Abstract
Here, we give a historical overview of the search for genetic variants that influence the susceptibility of an individual to a chronic disease, from RA Fisher's seminal work to the current excitement of whole-genome association studies (WGAS). We then discuss the concepts behind the identification of common variants as disease causal factors and contrast them to the basic ideas that underlie the rare variant hypothesis. The identification of rare variants involves the careful selection of candidate genes to examine, the availability of highly efficient resequencing techniques and the appropriate assessment of the functional consequences of the implicated variant. We believe that this strategy can be successfully applied at present in order to unravel the contribution of rare variants to the multifactorial inheritance of common diseases, which could lead to the implementation of much needed preventative screening schemes.
The study of ‘quantitative’ inheritance based on mendelian principles was pioneered by R.A. Fisher in 1918 (ref. 1). His paper first introduced the term ‘variance’ in its modern sense, as well as the analysis of variance. However, after one further key paper extending these ideas2, he wrote in a letter in 1932, referring to the potential for serological studies and their likely ability to detect gene products, that “…such work is going to lead to a greater advance, both theoretical and practical, in the problems of human genetics than can be expected from any further work on biometrical or genealogical lines.” John Thoday, who succeeded Fisher as Professor of Genetics at Cambridge in 1959, introduced the idea in 1961 of what are now called ‘QTLs’ (quantitative trait loci)—namely, of using genetic mapping techniques to identify specific genes affecting a quantitative trait, in his case, Drosophila bristle number3. It is that idea, applied to human genetics for the identification of distinct genes affecting disease susceptibility, which underlies the present enormous flurry of activity in whole-genome association studies. The aim of this review is to describe the historical background of the ideas behind such studies, and then to provide an overview and critical interpretation of the many recent studies identifying common variants influencing the incidence of common multifactorial diseases, and to contrast these data with the evidence for the substantial contribution of rare variants.
Historical background
ABO and disease associations
E.B. Ford was a close associate of Fisher and a pioneer of what he called ‘ecological genetics’, particularly the study of natural selection in natural populations. In 1945, Ford urged a search for associations between the ABO blood groups and disease in order to explain the selection he assumed was needed for the maintenance of the ABO polymorphism. The first such association, described in 1953 (ref. 4), was between ABO types and stomach cancer. A 1961 (ref. 5) summary of data on ABO and disease associations is shown in Table 1 (Table 5.4 in ref. 6). Several of the odds ratios (ORs) listed are on the high side of those now being found by WGAS for a variety of common chronic diseases, with similarly low probabilities. The genes determining ABO types were effectively the first candidate genes, but there is so far no convincing explanation for these associations. Indeed, they are almost forgotten, perhaps because of the much larger ORs later found for associations between HLA types and certain diseases.
Table 1.
ABO and disease association
| Disease | ABO allele | OR | χ 2 |
|---|---|---|---|
| Duodenal ulcer | O | 1.40 | 200 |
| Stomach cancer | A | 1.25 | 49 |
| Stomach ulcer | O | 1.82 | 37 |
| Pernicious anemia | A | 1.50 | 17 |
| Pancreas cancer | A | 1.27 | 8 |
As reported in ref. 5.
HLA and disease associations: the importance of linkage disequilibrium
The idea of doing studies on the association between HLA types and disease was first discussed around the mid-1960s, largely stimulated by Ruggero Ceppellini (a pioneer of early HLA studies who coined the word ‘haplotype’ in 1967) and based on the association between inherited blood disorders and malaria, a suggestion made by J.B.S. Haldane in 1949 (ref. 7). The first published study of an HLA and disease association was on Hodgkin's disease in 1967 (ref. 8). The claimed association was with an antigen then called ‘4c’. Even with the few antigens (about five) then ascertainable, the association (OR = 2.8, χ2 = 5.06) was not considered significant because of the problem of multiple comparisons. The study was, however, based on a good rationale—that is, on the association between Gross virus–induced leukemia and H-2 in the mouse9. The discovery of H-2–linked immune response genes by McDevitt and others soon provided the best explanation for such associations10. Although the association between Hodgkin's disease and HLA turned out to be relatively weak, with ORs < 1.5, it has been abundantly confirmed. Nevertheless, in spite of the likely explanation in terms of immune response differences between HLA types, this pioneering association has never been properly explained at a functional level.
The first suggestion that linkage disequilibrium could account for associations between a genetic variant and a disease was made in 1972 in the context of the HLA association with Hodgkin's disease11. The overall data on the HLA and Hodgkin's disease association were already then—and remain—significant, although with low ORs. These data led to the explanation of how genetic marker associations with a disease could be due to variation in a gene closely linked to that giving rise to the observed disease association, by linkage disequilibrium. This was the origin of the idea of genetic marker and disease association studies, which have now become feasible on a large scale because of the huge range of SNPs now available at the DNA level, and because of the associated development of high-throughput technology.
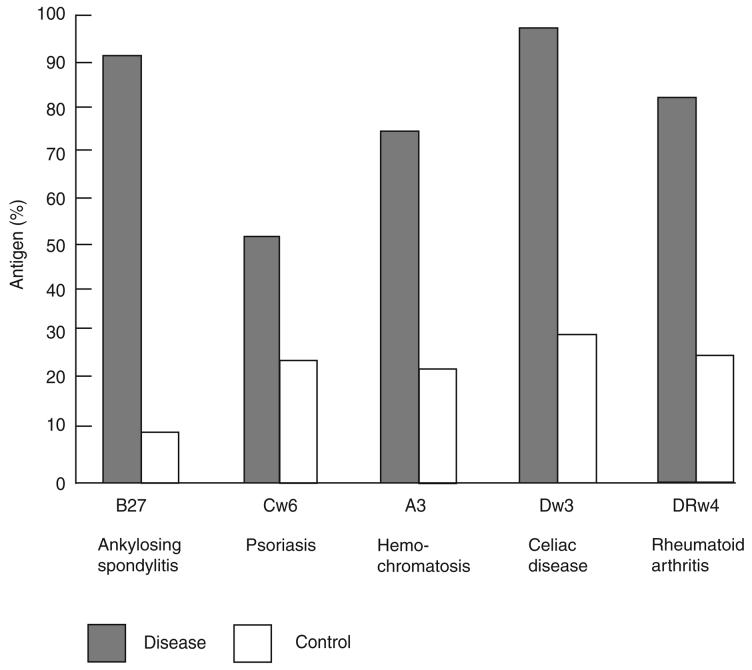
On the basis of the associations between mouse H-2 types and immune response, many studies were carried out on the associations between HLA types and diseases with a possible immune etiology. Early data are summarized in Figure 1. These studies were simple case–control comparisons of the frequencies of different HLA types in disease as compared to control populations. The most notable early result was the association between HLA-B27 and ankylosing spondylitis. Of the diseases shown in Figure 1, the only one with no connection with an immune etiology is hemochromatosis, which became the first and possibly still the best example of finding, by LD, a previously unknown functional gene for a relatively common disease12. The ORs for most of the ten or more diseases that had been investigated in several different studies by 1974 were above 5, with that for ankylosing spondylitis being over 100. The corresponding χ2 values were nearly all at least 15, and many were much greater. The exceptions to high ORs were those for multiple sclerosis (OR = 1.7), acute lymphatic leukemia (OR = 1.7) and Hodgkin's disease (ORs = 1.3–1.7). The multiple sclerosis association became stronger with the discovery of the HLA class II antigens, and later data suggested an OR of about 2 for the association between Hodgkin's disease and HLA-DP13.
Figure 1.
HLA and disease associations. Association of HLA alleles and disease as originally reported in ref. 34.
Notably absent from Figure 1 is the association between HLA and type 1 diabetes (T1D). This was first described as an association with ‘B15’ in 1973 (ref. 14), later also with B8, and then, in 1975 (ref. 15), as an association with Dw3 and Dw4 defined by mixed lymphocyte culture typing. The latter became an association with the HLA-DR3 and HLA-DR4 serological determinants in 1977. Winearls et al.16 observed that the association between B15 and DR4 was much stronger in individuals with T1D than in controls, in contrast to the association between B8 and DR3, which was the same in both affected individuals and controls. This, together with the observation that the T1D association was strongest in DR3/DR4 heterozygotes, suggested that the association was most probably with DQ, as this was the only product both of whose chains were polymorphic, thus allowing the possibility of association with a particular heterozygous combination at the DQ locus. The association with DQ was subsequently established in 1987 (ref. 17).
The rare variant hypothesis: colorectal cancer as a model
About 5% of cases of colorectal cancer (CRC) are associated with inherited, dominant, familial mendelian susceptibility, especially FAP (familial adenomatous polyposis), caused by severely deleterious highly penetrant mutations in the APC gene, and HNPCC (hereditary nonpolyposis colorectal cancer), caused by mutations in mismatch repair genes (see ref. 18 for an example). Another 20–30% of cases are thought to be due to inherited susceptibility that is ‘multifactorial’, namely, associated with much lower penetrance variants that do not give rise to clear-cut familial patterns of inheritance. An important role for rare variants in inherited multifactorial susceptibility to colorectal cancer was first suggested by the effects of rare missense variants in APC19,20. The biggest gap in our knowledge of the inherited susceptibility to colorectal cancer—as also for essentially all the relatively common chronic diseases—concerns the 20–30% of cases that are multifactorial. It is that gap which WGAS and rare variant studies aim to fill.
The ‘rare variant hypothesis’20,21 proposes that a significant proportion of the inherited susceptibility to relatively common human chronic diseases may be due to the summation of the effects of a series of low frequency dominantly and independently acting variants of a variety of different genes, each conferring a moderate but readily detectable increase in relative risk. Such rare variants will mostly be population specific because of founder effects resulting from genetic drift.
Further evidence for the hypothesis was obtained by screening DNA from 124 individuals with multiple (from 3 to 100) colorectal adenomatous polyps for germline variants in a variety of genes involved in Wnt signaling (APC, AXIN1 and CTNNB1) and mismatch repair (MLH1 and MSH2)22. The overall frequency of variants in the individuals with adenoma was 24.9%, significantly higher than that of 11.5% in the controls. Each variant was also assessed for its possible functional effect, and essentially all satisfied the criteria one might expect22,23, as discussed later. Very similar overall results to those described above for colorectal adenomas have been found in a systematic study of the control of plasma levels of HDL cholesterol24.
The search for common or rare variants
Common variants
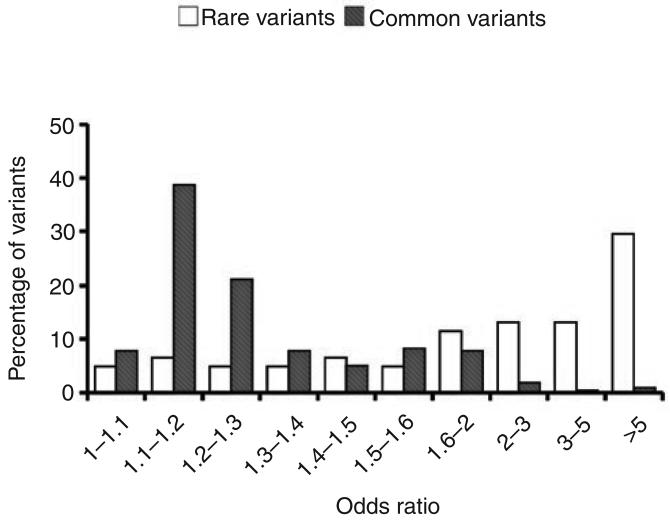
The search for common variants affecting the incidence of a disease has now become possible without making any prior assumptions as to the nature of the variants involved, through the ability to screen a sufficiently large number of well-spaced SNPs providing almost complete genomic coverage. It should then, in principle, be possible to identify the real disease-associated variant by scanning nearby genes for variants that plausibly satisfy the requirement for having an effect on the disease. Most of the common variants found so far in the recent enormous accumulation of new data on WGAS for a wide range of diseases are, however, associated with ORs of only between about 1.2 and 1.5 (Fig. 2). The main challenge to their identification has been to do large enough studies, with replication, to achieve unequivocal statistical significance. The studies must also take into account (see ref. 25 for an example) small overall effects needing large studies for their detection, the potential confounding effects of hidden population substructure, and multiple comparisons, namely the testing of very large numbers of SNPs, which entails using very stringent significance levels—often down to 10−7—to avoid large numbers of false positives.
Figure 2.
Distribution of odds ratios for common and rare variants. Odds ratios were obtained from the literature (Supplementary Note). We included 61 rare variants and 217 common variants in this analysis.
Rare variants
Because of their low frequency and individually small contributions to the overall inherited susceptibility of a disease, rare variants will not be detectable by population association studies based on the use of linked polymorphic markers, even very large WGAS. Their discovery depends on the strategy used in the search for variants influencing colorectal adenomas22,23 and HDL cholesterol levels24. Candidate genes are first sequenced in each member of the chosen disease group. Variants considered to be rare—that is, those not obviously polymorphic but not as rare as obviously deleterious mutations—are then assessed for their frequency in an appropriate control population. Variants are also assessed for their potential consequences to the function of the relevant gene product by criteria such as occurrence in conserved regions, charge changes, and bulky changes likely to affect protein structure and thus function, and also by direct biochemical or functional assays. A variant is considered a good candidate for an effect on inherited susceptibility if it shows a significant difference in frequency between disease and control groups either singly or, more often, as a member of a group of variants affecting the same gene or a set of genes with related functions, and it is assessed to have a substantial probability of affecting the function of the relevant gene product. The challenges of such studies are the choice of candidate genes, the choice of appropriate case groups, the need for extensive DNA resequencing of many genes in comparatively large numbers of individuals, and the assessment of the functional consequences of variants. Most critical of these is the choice of candidate genes made by two main criteria: (i) genes in which obviously severe disruption of function gives rise to a severe, usually clearly familial, version of the disease being studied and (ii) genes known to be involved in the biology of the disease based on biochemical and physiological studies. For example, for cancer, the most obvious candidates are genes that are mutated somatically or epigenetically changed in their expression in a significant proportion of cancers. Case groups should be chosen to be enriched for the presence of rare variants. Generally these will include cases with one or more close relatives affected, but which are not clearly familial, and, especially for cancer, with an early age of onset. Control populations should ideally consist of individuals known to be free of the disease. Selection of large numbers of controls whose provenance is known will help to minimize population stratification effects.
Common and rare variants compared
Common and rare variant frequencies
Given that there is a huge amount of variation at the molecular level which has no obvious functional relevance and that there must therefore be many neutral variants that will achieve significant frequencies simply by chance, a more or less arbitrary lower threshold of 1% has been proposed as the definition of polymorphic variation6. This value is mostly well above that attained by a deleterious mutation maintained in the population by mutation-selection balance. Even for completely recessive deleterious mutations, the corresponding maximum expected incidence is probably only just over 3%.
So far, WGAS have been limited to SNPs with minor allele frequencies (MAF) greater than about 5%. Rare variants, being mostly neutral or nearly neutral, will often be founders and so relatively population specific. They are distinguished from clearly deleterious mutations by having frequencies that lie somewhere between ∼0.1%, the upper limit for deleterious mutations, and ∼1%, the lower limit of polymorphic variation. These frequency boundaries are, however, not absolutely defined, so there is likely to be some overlap at the margins between low-frequency common variants and high-frequency rare variants.
Neither common nor rare variants are familial
A critical feature shared by common and rare variants is that they do not give rise to a familial concentration of cases. This is because the penetrance of such variants, namely, the probability of a given genotype having the disease in question, is low. Assuming, for example, that the penetrance of the heterozygote for a disease susceptibility allele Dd is 10%, it can be shown that for matings Dd × dd, only 1.4% of families even with four offspring will include more than one affected offspring. For a penetrance of 20%, which, as discussed in Box 1 is high even for a variant with an OR of 3, this proportion is still only 5.2%. Only when penetrances are well above 50% does one approach a familial concentration that begins to look like a standard mendelian segregation. Family studies, therefore, are simply not relevant for the discovery and interpretation of either common or rare variants.
BOX 1 Population attributable risk and individual risk.
There are two important contrasting measures of the relative contributions of common and rare variants to multifactorial inherited disease susceptibility. The first, at the population level, is the contribution of a variant to the proportion of cases that comprise the multifactorial inherited component of the population incidence of a disease. The second, at the individual level, is the contribution of a variant to an individual's risk of getting a disease.
Population attributable risk (PAR) as a measure of the multifactorial inherited component of a disease
In this context, the PAR is defined by the relationship:
| (1) |
where K = observed disease incidence and y = disease incidence in the absence of the genetic variant. Although R is not itself additive or multiplicative with respect to combining the effects of different variants, the concept is readily generalizable by assuming that y is the disease incidence in the absence of all genetic variants that influence the probability of getting the disease. R is, therefore, an appropriate measure of the proportion of the disease incidence that can be attributed to genetic factors, at least with respect to those that increase the risk of disease.
Variant penetrance
Assuming a simple single-locus two-allele dominant model for a variant's mode of action, it can be shown (W.B., unpublished data) that the additional penetrance, f, due to the presence of a dominant variant is related to α, the corresponding OR, by the formula:
| (2) |
where y, as before, is the probability of getting the disease in the absence of the variant, and so f is independent of the gene frequency of the variant. Assuming that the relative effect of a single variant on disease incidence is small, y in equation (2) can be replaced by the observed disease incidence, K. For a rare variant, the frequency of homozygotes can be neglected, and so the assumption of dominance is appropriate. For a common variant, equation (2) can be used separately to relate the ORs for the disease variant heterozygotes and homozygotes to their corresponding penetrances. When either, or both, of y and (α − 1) are small, the penetrance, f, is approximately just K(α − 1).
The relationship between the OR, which is observed, and the penetrance, which must be estimated from the OR, is fundamental to any consideration of the practical application of an intervention strategy based on the presence of one or more disease susceptibility variants in an individual. For example, for T1D, assuming a population incidence of 0.005 and additive penetrances, even with ten common variants all with gene frequencies of 0.5 and ORs of 1.4, only about 1/1000 individuals would approach having an increased risk of 2%, which hardly would justify any individual interventional strategy. On the other hand, the data for colorectal cancer suggest the existence of a substantial number of rare variants with ORs of at least 2–3, which corresponds to penetrances of 9–17%, assuming a population incidence of 10%. These penetrances approach a level where individual intervention may be justified.
Comparison of PARs for common and rare variants
The PAR for any given variant can be calculated, at least approximately, from equations (1) and (2), assuming known ORs, α and population disease incidence K. For an individual rare variant and a disease with an incidence of, say, 0.1 or less, the PAR is approximately 2fp/K, where p is the frequency of the rare disease-associated variant. For a number of such variants, the overall PAR will be the sum of these individual contributions, as the probability of an individual having more than one variant will be very small.
For common variants that are not simply dominant, values of the penetrance, f, assuming dominance can be chosen which lie between the separate estimates from equation (1) for the disease variant heterozygotes and the homozygotes, and give a result equivalent to assuming different penetrances for the hetero- and homozygotes. A single common variant with a modest OR can make a substantial contribution to the PAR because of its relatively high frequency. Estimating the combined PAR for a set of common variants is more complicated than for rare variants, as the joint occurrence of several variants in an individual must be taken into account. Assuming n1 loci, each with variants at a frequency of 0.5 and ORs of α1 acting independently, a rough approximation suggests an overall contribution to the PAR of n1(α1 − 1)y/K. The overall approximate contribution to the PAR of n2 rare variants, each with frequency p and ORs of α2 is 2n2p(α2 − 1)y/K. On these model assumptions, the relative contributions of common as compared to rare variants are:
| (3) |
For example, if the number of common variants for a disease is 10 and their average OR is 1.4, whereas the number of rare variants is 200, their average frequency is 0.002 and their average OR is 3.5, then the ratio of common to rare variants, according to equation (3), would be 4:2, suggesting that rare variants can make a substantial contribution to the overall multifactorial inheritance of a disease. The relative balance of the two contributions offsets the higher frequency, but lower ORs and lower number of common variants, against the lower frequency but higher ORs and larger number of rare variants.
Odds ratio distributions for common and rare variants
A summary of the OR distributions for rare and common variants from a wide range of recent publications is shown in Figure 2 (see Supplementary Note online). The difference between the two distributions is quite striking. For common variants, relatively few have values above 2, and the mean OR is 1.36. For the rare variants, on the basis of a smaller set of observations but with many for which the OR could not be assessed because the variant was not observed in the controls, most have ORs above 2, and the mean OR is 3.74.
The overall picture is already reasonably clear. Most common disease-associated variants will have ORs of at most up to 2, with many between 1.1 and 1.4, whereas many, if not most, rare variants will have ORs greater than 2, with a significant number considerably greater than 2.
Functional assessment of common versus rare variants
The discovery of a variant that influences the probability of getting a disease can make a contribution to understanding the disease etiology only if the causal functionally relevant variation can be identified. There is, in this respect, a fundamental difference between the ability to identify the functional basis of common as compared to rare variants.
For rare variants, it will nearly always be the case that the functional effect is due to the variant itself. This is because of the choice of candidate gene, the assessment of the effect of the variant on the function of the gene product, and the extremely low probability of finding two rare variants with comparable functional effects in closely linked genes. Most rare variants are likely to be missense variants, and their functional effects may be expected to arise mostly from amino acid changes that affect protein–protein interactions and that can thus have mildly dominant or dominant-negative effects. Variants in promoter regions may also be relevant, through dominant effects on gene expression.
For common variants, in most cases, the disease-associated variant itself is unlikely to be functionally relevant. The whole premise of WGAS is that an association can uncover the effect of a closely linked functional variant that is in LD with the observed associated variant. However, when the OR is near 1, and so the effect of a variant is relatively small, it is likely to be very difficult to establish which of a set of closely linked variants in LD with each other is the one that is most relevant functionally.
The problem of identifying the functional variant is well illustrated by the extensive studies on the undoubtedly significant association of SNPs at 8q24 with both colorectal and prostate cancer26-28. For colorectal cancer, the highest overall OR was 1.22 and the estimated population attributable risk (PAR) around 20% (ref. 26). Nevertheless, extensive sequencing around the most associated SNPs has not yet given any real clues as to which is the causal variation. The causal basis for the rare variants described for colorectal adenomas23 was, on the other hand, quite unequivocal. However, highly suggestive causal common variants have been identified for both Crohn's disease29 and T1D (ref. 30). This is in keeping with the idea that common variants with higher ORs may be those that have been subject to comparatively recent natural selection, such as variants in HLA and other immune function genes in relation to infections, and perhaps the diabetes-associated variants in relation to available food supplies.
Conclusions
Family studies do not have a significant role in the discovery or analysis of either common or rare disease associated variants, both of which have relatively low penetrances at the individual level (Box 1 and Table 2). That is the basis for the need for quite different strategies for the discovery of either type of variant. Common variants depend on large-scale genotyping of large numbers of cases and controls to be sure of the statistical significance of a suspected SNP association. Rare variants depend on extensive resequencing of carefully selected candidate genes in relatively large numbers of carefully chosen cases, together with a thorough analysis of the functional effects of any suspected variants. Both types of studies assume that background genetic and environmental effects are averaged out, so that, in experimental design terminology, it is the ‘marginal’ effect of a variant that is being assessed.
Table 2.
Characteristics of common and rare disease variants compared
| Common disease variants | Rare disease variants |
|---|---|
| Discovery by population association, case-control studies, using genome-wide markers (WGA) |
Discovery by DNA resequencing of candidate genes, preferably in early onset cases with one or more relatives affected |
| Mostly MAF > 5% | MAF > 0.1% to 2–3% |
| Higher than rare familial mutations, lower than polymorphisms. Often population specific. | |
| Explained by LD with functional variant | Not detected by WGA |
| OR mostly between 1.2 and 1.5 | OR mostly ≥ 2 |
| Higher ORs could be due to recent natural selection | |
| No familial concentration | No familial concentration |
| Need large studies with control for ethnic heterogeneity to achieve statistical significance and minimize false positives |
Assess significance by increased frequencies in cases vs. controls and by functional analysis of variant |
| Make substantial contribution to PAR | Summation of effects of several variants make significant contribution to PAR |
| Low penetrance makes prophylactic intervention unlikely | Penetrance often high enough to justify prophylactic interventions |
| Hard to find functionally relevant variant | Variants identified are functionally relevant |
| Contribution to disease etiology questionable | Make a contribution to understanding disease etiology |
| May suggest candidates for rare variant search | Effect may be modified by common variants |
There is no doubt that WGAS have uncovered, and will continue to uncover, interesting and previously unknown polymorphic variants with measurable significant effects on a variety of common chronic diseases. Our analysis shows, however, that as the odds ratios for common variants will mostly be small, the penetrance of these variants will be very small, even though the contribution of an individual variant to the overall inherited susceptibility of a disease, as measured by the PAR, may be relatively large (Box 1). It is the penetrance, however, that determines the possibility of applying potential preventative approaches on the basis of whether an individual is a carrier of a variant. Small ORs make it very difficult to establish the functional basis for any particular association, and so to make a convincing contribution to understanding the etiology of the disease. Thus, whereas WGAS may make a major contribution to understanding the population genetic architecture of a disease, their practical applications in terms of understanding the etiology of a disease and in targeted prevention are likely to be very limited.
It seems likely that, considering the scale of studies so far carried out and the wide range of SNPs used, most of the associations with ORs around 1.2 or greater for the diseases so far studied may already have been found, at least in populations of European origin. There is always the possibility that positive interactions between one or more common variants may give rise to a much increased OR. This is, however, very difficult to test for, unless the marginal effects of the variants being tested for their interactions are themselves significant. Even then, the number of pairwise combinations to be assessed is likely to be prohibitive. Furthermore, it seems a priori unlikely that variants with small primary effects would give rise to significant interactions.
There remain two key questions. First, is there a long tail of low OR associations still to be found? Second, are there, as might be expected, different associations in non-European populations? The lower the OR, the larger the study needed to achieve statistical significance and the harder it will be to find an association against a background of inevitably increased environmental, and possibly ethnic, heterogeneity. There is a sort of uncertainty principle here, as variant effects merge into the effects of a variable background environment. Given the difficulty of applying even those results associated with larger ORs, it is a serious question as to whether it is cost effective to do larger and larger studies simply to try and find out in more detail the population specific genetic architecture of a disease. Genotype by environmental effects will only be found by very large WGAS in different well-controlled environments that are not confounded by ethnic differences. It may well be questioned whether such studies are, in general, even possible, let alone worthwhile. It must be expected that the smaller the OR, the more likely it will be that environmental factors predominate.
Our analysis suggests that rare variants may make a substantial contribution to the multifactorial inheritance of common chronic diseases and may often have penetrances large enough to justify preventative screening strategies (Box 1). Thus, even though individual rare variants may not contribute much to the overall inherited tendency of a disease, their discovery is likely to be much more rewarding than that of common variants in terms of practical applications, including understanding disease etiology.
In order to meet the challenge of finding rare variants, it is critical that the resources of the newer DNA sequencing technologies are made available for rare variant searches to at least the same extent as SNP typing resources have been made available for WGAS.
There are two important ways in which studies of rare and common variants might intersect. The first is the possibility that common variants may act as significant modifiers of the effects of rare variants (see ref. 31 for an example). This could be investigated, for example, by looking at the effects of established common variants influencing breast cancer susceptibility on the ORs for putative rare variants at the BRCA1 and BRCA2 loci (Box 2). The second point of interaction is that the genes for which common variants are found, or genes nearby that may contain the functionally relevant variant, could be considered candidates for the search for rare variants. They may also then help identify the functional variant associated with a common disease variant.
BOX 2 Rare variants in BRCA1 and BRCA2.
BRCA1 and BRCA2 mutations as listed in the Breast Cancer Information Core database are considered clinically significant if they are associated with a clear-cut familial pattern of disease incidence. These are predominantly frameshift or nonsense mutations with obviously disruptive effects on gene function, with just a small proportion of missense changes. Variants classified as of ‘unknown significance’ (VUS) or as ‘not clinically significant’, mainly because they do not show familial aggregation, have a notably different distribution of changes. These BRCA1 and BRCA2 variants are often hardly, if at all, referred to in reviews of breast cancer susceptibility (see ref. 32 for an example). A high proportion of the VUS are missense changes, with a very small proportion of frameshift or nonsense changes (Supplementary Table 1 online). The functional consequences of these missense changes can be assessed in the usual way, according to the probable severity of the effect of the amino acid change on the function of the gene product (see ref. 33 for an example). Intervening sequence changes are found relatively often in all three categories of BRCA1 and BRCA2 variants. The noteworthy feature of these data on types of mutations is the similarity of the distributions for VUS and ‘not clinically significant’ variants, if we ignore the synonymous changes, to the distribution expected for rare variants. This suggests that most, at least of the missense changes, in the VUS and ‘not clinically significant’ categories may actually be rare variants that do have some clinical significance. Assessment of function on the basis of familial aggregation will completely miss the potential pathological significance of these BRCA1 and BRCA2 categories of variants, because of their relatively low penetrances.
For the severe mutations, assuming a mutation rate per base pair of about 5 × 10−8 and, conservatively, a selective disadvantage of about 0.1, a penetrance of 1, and 1,000 mutations, their total contribution to the PAR per locus is about 2 × (5 × 10−8/ 0.1) × 1 × 1,000 = 0.001. For the missense mutations as rare variants, we can reasonably assume an OR of 2, an average frequency of 0.002, a population incidence for breast cancer of 0.1 and also 1,000 variants, giving a contribution to the PAR of 0.4. Thus, on the basis of these fairly conservative assumptions, the contribution of the VUS and ‘not clinically relevant’ missense variants to the overall inherited risk of breast cancer would be 400 times that of the usual familial mutations. Given that the increased breast cancer risk to variant carriers could be between 10% and 20%, there is a strong case for considering some sort of genetic screening program for these variants, coupled with a more intensive breast cancer screening protocol for the carriers, once identified.
How many rare variants does each of us carry? This is analogous to the classic question of genetic load and the average number of recessive lethals per individual. Given the likely average frequency of rare variants (though the frequency distribution is probably very skewed), and the many thousands of genes in which such variants could occur, it seems possible that the average number of rare variants per person could easily be ten or more. As it is almost only the rare variants that are associated with high enough penetrances to influence individual prophylactic decisions, it is this type of low frequency variation that may be much more likely to become the basis for some sort of personalized medicine, than that usually discussed in relation to common polymorphic variation.
ACKNOWLEDGMENTS
We are grateful to N. Fearnhead and B. Winney for their original contributions to the adenoma study and for many helpful discussions. We are also grateful to P. Goodfellow for comments that helped to clarify our presentation of concepts. C.B. is the CRUK Julia Bodmer Fellow, and the overall work was supported by a CRUK program grant to W.B.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Fisher R. The correlation between relatives on the supposition of Mendelian inheritance. Trans. Roy. Soc. Edinb. 1918;52:399–433. [Google Scholar]
- 2.Fisher R, Immer F, Tedin O. The genetical interpretation of statistics of the third degree in the study of quantitative inheritance. Genetics. 1932;17:107–124. doi: 10.1093/genetics/17.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoday J. Location of polygenics. Nature. 1961;191:368–370. [Google Scholar]
- 4.Aird I, Bentall H, Roberts J. A relationship between cancer of stomach and the ABO blood groups. BMJ. 1953;1:799–801. doi: 10.1136/bmj.1.4814.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke C. Blood groups and disease. In: Steinberg A, editor. Progress in Medical Genetics. Vol. 1. New York: Grune and Stratton; 1961. [Google Scholar]
- 6.Cavalli-Sforza L, Bodmer W. The Genetics of Human Populations. San Francisco: Freeman & Co.; New York: Dover Publications, Inc.; 1971. 1999. [Google Scholar]
- 7.Haldane J. Disease and evolution. Ricerca Sci. 1949;19:3–10. [Google Scholar]
- 8.Amiel J. Study of leucocyte phenotypes in Hodgkin's disease. In: Curtoni E, Mattiuz P, Tosi R, editors. Histocompatibility Testing 1967. Copenhagen: Munksgaard; 1967. pp. 79–81. [Google Scholar]
- 9.Lilly F, Boyse E, Old L. Genetic basis of susceptibility to viral leukaemogenesis. Lancet. 1964;2:1207–1209. doi: 10.1016/s0140-6736(64)91043-8. [DOI] [PubMed] [Google Scholar]
- 10.McDevitt H, Bodmer W. HL-A immune-response genes, and disease. Lancet. 1974;1:1269–1275. doi: 10.1016/s0140-6736(74)90021-x. [DOI] [PubMed] [Google Scholar]
- 11.Bodmer W. Genetic factors in Hodgkin's disease: association with a disease susceptibility locus (DS-A) in the HLA-region. Natl. Cancer Inst. Monogr. 1972;36:127–134. [PubMed] [Google Scholar]
- 12.Feder J, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat. Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 13.Bodmer J, Tonks S, Oza A, Lister T, Bodmer W. HLA-DP based resistance to Hodgkin's disease. Lancet. 1989;1:1455–1456. doi: 10.1016/s0140-6736(89)90166-9. [DOI] [PubMed] [Google Scholar]
- 14.Singal D, Blajchman M. Histocompatibility (HL-A) antigens, lymphocytotoxic antibodies and tissue antibodies in patients with diabetes mellitus. Diabetes. 1973;22:429–432. doi: 10.2337/diab.22.6.429. [DOI] [PubMed] [Google Scholar]
- 15.Thomsen M, Platz P, Andersen O, Christy M. MLC typing in juvenile diabetes mellitus and idiopathic Addison's disease. Transplant. Rev. 1975;22:125–147. doi: 10.1111/j.1600-065x.1975.tb01555.x. [DOI] [PubMed] [Google Scholar]
- 16.Winearls B, et al. A family study of the association between insulin dependent diabetes mellitus, autoantibodies and the HLA system. Tissue Antigens. 1984;24:234–246. doi: 10.1111/j.1399-0039.1984.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 17.Todd J, Bell J, McDevitt H. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 18.Bodmer W. Cancer genetics: colorectal cancer as a model. J. Hum. Genet. 2006;51:391–396. doi: 10.1007/s10038-006-0373-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laken S, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat. Genet. 1997;17:79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]
- 20.Frayling I, et al. The APC variants I1307K and E1317Q are associated with colorectal tumors, but not always with a family history. Proc. Natl. Acad. Sci. USA. 1998;95:10722–10727. doi: 10.1073/pnas.95.18.10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bodmer W. Familial adenomatous polyposis (FAP) and its gene, APC. Cytogenet. Cell Genet. 1999;86:99–104. doi: 10.1159/000015360. [DOI] [PubMed] [Google Scholar]
- 22.Fearnhead N, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc. Natl. Acad. Sci. USA. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fearnhead N, Winney B, Bodmer W. Rare variant hypothesis for multifactorial inheritance: susceptibility to colorectal adenomas as a model. Cell Cycle. 2005;4:521–525. doi: 10.4161/cc.4.4.1591. [DOI] [PubMed] [Google Scholar]
- 24.Cohen J, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 25.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 27.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 28.Freedman M, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathew C. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat. Rev. Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 30.Todd J, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felix R, et al. GSTM1 and GSTT1 polymorphisms as modifiers of age at diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC) in a homogeneous cohort of individuals carrying a single predisposing mutation. Mutat. Res. 2006;602:175–181. doi: 10.1016/j.mrfmmm.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Stratton M, Rahman N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 33.Lovelock P, et al. Identification of BRCA1 missense substitutions that confer partial functional activity: potential moderate risk variants? Breast Cancer Res. 2007;9:R82. doi: 10.1186/bcr1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bodmer W. HLA—The Major Human Tissue Typing System. In: Messel H, editor. The Biological Manipulation of Life. Sydney: Pergamon Press; 1981. pp. 217–244. [Google Scholar]