Abstract
The pathophysiology of endometriosis remains unclear but involves a complex interaction between ectopic endometrium and host peritoneal tissues. We hypothesized that disruption of this interaction would suppress endometriotic lesion formation. We hoped to delineate the molecular and cellular dialogue between ectopic human endometrium and peritoneal tissues in nude mice as a first step toward testing this hypothesis. Human endometrium was xenografted into nude mice, and the resulting lesions were analyzed using microarrays. A novel technique was developed that unambiguously determined whether RNA transcripts identified via microarray analyses originated from human cells (endometrium) or mouse cells (mesothelium). Four key pathways (ubiquitin/proteasome, inflammation, tissue remodeling/repair, and ras-mediated oncogenesis) were revealed, demonstrating communication between host mesothelial cells and ectopic endometrium. Morphometric analysis of nude mouse lesions confirmed that necrosis, inflammation, healing and repair, and cell proliferation occurred during xenograft development. These processes were entirely consistent with the molecular networks revealed by the microarray data. The transcripts detected in the xenografts overlapped with differentially expressed transcripts in a comparison between paired eutopic and ectopic endometria from human endometriotic patients. For the first time, components of the interaction between ectopic endometrium and peritoneal stromal tissues are revealed. Targeted disruption of this dialogue is likely to inhibit endometriotic tissue formation and may prove to be an effective therapeutic strategy for endometriosis.
Endometriosis is defined by the presence of endometrial glands and stroma outside the uterine cavity. This disease affects approximately 10 to 15% of women of reproductive age,1 causing painful menstrual periods (dysmenorrhea), chronic pelvic pain, painful intercourse (dyspareunia), and subfertility.2 Current evidence supports Sampson’s theory that endometriosis is caused by retrograde menstruation and the implantation of refluxed endometrial tissue within the pelvic cavity.3 It has been hypothesized that hormonal,4 immunological,5 genetic,6 and environmental7 factors contribute to the etiology of endometriosis. However, molecular interactions between the ectopic endometrial tissue, peritoneal tissues and infiltrating leukocytes, endothelial cells, and fibroblasts appear to underpin this disease.
Endometriotic lesion growth is supported by vascular endothelial growth factor (VEGF)-A secreted by the ectopic endometrium, acting on the endothelial cells within the peritoneum to induce angiogenesis.8 Blocking this interaction with anti-VEGF-A therapies reduces the number of endometriosis-like lesions.8,9 It is likely that additional interactions between ectopic endometrium and components of peritoneal tissues also play a role in the pathogenesis of endometriosis.
Transcripts that are differentially expressed in the endometrium of women with and without endometriosis have been identified,10 some of which appear to encode molecular signals mediating cross-talk between epithelial and stromal cells. Several groups have also used microarrays to compare eutopic and ectopic endometrium from women with endometriosis.11,12 In addition, laser capture microdissection of glandular epithelial cells from eutopic and ectopic sites identified differentially expressed transcripts associated with several biological processes in epithelial cells.13 Taken together, these studies have identified large numbers of endometriosis-associated RNA transcripts, and several appear to encode molecular signals mediating cross-talk between epithelial and stromal cells in endometriosis.
However, to fully understand the molecular interactions between the ectopic endometrial tissue and its site of attachment, transcripts expressed by ectopic endometrial cells need to be distinguished from those derived from the other cells in the lesions. The nude mouse xenograft model of endometriosis offers an opportunity to do this.14 Nude mice (nu/nu) have a congenital absence of the thymus gland and a resultant defect in T lymphocyte activity, although macrophage and natural killer cell activity is intact, albeit altered. Thus human endometrial tissue can be implanted into nude mice without generating a host-versus-graft response.15 The resulting lesions contain human endometrial glandular epithelium and stroma and mouse-derived mesothelial cells16 that can be distinguished on the basis of species.17
Techniques to analyze separately selected subsets of RNA transcripts from the human and mouse components of tumor xenografts using custom-made Affymetrix microarrays have been described18,19 but the number of genes in these analyses was limited. In the study described here, we have combined the xenograft model of endometriosis with novel microarray analysis methods to distinguish RNA transcripts derived from human and mouse cells. We then interpreted these transcript abundance patterns in the context of the histological events that occur during endometriotic lesion development and in the context of new transcript abundance profiles from endometriotic lesions in women with this disease.
Materials and Methods
Collection of the tissue used in this project was approved by the Cambridge Local Research Ethics Committee, Cambridge, UK, or the Canterbury Regional Ethics Committee, New Zealand. All patients gave written informed consent. All animal procedures were performed in accordance with the Animals Act 1986 (Scientific Procedures) and licensed by the Home Office of the United Kingdom.
Collection of Human Endometrial Tissue Samples for Xenograft Experiments
For the nude mouse model microarray and morphometric analyses, endometrial biopsies were collected using Pipelle suction curettes (Endocell, Wallach Surgical Devices Inc, Orange, CT). Each volunteer provided enough tissue for one nude mouse experiment. All women (n = 6) had regular menstrual cycles (28–30 days) and were not taking medications. The cycle phase of all biopsies was histologically confirmed using Noyes criteria.20 The cycle phase and disease status of the eutopic endometrial biopsies did not influence the number of lesions that developed in nude mice nor their gross and histological appearance in this study. Two of these samples were used for the microarray experiments whereas the remaining four were used for morphometric analysis of nude mouse lesions. All collected endometrial biopsies were immediately placed in prewarmed phenol red free DMEM/F12 culture media (Sigma-Aldrich Co. Ltd, Dorset, UK) for a maximum of 20 minutes. The nude mouse model of endometriosis was used as previously described.8,21
For the microarray experiments, endometrium was obtained on days 12 and 13 of the menstrual cycle. One donor had rAFS stage 1 endometriosis whereas the other had no detectable endometriosis at laparoscopy. The samples used for the morphometric analysis were collected on days 12, 14, 15, and 18 of the cycle. Disease status was surgically defined in one woman with rAFS stage 2 endometriosis and two disease-free women. The other volunteer had not had a surgical procedure but was asymptomatic.
Microarray Analysis of Nude Mouse Lesions
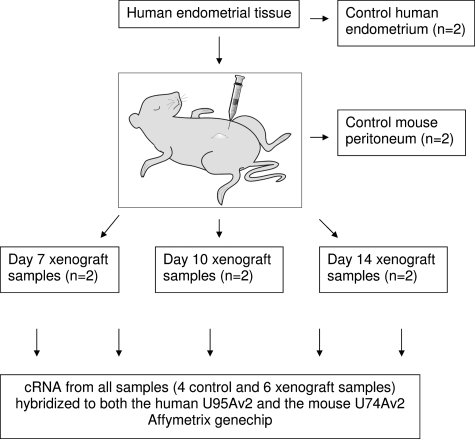
A portion of eutopic endometrium from the same samples used in the nude mouse model was snap frozen to provide control human endometrial tissue. Endometriosis-like lesions were harvested from the nude mice on days 7, 10, and 14 after tissue implantation. A fragment of tissue was removed from the middle third of each lesion for histological assessment and the remainder snap frozen for RNA extraction. Fragments from several nude mouse lesions were pooled (n = 3–5 lesions) based on the day of harvesting and the source of implanted endometrium. A single lesion that did not contain endometrial glands and stroma was excluded from the analysis. A peritoneal biopsy (3 × 3 mm2) was taken from a day 7 and a day 14 nude mouse in both experiments to serve as the mouse-only peritoneal controls (Figure 1).
Figure 1.
Diagram illustrating the source of tissue for all control and xenograft samples. Each xenograft sample was pooled from three to five nude mouse lesions at each time point. cDNA from each sample was hybridized to both the HU-95A (human) and MU-74-Av2 (murine) Affymetrix GeneChip.
Total RNA was isolated from the frozen xenograft tissue, human endometrium, and mouse peritoneum samples, using Trizol (Invitrogen Ltd, Paisley, UK) and Qiagen columns (Qiagen Ltd, Crawley, UK) and then assessed using an Agilent-2100 Bioanalyzer before labeling for Affymetrix microarray analysis as described by Affymetrix (Santa Clara, CA). Each cRNA sample from the mouse xenograft model was hybridized to both a MGU-74Av2 (mouse) and HU-95Av2 (human) Affymetrix GeneChip (Figure 1). Hybridization and data extraction was performed in the Pfizer Microarray Core Facility in Sandwich, Kent, UK. Arrays were analyzed for quality control only using the Affymetrix MAS 5.0 package. Quality control, background correction and normalization were performed using the statistical environment R and the Bioconductor affy package (http://www.R-project.org, April 2007).22
Hybridization of a mixture of cRNA derived from two different species to a GeneChip is only meaningful if the probe sets can be demonstrated to be species-specific. We used a conservative approach that involved the hybridization of control material (human-only endometrium and mouse-only peritoneum) to both human and mouse GeneChips.
To compare and combine data obtained from both human-specific GeneChips and mouse-specific GeneChips we identified orthologous genes and measured the signal derived from the same transcript on both GeneChips. Orthologous probes were defined by one-to-one mapping of the human probe sets on the U95Av2 chip to the mouse probe sets on the U74v2 GeneChip with information supplied by Affymetrix (Santa Clara, CA). The orthologous probes were cross-referenced through their Swiss-Prot ID, generating a many-to-many set of relationships. There were stored in an SQL database to simplify the data mining.
The assumptions about hybridization data, which are implicit in conventional normalization protocols (ie, that the vast majority of signals do not differ between the two populations), are not valid in this case. Normalization methods, which rely on scaling to the global median or a loess-based transformation on the entire data set are thus inappropriate. Therefore control probes homologous to cRNAs spiked into the hybridization mixture (BioB, BioC, BioD, and cre) were used as an invariant set to normalize the data (see Supplemental Table 1 at http://ajp.amjpathol.org). A transformation based on a regression line fitted through the average of the replicates of these invariant probes was used to normalize the entire data set at the probe level.
The 1.5-pmol BioB probe set is considered to be the limit of sensitivity of the assay and should be called as present 50% of the time (http://www.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual.pdf, Dec 2007). The other controls BioC, BioD, and cre should always be called present, with increasing signal values showing their relative frequencies. We used the median value of the 1.5 pmol of BioB control probe sets on each normalized array as the threshold for determining which targets were present.
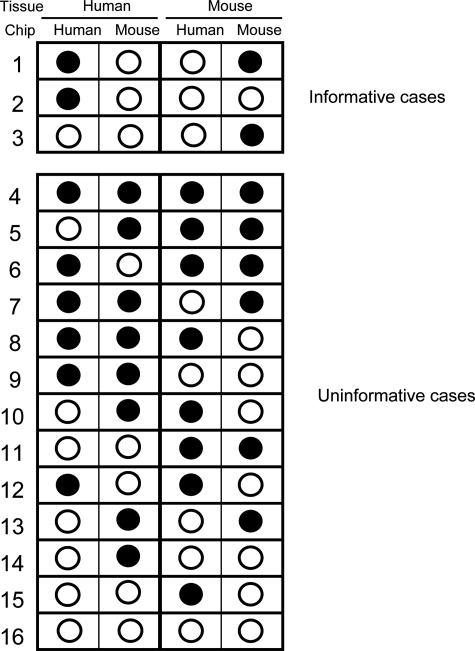
Given that there are four distinct types of control hybridization (mouse tissue on mouse chip, mouse tissue on human chip, human tissue on human chip, and human tissue on mouse chip), there are 16 possible combinations of different signals that can be obtained for every probe set. Only three of these scenarios are informative and these define the probe sets that do not cross-hybridize (Figure 2, upper panel). In case 1, the human probe set is unambiguously able to identify the human transcript without cross-hybridizing to the corresponding mouse RNA. Equally, this probe set on the mouse chip will detect the mouse-specific transcript, but not the human transcript. In case 2, the human probe set unambiguously detects the human transcript as there is no cross-hybridization of the human material to the mouse probe set, but since the mouse target RNA was not present in the mouse-only sample, it is not known whether the mouse RNA cross-reacts with the human probe set. The converse is true in case 3, where the mouse probe set will detect the mouse transcript and this RNA is not detected by the human probe set but it is uncertain whether the human RNA cross-reacts with the mouse probe set, as this transcript was not present in the human-only sample. Using this categorization, it is possible to determine on a gene-by-gene basis which case scenario describes the result for each probe set. Only probe sets within cases 1, 2, and 3 are informative.
Figure 2.
Schematic representation of the possible outcomes of hybridizing human cRNA and mouse cRNA to both human and mouse arrays. Each probe set on the array will produce one of the 16 possible hybridization patterns. Filled circles represent positive hybridization. Some probe sets hybridize with cRNAs derived from both species (ie, cross-hybridize, for example cases 9 and 11); others are uninformative as there is no signal from the hybridization with the correct species (for example cases 14–16).
We then hybridized the xenograft-derived cRNAs (which contain both human and mouse cRNA) to both the mouse U74v2 and human U95Av2 GeneChips. Using BioB-derived thresholds, we determined whether signal was present on the mouse array or the human array or on both. To conclude that a transcript was present in the xenograft the signal had to be above the BioB-derived threshold in five of the six individual arrays (ie, the replicate arrays collected on days 7, 10, and 14 after implantation). It was therefore possible to assign a present or absent value for each of the non-cross-hybridizing orthologous probe sets present on the GeneChip.
The xenograft-derived data sets were analyzed using Ingenuity Pathways Analysis (IPA, Ingenuity Systems, Redwood City, CA). Genes were overlaid onto global molecular networks that were generated using information primarily derived from the published literature stored in the Ingenuity Pathways knowledge base. Pathways of interest were selected based on P values calculated using a right-tailed Fisher’s exact test.
Morphometric Analysis of Nude Mouse Lesions
Mice were sacrificed at days 7 or 14 after injection of human tissue and the lesions collected, fixed in formalin, and embedded in paraffin wax. All day 7 (n = 9) and 14 (n = 11) lesions >2 mm in greatest dimension were included in the analysis. Serial 5-μm sections of the whole lesion were cut and every 13th tissue section dewaxed, rehydrated, and stained with hematoxylin and eosin. Five of these were randomly selected from each lesion for morphometric analysis.
The volume fraction of glands, stroma, necrosis, cysts, inflammation, and hemosiderin were determined in day 7 and day 14 xenografts using the computer-assisted stereological toolbox (CAST, Visiopharm Denmark) morphometric analysis system, essentially as described in Cheng et al.23 Additionally, in each field the glands were counted and an estimate of glandular size (using the two-dimensional nucleator function of the CAST system) was performed. As normality of the data could not be guaranteed, nonparametric statistical analysis was performed using Statview 5.0, and P < 0.05 was taken as significant.
Immunohistochemistry and Histochemistry
Antigen retrieval was performed using the rehydrated dewaxed sections by enzymatic digestion with 0.1% trypsin (DIFCO Laboratories, BD, Oxfordshire, UK) in 0.1% calcium chloride phosphate-buffered saline (pH 7.8) at 37°C for 10 minutes. Frozen sections (5 μm) were thawed on ice, fixed in ice-cold acetone for 5 minutes, air-dried, and placed in distilled water (dH2O). The avidin/biotin kit (Vector Laboratories Ltd, Cambridgeshire, UK) reduced nonspecific streptavidin binding. Endogenous peroxidases were quenched with 3% hydrogen peroxidase (DIFCO Laboratories) in methanol.
The sections were preincubated in goat serum in 0.1% bovine serum albumin (DIFCO Laboratories)/phosphate-buffered saline (F4/80 assay) or in serum from the mouse on mouse kit (Vector Laboratories). The mouse on mouse kit protocol was followed for all immunohistochemistry that involved mouse monoclonal antibodies including mouse anti-human CD 68 (1.25 μg/ml, DAKO, Cambridgeshire UK), mouse anti-human major histocompatibility complex (MHC) class I (2 μg/ml DAKO) and mouse anti-human α-smooth muscle actin (α-SMA) (Clone 1A4, 2.3 μg/ml, Sigma-Aldrich). Rat anti-human F4/80 antibodies (10 μg/ml, Serotec Ltd, Oxfordshire, UK) and rabbit anti-human von Willebrand factor antibodies (5.7 μg/ml DAKO) were incubated overnight at 4°C. Biotinylated goat anti-rat secondary antibodies (5 μg/ml, Serotec) and goat anti-rabbit (5 μg/ml, Serotec) were then applied to these sections, respectively, for 1 hour at 37°C. Isotype-specific mouse IgG1 and IgG2 (Harlan Sera-Lab Ltd., Loughborough, UK) and rat IgG1 (Serotec) and Rabbit IgG (DAKO) were used as negative controls. The antibodies were detected and visualized with the Vectastain ABC kit (Vector Laboratories) and diaminobenzidine (Sigma-Aldrich). All sections were counterstained with Carazzi’s hematoxylin. The sections were dehydrated and mounted in Depex (BDH Laboratory Supplies Ltd, Dorset, UK). Collagen was detected in dewaxed and rehydrated formalin-fixed 5-μm sections using Weigert’s iron hematoxylin and Van Gieson’s method.
Microarray Analysis of Paired Eutopic and Ectopic Human Endometrial Tissue
For microarray analysis, paired biopsies from eutopic endometrium and ectopic lesions were taken from nine women with endometriosis (rAFS stages 2–4). The median age of patients was 34 years (range, 20–46 years). Samples of eutopic endometrium and peritoneal ectopic endometriosis from either the broad ligament (visceral peritoneum) or parietal peritoneum were collected at The Oxford Clinic, Christchurch, New Zealand. Histological examination of a fragment of all endometriotic biopsies confirmed the presence of glandular epithelium and stroma. Eutopic endometrial samples were dated histologically using Noyes criteria.20 Of the nine patients, five were in the proliferative phase and four were in the secretory phase. All eutopic and ectopic tissue samples were immediately snap-frozen in liquid nitrogen and stored for RNA extraction.
The nine paired eutopic and ectopic endometrial samples were homogenized in 1 ml of Trizol (Invitrogen Ltd, Paisley, UK) and RNA extracted as described by the manufacturer. The RNA was further purified using Qiagen columns (Qiagen Ltd, Crawley, UK) and DNase (Invitrogen Ltd.). RNA integrity was assessed using an Agilent-2100 Bioanalyzer. cRNA for array analysis was prepared as described by Affymetrix (http://www.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual.pdf, Dec 2007). The paired ectopic and eutopic human samples (n = 18) were hybridized to U133A Affymetrix cDNA arrays containing ∼23,000 probe sets in total.
The raw data were normalized both within arrays and between arrays using the LIMMA software package.24 Transcript abundance data were compared between eutopic endometrium and endometriotic lesion from the same patient samples using the paired CyberT algorithm (version 3.70; experror = 0.25, winsize = 101, conf. = 30, minrep = 10, betafit = 3)25 (http://cybert.microarray. ics.uci.edu, May 2007). This algorithm is a paired t-test, modified by the inclusion of a Bayesian prior based on the variance of other transcripts with similar expression values in the data set.26 Transcripts with an absolute fold change of greater than 2, a Bayesian P < 0.0001 and posterior probability for differential expression (ppde) >0.99 were selected.25 In addition, the rank products method described by Breitling27 was used and an additional threshold of RankProd P < 0.001 imposed to select the final list of regulated transcripts. The transcripts finally selected had to meet all these criteria. Gene ontologies were identified using FatiGO28 (http://www.fatigo.org, Sept 2007). The transcripts identified as having significantly different abundance were analyzed using the Ingenuity Pathway Analysis package (Redwood City, CA).
The resulting list of transcripts present in the xenograft lesions was compared with that obtained by analyzing the eutopic and ectopic human tissue specimens. The probability of identifying overlapping sets of transcripts was determined using the hypergeometric distribution function dhyper in the statistical language R. The microarray data from the xenograft and eutopic/ectopic endometrial experiments have been submitted to Gene Expression Omnibus [http://www.ncbi.nlm.nih.gov/projects/ geo; accession numbers GSE11691 for eutopic and ectopic human endometrium (endometriosis) and GSE11768 for nude mouse model of endometriosis].
Results
Human and Mouse Microarray Analysis
In all, 5993 orthologous probe sets on the human (U95Av2) and the mouse (U74Av2) Affymetrix GeneChips were identified from information supplied by Affymetrix. As controls, human endometrium and mouse peritoneum were each hybridized to both human and mouse GeneChips, followed by normalization and thresholding based on the median of the 1.5-pmol BioB signals. This identified that, of the 5993 orthologous probe sets, 667 could be used to unambiguously identify both human and mouse transcripts (case 1 in Figure 2), 856 probe sets could be used to unambiguously identify human but not mouse transcripts (case 2 in Figure 2), and 582 probe sets could be used to unambiguously identify mouse but not human transcripts ie, case 3 in Figure 2. Thus, a total of 2105 probe sets could be used to reliably and specifically interrogate the transcript abundance profiles of the endometriosis xenografts.
Of the transcripts identified by these 2105 probe sets, 129 were present in the control tissues (ie, both human endometrium and mouse peritoneum) but not present in five of six nude mouse xenograft samples analyzed. These transcripts are listed in Supplemental Table 2 at http://ajp.amjpathol.org and are not associated with ectopic endometrial lesions. Furthermore, none of these 129 transcripts was identified as differentially expressed in the analysis of the eutopic/ectopic human tissues.
The presence or absence of the remaining 1976 transcripts in either human (endometrium), mouse (stroma), or both components of the xenografts was determined. The number of transcripts that showed consistent signal (ie, in five of the six pooled xenograft lesions analyzed) as unambiguously being present in the human and mouse transcriptomes is summarized in Table 1. Functional information about the proteins encoded by these transcripts is in Supplemental Table 3 at http://ajp.amjpathol.org.
Table 1.
Numbers of Mouse and Human Transcripts Unambiguously Identified in Nude Mouse Xenografts
| Both human and mouse transcripts absent from lesion | 129 |
| Both human and mouse transcripts present in lesion | 177 |
| Mouse transcript present, human transcript absent from lesion | 97 |
| Human transcript present, mouse transcript absent from lesion | 53 |
| Mouse transcript present in lesion | 190 |
| Human transcript present in lesion | 187 |
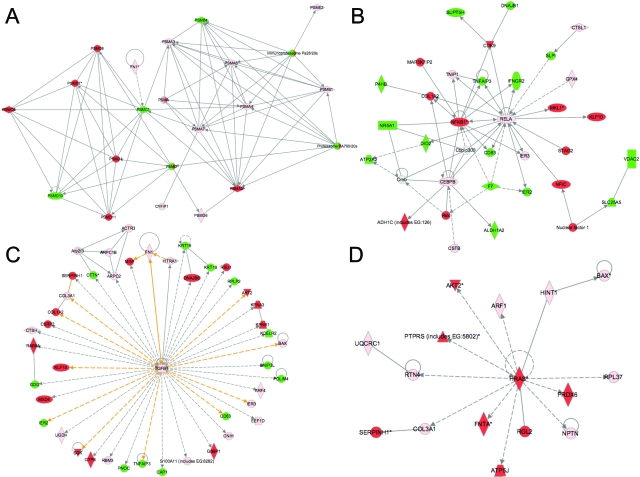
Analysis of the possible relationships among the proteins encoded by transcripts identified in the xenograft lesions was performed using IPA. This revealed four pathways relevant to endometriosis: cell injury and necrosis (Figure 3A), inflammation (Figure 3B), tissue remodeling and collagen deposition (Figure 3C), and tumorigenesis (Figure 3D).
Figure 3.
Four key Ingenuity Pathways Analyses (A–D) representing the molecular relationships between the unambiguously identified human and mouse transcripts in nude mouse xenografts. Each node represents a transcript: green, mouse-specific; red, human-specific; pink, represented in both human and mouse transcriptomes. Supplemental Table 3 at http://ajp.amjpathol.org lists the gene names for all abbreviations. The gene products are represented as nodes, and the biological relationship between two nodes is represented as a line. All lines are supported by at least one reference in literature, textbook, or from canonical information stored in the Ingenuity Pathways knowledge base.
Histology of Nude Mouse Lesions
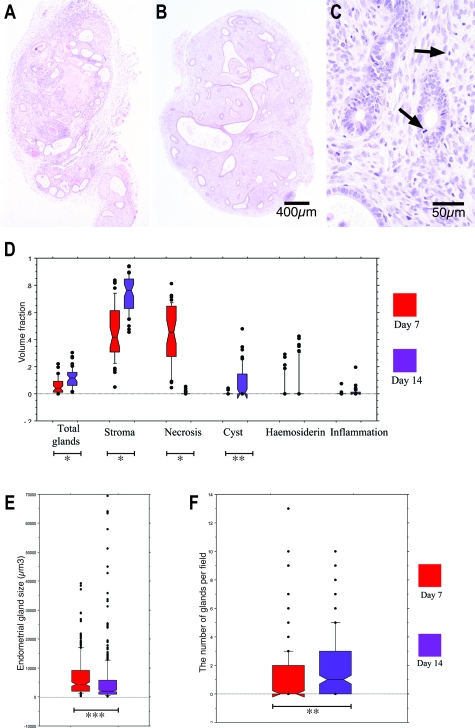
Day 7 lesions were characterized by a central necrotic area surrounded by stroma and dilated glands with flattened epithelium (Figure 4A). Epithelialized cystic structures were present at the core of most day 14 lesions and necrotic tissue was rarely seen. These cystic spaces were surrounded by large numbers of small pseudostratified glands located in stromal tissue (Figure 4B). Mitotic figures were noted in the glandular epithelium and in centrally located stromal cells in both day 7 and day 14 xenografts (Figure 4C).
Figure 4.
A–C: Formalin-fixed sections stained with hematoxylin and eosin from day 7 (A) and day 14 (B and C) nude mouse xenografts. A: Day 7 nude mouse xenografts exhibited central necrosis, glandular degeneration, and leukocyte infiltration. B: Day 14 implants contained an epithelialized centralized cystic space surrounded by multiple pseudostratified glands. C: Mitotic figures are identified in day 14 glandular and stromal cells (arrows). D: Volume fraction analysis of day 7 and day 14 nude mouse explants. Necrosis occupied a larger volume of day 7 lesions, whereas a higher volume fraction of total glands, stroma, and cysts was present in day 14 xenografts (*P < 0.0001; **P < 0.001). E: Two-dimensional nucleation endometrial gland size estimates from day 7 and day 14 nude mouse lesions. Endometrial gland size was significantly reduced in day 14 lesions (*P < 0.0001). F: The number of glands per field in day 7 and day 14 nude mouse xenografts. There were more glands per field in day 14 xenografts (***P = 0.002).
Morphometric Study
Histological differences between day 7 and day 14 xenografts were consistently seen in 20 lesions derived from four different individuals’ endometria (Figure 4D). Stroma and necrosis occupied the largest volume fraction of day 7 lesions. The volume fraction of stroma was higher (Mann-Whitney U test, P < 0.0001) and necrosis lower (P < 0.0001) in day 14 lesions. A larger volume fraction of day 14 lesions was occupied by glands (P < 0.0001) and cystic structures (P = 0.001) when compared to day 7 lesions. No statistically significant difference was seen in the low volume fractions of hemosiderin and inflammation between days 7 and 14 (P = 0.788 and P = 0.096, respectively) (Figure 4D).
Glands in day 14 lesions were more frequent (P = 0.002) but were smaller than those in day 7 lesions (P < 0.001). A few unusually large glands seen in the day 14 data set may represent a cut through the edge of a central cyst (Figure 4, E and F).
Identification of Human and Mouse Cells in Nude Mouse Lesions
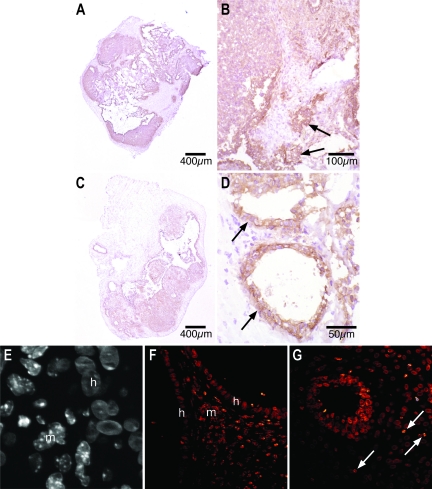
Human cells are anti-human MHC class I+ and show uniform nuclear Hoechst staining, whereas mouse cells are anti-human MHC class I− and show punctate nuclear Hoechst staining (Figure 5). In day 7 lesions, intense human MHC class I immunoreactivity was seen in centrally located endometrial glands and stroma. Unstained cells were seen in the outer two-thirds of the lesion and surrounding human endometrial glands (Figure 5, A and B). At day 7, murine cells with punctate Hoechst-stained nuclei were present in the stromal tissue at the periphery of the lesion, often immediately adjacent to human glandular epithelial cells (Figure 5E, arrow).
Figure 5.
A–D: Immunohistochemical localization of human MHC class I antigens in frozen sections from day 7 (A and B) and day 14 (C and D) nude mouse lesions. A and B: In day 7 lesions, centrally located glandular epithelium and stroma demonstrate intense brown human MHC class I immunoreactivity whereas unstained murine cells are seen around the circumference of the lesion (A) and in tongues extending into the center (B). C and D: In day 14 xenografts, anti-human MHC class I antibody staining is more diffuse (C) with significant numbers of unstained murine cells seen in central locations of the xenografts (D). The glandular epithelium maintains strong human MHC class I immunoreactivity in day 7 (B, arrows) and day 14 (D, arrows) lesions. E–G: Fluorescent staining of formalin-fixed nude mouse xenografts using the Hoechst reagent. E: A gray scale high-magnification view shows cells with uniformly Hoechst-stained nuclei (h, human) immediately adjacent to cells with speckled nuclei (m, murine). F: In a day 7 lesion, the glow over function demonstrates the punctate nuclei of murine stromal cells (m) between the homogeneously nucleated human cells of two glandular epithelia (h). G: Punctate solitary murine nuclei (arrows) can be seen completely surrounded by cells with homogenous human nuclei in the central part of a day 14 lesion.
Diffuse anti-human MHC class I staining was detected in central areas of day 14 xenografts. Unstained stromal cells were interspersed with human MHC class I+ stromal cells in more central locations. The glandular epithelium remained anti-human MHC class I+ in day 14 lesions (Figure 5, C and D). Similarly, large numbers of murine cells with punctate Hoechst-stained nuclei were in the periphery of day 14 nude mouse lesions but solitary, murine cells with speckled nuclei were also seen among clusters of cells with human-type nuclei in the core of day 14 lesions (Figure 5G, arrows). Both day 7 and day 14 xenografts displayed cells with human and murine Hoechst nuclear staining characteristics in close association (Figure 5, E–G).
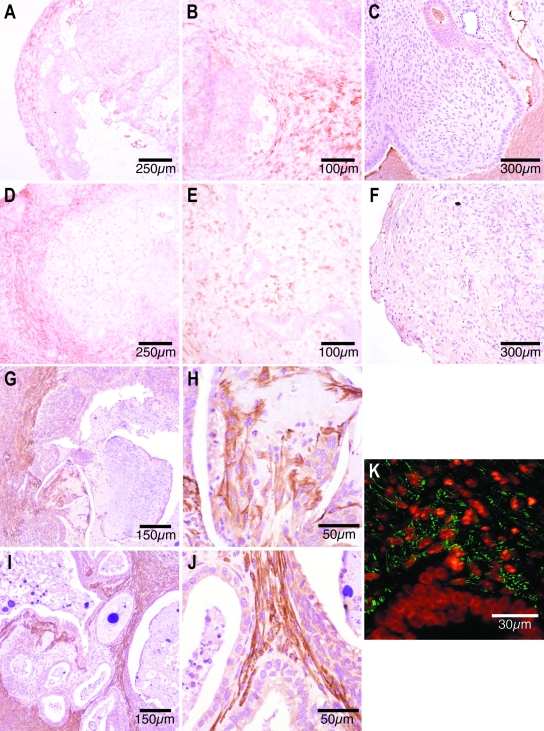
Macrophages
No human CD68-positive macrophages were detected in nude mouse lesions (Figure 6, C and F). In contrast, antibodies against the murine macrophage specific antigen, F4/80, revealed murine macrophage infiltration in nude mouse lesions. In day 7 xenografts, murine macrophages were only present at the edge of the lesion (Figure 6, A and B). In day 14 lesions, many isolated macrophages were also apparent in a central location, scattered in the stromal tissue (Figure 6, D and E).
Figure 6.
Antibody reactivity in frozen sections from day 7 (A–C) and day 14 (D–F) nude mouse xenografts. A and B: Many brown-stained, F4/80-positive murine macrophages are identified in the peripheral rim of frozen sections from day 7 xenografts (A) and between peripheral glandular epithelia (B). D and E: Large numbers of brown F4/80-positive murine macrophages are present in the periphery of day 14 nude mouse lesions (D). F4/80-positive macrophages are less densely scattered throughout the stroma of central portions of the day 14 xenografts (E). C and F: Anti-human CD68 immunoreactivity in paraffin-embedded nude mouse lesions. There were no CD68-positive cells in paraffin-embedded sections from day 7 (C) or day 14 (F) nude mouse xenografts. G–J: Immunohistochemical identification of α-SMA-positive cells in paraffin-embedded sections from day 7 (G and H) and day 14 (I and J) nude mouse xenografts. Large numbers of brown-stained, α-SMA-positive myofibroblasts are present in the outer third of paraffin-embedded sections from day 7 nude mouse lesions (G), and some are immediately adjacent to the necrotic core (H). Spindle-shaped α-SMA-positive myofibroblasts are seen subepithelially in the stromal tissue in central parts of the day 14 xenografts (I and J). K: Fluorescent colocalization of anti-α-SMA antibodies and the Hoechst nuclear stain in a formalin-fixed day 14 nude mouse lesion revealed green α-SMA-positive fibers in the cytoplasm of subepithelial cells that contain orange speckled Hoechst-stained (murine) nuclei. The glandular epithelium is made up of cells with a homogeneous (ie, human) nuclear appearance after Hoechst staining.
Smooth Muscle Actin
Immunohistochemical staining revealed large numbers of α-SMA containing myofibroblasts in the periphery of day 7 explants (Figure 6, G and H) and in more central locations at day 14 (Figure 6, I and J). Colocalization of the α-SMA antibody (green) to subepithelial cells that exhibited typically murine, punctate Hoechst-stained nuclei confirmed the murine origin of these α-SMA-containing cells (Figure 6K).
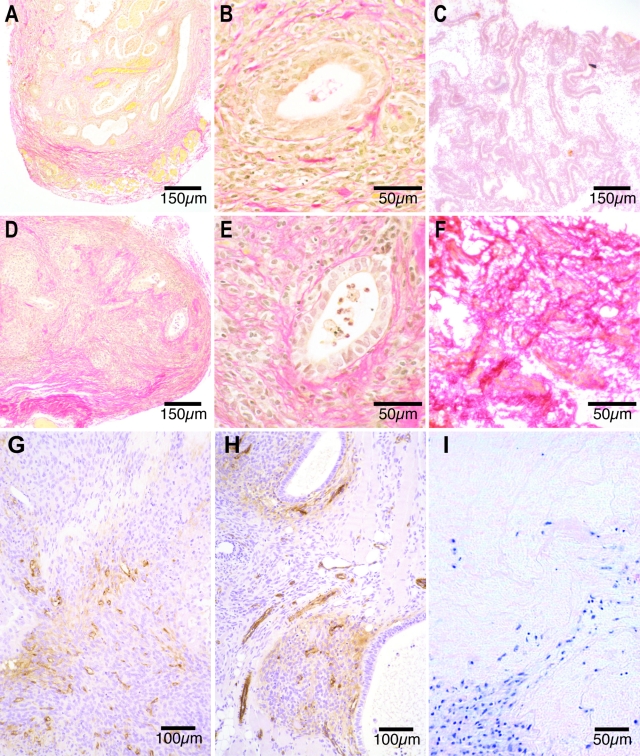
Collagen
Ectopic endometrium demonstrated stronger collagen staining than eutopic endometrium from the same and other women (Figure 7, C and F). Light peripheral collagen staining was apparent in the periphery of day 7 nude mouse lesions (Figure 7, A and B). Compared to day 7 xenografts, collagen staining in day 14 xenografts was increasingly intense and seen in progressively more central locations over time (Figure 7, D and E). At all time points collagen fibers were seen encircling but not penetrating the glandular epithelium (Figure 7, B and E). Hematoxylin and eosin-stained sections revealed cells lined up along extracellular matrix (ECM) fibers in nude mouse lesions (Figure 7I).
Figure 7.
A–F: Staining of collagen fibers using Weigert’s iron hematoxylin and Van Gieson’s method in formalin-fixed day 7 (A and B) and day 14 (D and E) nude mouse lesions and in frozen sections of human eutopic (C) and ectopic (F) endometrium from the same volunteer. Sparse red collagen fibers are present at the edges of formalin-fixed day 7 nude mouse lesions (A) and in more central locations by day 14 (D). The collagen fibers encircle the endometrial glands between day 7 (B) and 14 (E). Eutopic human endometrium (C) displays light red collagen staining, whereas ectopic endometrium (F) exhibits intense collagen staining. G and H: Von Willebrand factor (vWF) immunoreactivity in formalin-fixed sections from day 7 (G) and 14 (H) nude mouse lesions. Scattered brown vWF vascular endothelial cells are present in the periphery of day 7 xenografts (G), whereas small vessels made up of brown endothelial cells are present in central locations of day 14 lesions (H). I: Hematoxylin and eosin staining of a day 7 nude mouse lesion show purple-stained cells lined up along fibers of the extracellular matrix.
Endothelial Cells
Endothelial cells stained for von Willebrand factor were present in the periphery of day 7 nude mouse lesions in small unstructured clusters (Figure 7G). In day 14 nude mouse lesions von Willebrand factor-positive vascular endothelial cells were seen in discrete vessels in central locations of the xenograft in close proximity to the glandular epithelium (Figure 7H).
RNA Transcripts Differentially Expressed Between Eutopic and Ectopic Endometrium of Patients with Endometriosis
After normalization, the 18 paired eutopic and ectopic samples were compared using Cyber-T and RankProd, and 232 transcripts were found to be significantly up-regulated and 390 transcripts to be significantly down-regulated in ectopic lesions relative to eutopic endometrium. These transcripts are listed in Supplemental Table 4 at http://ajp.amjpathol.org. The regulated transcripts were assigned to functional subsets by ontology analysis of biological processes, molecular function, and cellular component (Table 2). The biological processes categories showing significant over- or under- representation included inflammatory response (n = 18), response to wounding (n = 22), cell adhesion (n = 35), calcium-independent cell adhesion (n = 5), primary metabolic process (n = 104), and DNA metabolic process (n = 2). The only ontology in the molecular function category showing a significant over- or under- representation was nucleic acid binding (n = 30). The cellular components identified by this analysis were extracellular space (n = 28), intercellular junction (n = 10), extracellular matrix (n = 17), integral to membrane (n = 85), and nuclear part (n = 4). Additional details relating to all of the regulated transcripts and their Gene Ontology (GO) categories are shown in Supplemental Table 5 at http://ajp.amjpathol.org.
Table 2.
Gene Ontology Analysis of Genes Differentially Expressed in Human Eutopic/Ectopic
| Gene ontology category | Level | Number of genes differentially expressed in microarray analysis | Percentage of genes differentially expressed in microarray analysis | Number of genes on U133A GeneChip | Percentage of genes on U113A GeneChip | Unadjusted P value | Adjusted P value | |
|---|---|---|---|---|---|---|---|---|
| Biological process | ||||||||
| Cell adhesion | 3 | 35 | 14.71 | 475 | 5.67 | 4.50E-07 | 3.59E-04 | |
| Defense response | 3 | 29 | 12.18 | 372 | 4.44 | 1.70E-06 | 9.03E-04 | |
| Primary metabolic process | 3 | 104 | 43.7 | 4830 | 57.62 | 2.57E-05 | 1.02E-02 | |
| Macromolecule metabolic process | 3 | 86 | 36.13 | 4162 | 49.65 | 4.20E-05 | 1.57E-02 | |
| Cellular metabolic process | 3 | 112 | 47.06 | 4953 | 59.09 | 2.32E-04 | 4.92E-02 | |
| Biopolymer metabolic process | 4 | 50 | 21.65 | 3154 | 38.33 | 1.30E-07 | 1.19E-04 | |
| Nucleobase, nucleoside, nucleotide, and nucleic acid process | 4 | 34 | 14.72 | 2359 | 28.67 | 1.19E-06 | 7.55E-04 | |
| Response to wounding | 4 | 22 | 9.52 | 303 | 3.68 | 7.48E-05 | 2.10E-02 | |
| Cell-cell adhesion | 4 | 14 | 6.06 | 153 | 1.86 | 1.83E-04 | 4.15E-02 | |
| Calcium-independent cell-cell adhesion | 5 | 5 | 2.3 | 11 | 0.14 | 4.68E-05 | 1.66E-02 | |
| Inflammatory response | 5 | 18 | 8.29 | 215 | 2.75 | 5.47E-05 | 1.76E-02 | |
| DNA metabolic process | 5 | 2 | 0.92 | 530 | 6.77 | 7.58E-05 | 2.10E-02 | |
| Molecular function | ||||||||
| Nucleic acid binding | 3 | 30 | 11.95 | 2001 | 22.91 | 1.62E-05 | 6.86E-03 | |
| Cellular component | ||||||||
| Extracellular space | 4 | 28 | 12.07 | 358 | 4.4 | 2.59E-06 | 1.27E-03 | |
| Intercellular junction | 8 | 10 | 7.94 | 93 | 1.59 | 5.53E-05 | 1.76E-02 | |
| Extracellular matrix(Sensu Metazoa) | 4 | 17 | 7.33 | 193 | 2.37 | 6.96E-05 | 2.10E-02 | |
| Integral to membrane | 7 | 85 | 52.47 | 2546 | 37.5 | 1.48E-04 | 3.94E-02 | |
| Nuclear part | 9 | 4 | 7.55 | 705 | 29.87 | 1.77E-04 | 4.15E-02 | |
The list of the all human and mouse transcripts that could be definitively identified in the xenograft lesions was compared with that derived from the analysis of the eutopic and ectopic tissues. Thirty-three unique transcripts were identified as being present in both lists, although there were several instances in which transcripts were detected by more than one probe set in each of the analyses. Analysis using a hypergeometric distribution indicated that the probability of at least this degree of overlap occurring by chance was 0.0007. These transcripts are listed in Table 3. It is noteworthy that 16 of 16 distinct transcripts identified as murine in the xenografts were up-regulated in the ectopic human tissue, and nine of 10 exclusively human transcripts were down-regulated in the ectopic tissue.
Table 3.
Transcripts That Are Present in Nude Mouse Xenografts and Differentially Regulated in Human Endometriotic Lesions Versus Eutopic Endometrium
| Official gene symbol | Title | Species specificity | Ratio of ectopic to eutopic samples |
|---|---|---|---|
| IL11RA | Interleukin 11 receptor, alpha | Mouse | 2.41 |
| LMNA | Lamin A/C | Mouse | 2.42 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | Mouse | 2.44 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | Mouse | 2.44 |
| FMOD | Fibromodulin | Mouse | 2.74 |
| CR1 | Complement component (3b/4b) receptor 1 (Knops blood group) | Mouse | 2.87 |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | Mouse | 3.07 |
| MCAM | Melanoma cell adhesion molecule | Mouse | 3.45 |
| MGP | Matrix Gla protein | Mouse | 3.67 |
| ITM2A | Integral membrane protein 2A | Mouse | 4.17 |
| AQP1 | Aquaporin 1 (Colton blood group) | Mouse | 4.39 |
| COL14A1 | Collagen, type XIV, alpha 1 (undulin) | Mouse | 4.63 |
| COL14A1 | Collagen, type XIV, alpha 1 (undulin) | Mouse | 4.63 |
| MNDA | Myeloid cell nuclear differentiation antigen | Mouse | 4.72 |
| CCL2 | Chemokine (C-C motif) ligand 2 | Mouse | 4.95 |
| LPL | Lipoprotein lipase | Mouse | 6.23 |
| FHL1 | Four and a half LIM domains 1 | Mouse | 6.69 |
| DPT | Dermatopontin | Mouse | 8.44 |
| DPT | Dermatopontin | Mouse | 8.44 |
| PAPSS1 | 3′-Phosphoadenosine 5′-phosphosulfate synthase 1 | Human and mouse | 0.21 |
| CSPG2 | Chondroitin sulfate proteoglycan 2 (versican) | Human and mouse | 0.32 |
| CSPG2 | Chondroitin sulfate proteoglycan 2 (versican) | Human and mouse | 0.32 |
| CSPG2 | Chondroitin sulfate proteoglycan 2 (versican) | Human and mouse | 0.32 |
| MSX1 | msh homeobox 1 | Human and mouse | 0.39 |
| MSX1 | msh homeobox 1 | Human and mouse | 0.39 |
| TUBB2A | Tubulin, beta 2A | Human and mouse | 2.98 |
| ACTA2 | Actin, alpha 2, smooth muscle, aorta | Human and mouse | 3.18 |
| S100A4 | S100 calcium binding protein A4 | Human and mouse | 3.23 |
| TPM1 | Tropomyosin 1 (alpha) | Human and mouse | 3.54 |
| DEFB1 | Defensin, beta 1 | Human | 0.13 |
| INDO | Indoleamine-pyrrole 2,3 dioxygenase | Human | 0.17 |
| HOMER2 | Homer homolog 2 (Drosophila) | Human | 0.26 |
| SPINT2 | Serine peptidase inhibitor, Kunitz type, 2 | Human | 0.28 |
| WFDC2 | WAP four-disulfide core domain 2 | Human | 0.31 |
| ARHGEF5 | Rho guanine nucleotide exchange factor (GEF) 5 | Human | 0.37 |
| DLX5 | Distal-less homeobox 5 | Human | 0.38 |
| SLPI | Secretory leukocyte peptidase inhibitor | Human | 0.46 |
| KRT19 | Keratin 19 | Human | 0.48 |
| MXRA5 | Matrix-remodeling associated 5 | Human | 4.31 |
Ratios in italics denote down-regulation in ectopic tissue.
Discussion
Drug development is hindered by a lack of knowledge about the pathogenesis of endometriosis. In particular, we do not yet understand the cross-talk signals that pass between the ectopic endometrium and the mesothelial cells derived from the peritoneum in the lesions. To address this, a unique cross-species microarray analysis of xenografts from the nude mouse model of endometriosis was performed. This analysis revealed for the first time separate molecular pathways that operate between the endometrial and peritoneal components of endometriotic lesions. These molecular changes were then linked to histological changes identified in developing endometriosis-like lesions and to gene signatures in human endometriotic tissues.
Nude mice are commonly used as models of endometriosis21,29 and cancer30 despite the species mismatch between some receptor-ligand pairs. Although these animals have a partial immune deficit they have nonetheless been widely used to investigate the effect of human therapeutic agents in endometriosis,8,9,31 Furthermore, we found a highly significant overlap between the transcripts present in nude mouse xenografts and those differentially regulated between human eutopic and ectopic endometrial tissue. This suggests that similar cellular processes occur in both human disease and nude mouse xenografts.
It would be desirable to quantitatively assess and compare the absolute level of transcripts in xenograft lesions. However, to correctly perform this quantitative determination, it would be essential to identify polymerase chain reaction primers for both the transcript to be quantified and for a normalizing “housekeeping” transcript. However, all commonly used “housekeeping” transcripts are highly conserved between man and mouse therefore normalization is technically difficult. In addition, normalization using house keeping transcripts would not account for the variable proportion of human cells in xenograft lesions. Thus, changes in xenograft composition could erroneously be interpreted as changes in transcript level. To ensure our data were not confounded by differences in tissue composition between xenografts, we chose to nonquantitatively identify the tissue of origin of transcripts present in the xenografts.
Robust methods were used to exclude cross-species hybridization and, although some transcripts may be excluded that are truly present in nude mouse lesions, this conservative approach is preferable to incorrectly including data from cross-hybridizing transcripts. Due to the small number of replicate xenografts analyzed in this study and because of the possibility of confounding due to differences in xenograft tissue composition, transcript abundance was not quantified, rather the transcripts were identified as either present or absent in the human or mouse compartments of the lesions. This approach provided valuable information about the molecular pathways involved in the dialogue between endometrial and peritoneal tissues in endometriotic lesions.
Cellular Injury
One pathway identified in nude mouse lesions contained numerous transcripts encoding endometrial and host-derived proteasome subunits (Figure 3A); for example five of the seven α-type subunits (PSMA2, PSMA3, PSMA4, PSMA6, and PSMA7) and six components of the 19S regulatory particle (PSMD1, PSMD6, PSMD7, PSMD8, PSMD10, and PSMD11) were present. Proteasomes are multi-subunit complexes that degrade ubiquitin-tagged proteins in a highly regulated ATP-dependent manner. The identification of transcripts encoding these subunits suggests that protein recycling and turnover is occurring in the xenografts. This is borne out by the histological observation that a large necrotic core was present in the day 7 nude mouse lesions. Becker et al demonstrated increased staining density of the hypoxia probe, pimonidazole hydrochloride, in central locations of early xenografts from the nude mouse model of endometriosis.32 These findings suggest that ischemia-mediated cell death in the inner poorly vascularized core of day 7 nude mouse lesions may cause central necrotic changes.
Inflammation
Tissue damage activates the innate immune system and triggers an inflammation and repair response.33 Nuclear factor κB1 (NF-κB1) and RELA were central components of another ingenuity pathway (Figure 3B) describing relationships between transcripts derived from endometrial and host site tissues in nude mice. The NF-κB transcription factor system up-regulates the transcription of proinflammatory mediators such as cyclooxygenase-2, MCP-1, regulated on activation normal T cell expressed and secreted (RANTES), granulocyte macrophage–colony-stimulating factor (Gm-CSF), interleukin (IL)-6, IL-8, MHC class I, and matrix metalloproteinase (MMP) 7,33 all of which are associated with endometriotic disease (Table 3). Tumor necrosis factor (TNF)-α and IL-1β, which up-regulate the inflammatory NF-κB pathway, have been found at high levels in the peritoneal fluid from women with endometriosis.34,35 NF-κB was shown to be activated in endometriotic lesions29 and in peritoneal macrophages from women with endometriosis.36 Moreover, two inhibitors of the NF-κB pathway significantly reduced xenograft development in the nude mouse model of endometriosis.37
The presence of numerous leukocytes in the xenografts38(Figure 5) and an elevation of activated macrophages in the peritoneal environment of women with endometriosis39 are further indicators of the inflammatory response in both the animal model and human disease. In the human microarray analysis, transcripts in GO categories “inflammatory response” and “response to wounding” were significantly over-represented (Table 3). In addition, several inflammation-related transcripts were differentially expressed in human eutopic and ectopic endometrial tissue and present in the xenograft model (for example CXCL12, CCL2, and IL11RA; Table 4).
Table 4.
Summary of Correlation between Ingenuity Pathways, Histology of Xenografts, Other Transcripts Present in Xenografts, Human Eutopic/Ectopic GO-Ontology Analysis, and Findings in the Literature
| Cellular events | Ingenuity pathway | Microscopic findings in xenografts | Transcript associations in xenografts | GO ontology analysis in human endometrial tissues (P < 0.05) | Literature citations |
|---|---|---|---|---|---|
| Cellular injury | Proteasome pathway | Central necrosis in day 7 lesions | Ubiquitin/proteasome pathway | Oxidative damage58 | |
| Breakdown | Hypoxia32 | ||||
| Inflammation | NF-κB pathway | Infiltration of host-derived macrophages | Macrophages | Response to wounding (n = 22) | TNF-α29 |
| TNF-α and IL-1β | NFKB,59 COX-2,60 MHC class I61 | ||||
| NF-κB | Inflammatory response (n = 18) | RANTES,62 MCP-1,63 GM-CSF,64 macrophages65,66 | |||
| Chemokines | |||||
| Tissue repair and remodeling | TGF-β pathway | Morphometric evidence of glandular remodeling | Myofibroblasts, actin cytostructure | Cell adhesion (n = 35) | TGF-β,67,68 α-SMA,69 Thy243,44 |
| Infiltration of myofibroblasts | Integrins and cell adhesion molecules | Calcium-independent cell adhesion (n = 5) | Interleukin-6,70 interleukin 8,71 interleukin 10,70 integrins,72,73 MMP-7,74 fibrinogen,75 tenascin,76 collagen77 | ||
| Collagen and other ECM laid down | Matrix metalloproteinases | Extracellular matrix (n = 17) | |||
| Cell matrix interaction | Extracellular matrix | Intracellular junction (n = 10) | |||
| Cellular proliferation | KRAS pathway | Increased volume fraction of glands and stroma over time | Oncogenes | DNA metabolic process (N = 2) | KRAS,48 VEGF-A,8 proliferation,78 cell survival79,80 |
| Mitotic figures in glands and stromal cells | Cell survival | Nucleic acid binding (n = 30) |
Tissue Repair and Remodeling
In tissue injury, inflammatory processes are self-limited and are followed by healing and repair.40 Transforming growth factor (TGF)-β1 promotes Thy2+ interleukin activity, inhibiting macrophage activation and inducing cellular proliferation and extracellular matrix formation.41 TGF-β1 was a central factor in another xenograft-derived ingenuity pathway (Figure 3C), and several ECM transcripts (COL1A2, COL3A1, FN1, and MGP) were downstream of TGF-β1 in the IPA networks generated from nude mouse lesions. Furthermore, elevated levels of TGF-β1 are present in the peritoneal fluid from women with endometriosis,42 and endometriosis is associated with a Thy2 immune response pattern.43,44
Myofibroblast-associated transcripts were present in both human and nude mouse microarray gene lists, for example α-SMA (ACTA2). Myofibroblasts attracted to inflammatory sites, deposit collagen and other ECM proteins that provide structure for proliferating stromal and epithelial cells.45 In addition to the ECM components in the IPA network mentioned above, transcripts encoding other matrix components or matrix-modifying factors were identified in both the human and xenograft arrays (CSPG2, COL14A1, MXRA5, SLPI, MCAM, DPT). Staining of nude mouse lesions for collagen showed a progressively central and more intense staining pattern that encircled the epithelial glands over time (Figure 7, B and E).
In the nude mouse, host myofibroblasts (identified by staining for α-SMA; Figure 6, H–K) were in the periphery of day 7 lesions, whereas by day 14 they were located centrally. Cellular motility is achieved by restructuring the actin cytoskeleton,46 and several actin-associated proteins were present in both species compartments from nude mouse explants (for example CTTN, DSTN, ACTB ACTR3, ARPC2, and ARPC1B). Other transcripts encoding proteins involved in cell motility are also present in both xenografts and human tissue (TUBBA2, TPM1, and S100A4). Integrins mediate cell matrix signaling and provide anchorage during cell migration and several integrin transcripts were present in the xenografts (ITGB1, ITGB5, and ITGAV). MMPs and other proteases degrade proteins in the pathway of motile cells and cleave integrin-mediated anchoring interactions between cells and the ECM,46 and both mouse MMP7 and MMP11 transcripts were identified in the lesions.
In healing intestinal glandular tissue, actin fibers were seen connecting subepithelial myocytes and epithelial cells. The contraction of myocytes promoted epithelial cell-to-cell contact and re-epithelialization of damaged intestinal glands.47 A similar interaction may occur between human endometrial glands and host derived myocytes in endometriotic tissues as murine myofibroblasts were seen encircling human ectopic endometrial glandular cells in nude mouse lesions (Figure 6, I and J).
Cellular Proliferation
Ectopic regeneration of the endometrial glandular epithelium was evidenced by several epithelial specific transcripts in the human nude mouse-derived transcriptome (KRT18 and KRT19). Additionally, the volume fraction, size, and number of endometrial glands were higher in day 14 nude mouse xenografts when compared to day 7 lesions, whereas mitotic figures were seen in both glandular epithelial and stromal cells.
The KRAS transcript was a central transcript in the last IPA network (Figure 3D) derived from nude mouse xenografts. KRAS participates in cell proliferation, regulating the cell cycle through its GTPase activity. In a mouse model, activation of KRAS activity was associated with the development of endometriosis and endometrioid ovarian cancer,48underlining its importance in the endometriotic disease process. Other tumor-associated transcripts were present in the endometrial component of nude mouse xenografts (for example ABL1 and AKT) suggesting that similar proteins direct cell proliferation in both tumor cells and grafted ectopic human endometrium. Furthermore, epidemiological studies that show an increased incidence of ovarian cancer in patients diagnosed with endometriosis.49 The molecular studies reported here and those of Dinulescu et al48 may suggest a mechanism behind the population-based studies but this warrants further investigation.
Other Cellular Processes
Like tumors, ectopic endometrium must attract a vascular supply to survive. In our study complexes of small blood vessels and endothelial cells were seen at the edge of day 7 nude mouse lesions, whereas by day 14 the endothelial cells were centrally located (Figure 7, G and H). We have previously determined that these endothelial cells are mouse-derived,8 and it may be that hypoxia inducible factor up-regulates VEGF-A50 in the center of the nude mouse xenografts, attracting host endothelial cells to the lesion to form blood vessels. This would account for the success of antiangiogenic agents in suppressing lesion formation in in vivo models of endometriosis.8,9 In our study, mouse VEGF-A was present in the nude mouse xenografts, suggesting secretion of this factor by infiltrating host cells such as macrophages,39 although previous data suggested that inhibition of human VEGF-A was critical to lesion suppression.8
Inflammatory cell infiltration, extracellular matrix remodeling, cellular proliferation, and vascular formation are integral to the development of both tumors and endometriosis.51 In a similar fashion to cancer, endometriotic cellular interactions were more complex than a mere host-graft dialogue in ectopic endometrial lesions.52,53 Three different host-derived cell types (macrophages, myofibroblasts, and vascular endothelial cells) were identified in the stroma of nude mouse xenografts, and stromal cells derived from the endometrium were also present. Furthermore, an interaction between the ECM and cells in nude mouse lesions was apparent, with cells directly visualized lying along fibers of the ECM (Figure 7E).
The xenograft model of endometriosis we used has two great advantages over other methods. First, it incorporates human endometrium, which, if the retrograde menstruation theory is correct, is the ultimate source of the diseased tissue. Second, it allows us to dissect the epithelial and stromal components of the disease using species-specific microarray analysis. However, in xenograft models the molecular cross-talk between endometrium and stromal cells is potentially reduced by species-specific differences in receptor-ligand affinities and the immunosuppressed nature of the mice. Therefore, we have supplemented our xenograft analysis with analysis of transcript abundance in human patients with this disease. The nude mouse model of endometriosis is likely to be a good representation of human disease, as highly significant numbers of transcripts were present in both nude mouse xenografts and in the differentially regulated gene lists from the human eutopic/ectopic microarray analysis (Table 3).
Two groups have performed a microarray analysis of endometriosis-like lesions after autologous transplantation of uterine fragments in a rat endometriosis model. Several transcripts were revealed that were present in the nude mouse xenograft transcriptomes.54,55 Although both the rat and nude mouse model of endometriosis provide a similar picture of the longitudinal development of an ectopic endometrial lesion,54 the interaction between ectopic endometrial tissue and its site of attachment can only be explored in detail in nude mouse xenografts.
Although this study revealed many components in the dialogue between ectopic endometrial tissue and its peritoneal site of attachment, several transcripts that were anticipated to be differentially regulated were not present in our human eutopic/ectopic analysis (for example aromatase4). Similarly Wren et al found several molecular associations in the endometriosis literature that were not apparent in published endometriosis microarray studies.56 These findings point to other mechanisms particpating in the disease process for example post-translational protein modification or post-transcriptional regulation of protein production by microRNAs.57
Unique bioinformatics methods were used to characterize cellular and molecular changes in lesions from the nude mouse model of endometriosis and link them to gene signatures in human disease (Table 4). Several networks were identified that broached endometrial and host site tissue compartments in ectopic endometrial lesions. We believe that disruption of the dialogue between peritoneum and ectopic endometrium is likely to inhibit the cellular interactions necessary for endometriotic lesion development. Delineating the interactions between endometrial tissue and its ectopic environment may be the first step in the development of novel pharmacological therapies for this disabling condition.
Acknowledgments
We thank patients at The Rosie Hospital, Cambridge, UK, and at The Oxford Clinic, Christchurch, New Zealand, who participated in this study. We are also grateful to Barry Potter, Adrian Neuman, John Bashford, and Diana License for expert technical assistance.
Footnotes
Address reprint requests to Louise Hull, Research Centre for Reproductive Health, School of Paediatrics and Reproductive Health, Discipline of Obstetrics and Gynaecology, Level 6 Medical School North, University of Adelaide, South Australia, 5006, Australia. E-mail: louise.hull@adelaide.edu.au.
Supported by Pfizer Central Research, Sandwich, Kent, UK, and Medical Research Council, UK (MRC) grant G9623012 and the Cambridge National Institute for Health Research (NIHR) Biomedical Research Centre.
Supplemental material for this article can be found on http:// ajp.amjpathol.org.
References
- Strathy JH, Molgaard CA, Coulam CB, Melton LJ., 3rd Endometriosis and infertility: a laparoscopic study of endometriosis among fertile and infertile women. Fertil Steril. 1982;38:667–672. doi: 10.1016/s0015-0282(16)46691-4. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- Sampson JA. Peritoneal endometriosis due to menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14:442–469. [Google Scholar]
- Bulun SE, Yang S, Fang Z, Gurates B, Tamura M, Sebastian S. Estrogen production and metabolism in endometriosis. Ann NY Acad Sci. 2002;955:75–85. doi: 10.1111/j.1749-6632.2002.tb02767.x. [DOI] [PubMed] [Google Scholar]
- Ho HN, Wu MY, Yang YS. Peritoneal cellular immunity and endometriosis. Am J Reprod Immunol. 1997;38:400–412. doi: 10.1111/j.1600-0897.1997.tb00319.x. [DOI] [PubMed] [Google Scholar]
- Simpson JL, Bischoff F. Heritability and candidate genes for endometriosis. Reprod Biomed Online. 2003;7:162–169. doi: 10.1016/s1472-6483(10)61746-4. [DOI] [PubMed] [Google Scholar]
- Rier S, Foster WG. Environmental dioxins and endometriosis. Semin Reprod Med. 2003;21:145–154. doi: 10.1055/s-2003-41321. [DOI] [PubMed] [Google Scholar]
- Hull ML, Charnock-Jones DS, Chan CL, Bruner-Tran KL, Osteen KG, Tom BD, Fan TP, Smith SK. Antiangiogenic agents are effective inhibitors of endometriosis. J Clin Endocrinol Metab. 2003;88:2889–2899. doi: 10.1210/jc.2002-021912. [DOI] [PubMed] [Google Scholar]
- Nap AW, Griffioen AW, Dunselman GA, Bouma-Ter Steege JC, Thijssen VL, Evers JL, Groothuis PG: Antiangiogenesis therapy for endometriosis. J Clin Endocrinol Metab 2004, 1089–1095 [DOI] [PubMed] [Google Scholar]
- Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, Osteen K, Lessey BA, Giudice LC. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144:2870–2881. doi: 10.1210/en.2003-0043. [DOI] [PubMed] [Google Scholar]
- Eyster KM, Boles AL, Brannian JD, Hansen KA. DNA microarray analysis of gene expression markers of endometriosis. Fertil Steril. 2002;77:38–42. doi: 10.1016/s0015-0282(01)02955-7. [DOI] [PubMed] [Google Scholar]
- Arimoto T, Katagiri T, Oda K, Tsunoda T, Yasugi T, Osuga Y, Yoshikawa H, Nishii O, Yano T, Taketani Y, Nakamura Y. Genome-wide cDNA microarray analysis of gene-expression profiles involved in ovarian endometriosis. Int J Oncol. 2003;22:551–560. [PubMed] [Google Scholar]
- Wu Y, Kajdacsy-Balla A, Strawn E, Basir Z, Halverson G, Jailwala P, Wang Y, Wang X, Ghosh S, Guo SW. Transcriptional characterizations of differences between eutopic and ectopic endometrium. Endocrinology. 2006;147:232–246. doi: 10.1210/en.2005-0426. [DOI] [PubMed] [Google Scholar]
- Zamah NM, Dodson MG, Stephens LC, Buttram VC, Jr, Besch PK, Kaufman RH. Transplantation of normal and ectopic human endometrial tissue into athymic nude mice. Am J Obstet Gynecol. 1984;149:591–597. doi: 10.1016/0002-9378(84)90240-0. [DOI] [PubMed] [Google Scholar]
- Bergqvist A, Jeppsson S, Kullander S, Ljungberg O. Human uterine endometrium and endometriotic tissue transplanted into nude mice: morphologic effects of various steroid hormones. Am J Pathol. 1985;121:337–341. [PMC free article] [PubMed] [Google Scholar]
- Tabibzadeh S, Miller S, Dodson WC, Satyaswaroop PG. An experimental model for the endometriosis in athymic mice. Front Biosci. 1999;4:C4–9. doi: 10.2741/tabibzad. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Vanderslice KD. Identification in histological sections of species origin of cells from mouse, rat and human. Stain Technol. 1984;59:7–12. doi: 10.3109/10520298409113823. [DOI] [PubMed] [Google Scholar]
- Schwartz DR, Moin K, Yao B, Matrisian LM, Coussens LM, Bugge TH, Fingleton B, Acuff HB, Sinnamon M, Nassar H, Platts AE, Krawetz SA, Linebaugh BE, Sloane BF. Hu/Mu ProtIn oligonucleotide microarray: dual-species array for profiling protease and protease inhibitor gene expression in tumors and their microenvironment. Mol Cancer Res. 2007;5:443–454. doi: 10.1158/1541-7786.MCR-06-0337. [DOI] [PubMed] [Google Scholar]
- Acuff HB, Sinnamon M, Fingleton B, Boone B, Levy SE, Chen X, Pozzi A, Carbone DP, Schwartz DR, Moin K, Sloane BF, Matrisian LM. Analysis of host- and tumor-derived proteinases using a custom dual species microarray reveals a protective role for stromal matrix metalloproteinase-12 in non-small cell lung cancer. Cancer Res. 2006;66:7968–7975. doi: 10.1158/0008-5472.CAN-05-4279. [DOI] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Fertil Steril. 1975;1:3–25. doi: 10.1016/j.fertnstert.2019.08.079. [DOI] [PubMed] [Google Scholar]
- Hull ML, Prentice A, Wang DY, Butt RP, Phillips SC, Smith SK, Charnock-Jones DS. Nimesulide, a COX-2 inhibitor, does not reduce lesion size or number in a nude mouse model of endometriosis. Hum Reprod. 2005;20:350–358. doi: 10.1093/humrep/deh611. [DOI] [PubMed] [Google Scholar]
- R Development Core Group (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-90051-07-01 [Google Scholar]
- Cheng CW, Bielby H, Licence D, Smith SK, Print CG, Charnock-Jones DS. Quantitative cellular and molecular analysis of the effect of progesterone withdrawal in a murine model of decidualization. Biol Reprod. 2007;76:871–883. doi: 10.1095/biolreprod.106.057950. [DOI] [PubMed] [Google Scholar]
- Smyth GK. New York: Springer; LIMMALinear Models for Microarray Data. 2005:pp 397–420. [Google Scholar]
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework: analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ramos R, Van Langendonckt A, Defrere S, Lousse JC, Mettlen M, Guillet A, Donnez J. Agents blocking the nuclear factor-kappaB pathway are effective inhibitors of endometriosis in an in vivo experimental model. Gynecol Obstet Invest. 2007;65:174–186. doi: 10.1159/000111148. [DOI] [PubMed] [Google Scholar]
- Rizki A, Weaver VM, Lee SY, Rozenberg GI, Chin K, Myers CA, Bascom JL, Mott JD, Semeiks JR, Grate LR, Mian IS, Borowsky AD, Jensen RA, Idowu MO, Chen F, Chen DJ, Petersen OW, Gray JW, Bissell MJ. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68:1378–1387. doi: 10.1158/0008-5472.CAN-07-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner-Tran KL, Eisenberg E, Yeaman GR, Anderson TA, McBean J, Osteen KG. Steroid and cytokine regulation of matrix metalloproteinase expression in endometriosis and the establishment of experimental endometriosis in nude mice. J Clin Endocrinol Metab. 2002;87:4782–4791. doi: 10.1210/jc.2002-020418. [DOI] [PubMed] [Google Scholar]
- Becker CMB, Cramer T, Funakoshi T, Rohwer N, Bernhart W, Treston AM, Folkman J, D'Amato RJ. Hypoxia may be an important regulator of disease progression in a mouse model of endometriosis. Obstet Gynecol. 2005;123:S19–S20. [Google Scholar]
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Overton C, Fernandez-Shaw S, Hicks B, Barlow D, Starkey P. Peritoneal fluid cytokines and the relationship with endometriosis and pain. Hum Reprod. 1996;11:380–386. doi: 10.1093/humrep/11.2.380. [DOI] [PubMed] [Google Scholar]
- Mori H, Sawairi M, Nakagawa M, Itoh N, Wada K, Tamaya T. Peritoneal fluid interleukin-1 beta and tumor necrosis factor in patients with benign gynecologic disease. Am J Reprod Immunol. 1991;26:62–67. doi: 10.1111/j.1600-0897.1991.tb00972.x. [DOI] [PubMed] [Google Scholar]
- Lousse JC, Van Langendonckt A, Gonzalez-Ramos R, Defrere S, Renkin E, Donnez J. Increased activation of nuclear factor-kappa B (NF-kappaB) in isolated peritoneal macrophages of patients with endometriosis. Fertil Steril. 2007;1:217–220. doi: 10.1016/j.fertnstert.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ramos R, Donnez J, Defrere S, Leclercq I, Squifflet J, Lousse JC, Van Langendonckt A. Nuclear factor-kappa B is constitutively activated in peritoneal endometriosis. Mol Hum Reprod. 2007;13:503–509. doi: 10.1093/molehr/gam033. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Lai MD, Lei HY, Wing LY. Neutrophils and macrophages promote angiogenesis in the early stage of endometriosis in a mouse model. Endocrinology. 2006;147:1278–1286. doi: 10.1210/en.2005-0790. [DOI] [PubMed] [Google Scholar]
- McLaren J, Prentice A, Charnock-Jones DS, Smith SK. Vascular endothelial growth factor (VEGF) concentrations are elevated in peritoneal fluid of women with endometriosis. Hum Reprod. 1996;11:220–223. doi: 10.1093/oxfordjournals.humrep.a019023. [DOI] [PubMed] [Google Scholar]
- Frangogiannis NG. The role of the chemokines in myocardial ischemia and reperfusion. Curr Vasc Pharmacol. 2004;2:163–174. doi: 10.2174/1570161043476375. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Oosterlynck DJ, Meuleman C, Lacquet FA, Waer M, Koninckx PR. Flow cytometry analysis of lymphocyte subpopulations in peritoneal fluid of women with endometriosis. Am J Reprod Immunol. 1994;31:25–31. doi: 10.1111/j.1600-0897.1994.tb00843.x. [DOI] [PubMed] [Google Scholar]
- Podgaec S, Abrao MS, Dias JA, Jr, Rizzo LV, de Oliveira RM, Baracat EC. Endometriosis: an inflammatory disease with a Th2 immune response component. Hum Reprod. 2007;22:1373–1379. doi: 10.1093/humrep/del516. [DOI] [PubMed] [Google Scholar]
- Akoum A, Lawson C, Herrmann-Lavoie C, Maheux R. Imbalance in the expression of the activating type I and the inhibitory type II interleukin 1 receptors in endometriosis. Hum Reprod. 2007;22:1464–1473. doi: 10.1093/humrep/dem021. [DOI] [PubMed] [Google Scholar]
- Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- Small JV, Rottner K, Kaverina I, Anderson KI. Assembling an actin cytoskeleton for cell attachment and movement. Biochim Biophys Acta. 1998;1404:271–281. doi: 10.1016/s0167-4889(98)00080-9. [DOI] [PubMed] [Google Scholar]
- Andoh A, Fujino S, Okuno T, Fujiyama Y, Bamba T. Intestinal subepithelial myofibroblasts in inflammatory bowel diseases. J Gastroenterol. 2002;37(Suppl 14):33–37. doi: 10.1007/BF03326410. [DOI] [PubMed] [Google Scholar]
- Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- Melin A, Sparen P, Persson I, Bergqvist A. Endometriosis and the risk of cancer with special emphasis on ovarian cancer. Hum Reprod. 2006;21:1237–1242. doi: 10.1093/humrep/dei462. [DOI] [PubMed] [Google Scholar]
- Sharkey AM, Day K, McPherson A, Malik S, Licence D, Smith SK, Charnock-Jones DS. Vascular endothelial growth factor expression in human endometrium is regulated by hypoxia. J Clin Endocrinol Metab. 2000;85:402–409. doi: 10.1210/jcem.85.1.6229. [DOI] [PubMed] [Google Scholar]
- van Kempen LC, de Visser KE, Coussens LM. Inflammation, proteases and cancer. Eur J Cancer. 2006;42:728–734. doi: 10.1016/j.ejca.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Framson PE, Sage EH. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem. 2004;92:679–690. doi: 10.1002/jcb.20091. [DOI] [PubMed] [Google Scholar]
- Cheon YP, Li Q, Xu X, DeMayo FJ, Bagchi IC, Bagchi MK. A genomic approach to identify novel progesterone receptor regulated pathways in the uterus during implantation. Mol Endocrinol. 2002;16:2853–2871. doi: 10.1210/me.2002-0270. [DOI] [PubMed] [Google Scholar]
- Flores I, Rivera E, Ruiz LA, Santiago OI, Vernon MW, Appleyard CB. Molecular profiling of experimental endometriosis identified gene expression patterns in common with human disease. Fertil Steril. 2007;87:1180–1199. doi: 10.1016/j.fertnstert.2006.07.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno R, Fujiwara H, Netsu S, Odagiri K, Shimane M, Nomura H, Suzuki M. Gene expression profiling of the rat endometriosis model. Am J Reprod Immunol. 2007;58:330–343. doi: 10.1111/j.1600-0897.2007.00507.x. [DOI] [PubMed] [Google Scholar]
- Wren JD, Wu Y, Guo SW. A system-wide analysis of differentially expressed genes in ectopic and eutopic endometrium. Hum Reprod. 2007;22:2093–2102. doi: 10.1093/humrep/dem129. [DOI] [PubMed] [Google Scholar]
- Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA: endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- Slater M, Quagliotto G, Cooper M, Murphy CR. Endometriotic cells exhibit metaplastic change and oxidative DNA damage as well as decreased function, compared to normal endometrium. J Mol Histol. 2005;36:257–263. doi: 10.1007/s10735-005-3802-9. [DOI] [PubMed] [Google Scholar]
- Nasu K, Nishida M, Ueda T, Yuge A, Takai N, Narahara H. Application of the nuclear factor-kappaB inhibitor BAY 11–7085 for the treatment of endometriosis: an in vitro study. Am J Physiol Endocrinol Metab. 2007;293:E16–E23. doi: 10.1152/ajpendo.00135.2006. [DOI] [PubMed] [Google Scholar]
- Ota H, Igarashi S, Sasaki M, Tanaka T. Distribution of cyclooxygenase-2 in eutopic and ectopic endometrium in endometriosis and adenomyosis. Hum Reprod. 2001;16:561–566. doi: 10.1093/humrep/16.3.561. [DOI] [PubMed] [Google Scholar]
- Semino C, Semino A, Pietra G, Mingari MC, Barocci S, Venturini PL, Ragni N, Melioli G. Role of major histocompatibility complex class I expression and natural killer-like T cells in the genetic control of endometriosis. Fertil Steril. 1995;64:909–916. doi: 10.1016/s0015-0282(16)57901-1. [DOI] [PubMed] [Google Scholar]
- Hornung D, Klingel K, Dohrn K, Kandolf R, Wallwiener D, Taylor RN. Regulated on activation, normal T-cell-expressed and -secreted mRNA expression in normal endometrium and endometriotic implants: assessment of autocrine/paracrine regulation by in situ hybridization. Am J Pathol. 2001;158:1949–1954. doi: 10.1016/S0002-9440(10)64664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolicoeur C, Boutouil M, Drouin R, Paradis I, Lemay A, Akoum A. Increased expression of monocyte chemotactic protein-1 in the endometrium of women with endometriosis. Am J Pathol. 1998;152:125–133. [PMC free article] [PubMed] [Google Scholar]
- Sharpe-Timms KL, Bruno PL, Penney LL, Bickel JT. Immunohistochemical localization of granulocyte-macrophage colony-stimulating factor in matched endometriosis and endometrial tissues. Am J Obstet Gynecol. 1994;171:740–745. doi: 10.1016/0002-9378(94)90091-4. [DOI] [PubMed] [Google Scholar]
- McLaren J, Prentice A, Charnock-Jones DS, Millican SA, Muller KH, Sharkey AM, Smith SK. Vascular endothelial growth factor is produced by peritoneal fluid macrophages in endometriosis and is regulated by ovarian steroids. J Clin Invest. 1996;98:482–489. doi: 10.1172/JCI118815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme J, Becker S, Wing R. Accentuated cyclic activation of peritoneal macrophages in patients with endometriosis. Am J Obstet Gynecol. 1984;148:85–90. doi: 10.1016/s0002-9378(84)80037-x. [DOI] [PubMed] [Google Scholar]
- Chegini N, Gold LI, Williams RS. Localization of transforming growth factor beta isoforms TGF-beta 1. TGF-beta 2, and TGF-beta 3 in surgically induced endometriosis in the rat. Obstet Gynecol. 1994;83:455–461. [PubMed] [Google Scholar]
- Komiyama S, Aoki D, Komiyama M, Nozawa S. Local activation of TGF-beta1 at endometriosis sites. J Reprod Med. 2007;52:306–312. [PubMed] [Google Scholar]
- Anaf V, Simon P, Fayt I, Noel J. Smooth muscles are frequent components of endometriotic lesions. Hum Reprod. 2000;15:767–771. doi: 10.1093/humrep/15.4.767. [DOI] [PubMed] [Google Scholar]
- Punnonen J, Teisala K, Ranta H, Bennett B, Punnonen R. Increased levels of interleukin-6 and interleukin-10 in the peritoneal fluid of patients with endometriosis. Am J Obstet Gynecol. 1996;174:1522–1526. doi: 10.1016/s0002-9378(96)70600-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Velasco JA, Arici A. Interleukin-8 stimulates the adhesion of endometrial stromal cells to fibronectin. Fertil Steril. 1999;72:336–340. doi: 10.1016/s0015-0282(99)00223-x. [DOI] [PubMed] [Google Scholar]
- Beliard A, Donnez J, Nisolle M, Foidart JM. Localization of laminin, fibronectin, E-cadherin, and integrins in endometrium and endometriosis. Fertil Steril. 1997;67:266–272. doi: 10.1016/S0015-0282(97)81909-7. [DOI] [PubMed] [Google Scholar]
- Giannelli G, Sgarra C, Di Naro E, Lavopa C, Angelotti U, Tartagni M, Simone O, Trerotoli P, Antonaci S, Loverro G. Endometriosis is characterized by an impaired localization of laminin-5 and alpha3beta1 integrin receptor. Int J Gynecol Cancer. 2007;17:242–247. doi: 10.1111/j.1525-1438.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- Uzan C, Cortez A, Dufournet C, Fauvet R, Siffroi JP, Darai E. Eutopic endometrium and peritoneal, ovarian and bowel endometriotic tissues express a different profile of matrix metalloproteinases-2, -3 and -11, and of tissue inhibitor metalloproteinases-1 and -2. Virchows Arch. 2004;445:603–609. doi: 10.1007/s00428-004-1117-y. [DOI] [PubMed] [Google Scholar]
- Kauma S, Clark MR, White C, Halme J. Production of fibronectin by peritoneal macrophages and concentration of fibronectin in peritoneal fluid from patients with or without endometriosis. Obstet Gynecol. 1988;72:13–18. [PubMed] [Google Scholar]
- Harrington DJ, Lessey BA, Rai V, Bergqvist A, Kennedy S, Manek S, Barlow DH, Mardon HJ. Tenascin is differentially expressed in endometrium and endometriosis. J Pathol. 1999;187:242–248. doi: 10.1002/(SICI)1096-9896(199901)187:2<242::AID-PATH221>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Matsuzaki S, Canis M, Darcha C, Dechelotte P, Pouly JL, Bruhat MA. Fibrogenesis in peritoneal endometriosis: a semi-quantitative analysis of type-I collagen. Gynecol Obstet Invest. 1999;47:197–199. doi: 10.1159/000010094. [DOI] [PubMed] [Google Scholar]
- Toki T, Nakayama K. Proliferative activity and genetic alterations in TP53 in endometriosis. Gynecol Obstet Invest. 2000;50(Suppl 1):33–38. doi: 10.1159/000052876. [DOI] [PubMed] [Google Scholar]
- Johnson MC, Torres M, Alves A, Bacallao K, Fuentes A, Vega M, Boric MA. Augmented cell survival in eutopic endometrium from women with endometriosis: expression of c-myc. TGF-beta1 and bax genes. Reprod Biol Endocrinol. 2005;3:45. doi: 10.1186/1477-7827-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumenou A, Panayiotides I, Matalliotakis I, Vlachonikolis I, Tzardi M, Koumantakis E. Bcl-2 and Bax expression in human endometriotic and adenomyotic tissues. Eur J Obstet Gynecol Reprod Biol. 2001;99:256–260. doi: 10.1016/s0301-2115(01)00381-5. [DOI] [PubMed] [Google Scholar]