Abstract
Type 4 phosphodiesterase (PDE4) inhibitors are emerging new treatment for a number of disorders including asthma and chronic obstructive pulmonary disease. Here we report the biochemical characterization on the second generation inhibitor (+)-1 (L-869298, IC50 = 0.4 nM) and its enantiomer (−)-1 (L-869299, IC50 = 43 nM), and their co-crystal structures with PDE4D at 2.0 Å resolution. In spite of the 107-fold affinity difference, both enantiomers interact with the same sets of residues in the rigid active site. The weaker (−)-1 adopts an unfavorable conformation in order to preserve the pivotal interactions between the Mg-bound waters and N-oxide of pyridine. These structures support a model in which inhibitors are anchored by the invariant glutamine at one end and the metal-pocket residues at another end. This model provides explanation for most of the observed structure-activity relationship and the metal ion dependency of the catechol-ether based inhibitors and should facilitate their further design.
Keywords: L-869298, PDE4, enantiomer, cAMP, crystal structure
Introduction
Cyclic nucleotide phosphodiesterases (PDEs) are binuclear metallohydrolases that catalyze the hydrolysis of second messengers cAMP and cGMP. Eleven families of PDEs, encoded by twenty one genes and sharing a conserved catalytic domain of about 300 amino acids, are differentially expressed in various human tissues.1–5 Over the last two decades, a number of PDE inhibitors have been developed for therapeutic usage. For examples, cilostazol, a PDE3 inhibitor, is beneficial in the treatment of macrovascular disorders such as intermittent claudicating.6 Ibudilast, an effective treatment for asthma, cerebrovascular disorders, and allergic conjunctivitis, is a PDE4-preferred inhibitor.7 The PDE5 inhibitors, sildenafil, vardenafil, and tadalafil, are widely used in the treatment of male erectile dysfunction.8
The cAMP-specific type 4 phosphodiesterases are encoded by 4 genes (PDE4A to PDE4D) and are particularly abundant in inflammatory cells, immune cells, sensory neurons, airway smooth muscles, and airway epithelium. Inhibition of PDE4 will elevate cAMP and initiate a series of biological responses, including blockage of cell trafficking and proliferation, attenuation of the production of inflammatory cytokines and reactive oxygen species, and enhancement of the mucociliary clearance through CFTR activation.9 PDE4 inhibitors are clinically efficacious in treatment of asthma and chronic obstructive pulmonary disease (COPD) and in models of rheumatoid arthritis, multiple sclerosis, septic shock and neurological disorders.10–18 They may emerge as new therapeutics for a number of disorders in the near future.13
The promise of identifying additional novel therapeutics among family-specific PDE inhibitors has led to an intense effort to study their interaction at the atomic level. The understanding of ligand binding and PDE functions has been greatly enriched by the crystal structures of the catalytic domains of PDE1B,19 PDE2A,20 PDE3B,21 PDE4B and PDE4D,19, 22–29 PDE5A,19,26,30 PDE7A,31 and PDE9A.32 In addition to solidifying the bimetallic catalytic machinery hypothesis of PDEs,9 these structures have identified a number of interactions such as the glutamine-mediated hydrogen bond for nucleotide differentiation and shed light on the common and selective recognition of various inhibitors.19,25,28 However, it remains unclear how PDE recognizes enantiomeric inhibitors that often show over ten fold affinity difference. Here we report the kinetic and structural studies on two PDE4 enantiomeric inhibitors, (+)-1 and (−)-1, Fig. 1), and their complexes with PDE4D. Comparison of the cocrystal structures reveals that this pair of enantiomers interacts with the similar PDE4 residues, but the weaker inhibitor (−)-1 adopts a restrained conformation in order to preserve the other key interactions and to fit into the rigid pocket.
Fig. 1.
Chemical structures of PDE4 selective inhibitors. (+)-1 or (−)-1 has the chiral center at carbon-9, and can be divided into five subgroups (CORE and R1 to R4) to facilitate discussion.
Results and Discussion
Biochemical characteristics of the enantiomeric PDE4 inhibitors
The more potent enantiomer (+)-1, with its absolute configuration assigned previously by x-ray crystallographic analysis (Fig. 1), is a non-emetic 2nd generation PDE4-specific inhibitor exhibiting good in vivo efficacy in several animal models of pulmonary function with a wide therapeutic index on emesis and prolongation of the QTc interval.33 It potently inhibited all four PDE4 subtypes with the IC50 values of 0.4, 0.4, 1.1 and 0.3 nM against the catalytic domains of PDE4A, PDE4B, PDE4C and PDE4D, respectively. The less potent isomer (−)-1, with an enantiomeric purity at 99.7%, has the corresponding IC50 values of 56, 40, 135, and 43 nM, respectively (Table 1). The potencies of (+)-1 and (−)-1 against four representative full-length PDE4 variants of PDE4A4, PDE4B2, PDE4C2 and PDE4D3 are similar to those of the catalytic domains (data not shown). A linear dependence of the IC50 values for (+)-1 inhibition of PDE4A on cAMP concentration was obtained, indicating that it is an active site-directed competitive inhibitor (Fig. 2a). The linear plot yielded an apparent Ki value of 0.43 ± 0.07 nM. In addition, the activity of (+)-1-inhibited PDE4A was completely recovered within 90s after a 120-fold dilution, indicating that it is a rapidly reversible inhibitor (Fig. 2b). These data support that they are active site-directed and rapidly reversible inhibitors with comparable potency against the four PDE4 isoforms.
Table 1.
PDE4A, 4B, 4C and 4D Activity Inhibition by (+)-1 and (−)-1 (IC50, nM)
| Stereochemistry | Enantiomeric purity (ee*) | PDE4A | PDE4B | PDE4C | PDE4D | |
|---|---|---|---|---|---|---|
| (+)-1 (n=9) | (S)-(+) | 99.0% | 0.4 ± 0.2 | 0.4 ± 0.2 | 1.0 ± 0.2 | 0.4 ± 0.1 |
| (−)-1 (n=3) | (R)-(−) | 99.7% | 56 ±14 | 40 ±9 | 135 ±32 | 43 ±21 |
| IC50 Ratio((+)-1/(−)-1) | 140 | 100 | 135 | 107 |
IC50 values represent mean (± SD) using 0.1 μM cAMP as substrate concentration (see method for details). Under the conditions, they are close to their apparent Ki values.
ee, enantiomeric purity in excess.
Fig. 2.
Kinetic properties of (+)-1. (A) Inhibition of PDE4A by (+)-1 with respect to increased cAMP concentration. The linear response has an intercept of 0.43 ± 0.07 nM (apparent Ki) and a slope of 0.19 ± 0.02 nM/μM cAMP. (B). Rapidly reversible PDE4A inhibition by (+)-1. The activity (mean ± se, n = 3) of PDE4A inhibited with 15 nM (+)-1 was determined at 30, 60, 120, and 180 seconds following a 120-fold volume dilution. 100% activity represents the average of the corresponding controls (with DMSO as vehicle). Recovery time represents the dilution time plus the assay duration (60s) except for the 0s point for (+)-1, which represents the enzyme activity in the presence of 15 nM (+)-1.
Conformational conservation of the PDE4D catalytic domain
The cocrystals of PDE4D2 catalytic domain (amino acids 79–438) in complex with (+)-1 and (−)-1 contain four PDE4D2-inhibitor complexes in the asymmetric unit, which are apparently associated into a tetramer in the crystal state. The monomeric PDE4D2 molecule consists of 16 helices and two divalent metal ions (Fig. 3), as previously reported.25 A superposition between the monomers of the tetramers yields root-mean-squared (RMS) deviations of 0.38 and 0.32 Å for the backbone atoms of residues 79–412 in the PDE4D2-(+)-1 and PDE4D2-(−)-1 crystals, suggesting a conformational similarity within the tetramer. The superposition of the monomers of the PDE4D2-(−)-1 tetramer over the corresponding ones of the unliganded PDE4D2 or the PDE4D2-(+)-1 complex yields the RMS deviations of 0.21 and 0.28 Å for backbone atoms in the catalytic domains, respectively. These comparisons suggest no significant changes of the overall conformation of the catalytic domains upon enantiomer binding, and the rigidity of the PDE4D2 active site.
Fig. 3.
PDE4D2-inhibitor structures. (A) Ribbon diagram of monomeric PDE4D2 catalytic domain. (+)-1 and (+)-1 are shown as the green and golden sticks, respectively. Divalent metal Zn is drawn as a red ball and Mg as a purple ball. (B) Binding of (+)-1 (green sticks and balls) and (+)-1 (golden sticks) at the active site of PDE4D2. The residues of PDE4D2, which are involved in binding with the inhibitors, are labeled. The four metal binding residues are shown in light blue.
Binding of the enantiomeric inhibitors
In spite of the 107-fold affinity difference, (+)-1 and (−)-1 bind to the active site of PDE4D2 with similar orientations and interact with the same residues (Fig. 3, Table 2). The structures of the two enantiomers are divided into five subgroups and labeled as CORE and R1 to R4 in Fig. 1 to facilitate discussion. Since most subgroups of the two inhibitors interact with similar residues of PDE4D2, the discussion below will not differentiate them unless different contacts are observed. The catechol-ether group (CORE in Fig. 1) of both enantiomers forms two hydrogen bonds with the side chain of Gln369, an invariant residue among all PDEs (Table 2). In addition, its phenyl ring stacks against Phe372 and also contacts with Tyr159, Asn321, Ile336, and Gln369 via van der Waals’ forces. The difluoromethyl group (R1) is located in a small pocket interacting with Asn321, Pro322, Tyr329, Trp332, Thr333, Ile336, and Gln369. The cyclopropyl group (R2) sits in a hydrophobic pocket, interacting with residues Met357, Gln369 and Phe372. The hexafluoro-propyloxy group and the thiazole ring (R3) are surrounded by Met273, Phe340, Met357, Phe372, and Ile376.
Table 2.
Interactions of (+)-1 and (−)-1 with PDE4D2
| Hydrogen bonds | |||
|---|---|---|---|
| Inhibitor atom | Protein atom | Average distance (Å) | |
| (+)-1 | (−)-1 | ||
| O1 … | Gln369 NE2 | 3.13 ± 0.05 | 3.10 ± 0.04 |
| O2 … | Gln369 NE2 | 3.02 ± 0.08 | 2.99 ± 0.06 |
| O4 … | H2O (W7) | 2.95 ± 0.11 | 2.56 ± 0.06 |
| O4 … | H2O (W8) | 2.93 ± 0.12 | 3.43 ± 0.08 |
| Van der Waals’ interactions of (+)-1 and (+)-1 | |||
| difluoromethy1 | Asn321, Pro322, Tyr329, Trp332, Thr333, Ile336, Gln369 | ||
| Propyl ring | Met357, Gln369, Phe372 | ||
| phenyloxy | Tyr159, Asn321, Ile336, Gln369, Phe372 | ||
| pentyl ring | Met273, Phe340, Phe372 | ||
| F2 group | Met273, Met357, Ile376 | ||
| Pyridine-N-oxide | Thr271, Met273, Asp318, Leu319 | ||
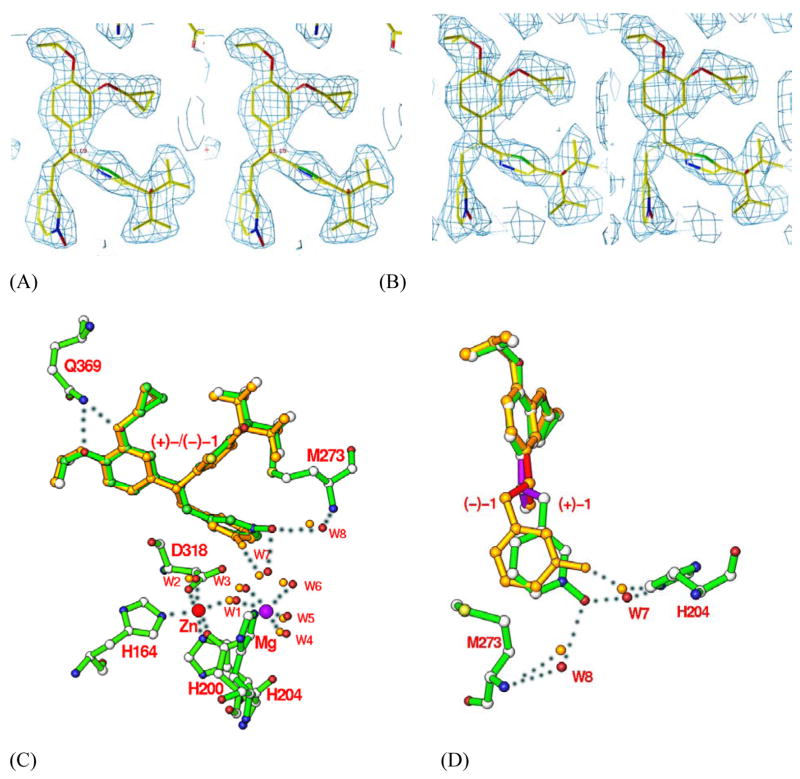
The pyridine-N-oxide (R4) orients towards the metal binding pocket and interact with residues Thr271, Met273, Asp318, and Leu319 (Figs. 3 & 4). The oxygen of the N-oxide in (+)-1 (O4 in Fig. 1 and Table 2) forms two hydrogen bonds with water molecules W7 and W8 that are bound respectively to His204 and Met273. However, only one hydrogen bond between W7 and the oxygen O4 of pyridine-N-oxide is found in the PDE4D2-(−)-1 complex. The hydrogen bond (2.9 Å) between pyridine-N-oxide and W8 in (+)-1 became a polar interaction with an average distance of 3.4 Å in the PDE4D2-(−)-1 structure (Fig. 4). The R4 group interacts indirectly with the divalent metals via a network of bound water molecules. There are three waters (W1, W2 and W3) that form van der Waals’ interactions with the pyridine ring. A Mg-bound W3 is 3.5 Å to the nitrogen of N-oxide in (+)-1 and 3.1 Å in (−)-1. Water W3 also impacts the inhibitor binding via a hydrogen bond relay through W7 (Fig. 4). These indirect interactions imply a role of the metal ions in the inhibitor binding.
Fig. 4.
Binding of (+)-1 and (−)-1 (L869298 and L869299) to the active site of PDE4D2. Stereoview of electron density for (+)-1 (A) and (−)-1 (B). The omitted (2Fo-Fc) maps were contoured at 1.5σ and 1.0σ for (+)-1 and (−)-1, respectively. (C) Superposition of (+)-1 (green) over (−)-1 (golden). The oxygen of pyridine-N-oxide of (+)-1 forms two hydrogen bonds with waters W7 and W8 that bound to His204 and Met273 (dotted lines), respectively. However, (−)-1 forms only one hydrogen bond with water W7 and has a distance of 3.4 Å to water W8. (D) Another view of the superposition of (+)-1 over (−)-1. The three bonds around the chiral center are related by a mirror symmetry and shown in purple color for (+)-1 and red for (−)-1. The R3 groups in (+)-1 and (−)-1 are omitted for a clear view of the enantiomeric configuration.
On the basis of the 107-fold affinity difference, one might suspect that (+)-1 and (−)-1 would interact with somewhat different residues. However, the cocrystal structures revealed that all subgroups of the inhibitors are bound similarly (Figs. 3 & 4, Table 2). The conformational difference of the bound enantiomers is limited to the arrangement of the four single bonds around the chiral center, which is in a proximate mirror-image relationship (Fig. 4d). Among the possible reasons, the change of the hydrogen bond between pyridine-N-oxide and water W8 in (+)-1 to a polar interaction in (−)-1 (Fig. 4) may account for a portion of the affinity difference. Another difference lies at the conformation of the bound enantiomers. While most subgroups of (+)-1 and (−)-1 are superimposable to each other, the dihedral angle around the C5-C9 bond (Fig. 1) is very different. This dihedral angle is about −70° relative to R4 in (+)-1, which is closer to the energetically favored “staggered” conformation of −60°. In comparison, the dihedral angle in (−)-1 is about 20°, which is closer to the unfavorable “eclipsed” conformation of 0°. It appears that, in order to preserve the favorable pyridine-N-oxide interaction at the metal pocket and, in the meantime, to accommodate the chirality, (−)-1 is forced into a distorted conformation in the rigid PDE4D active site.
Metal pocket serves as an anchor for high affinity binding of inhibitors
Previous structural studies have established that the common catalytic machinery of PDEs comprises two divalent cations with different affinities. The anomalous scattering of the PDE crystal at the absorption edge of zinc supports that the tightly bound cation is a Zn2+ ion.22 Even though a number of divalent cations including Mg2+, Co2+, Zn2+ and Ni2+ can support PDE catalysis in vitro, the loosely bound metal ion is likely a Mg2+ ion in vivo, since only its EC50 value at activating catalysis is within its free intracellular concentration range.34 The structures of PDEs in complex with various inhibitors have revealed two common factors for binding of most PDE inhibitors: the hydrophobic clamp for the catechol-ether ring of rolipram and the hydrogen bonds between catechol-ether oxygen and Gln369.25,27–29,32 The structures of PDE4D in complex with (+)-1 and (−)-1 support the early observation, but also suggest that hydrogen-bond network near the metal center serves as the third element for high affinity binding of PDE inhibitors. The hydrogen bonds with Gln369 and the hydrophobic clamp interaction alone appear to be insufficient for securing a high affinity inhibitor interaction. Earlier binding studies have uncovered that the Mg ion is responsible for eliciting the cAMP binding and the high affinity interaction of many catechol-ether based PDE4 inhibitors (Fig. 1).9,35 For example, the productive and high affinity cAMP binding decreased from 2 μM to a nonproductive binding of 179 μM, and the binding of cilomilast decreased from a Kd of 42 nM to > 4000 nM upon the removal of the Mg. The molecular origin of this Mg-dependency for cilomilast (Fig. 1) has now been resolved by the presence of two hydrogen-bonds between the oxygens of its carboxylate and Mg-bound waters in its cocrystals.28 Upon the removal of Mg, interaction of PDE4A with (+)-1 decreased by approximately 275-fold to a Kd of ~ 110 nM in comparison to its original Ki of 0.4 nM (data not shown). The interactions of the Mg-bound water (W3) directly and indirectly (through W7) with the N-oxide likely contribute significantly to this affinity difference. In short, this study supports a coordinated “two point anchoring model”, in which the hydrogen bonding to the glutamine at one end and the hydrophilic interactions in the metal pocket at the other end act as the pivotal factors in eliciting the high affinity binding of catechol-ether based PDE4 inhibitors.
Implication on PDE4 inhibitor optimization
Numerous PDE4 inhibitors with diverse structures have been synthesized over the past years, as exemplified in the earlier comprehensive review.11 The structures of PDE4B and PDE4D in complex with a number of more advanced inhibitors sharing the catechol-ether scaffold, including rolipram, cilamilast, roflumilast, mesopram, and piclamilast (Fig. 1), have been reported.23,25,28 Superposition of these structures revealed several common interacting features. The catechol-ether group (CORE in Fig. 1) appears to serve as a common element in the formation of the hydrogen bonds with the invariant glutamine, which likely contributes partially to the binding affinity.19,23–29,32 Other components for inhibitor binding in this area are the stacking against a conserved phenylalanine and the hydrophobic interactions with Ile336 and Phe340 in PDE4D2.25,28 It is interesting to note that the distance between (+)-1 (or (−)-1) and Phe340 is slightly longer than 4 Å, thus implying a general hydrophobic environment for the interactions.
The residues interacting with the R1 group vary significantly across different PDE families and may be, in principle, useful for improving family-specificity of inhibitors. However, this sub-pocket is relatively small and accommodates only three or so atoms such as difluoromethyl group. The R2 group sits in a hydrophobic pocket, and thus hydrophobic replacement of R2 group, such as replacing the cyclic propyl with the cyclic butyl, would not impact significantly the binding affinity. On the other hand, the introduction of polar atoms into the pocket is unfavorable, as reflected by ~ 70-fold decreased affinity when R2 is CF3.33 The different amino acids and volume of this sub-pocket among PDE4, 5, 7 and 927,31,33 offer the opportunity to improve family-specificity of inhibitors through R2 modifications. The R3 subgroup orients to an open space which is surrounded by the hydrophobic residues Met273, Phe340, Met357, Phe372 and Ile376. A hydrophobic group, such as the phenyl in CDP-840 (5 nM, PDE4D, Fig. 1), bis-trifluoromethyl-phenyl in L-826141 (0.3 nM) and bis-trifluoromethyl-thiazole in (+)-1 (0.4 nM), significantly enhances the affinities of these inhibitors, likely through desolvation contribution.36 The R4 group sits next to the central catalytic machinery, the bimetallic ion center. This sub-pocket is formed by a set of highly conserved residues across PDE families. The partial and/or rapidly reversible occupancy of the weaker Mg site leads to the differential binding of inhibitors, including the high and low affinity binding of rolipram.9 Therefore, the modification of R4, which is pivotal in eliciting and coordinating the high affinity interaction of catechol-ether based inhibitors, might not lead to selectivity improvement.
Experimental section
Chemicals
(S)-(+)-3-{2-[(3-Cyclopropyloxy-4-difluromethoxy)phenyl]-2-[5-(2-(1-hydroxy-1-trifluoromethyl-2,2,2-trifluoro)ethyl)thiazolyl]ethyl}pyridine N-Oxide ((+)-1) and (R)-(−)-3-{2-[(3-Cyclopropyloxy-4-difluromethoxy)phenyl]-2-[5-(2-(1-hydroxy-1-trifluoromethyl-2,2,2-trifluoro)ethyl)thiazolyl]ethyl}pyridine N-Oxide ((−)-1) were prepared as described previously.33 The enantiomeric purity of (+)-1 was approximately at 99%. Higher enantiomeric purity was required for (−)-1 in forming the PDE4D cocrystal. The starting (−)-1 with a 98.0% was further purified by HPLC using a CHIRALPAK AD column (eluting with 10% ethanol in hexanes) and by crystallization from ethanol/pentane to give a white solid. Analytical HPLC using the same conditions indicated that the enantiomeric purity of the final (−)-1 was at 99.7%.
PDE4A, PDE4B, PDE4C and PDE4D activity assays
The hydrolysis of cAMP by the four PDE4 isoforms were monitored as previously described.35 The enzymes used are the recombinant Gln-Thr (QT) versions of PDE4A, 4B, 4C and 4D, which encode the catalytic domains of PDE4s and have been shown to be in a fully activated conformation.9 They were generated by creating glutathione fusion constructs in frame with the QT located within UCR2 of PDE4A4, PDE4B2, PDE4C2 and PDE4D3 sequences. These QT constructs, encoding approximately the maximal common regions of all PDE4 variants, were expressed and purified to homogeneity from Sf9 cells. Their PDE activity was monitored using 0.1 μM [3H]-cAMP in a buffer containing 10 mM MgCl2, 1 mM EDTA, 100 mM KCl and 20 mM HEPES (pH 7.5). Compounds were introduced via 2 μL of DMSO as vehicle which exerted minimal perturbation on PDE activity. The IC50 values were similar to their apparent Ki values under the conditions from the use of 0.1 μM cAMP in comparison with their similar Km of ~ 2 μM.
Crystallization and data collection
The protein expression and purification of the catalytic domain of PDE4D2 (amino acids 79–438) have been described previously.25 The protein-inhibitor complexes were prepared by mixing 2 mM (+)-1 (or (−)-1) with 10 mg/mL PDE4D2 in a storage buffer of 50 mM NaCl, 20 mM Tris.HCl (pH 7.5), 1 mM β-mercaptoethanol, and 1 mM EDTA. The crystals of the catalytic domain of PDE4D2 in complex with (+)-1 were grown by vapor diffusion against a well buffer of 0.1 M HEPES (pH 7.5), 15% PEG3350, 25% ethylene glycol, 5% isopropanol, and 5% glycerol at 4°C. The catalytic domain of PDE4D2 in complex with (−)-1 was crystallized against a well buffer of 0.05 M HEPES (pH 7.5), 15% PEG3350, 25 % ethylene glycol, and 5% methanol at 4°C. Both crystals have the space group P212121. The diffraction data were collected on beamlines X26C ((+)-1) and X25 ((−)-1) at Brookhaven National Laboratory (Table 3), and processed with program HKL2000.37
Table 3.
Statistics on diffraction data and structure refinement of PDE4D-inhibitors
| Data collection | (+)-1 | (−)-1 |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit cell (a, b, c, Å) | 99.5, 112.2, 160.2 | 99.4, 112.8, 161.1 |
| Resolution (Å) | 2.0 | 2.03 |
| Total measurements | 836,316 | 499,063 |
| Unique reflections | 121,450 | 112,573 |
| Completeness (%) | 99.8 (99.0)* | 96.4 (84.9) |
| Average I/σ | 12.5 (3.5)* | 11.1 (3.0) |
| Rmerge | 0.080 (0.49)* | 0.092 (0.44) |
| Structure Refinement | ||
| R-factor | 0.223 | 0.231 |
| R-free | 0.245 | 0.260 |
| Resolution | 50-2.0 Å | 50-2.03 |
| Reflections | 116,679 | 106,847 |
| RMS deviation for | ||
| Bond (Å) | 0.0057 | 0.0064 |
| Angle | 1.19° | 1.20° |
| Average B-factor (Å2) | ||
| All atoms | 33.8 | 30.0 |
| Protein | 33.9 | 30.1 |
| Inhibitor | 48.8 | 40.1 |
| Zn | 26.6 | 23.2 |
| Mg | 24.4 | 20.7 |
| Water | 31.4 | 26.0 |
The numbers in parentheses are for the highest resolution shell.
Structure determination
The crystals of PDE4D2 in complex with inhibitors (+)-1 and (−)-1 contain a tetramer in the crystallographic asymmetric unit. The structures were solved by molecular replacement program AMoRe,38 using the catalytic domain of unliganded PDE4D2 as the initial model.25 The tetramer of PDE4D2-(+)-1 and PDE4D2-(−)-1 were optimized by rigid-body refinement of CNS.39 The electron density maps were improved by the density modification package of CCP4.40 The atomic model was rebuilt by program O41 and refined by CNS (Table 3).
Acknowledgments
We thank Beamlines X25 and X26C at NSLS for collection of diffraction data, Drs. Robert N. Young, Yves Girard and Richard W. Friesen at Merck Frosst for many helpful discussions. This work was supported in part by NIH (GM59791 to H.K).
Footnotes
Protein Data Bank accession codes
The atomic coordinates and diffraction data have been deposited into the RCSB Protein Data Bank with accession codes 2FM0 and 2FM5
References
- 1.Soderling SH, Beavo JA. Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr Opin Cell Biol. 2000;12:174–179. doi: 10.1016/s0955-0674(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 2.Conti M, Richter W, Mehats C, Livera G, Park JY, Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J Biol Chem. 2003;278:5493–5496. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- 3.Houslay MD, Adams DR. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maurice DH, Palmer D, Tilley DG, Dunkerley HA, Netherton SJ, Raymond DR, Elbatarny HS, Jimmo SL. Cyclic nucleotide phosphodiesterase activity, expression, and targeting in cells of the cardiovascular system. Mol Pharmacol. 2003;64:533–546. doi: 10.1124/mol.64.3.533. [DOI] [PubMed] [Google Scholar]
- 5.Goraya TA, Cooper DM. Ca2+-calmodulin-dependent phosphodiesterase (PDE1): current perspectives. Cell Signal. 2005;17:789–797. doi: 10.1016/j.cellsig.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Chapman TM, Goa KL. Cilostazol: a review of its use in intermittent claudication. Am J Cardiovasc Drugs. 2003;3:117–138. doi: 10.2165/00129784-200303020-00006. [DOI] [PubMed] [Google Scholar]
- 7.Huang Z, Liu S, Zhang L, Salem M, Greig GM, Chan CC, Natsumeda Y, Noguchi K. Preferential inhibition of human phosphodiesterase 4 by ibudilast. Life Science. 2005 doi: 10.1016/j.lfs.2005.10.026. in press. [DOI] [PubMed] [Google Scholar]
- 8.Corbin JD, Francis SH. Pharmacology of phosphodiesterase-5 inhibitors. Int J Clin Pract. 2002;56:453–459. [PubMed] [Google Scholar]
- 9.Liu S, Laliberte F, Bobechko B, Bartlett A, Lario P, Gorseth E, Van Hamme J, Gresser MJ, Huang Z. Dissecting the cofactor-dependent and independent bindings of PDE4 inhibitors. Biochemistry. 2001;40:10179–10186. doi: 10.1021/bi010096p. [DOI] [PubMed] [Google Scholar]
- 10.Barnette MS, Underwood DC. New phosphodiesterase inhibitors as therapeutics for the treatment of chronic lung disease. Curr Opin Pulmonary Med. 2000;6:164–169. doi: 10.1097/00063198-200003000-00014. [DOI] [PubMed] [Google Scholar]
- 11.Piaz VD, Giovannoni P. Phosphodiesterase 4 inhibitors, structurally unrelated to rolipram, as promising agents for the treatment of asthma and other pathologies. Eur J Med Chem. 2000;35:463–480. doi: 10.1016/s0223-5234(00)00179-3. [DOI] [PubMed] [Google Scholar]
- 12.Souness JE, Aldous D, Sargent C. Immunosuppressive and anti-inflammatory effects of cyclic AMP phophodiesterase (PDE) type 4 inhibitors. Immunopharmacol. 2000;47:127–162. doi: 10.1016/s0162-3109(00)00185-5. [DOI] [PubMed] [Google Scholar]
- 13.Huang Z, Ducharme Y, Macdonald D, Robichaud A. The next generation of PDE4 inhibitors. Curr Opin Chem Biol. 2001;5:432–438. doi: 10.1016/s1367-5931(00)00224-6. [DOI] [PubMed] [Google Scholar]
- 14.Giembycz MA. Development status of second generation PDE 4 inhibitors for asthma and COPD: the story so far. Monaldi Arch Chest Dis. 2002;57:48–64. [PubMed] [Google Scholar]
- 15.Sturton G, Fitzgerald M. Phosphodiesterase 4 inhibitors for the treatment of COPD. Chest. 2002;121:192s–196s. doi: 10.1378/chest.121.5_suppl.192s. [DOI] [PubMed] [Google Scholar]
- 16.Banner KH, Trevethick MA. PDE4 inhibition: a novel approach for the treatment of inflammatory bowel disease. Trends Pharmacol Sci. 2004;25:430–436. doi: 10.1016/j.tips.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Castro A, Jerez MJ, Gil C, Martinez A. Cyclic nucleotide phosphodiesterases and their role in immunomodulatory responses: advances in the development of specific phosphodiesterase inhibitors. Med Res Rev. 2005;25:229–244. doi: 10.1002/med.20020. [DOI] [PubMed] [Google Scholar]
- 18.Lipworth BJ. Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet. 2005;365:167–175. doi: 10.1016/S0140-6736(05)17708-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang KY, Card GL, Suzuki Y, Artis DR, Fong D, Gillette S, Hsieh D, Neiman J, West BL, Zhang C, Milburn MV, Kim SH, Schlessinger J, Bollag G. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol Cell. 2004;15:279–286. doi: 10.1016/j.molcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Iffland A, Kohls D, Low S, Luan J, Zhang Y, Kothe M, Cao Q, Kamath AV, Ding YH, Ellenberger T. Structural determinants for inhibitor specificity and selectivity in PDE2A using the wheat germ in vitro translation system. Biochemistry. 2005;44:8312–8325. doi: 10.1021/bi047313h. [DOI] [PubMed] [Google Scholar]
- 21.Scapin G, Patel SB, Chung C, Varnerin JP, Edmondson SD, Mastracchio A, Parmee ER, Singh SB, Becker JW, Van der Ploeg LH, Tota MR. Crystal structure of human phosphodiesterase 3B: atomic basis for substrate and inhibitor specificity. Biochemistry. 2004;43:6091–6100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 22.Xu RX, Hassell AM, Vanderwall D, Lambert MH, Holmes WD, Luther MA, Rocque WJ, Milburn MV, Zhao Y, Ke H, Nolte RT. Atomic structure of PDE4: Insight into phosphodiesteras mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 23.Xu RX, Rocque WJ, Lambert MH, Vanderwall DE, Luther MA, Nolte RT. () Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J Mol Biol. 2004;337:355–365. doi: 10.1016/j.jmb.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 24.Lee ME, Markowitz J, Lee JO, Lee H. Crystal structure of phophodiesterase 4D and inhibitor complex. FEBS Lett. 2002;530:53–58. doi: 10.1016/s0014-5793(02)03396-3. [DOI] [PubMed] [Google Scholar]
- 25.Huai Q, Wang H, Sun Y, Kim HY, Liu Y, Ke H. Three dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 26.Huai Q, Colicelli J, Ke H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- 27.Huai Q, Liu Y, Francis SH, Corbin JD, Ke H. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor IBMX suggest a conformation determinant of inhibitor selectivity. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 28.Card GL, England BP, Suzuki Y, Fong D, Powell B, Lee B, Luu C, Tabrizizad M, Gillette S, Ibrahim PN, Artis DR, Bollag G, Milburn MV, Kim SH, Schlessinger J, Zhang KY. Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure. 2004;12:2233–2247. doi: 10.1016/j.str.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Card GL, Blasdel L, England BP, Zhang C, Suzuki Y, Gillette S, Fong D, Ibrahim PN, Artis DR, Bollag G, Milburn MV, Kim SH, Schlessinger J, Zhang KY. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat Biotechnol. 2005;23:201–207. doi: 10.1038/nbt1059. [DOI] [PubMed] [Google Scholar]
- 30.Sung BJ, Yeon HK, Ho JY, Lee JI, Heo YS, Hwan KJ, Moon J, Min YJ, Hyun YL, Kim E, Jin ES, Park SY, Lee JO, Gyu LT, Ro S, Myung CJ. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature. 2003;425:98–102. doi: 10.1038/nature01914. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Liu Y, Chen Y, Robinson H, Ke H. Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J Biol Chem. 2005;280:30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 32.Huai Q, Wang H, Zhang W, Colman R, Robinson H, Ke H. Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor 3-isobutyl-1-methylxanthine binding. Proc Natl Acad Sci USA. 2004;101:9624–9629. doi: 10.1073/pnas.0401120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friesen RW, Ducharme Y, Ball RG, Blouin M, Boulet L, Cote B, Frenette R, Girard M, Guay D, Huang Z, Jones TR, Laliberte F, Lynch JJ, Mancini J, Martins E, Masson P, Muise E, Pon DJ, Siegl PK, Styhler A, Tsou NN, Turner MJ, Young RN, Girard Y. Optimization of a tertiary alcohol series of phosphodiesterase-4 (PDE4) inhibitors: structure-activity relationship related to PDE4 inhibition and human ether-a-go-go related gene potassium channel binding affinity. J Med Chem. 2003;46:2413–2426. doi: 10.1021/jm0204542. [DOI] [PubMed] [Google Scholar]
- 34.Liu S, Laliberte F, Bobechko B, Bartlett A, Lario P, Gorseth E, Van Hamme J, Gresser MJ, Huang Z. Dissecting the cofactor-dependent and independent bindings of PDE4 inhibitors. Biochemistry. 2001;40:10179–10186. doi: 10.1021/bi010096p. [DOI] [PubMed] [Google Scholar]
- 35.Laliberte F, Han Y, Govindarajan A, Giroux A, Liu S, Bobechko B, Lario P, Bartlett A, Gorseth E, Gresser M, Huang Z. Conformational difference between PDE4 apoenzyme and holoenzyme. Biochemistry. 2000;39:6449–6458. doi: 10.1021/bi992432w. [DOI] [PubMed] [Google Scholar]
- 36.Frenette R, Blouin M, Brideau C, Chauret N, Ducharme Y, Friesen RW, Hamel P, Jones TR, Laliberte F, Li C, Masson P, McAuliffe M, Girard Y. Substituted 4-(2,2-diphenylethyl)pyridine-N-oxides as phosphodiesterase-4 inhibitors: SAR study directed toward the improvement of pharmacokinetic parameters. Bioorg Med Chem Lett. 2002;12:3009–3013. doi: 10.1016/s0960-894x(02)00615-7. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Navaza J, Saludjian P. AMoRe: an automated molecular replacement program package. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 39.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]