Abstract
Background
Lamin A/C mutations are a well established cause of dilated cardiomyopathy (DCM), although their frequency has not been examined in a large cohort of patients.We sought to examine the frequency of mutations in LMNA, the gene encoding lamin A/C, in patients with idiopathic (IDC) or familial dilated cardiomyopathy (FDC).
Methods
Clinical cardiovascular data, family histories, and blood samples were collected from 324 unrelated IDC probands, of whom 187 had FDC. DNA samples were sequenced for nucleotide alterations in LMNA. Likely protein-altering mutations were followed up by evaluating additional family members, when possible.
Results
We identified 18 protein-altering LMNA variants in 19 probands, or 5.9% of all cases (7.5% of FDC; 3.6% of IDC). Of the 18 alterations, 11 were missense (one present in two kindreds), three were nonsense, three were insertion/deletions, and one was a splice site alteration. Conduction system disease and DCM were common in carriers of LMNA variants. Unexpectedly, in six of the 19 kindreds with a protein-altering LMNA variant (32%), at least one affected family member was negative for the LMNA variant.
Conclusions
Lamin A/C variants were observed with a frequency of 5.9% in probands with DCM. The novel observation of FDC pedigrees in which not all affected individuals carry the putative disease-causing LMNA mutation suggests that some protein-altering LMNA variants are not causative or that some proportion of FDC may be due to multiple causative factors. These findings warrant increased caution in FDC research and molecular diagnostics.
Keywords: dilated cardiomyopathy, heart failure, genetics, lamin A/C, bilineal inheritance
Introduction
Dilated cardiomyopathy (DCM) is the most common cause of heart failure, resulting in considerable morbidity and mortality. Idiopathic dilated cardiomyopathy (IDC) is DCM of unknown cause, in which the common discernable causes (such as coronary artery disease) have been excluded. Familial dilated cardiomyopathy (FDC) describes families with two or more members with IDC. Recent studies utilizing echocardiographic screening of relatives have shown that one-quarter to one-half of IDC cases are familial (FDC) (reviewed in1).
FDC is largely a genetic disease.1, 2 More than 20 known genes cause autosomal dominant FDC,1, 2 each accounting for only a few percent of cases.1 One of the more common genes identified in FDC is LMNA, which codes for lamin A/C proteins. Lamins A and C function within the nuclear lamina. Mutations in LMNA can cause dilated cardiomyopathy with conduction system disease,3-8 muscular dystrophy, lipodystrophy, progeria and several other disorders.9 LMNA may be among the leading causes of FDC,1 however, the proportion of IDC and FDC due to LMNA mutations remains unclear.
The purpose of this study was twofold: to better define the proportion of FDC due to LMNA, and to compare the role of LMNA in FDC and in non-familial cases (IDC).
Methods
Patient Population
Informed consent was obtained from all subjects for this IRB approved project. The study included 324 unrelated IDC probands, and used methods of clinical categorization previously described.10 For this study, families with confirmed and probable familial disease10 were classified as FDC; those with a negative family history or possible FDC10 were classified as IDC. In FDC cases, the patient and at least one first or second degree relative had confirmed IDC, defined as left ventricular enlargement (LVE) with systolic dysfunction after the exclusion of other detectable causes of DCM including coronary artery disease (CAD).10 Conduction system disease was first, second or third degree heart block identified by resting ECG, or the presence of a permanent pacemaker. Cardiac arrhythmias were defined as sustained atrial flutter or fibrillation, documented history (ECG, 24 hour Holter monitoring, or multiple event recordings) of recurrent, frequent paroxysmal supraventricular arrhythmias or atrial flutter or fibrillation, symptomatic brady-tachy syndrome, documented history of ventricular tachycardia or ventricular fibrillation, or one or more episodes of sudden cardiac death. No elevations in creatine kinase were observed in any subjects studied.
Genetic analysis
Genomic DNA was extracted from whole blood by a standard procedure, and from paraffin embedded lung tissue obtained at autopsy from subject P.23 using QIAamp DNA Mini Kit (Qiagen, Valencia, CA). For proband samples, each exon of LMNA (NM_170707) was PCR amplified by standard methods. Purified PCR products (individual exons and 20−40 nucleotides of 5’ and 3’ introns) were sequenced in both directions using BigDye v3.1 (Applied Biosystems Inc, Foster City, CA) and analyzed by capillary electrophoresis on an ABI 3100. Sequencher software (Gene Codes, Ann Arbor, MI) was used to visually inspect each sequence. Nucleotide changes were considered protein-altering if they predicted an amino acid change, frameshift, premature truncation, or mis-splicing event. None of the protein-altering variants were identified in 150 unrelated Caucasian control individuals (300 chromosomes), nor previously published in the Lamin A/C literature as benign polymorphisms, indicating they are likely not common variants. DNA samples from relatives of LMNA mutation carriers were sequenced bidirectionally for the exon containing the proband's mutation, with the exception of family N, whose members were evaluated by a combination of sequencing and allele-specific oligonucleotide hybridization. Clinically affected relatives who were negative for the proband's mutation (incomplete segregation) were then sequenced for the entire LMNA coding region. Primer sequences and PCR conditions are available at www.fdc.to/laminprimers.
Total RNA was obtained from the explanted heart of subject C.6 using the Qiagen RNeasy kit and RT-PCR was performed by standard methods. A minor band was excised from the agarose gel, purified using the Qiagen QIAquick gel extraction kit and sequenced using primers specific to exons 1 and 3.
All mutations resulting in insertions, deletions, altered splicing, or incomplete segregation were confirmed by an alternative molecular technique such as restriction fragment length polymorphism (RFLP), allele-specific PCR, RT-PCR, allele-specific oligonucleotide hybridization (ASO), or temperature gradient capillary electrophoresis (TGCE). All six kindreds demonstrating incomplete segregation were further analyzed by DNA sequencing of all coding exons and flanking intronic regions of six additional FDC genes: β-myosin heavy chain (MYH7), cardiac troponin T (TNNT2), cardiac sodium channel (SCN5A), titin-cap or telethonin (TCAP), LIM domain binding 3 (LDB3), muscle LIM protein (CSRP3).
Results
Molecular analysis of the FDC cohort
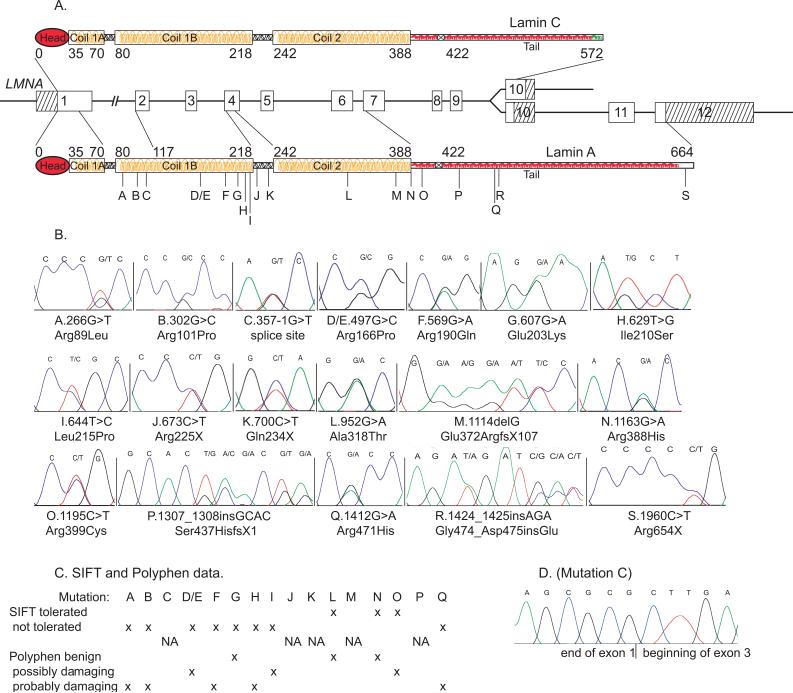
The entire coding region of LMNA (Figure 1) was sequenced in 324 unrelated IDC probands (187 with FDC). We identified 18 (three previously published by our group6, 7) protein-altering variants in 19 of the 324 subjects (5.9%) (Table 1), all of whom were Caucasian. Fourteen of 187 FDC probands carried a LMNA variant (7.5%), compared to five of 137 IDC cases (3.6%). There were 11 missense (one identified twice), three nonsense, two insertion, one splice site, and one deletion variant (Table 1). Seventeen variants in 18 probands were predicted to affect both lamin A and lamin C proteins (Figure 1). Fourteen of the 18 variants were novel; reports of previously observed relevant sequences are provided3, 8, 11-16 (Table 1). The 11 missense alterations changed conserved amino acids, and most were predicted to be not tolerated by SIFT17 or possibly or probably damaging by Polyphen18 prediction algorithms (Figure 1).
Figure 1. Mutations in LMNA and the lamin A and C proteins and amino acid conservation.
Panel A. The LMNA gene is shown. Lamin A (shown below) is coded by exons 1−12 and is a total of 664 amino acids in length. Lamin C (above) is coded by exons 1−9 and an alternatively spliced exon 10, and is 572 amino acids long. The locations of the 18 mutations are labeled with their respective letters. Panel B. Sequencing electropherograms show heterozygosity for each mutation labeled A through S. The nucleotide changes and their effects on amino acid sequence are indicated. Abbreviations are as follows: X, stop; fs, frame shift; del, deletion; ins, insertion. All amino acids changed by mutations were conserved across mouse, rat, chicken, and zebrafish except for mutation D/E and R (mouse, rat and chicken) and mutation O (mouse, rat).Panel C. SIFT17 or polyphen18 data are shown for missense mutations. Panel D. Sequence data shows skipping of LMNA exon 2 for Mutation C. The 357−1G>T splice site mutation results in a minor PCR product representing a transcript in which exon 3 follows immediately after exon 1.
Table 1.
LMNA mutations identified in this study
| Pedigree | Exon | Mutation - Amino Acid | Mutation Nucleotide | Novel Mutation? | Phenotype, Related Reports |
|---|---|---|---|---|---|
| A | 1 | Arg89Leu | 266G>T | no | same mutation, DCM11 |
| B | 1 | Arg101Pro | 302G>C | yes | |
| C | 2 | splice site | 357−1G>T | yes | |
| D/E | 2 | Arg166Pro | 497G>C | yes | |
| F | 3 | Arg190Gln | 569G>A | yes | DCM, Arg190Trp, 568C>T,8, 12 |
| G | 3 | Glu203Lys | 607G>A | yes*6 | DCM, Glu203Gly, 608A>G3 |
| H | 3 | Ile210Ser | 629T>G | yes | |
| I | 4 | Leu215Pro | 644T>C | yes*7 | |
| J | 4 | Arg225X | 673C>T | no*6 | same mutation, DCM13 |
| K | 4 | Gln234X | 700C>T | yes | |
| L | 6 | Ala318Thr | 952G>A | yes | |
| M | 6 | Glu372ArgfsX107 | 1114delG | yes | |
| N | 7 | Arg388His | 1163G>A | yes | |
| O | 7 | Arg399Cys | 1195C>T | no | same mutation, FPLD14 |
| P | 7 | Ser437HisfsX1 | 1307_1308insGCAC | yes | |
| Q | 8 | Arg 471His | 1412G>A | yes | |
| R | 8 | Gly474_Asp475insGlu | 1424_1425insAGA | yes | |
| S | 11 | Arg654X | 1960C>T | no | same mutation, progeria with homozygous ZMPSTE24 mutation;15 FPLD and DCM, 1961 duplication16 |
Previously published by the authors; no other reports unless noted. FPLD is familial partial lipodystrophy.
Clinical Analysis of Pedigrees
Clinical data are provided for the 16 pedigrees not previously published (Figures 2 and 3). Affected status was only assigned when stringent DCM criteria were met (systolic dysfunction and LVE) after all other causes of DCM (including CAD) had been excluded.10 Family members with documented DCM but without documented exclusion of CAD were given ‘unknown’ status. Others given ‘unknown’ status were those with either LVE or systolic dysfunction (but not both), or conduction system disease or arrhythmias without DCM.
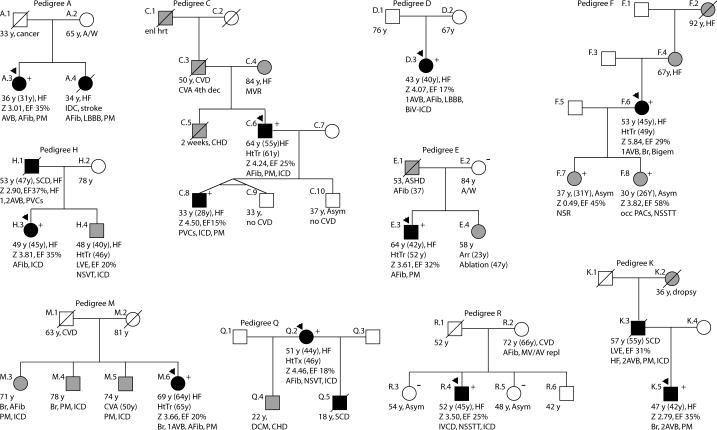
Figure 2. Pedigrees A, C, D, E, F, H, K, M, Q, R.
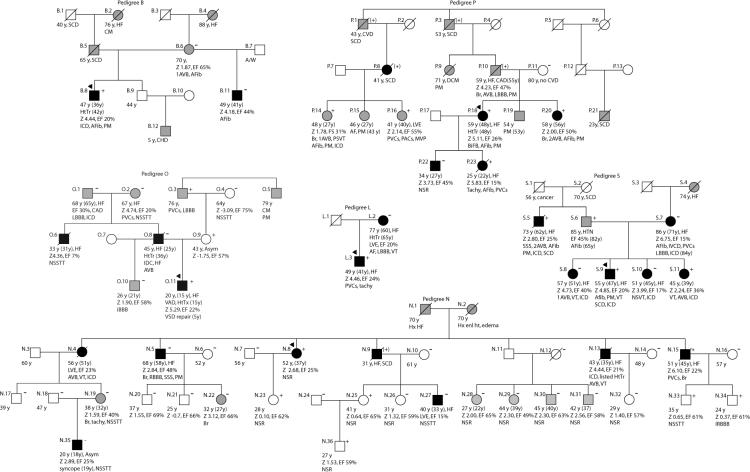
Pedigrees have been labeled by letter, which correspond to their respective mutation as shown in Figure 1. Squares represent males, circles females. An arrowhead denotes the proband. A diagonal line marks deceased individuals. Solid symbols indicate idiopathic dilated cardiomyopathy with or without heart failure, shaded symbols represent any cardiovascular abnormality. Open symbols represent unaffected individuals. The presence or absence of the familial LMNA mutation is indicated by a + or − symbol, respectively. Obligate carriers are noted in parenthesis, (+). Available clinical data are provided under the symbols. The first line is the current age or age of death, followed by the age of onset in parenthesis, and the current major medical diagnosis if alive, or the cause of death if known. Age at cardiac transplantation, when applicable, is the second line. The next line describes LV size and function; Z score is the standard deviation from the mean of the left ventricular end-diastolic dimension measured by echocardiography. The ejection fraction follows; where values are not available, descriptive terms are provided. The last line describes arrhythmias and/or conduction system disease. Abbreviations are as follows: Afib, atrial fibrillation; Asym, asymptomatic; Arr, arrhythmia; ASHD, atherosclerotic heart disease; AV, atrioventricular; AVB atrioventricular block; 1AVB, 2AVB, first or second degree AVB, respectively; A/W, alive and well; bigem, bigeminy; Bi-F, bi-fascicular block; BiV-ICD; biventricular pacemaker with implantable cardiac defibrillator; Br, bradycardia; CHD, congenital heart disease; CM, cardiomyopathy, unknown type; CVA, cerebrovascular accident; CVD, cardiovascular disease; EF, ejection fraction; enl hrt, enlarged heart; Htn, hypertension; HtTr, heart transplant; ICD, implantable cardiac defibrillator; IRBBB, incomplete right bundle branch block; irreg, irregular; HB, heart beat; LBBB, left bundle branch block; Nl scr, normal cardiovascular screening; NSR, normal sinus rhythm; NSSTT, non-specific ST T changes on ECG; NSVT, nonsustained ventricular tachycardia; PM, pacemaker; occ, occasional; PAC, paroxysmal atrial contractions; PVCs, premature ventricular contractions; RBBB, right bundle branch block; SCD, sudden cardiac death; tachy, sinus tachycardia; SSS, sick sinus syndrome; VT, ventricular tachycardia; Z, z-score.
Figure 3. Pedigrees with incomplete segregation.
Refer to Figure 2 legend.
FDC Pedigrees A, C, D, E, F, H, K, M, Q, R (Figure 2)
All affected members of the 10 pedigrees had conduction system disease and/or arrhythmias, as with the previously published pedigrees (G,6 I,7 and J6), consistent with the cardiovascular phenotype associated with lamin A/C mutations.3-8, 11, 12, 19, 20 Atrial fibrillation and pacemakers were common. Five of the 10 pedigrees (A, C, H, K, Q) had FDC, along with the previously published G,6 I7 and J6. Four of the FDC pedigrees (C, and the previously published G,6 I7 and J6) had DNA available from multiple affected individuals, all of whom carried the familial LMNA mutation. In family C, the identified mutation affected the splice recognition site21 and altered mRNA splicing (Figure 1.D).
IDC Pedigrees
Five IDC subjects (Pedigrees D, E, F, M and R) carried LMNA mutations. Four of the five (all but Pedigree D) had at least one first-degree relative with cardiovascular disease. In families E and R the relatives were mutation-negative and clinically unaffected. In Family F, the proband's daughters (ages 30 and 36) carried the mutation and showed mild disease that had not yet met the criteria for DCM. The probands of Pedigrees D and E, not known to be related, carried a 497G>C (Arg166Pro) mutation.
FDC Pedigrees with Incomplete Segregation
Six pedigrees (Figure 3) showed incomplete segregation of the identified LMNA mutation. In these pedigrees, at least one family member met formal criteria for IDC affected status (DCM with exclusion of all detectable causes) but did not carry the LMNA variant found in the proband.
Pedigree B
The proband's half brother (B.11) who was also affected with IDC, was mutation negative, as was their mother (B.6). The mutation may have been de novo or inherited from the father (B.5), who died of sudden cardiac death. The paternal grandfather (B.1) and grandmother (B.2), demonstrated a family history consistent with familial cardiomyopathy. No genetic material or medical records were available from the father or paternal grandparents.
Pedigree L
The proband's affected mother does not carry the 952G>A (Ala318Thr) mutation. No medical history or DNA was available from the proband's father.
Pedigree N
This multigenerational family has several affected members, only some of whom harbor LMNA 1163G>A (Arg388His). Four affected siblings in the second generation had ascertainable genotypes; of these, two were mutation positive (N.8, N.15), one was an obligate carrier (N.9), and one (N.5) was mutation negative. Family member N.27 had IDC and did not carry the LMNA mutation, although his similarly affected father was an obligate carrier. Similarly, N.5 and N.35 were phenotype-positive but genotype-negative for the familial LMNA mutation.
Pedigree O
The diagnosis of FDC was based upon advanced DCM requiring cardiac transplantation in the proband at 15 years and his father at 36 years. The 1195C>T (Arg399Cys) variant was identified in the proband, but absent in his affected father. We therefore obtained a DNA specimen from the proband's asymptomatic mother (O.9), who did carry the variant. Her cardiovascular evaluation was negative. However, her family history included arrhythmia, cardiomyopathy and a pacemaker in her father, who was positive for the variant, and a paternal uncle from whom a DNA specimen and medical records were not available.
Pedigree P
The proband (P.18) harbors an insertion (c1307_1308insGCAC) within exon 7. The resulting frameshift creates a stop at codon 438, truncating lamin A and C proteins by 227 and 135 amino acids, respectively. The four nucleotide GCAC motif is found three times in the wildtype sequence but four times in the variant allele, indicating a likely duplication event. The mutation was identified in autopsy material from the proband's affected daughter (P.23), who died waiting for a cardiac transplant. It was also identified in the proband's sister (P.20) and second cousin (P.14), both of whom have significant conduction system disease requiring pacemakers, as well as in another second cousin (P.16), who is clinically normal. The father (P.10) and three other family members were obligate carriers. The proband's affected son (P.22) did not carry the familial mutation (incomplete segregation). Little information is available on his father (P.17). The clear IDC diagnosis in the proband's mutation-negative son, indicates that the LMNA alteration is not responsible for all cases of IDC in the family (incomplete segregation).
Pedigree S
The diagnosis of FDC was based upon data from four affected siblings (S.8, S.9, S.10, S.11) and their mother (S.7), however the mother and two of the siblings (S.8 and S.10) were LMNA mutation-negative. Upon further investigation, we found that the father (S.6), who had been reported as unaffected, had experienced atrial fibrillation for over 20 years, and his brother (S.5) carried a diagnosis of IDC. We obtained samples from S.5 and S.6, both of whom carried the LMNA truncation variant.
SIFT17 and polyphen18 were developed to predict whether missense mutations were injurious using biochemical and protein information when clinical information was not available. Although agreeing with one another in 9 of the 11 missense mutations in this study (Figure 1), in two cases the predictions were contradictory: polyphen yielded a benign prediction for mutation G, but that mutation was highly likely to be pathologic as shown by segregation of the mutation with disease in a large multigenerational family.6 The other mismatch occurred for family O, where the SIFT predicted a tolerated mutation. However, late onset conduction system disease suggests that this mutation was pathogenic. The utility of algorithms such as these remains uncertain for studies such as this one when clinical and family information are available.
In an effort to identify an additional disease-causing mutation in the cases with incomplete segregation, the proband and one genotype-negative, phenotype-positive individual were analyzed for six additional FDC genes: β-myosin heavy chain (MYH7), cardiac troponin T (TNNT2), cardiac sodium channel (SCN5A), titin-cap or telethonin (TCAP), LIM domain binding 3 (LDB3), and muscle LIM protein (CSRP3). All coding exons and flanking intronic sequences of each gene were sequenced. No likely disease-causing mutations were identified.
Age of Onset
Within the 19 families, the average age of onset for 37 patients with DCM was 42.8±8.7 years, with a median of 42 years. The mean age of onset of conduction system disease in 56 patients was 40.8±9.6 years, with a median age of 40. In the 65 patients with any type of cardiovascular disease the average onset was 39.6± 9.8 years, with a median age of 40.
Discussion
In 324 unrelated patients with IDC or FDC we identified 19 with protein-altering LMNA nucleotide variants (5.9%) of which three were previously published by our group.6, 7 We were interested in the relative risk of lamin A/C cardiomyopathy in patients with FDC versus those with apparent non-familial IDC. Notably, 14 of 187 FDC probands (7.5%) carried LMNA mutations compared to five of 137 IDC cases (3.6%). In addition to dilated cardiomyopathy, 18 of the 19 probands (95%) also had conduction system disease, which has been extensively documented in association with LMNA mutations.3-8, 11, 12, 19, 20 The finding that LMNA mutations are relatively common in IDC (3.6%, or 1 in 28) suggests that regardless of family history, it may be reasonable to consider LMNA mutation analysis in assessing the cause of IDC.
To date this is the largest cohort to be sequenced for LMNA gene mutations. The 5.9% frequency of LMNA mutations can only be considered a general estimate of the burden of lamin A/C-associated genetic disease in the IDC/FDC population. Our 324 index patients came from OHSU, outside professional referrals and self-referral.10 However, we cannot stipulate that the patients in this study represent an unselected cohort of IDC patients that might be seen in community practice, as our research recruitment efforts have emphasized familial disease. These general estimates are in reasonable agreement with prior studies that conducted systematic sequencing of FDC or IDC subjects, in which estimates ranged from 3 of 47 FDC probands (6.4%),19 5 of 75 (6.8%)8 4 of 49 (40 familial) (8.2%),11 to 6 of 66 heart transplant recipients (9.1%).20
Unexpectedly, we found that in 6 of the 19 pedigrees (32%) with a putative disease-causing LMNA mutation not all affected family members carried the alteration. It is not surprising in an adult-onset disease with age-dependent penetrance to find genotype-positive, phenotype-negative individuals (e.g., Subjects N.23, N.25, N33, N34, etc). However, in our cohort we found genotype-negative, phenotype-positive individuals in families with a LMNA alteration of the type currently understood to be likely disease-causing. We note that in four families, (A, H, K and Q) no genetic material was available from the additionally affected family members; therefore segregation could not be assessed.
There are two possible explanations for the patterns in the six incompletely segregating pedigrees. The first is that the observed LMNA alterations are not responsible for DCM in these families. This is unlikely, as extensive data from previous reports (summarized at http://www.dmd.nl) have shown that many other similar LMNA missense mutations can cause FDC (and other diseases), none of these sequence variants were found in our control DNAs or have been reported as common variants in extensive LMNA mutation databases (http://www.dmd.nl and http://www.umd.be:2000/IFAM.shtml), and the molecular genetic and clinical evidence suggest that the LMNA mutations identified here may be pathogenic. The alternative explanation is that the LMNA mutations are contributing to DCM in the genotype-positive individuals and some other causative factor is involved in the phenotype-positive, LMNA genotype-negative members of these kindreds. The nature of the possible cause of DCM in the mutation-negative family members remains unknown. Common causes of DCM (ischemic disease, cardiotoxic drugs, valvular or congenital heart disease) were excluded in all cases by medical records and cardiovascular evaluation. Remaining possible causes include subtle environmental influences (toxins, viruses, etc) not detected by our study, or genetic factors, such as a mutation in an additional DCM gene. That a second DCM gene mutation may be present in the LMNA mutation-negative affected family members is supported by the multigenerational heritability of DCM in Families N and S and a positive family history in both maternal and paternal lineages, as in Family S. If one argues that LMNA is unrelated to the DCM in Family S, then there must be two unknown factors contributing to the bilineal inheritance of DCM in this kindred. The more likely explanation seems to be that LMNA is at least partially responsible for the DCM in those individuals carrying a LMNA mutation, and another gene is responsible for the DCM in the LMNA mutation-negative relatives. Compound heterozygosity in cardiovascular disease is not new, as the combined phenotypic effects of multiple mutations have been observed in other cardiovascular genetic diseases. Keating and Sanguinetti have suggested a ‘multihit hypothesis’ for the long QT syndrome (LQTS),22 in which the presence of more than one mutation caused earlier onset and more severe disease. Compound mutations and compound heterozygosity were identified in 7.9%23 and 4.6%24 of LQTS index patients, and were associated with earlier and more aggressive disease. Compound mutations and accelerated disease have also been reported in 2.5%,25 3.5%,26 and 5%27 of hypertrophic cardiomyopathy cases. Thus, these findings suggest that compound heterozygosity may be more common in IDC/FDC than previously appreciated.
We note that this FDC/IDC cohort is larger by four-fold than any other published to date. The size of this cohort may have made the identification of this pattern of incomplete segregation more probable. Also, we actively recruit kindreds with a history of familial disease, which may have created an ascertainment bias toward subjects with particularly strong family histories, possibly due to the presence of two disease-causing factors, thereby yielding these incomplete segregation patterns.
These findings have important implications for researchers, providers, and patients. For scientists, these FDC pedigrees challenge an assumption that identification of a single gene mutation is generally sufficient to explain FDC, and could greatly confound FDC gene discovery using one-locus linkage analysis or candidate gene approaches. For providers, genetic counseling and/or referral to a specialty center expert in genetic cardiomyopathies becomes critical. LMNA genetic testing is now clinically available in several U.S. laboratories, and FDC pedigrees with two causative factors is particularly important when considering genetic testing for presymptomatic at-risk relatives. Relatives who test negative for a known family mutation may still be at risk to develop FDC. We therefore urge caution in the interpretation of clinical molecular results, and we emphasize the importance of eliciting a detailed family history on both maternal and paternal sides of a family regardless of prior assumptions.
Limitations
Physiological data demonstrating altered cellular function establishing causation of the LMNA alterations in these families is not available. No second genetic factors contributing to FDC have yet been identified in the incompletely segregating kindreds. Also, mutations involving exon splice enhancers or exon splice silencers, or deletions of multiple exons would not have been readily identified.
Conclusion
The incidence of LMNA mutations in FDC is approximately 7.5% compared to 3.6% in IDC. This is the first description of FDC pedigrees in which apparently pathogenic LMNA mutations are identified but which do not account for all cases of IDC in the family, suggesting that more than one factor may be responsible for FDC in some kindreds. Collectively these findings suggest a more complex basis of genetic dilated cardiomyopathy than previously appreciated. We urge caution in the interpretation of clinical molecular genetic test results, particularly when a presymptomatic family member tests negative for a putative familial mutation.
Acknowledgements
We thank the many families and referring physicians for their participation in the OHSU Familial Dilated Cardiomyopathy Research Program, without whom these studies would not have been possible. This work was supported by NIH awards RO1-HL58626 (Dr Hershberger) and 5 M01 RR000334.
Supported by NIH awards RO1-HL58626 and 5 M01 RR000334
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: No conflicts.
References
- 1.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45(7):969–81. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 2.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J Clin Invest. 2005;115(3):518–26. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fatkin D, MacRae C, Sasaki T, Wolff M, Porcu M, Frenneaux M, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–24. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky G, Muntoni F, Miocic S, Sinagra G, Sewry C, Mestroni L. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circ. 2000;101:473–476. doi: 10.1161/01.cir.101.5.473. [DOI] [PubMed] [Google Scholar]
- 5.Becane HM, Bonne G, Varnous S, Muchir A, Ortega V, Hammouda EH, et al. High incidence of sudden death with conduction system and myocardial disease due to lamins A and C gene mutation. Pacing Clin Electrophysiol. 2000;23(11 Pt 1):1661–6. doi: 10.1046/j.1460-9592.2000.01661.x. [DOI] [PubMed] [Google Scholar]
- 6.Jakobs PM, Hanson E, Crispell KA, Toy W, Keegan H, Schilling K, et al. Novel lamin A/C mutations in two families with dilated cardiomyopathy and conduction system disease. J Card Fail. 2001;7:249–256. doi: 10.1054/jcaf.2001.26339. [DOI] [PubMed] [Google Scholar]
- 7.Hershberger RE, Hanson E, Jakobs PM, Keegan H, Coates K, Bousman S, et al. A novel lamin A/C mutation in a family with dilated cardiomyopathy, prominent conduction system disease, and need for permanent pacemaker implantation. Am Heart J. 2002;144:1081–6. doi: 10.1067/mhj.2002.126737. [DOI] [PubMed] [Google Scholar]
- 8.Arbustini E, Pilotto A, Repetto A, Grasso M, Negri A, Diegoli M, et al. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 2002;39(6):981–90. doi: 10.1016/s0735-1097(02)01724-2. [DOI] [PubMed] [Google Scholar]
- 9.Worman HJ, Courvalin JC. How do mutations in lamins A and C cause disease? J Clin Invest. 2004;113(3):349–51. doi: 10.1172/JCI20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kushner JD, Nauman D, Burgess D, Ludwigsen S, Parks S, Pantely G, et al. Clinical characteristics of 304 kindreds evaluated for familial dilated cardiomyopathy. J Cardiac Failure. 2006;12:422–29. doi: 10.1016/j.cardfail.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Taylor MR, Fain PR, Sinagra G, Robinson ML, Robertson AD, Carniel E, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41(5):771–80. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 12.Pethig K, Genschel J, Peters T, Wilhelmi M, Flemming P, Lochs H, et al. LMNA mutations in cardiac transplant recipients. Cardiology. 2005;103(2):57–62. doi: 10.1159/000082048. [DOI] [PubMed] [Google Scholar]
- 13.van Tintelen JP, Hofstra RM, Katerberg H, Rossenbacker T, Wiesfeld AC, du Marchie Sarvaas GJ, et al. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am Heart J. 2007;154(6):1130–9. doi: 10.1016/j.ahj.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 14.Lanktree M, Cao H, Rabkin SW, Hanna A, Hegele RA. Novel LMNA mutations seen in patients with familial partial lipodystrophy subtype 2 (FPLD2; MIM 151660). Clin Genet. 2007;71(2):183–6. doi: 10.1111/j.1399-0004.2007.00740.x. [DOI] [PubMed] [Google Scholar]
- 15.Denecke J, Brune T, Feldhaus T, Robenek H, Kranz C, Auchus RJ, et al. A homozygous ZMPSTE24 null mutation in combination with a heterozygous mutation in the LMNA gene causes Hutchinson-Gilford progeria syndrome (HGPS): insights into the pathophysiology of HGPS. Hum Mutat. 2006;27(6):524–31. doi: 10.1002/humu.20315. [DOI] [PubMed] [Google Scholar]
- 16.Decaudain A, Vantyghem MC, Guerci B, Hecart AC, Auclair M, Reznik Y, et al. New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J Clin Endocrinol Metab. 2007;92(12):4835–44. doi: 10.1210/jc.2007-0654. [DOI] [PubMed] [Google Scholar]
- 17.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebillon P, Bouchier C, Bidot LD, Bonne G, Ahamed K, Charron P, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40(8):560–7. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karkkainen S, Reissell E, Helio T, Kaartinen M, Tuomainen P, Toivonen L, et al. Novel mutations in the lamin A/C gene in heart transplant recipients with end stage dilated cardiomyopathy. Heart. 2006;92(4):524–6. doi: 10.1136/hrt.2004.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mount SM. Genomic sequence, splicing, and gene annotation. Am J Hum Genet. 2000;67(4):788–92. doi: 10.1086/303098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104(4):569–80. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 23.Westenskow P, Splawski I, Timothy KW, Keating MT, Sanguinetti MC. Compound mutations: a common cause of severe long-QT syndrome. Circulation. 2004;109(15):1834–41. doi: 10.1161/01.CIR.0000125524.34234.13. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz PJ, Priori SG, Napolitano C. How really rare are rare diseases?: the intriguing case of independent compound mutations in the long QT syndrome. J Cardiovasc Electrophysiol. 2003;14(10):1120–1. doi: 10.1046/j.1540-8167.2003.03339.x. [DOI] [PubMed] [Google Scholar]
- 25.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(9):1903–10. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 26.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–32. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 27.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42(10):e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]