice for 15 min each time and a neutralization solution (3 M NaCl, 0.5 M Tris-HCl) for 30 min with continuous agitation at room temperature. The DNA was then transferred onto a nylon membrane by a vacuum blotter (trans-Vac, Hoefer, San Francisco, CA) with 10 × SSC and cross-linked to the membrane by exposure to short-wavelength UV light from a UV Spectrolinker (Spectronics, Westbury, NY). The 1.3-kb kanamycin resistance cassette-containing DNA fragment was cleaved by Hind III from the pUC4K plasmid and isolated from the agarose gel using a Montage Gel Extraction Kit (Millipore, Bedford, MA). Probe labeling and hybridization were performed using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Mannheim, Germany) according to the protocols provided by the manufacturer. The 1.3-kb DNA probe was labeled by the randomly primed incorporation of digoxigenin-labeled dUTP. Briefly, hybridization to the blot was performed at 68°C in a standard buffer (5 × SSC, 0.1% N-lauroylsarcosine, 0.02% SDS, and 1% blocking reagent) for 16 h. Two 5-min washes were performed at room temperature in a 2 × SSC (1 × SSC is comprised of 0.15 M NaCl and 0.015 M sodium citrate)-0.1% SDS solution. The blot was then washed with two 15-min washes in 0.1 × SSC-0.1% SDS solutions at 68°C. Hybrids were detected using an antibody conjugate (anti-digoxigenin-alkaline phosphate conjugate) and NBT/BCIP (18.75 mg/ml nitrobluetetrazolium chloride and 9.4 mg/ml of a 5-bromo-4-chloro-3-indolyl-phosphate in 67% [vol/vol] DMSO) solution.
Identification of transposon insertion sites
The transposon insertion sites in the mutants of interest were identified by a three-step PCR using a DNA Walking SpeedUp Premix Kit (Seegene, Seoul, Korea). This kit uses annealing control primer (ACP) technology [60,61]. The DW-ACP primers provided by the kit are designed to capture unknown target sites, and with primers internal to the known sequence, will amplify DNA fragments possessing the junction region of known and adjacent sequences. The first PCR reaction was performed independently in four individual PCR tubes using the kan-5 primer internal to the kanamycin resistance cassette with one of the DW-ACP 1, 2, 3, or 4 primers. The total 50 μl of the PCR contained genomic DNA of the mutant under study, 4 μl of the DW-ACP primer, the kan-5 primer, distilled water, and 25 μl of 2 × SeeAmp ACP Master Mix II. The PCR program was set as follows: one cycle at 94°C for 5 min, 42°C for 1 min, and 72°C for 2 min; followed by 30 cycles of 94°C 30 s, 55°C 30 s, and 72°C for 100 s. Finally the PCR was extended at 72°C for 7 min. The product from the first PCR was purified using Montage PCR Filter Units (Millipore) and was used as the template for the second PCR. The second PCR with a final volume of 20 μl contained the first PCR product as the template, 1 μl DW-ACPN, a kanamycin resistance cassette-specific kan-6 primer upstream from kan-5, distilled water, and 10 μl of 2 × SeeAmp ACP Master MixII. The PCR program was set as follows: denaturation at 94°C for 3 min; 35 cycles of 94°C 30 s, 56°C 30 s, and 72°C 100 s; then the PCR was extended at 72°C for 7 min. The third PCR used 2 μl of the second PCR product (without purification) as the template. The remaining components consisted of 1 μl universal primer, 1 μl of the kan-7 primer upstream from kan-6, distilled water, and 10 μl of 2 × SeeAmp ACP Master MixII. The third PCR reaction consisted of 94°C for 3 min; followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 100 s; then the PCR was extended at 72°C for 7 min. The products from the third PCR were purified from the agarose gel and sequenced by using the kan-7 primer to determine the junction sequence where the transposon was inserted into the genomic DNA. The sequence of the junction region of the transposon was compared against the S. Typhimurium LT2 genome sequence [GenBank: NC 003197].
Complementation Test
Primers used for complementation test (cafA-F, cafA-R, res-F, res-R, miaA-F, miaA-R, nusA-F, nusA-R, ubiB-F and ubiB-R) were listed in Table 3 and were used to amplify genomic DNA of S. Typhimurium LB5010. The PCR product that contained the full coding sequence of the respective gene was cloned into the yT&A vector (Yeasten Biotech, Taipei, Taiwan) according to the protocol provided by the manufacturer. The resulting plasmid was transformed into the corresponding mutant strain by electroporation. The complemented strains were tested for type 1 fimbrial expression in static broth and on solid agar medium by yeast agglutination test.
Reverse transcription polymerase chain reaction (RT-PCR) analysis
The bacteria were stabilized by adding RNAprotect bacteria reagent (Qiagen, Valencia, CA). Total RNA was prepared by using an RNeasy Mini Kit (Qiagen) and RNase-free DNase (1 unit/1 μg RNA) according to the manufacturer's protocol. RT-PCR was performed using a Fast-Run HotStart RT-PCR (AMV) kit (Protech, Taipei, Taiwan). Briefly, RNA was denatured at 58°C for 5 min, followed by cDNA synthesis at 42°C for 30 min and inactivation at 94°C for 2 min. The following PCR conditions are: 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s. An additional extension was performed at 72°C for 5 min. Primers fimA-F and fimA-R were used to detect the mRNA expression of fimA, while 16S-F and 16S-R primers were used to detect the mRNA expression of 16S rRNA as a control. Densitometry was performed using the Image Quant version 5.2 image analysis software (Molecular Dynamics, GE Healthcare, UK).
Authors' contributions
Y-CC drafted the manuscript. K-CW participated in the sequence alignment and molecular cloning experiments. Y-TC, C-HY, S-CM, and C-CF were responsible for the library screening, molecular cloning, and hybridization experiments. L-HC carried out the complementation tests. K-SY conceived and coordinated the study, and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was sponsored by the Chi Mei Medical Center, Tainan, Taiwan and Taipei Medical University, Taipei, Taiwan under the contract number 94CM-TMU-05. We would like to acknowledge Mr. Nan Jung Li from the Division of Animal Medicine, Animal Technology Institute Taiwan for assistance with electron microscopy. The authors would also like to thank Dr. Steve Clegg from the University of Iowa, Iowa City, U.S.A. for his critical comments on this manuscript.
Contributor Information
Yin-Ching Chuang, Email: chuangkenneth@hotmail.com.
Ke-Chuan Wang, Email: d102094010@tmu.edu.tw.
Yi-Tseng Chen, Email: m102093024@tmu.edu.tw.
Chia-Huei Yang, Email: m102094027@tmu.edu.tw.
Shang-Chin Men, Email: m102094019@tmu.edu.tw.
Chia-Chun Fan, Email: m102094017@tmu.edu.tw.
Li-Huan Chang, Email: m102095020@tmu.edu.tw.
Kuang-Sheng Yeh, Email: ksyeh@tmu.edu.tw.
References
- Todd EC. Epidemiology of food-borne disease: a worldwide review. World Health Stat Q. 1997;50:30–50. [PubMed] [Google Scholar]
- Jones GW, Isaacson RE. Proteinaceous bacterial adhesins and their receptors. Crit Rev Microbiol. 1983;10:229–260. doi: 10.3109/10408418209113564. [DOI] [PubMed] [Google Scholar]
- Duguid JP, Smith IW, Dempster G, Edmunds PN. Non-flagellar filamentous appendages ("fimbriae") and haemagglutinating activity in Bacterium coli. J Pathol Bacteriol. 1995;70:335–348. doi: 10.1002/path.1700700210. [DOI] [PubMed] [Google Scholar]
- Duguid JP, Anderson ES, Campbell I. Fimbriae and adhesive properties in Salmonella. J Pathol Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- Brinton CC., Jr. The structure, function, synthesis and genetic control of bacterial pili and a molecular model of DNA and RNA transport in gram negative bacteria. Trans N Y Acad Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- Duguid JP, Gillies RR. Fimbriae and adhesive properties in dysentery bacilli. J Pathol Bacteriol. 1957;74:397–411. doi: 10.1002/path.1700740218. [DOI] [Google Scholar]
- Althouse C, Patterson S, Fedorka-Cray P, Isaacson RE. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect Immun. 2003;71:6446–6452. doi: 10.1128/IAI.71.11.6446-6452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thankavel K, Shah AH, Cohen MS, Ikeda T, Lorenz RG, Curtiss III R, Abraham SN. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J Biol Chem. 1999;274:5797–5809. doi: 10.1074/jbc.274.9.5797. [DOI] [PubMed] [Google Scholar]
- van der Velden AWM, Bäumler AJ, Tsolis RM, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S, Swenson DL. Salmonella fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, FL , CRC Press; 1994. pp. 105–114. [Google Scholar]
- Old DC., I. Corneil, L. F. Gibson, A. D. Thomson, and J. P. Duguid Fimbriation, pellicle formation and the amount of growth of salmonellas in broth. J Gen Microbiol. 1968;51:1–16. doi: 10.1099/00221287-51-1-1. [DOI] [PubMed] [Google Scholar]
- Old DC, Duguid JP. Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol. 1970;103:447–456. doi: 10.1128/jb.103.2.447-456.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KS, Tinker JK, Clegg S. FimZ binds the Salmonella typhimurium fimA promoter region and may regulate its own expression with FimY. Microbiol Immunol. 2002;46:1–10. doi: 10.1111/j.1348-0421.2002.tb02670.x. [DOI] [PubMed] [Google Scholar]
- Yeh KS, Hancox LS, Clegg S. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J Bacteriol. 1995;177:6861–6865. doi: 10.1128/jb.177.23.6861-6865.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker JK, Hancox LS, Clegg S. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2001;183:435–442. doi: 10.1128/JB.183.2.435-442.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker JK, Clegg S. Control of FimY translation and type 1 fimbrial production by the arginine tRNA encoded by fimU in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2001;40:757–768. doi: 10.1046/j.1365-2958.2001.02430.x. [DOI] [PubMed] [Google Scholar]
- Swenson DL, Kim KJ, Six EW, Clegg S. The gene fimU affects expression of Salmonella typhimurium type 1 fimbriae and is related to the Escherichia coli tRNA gene argU. Mol Gen Genet. 1994;244:216–218. doi: 10.1007/BF00283525. [DOI] [PubMed] [Google Scholar]
- Swenson DL, Clegg S. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J Bacteriol. 1992;174:7697–7704. doi: 10.1128/jb.174.23.7697-7704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker JK, Clegg S. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect Immun. 2000;68:3305–3313. doi: 10.1128/IAI.68.6.3305-3313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland KA, Lucchin S, Hinton JCD, Dorman CJ. The leucine-responsive regulatory protein, Lrp, activates transcription of the fim operon in Salmonella enterica serovar Typhimurium via the fimZ regulatory gene. J Bacteriol. 2008;190:602–612. doi: 10.1128/JB.01388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker JD, Ledeboer NA, Jagnow J, Jones BD, Clegg S. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol Microbiol. 2002;45:1255–1265. doi: 10.1046/j.1365-2958.2002.03121.x. [DOI] [PubMed] [Google Scholar]
- Baumler AJ, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich MJ, Kinsey NE, Vila J, Kadner RJ. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Saulino ET, Hultgren SJ. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr Opin Microbiol. 1998;1:223–231. doi: 10.1016/S1369-5274(98)80015-5. [DOI] [PubMed] [Google Scholar]
- Rouviere-Yaniv J, Gros F. Characterization of a novel, low-molecular weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci USA. 1975;72:3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatwole VM, Somerville RL. Cloning, nucleotide sequence, and characterization of mtr, the structural gene for a tryptophan-specific permease of Escherichia coli K-12. J Bacteriol. 1991;173:108–115. doi: 10.1128/jb.173.1.108-115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J, Reizer A, Saier MHJ. A functional superfamily of sodium/solute symporters. Biochim Biophys Acta. 1994;1197:133–166. doi: 10.1016/0304-4157(94)90003-5. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature (London) 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a "ribosome modulation factor" associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci USA. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Coissac E, Netter P, Bossi L. Unsuspected prophage-like elements in Salmonella typhimurium. Mol Microbiol. 1997;25:161–173. doi: 10.1046/j.1365-2958.1997.4451807.x. [DOI] [PubMed] [Google Scholar]
- Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- Greenblatt J, Li J. Interaction of the sigma factor and the NusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981;24:421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- Blomfield I, van der Woude M. Regulation and function of phase variation in Escherichia coli. In: Wilson W, editor. Bacterial Adhesion to Host Tissues Mechanisms and Consequences. Cambridge, United Kingdom , Cambridge University Press; 2002. pp. 89–113. [Google Scholar]
- Abraham JM, Fretiag CS, Clements JR, Eisenstein BI. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawula TH, Orndorf PE. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histone-like protein H-NS. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield IC, Kulasekara DH, Eisenstein BI. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol Microbiol. 1997;23:705–717. doi: 10.1046/j.1365-2958.1997.2241615.x. [DOI] [PubMed] [Google Scholar]
- Gally DL, Rucker TJ, Blomfield IC. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli. J Bacteriol. 1994;176:5665–5672. doi: 10.1128/jb.176.18.5665-5672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, HKosuge T, Mori H, Perna NT, Plunkett III G, Rudd KE, Serres MH, Thomas GH, Thomason NR, Wishart D, Wanner BL. Escherichia coli K-12: a cooperatively developed annotation snapshot-2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S, Hancox LS, Yeh KS. Salmonella typhimurium fimbrial phase variation and FimA expression. J Bacteriol. 1996;178:542–545. doi: 10.1128/jb.178.2.542-545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend S, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Ougan G, Baumler AJ. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichoison B, Low D. DNA methylation-dependent regulation of Pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- Klemm P, Christiansen G, Kreft B, Marre R, Bergmans H. Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J Bacteriol. 1994;176:2227–2234. doi: 10.1128/jb.176.8.2227-2234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden NJ, Totsika M, Mahler E, Roe J, Catherwood K, Lindner K, Dobrindt U, Gally DL. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiol-SGM. 2006;152:1143–1153. doi: 10.1099/mic.0.28677-0. [DOI] [PubMed] [Google Scholar]
- Nuccio SP, Chessa D, Weening EH, Raffatellu M, Clegg S, Baumler AJ. SIMPLE approach for isolating mutants expressing fimbriae. Appl Environ Microbiol. 2007;73:4455–4462. doi: 10.1128/AEM.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella enterica Sv. Typhimurium. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Umitsuki G, Wachi M, Takada A, Hikichi T, Nagai K. Involvement of RNase G in in vivo mRNA metabolism in Escherichia coli. Genes Cells. 2001;6:403–410. doi: 10.1046/j.1365-2443.2001.00430.x. [DOI] [PubMed] [Google Scholar]
- Morgenroth A, Duguid JP. Demonstration of different mutational sites controlling rhamnose fermentation in FIRN and non-FIRN rha-strains of Salmonella typhimurium: an essay in bacterial archaeology. Genet Res. 1968;11:151–169. doi: 10.1017/s0016672300011320. [DOI] [PubMed] [Google Scholar]
- Barak JD, Gorski L, Naraghi-Arani P, Charkowski AO. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl Environ Microbiol. 2005;71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CH, Friedman DI. Reduced Rho-dependent termination permits NusA independent growth of E. coli. Proc Natl Acad Sci USA. 1994;91:7543–7547. doi: 10.1073/pnas.91.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson JU, Bjork GR. Pleiotropic effects induced by modification deficiency next to the anticodon of tRNA from Salmonella typhimurium LT2. J Bacteriol. 1986;166:1013–1021. doi: 10.1128/jb.166.3.1013-1021.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/JB.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaca: evolution of the "eukaryotic" protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- Winfield MD, Groisman EA. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol. 2003;69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullas LR, Ryu JI. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanish-Perron B, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Galbraith NS. Studies of human salmonellosis in relation to infection in animals. Vet Res. 1961;73:1296–1303. [PubMed] [Google Scholar]
- Terauchi R, Kahl G. Rapid isolation of promoter sequences by TAIL-PCR: the 5'-flanking regions of pal and pgi genes from yams (Dioscorea) Mol Gen Genet. 2000;263:554–560. doi: 10.1007/s004380051201. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-J. [DOI] [PubMed] [Google Scholar]


