Abstract
In liver wound healing, transforming growth factor-β (TGF-β) plays a critical role in stellate cell activation as well as signaling cascades in the fibrogenic response to injury. We postulate that the TGF-β-dependent downstream signaling pathway may vary according to the mechanism of stellate cell activation; this study was undertaken to ascertain whether the downstream signaling pathways mediated by TGF-β vary in different liver injury models. We measured Smad3 and MAP kinase activation after isolating stellate cells from rat livers injured by either bile duct ligation (BDL) or repeated carbon tetrachloride (CCl4) administration. Phospho-Smad3 was dramatically up-regulated in stellate cells after CCl4 injury, but not after BDL-induced injury. TGF-β signaling in stellate cells activated after BDL was mediated prominently through ERK activation, whereas activation induced by CCl4 injury or culture led to a cross-signaling mechanism involving both Smad3 and p38. The divergent Smad signaling pathways observed appeared to be attributable to the differential regulation of the early growth response gene-1 (Egr-1), an apparent negative transcriptional factor for Smad3 in our system. In addition, inhibition of ERK activation in stellate cells from BDL-injured liver led to a decrease in expression of endothelin-converting enzyme-1, a critical regulator of endothelin-1. We speculate that TGF-β signaling proceeds through differential signaling pathways depending on the mechanism of liver injury that leads to stellate cell activation.
Transforming growth factor-β (TGF-β) is a multifunctional cytokine involved in controlling cell proliferation, cell differentiation, cell adhesion, and extracellular matrix production. TGF-β is also commonly accepted as a central component of fibrogenic response to wounding and is up-regulated in a variety of different diseases such as those involving the skin, pancreas, liver, and others.1,2 In murine hepatic wounding, TGF-β1 (henceforth referred to as TGF-β) mRNA and (active) protein levels are elevated and correlate closely with collagen gene transcription as well as its accumulation and scar formation.3 A major cellular target of TGF-β’s effect is the hepatic stellate cell, a critical effector of the hepatic wounding response4; evidence indicates that TGF-β exerts direct effects on hepatic stellate cells, specifically leading to their activation with concomitant extracellular matrix production. In aggregate, the data point to TGF-β as a critical modulator of the wounding response.
Extensive investigation has begun to elucidate the mechanism(s) by which TGF-β exerts its effect on gene expression. In this regard, the Smads have received great attention as important TGF-β signaling effectors.5 In most cell types, TGF-β binds to the constitutively active type II receptor, which then recruits and phosphorylates type I receptor. Subsequent to receptor phosphorylation is stimulation of activin-like kinase (ALK)-5 and phosphorylation of Smad2 and Smad3. Smad1 and Smad5 are activated by bone morphogenetic protein type I receptors in a similar pathway.6 In endothelial cells and stellate cells, TGF-β exerts its effects through activation of ALK-1 and the Smad1/5 pathway in addition to activation of ALK-5 and Smad2/3 cascade.7,8 Smad4 is a co-Smad (ie, requiring binding with another Smad, forming a heteromeric complex). After oligomerization, Smad4-containing heteromers enter the nucleus where they exert effects on gene transcription.9 Smad6 and Smad7 are inhibitory Smads that antagonize signaling by other Smads.10 In liver wound healing; Smad3 is required for hepatic stellate cell matrix production and matrix interactions11 and maximal type I collagen induction12 whereas Smad2 on the other hand, plays an antiproliferative role without an apparent function in fibrogenesis.11 The mitogen-activated protein kinase (MAPK) family also appears to be crucial in TGF-β-mediated signal transduction.13 Three distinct members of the MAPK family including ERK1/2, p38, and JNK may be activated by TGF-β stimulation.14,15 ERKs are central elements in mitogenic signal transduction downstream of Ras, p38 appears to be important in pro-inflammatory cytokine and environmental stress signaling, and JNK plays a key role in the cellular response to pro-inflammatory cytokines and apoptotic agents. In quiescent hepatic stellate cells16 and in pancreatic stellate cells,17 p38 MAPK activity influences cell activation. TGF-β itself increases α1(1) collagen mRNA stability through p38 MAPK.18,19
Not only have the Smads been shown to be important regulators of gene transcription, but the early growth response gene-1 (Egr-1) has likewise emerged as an important putative transcriptional regulator.20 In cultured hepatic stellate cells, Egr-1 has been shown to be up-regulated in response to physiologically relevant concentrations of bile acids indicating its role in perpetuation of hepatic fibrosis.19 Also, Egr-1 is up-regulated in endothelial and smooth muscle cells after vascular injury21 and it is induced by extracellular stimuli such as growth factors and cytokines,20 including TGF-β. For example, expression of this DNA-binding protein is induced by TGF-β in fibroblasts in vitro and in vivo.22 Studies have also shown that Egr-1 stimulates TGF-β synthesis in a variety of mesenchymal cell types and enhances their sensitivity to TGF-β.23
TGF-β in hepatic wound healing has also been linked to the regulation of endothelin-1 (a key vasoactive compound) in stellate cells, Endothelin-1 is a key peptide important in the fibrogenic response24 and may contribute to abnormal vascular responses and possibly wound contraction. This latter process has been linked to enhanced expression of endothelin-converting enzyme (ECE-1). ECE-1 is a neutral membrane-bound metalloprotease with Mr = 120 kDa, belonging to the endo-peptidase-24.11 family found in brain.25 ECE-1 is responsible for specific cleavage of an intermediate endothelin-1 precursor (big endothelin)26 into biologically active peptides. The mechanism by which TGF-β-regulated endothelin-1 overproduction in the injured livers occurs is linked to increased ECE-1 expression and posttranscriptional modulation of ECE-1 mRNA.27
TGF-β is prominent in several mechanistically distinct experimental models of hepatic wound healing.28 We hypothesize that at the cellular level, specific TGF-β signaling pathways were likely to be present. In this study, we have taken advantage of our ability to isolate populations of stellate cells after induction of in vivo injury and hepatic wound healing and using this model, we have carefully examined putative TGF-β signaling. We reveal differential Smad3 and MAPK expression, and moreover that TGF-β signaling proceeds through divergent Smad and MAPK pathways after different types of liver injury and this has a functional effect on the expression of endothelin-converting enzyme-1. Further, we show that Egr-1 appears to be a critical factor in the regulation of Smad3 expression in vivo during hepatic wound healing. The data indicate that TGF-β utilizes diverse signaling pathways in hepatic wound healing and has important implications for targeting compounds that may interrupt this process.
Materials and Methods
Materials
Recombinant human TGF-β1 was from R&D Systems (Minneapolis, MN); polyclonal anti-Smad3 antibody was from Zymed Laboratories Inc. (South San Francisco, CA); anti-ECE-1 primary antibody B61/104 was kindly provided by Dr. Thomas Subkowski, BASF, Ludwigshafen, Germany; desmin, anti-β-actin, and smooth muscle α-actin antibodies were from Sigma Biochemicals (St. Louis, MO); monoclonal collagen 1 antibody was from Abcam (Cambridge, MA); Egr-1 and CRBP-1 were from Santa Cruz Biotechnology (Santa Cruz, CA); monoclonal phospho-p38 MAPK, phospho-SAPK/JNK MAPK, phospho-p44/42 MAPK (to detect phospho-ERK1/2), p38, SAPK/JNK, p44/42 (to detect ERK1/2), and phospho-Smad3 antibodies were from Cell Signaling Technology (St. Louis, MO) and Epitomics (Burlingame, CA). SAPK/JNK, p38, and p44/42 MAPK activity assay kits were purchased from Cell Signaling Technology, Inc. (Beverly, MA). A soluble TGF-β type II receptor (STR), generated by fusion of rabbit type II TGF-β receptor with Fc region of human IgG1 (this construct inhibits biological activity of TGF-β1 and TGF-β3), from Phillip Gotwals (Biogen Inc., Cambridge, MA) was synthesized as described.29
Cell Isolation and Culture
Hepatic stellate cells were isolated from male Sprague-Dawley rats (450 to 500 g; Harlan Sprague Dawley, Indianapolis, IN) as described previously.30 In brief, after in situ perfusion of the liver with 0.20 mg/100 ml of pronase (Roche Molecular Biochemicals, Indianapolis, IN), followed by 0.013 mg/100 ml of collagenase (Crescent Chemical, Hauppauge, NY), dispersed cell suspensions were layered on a discontinuous density gradient of 8.2% and 15.6% Accudenz (Accurate Chemical and Scientific, Westbury, NY). The resulting upper layer consisted of more than 95% stellate cells (data not shown). Cell purity was assessed microscopically and by examining vitamin A autofluorescence and desmin staining, which was established to be 90 to 95% for all experimental samples studied. Cells were suspended in modified medium 199, containing 20% serum (10% horse serum and 10% calf serum; Life Technologies, Inc., Gaithersburg, MD) at a density of ∼1 × 106 cells/ml. Cultures were incubated in a humidified incubator containing 95% O2/2.5% CO2. Cell viability was greater than 95% in all of the cultures used for study. Cells isolated from normal rat livers and harvested immediately for total proteins were considered to be quiescent, whereas cells grown for 5 or more days in the presence of serum-containing medium were considered to be activated in vitro. Cells isolated from injured rat livers were assayed immediately after isolation or in some situations were cultured very short periods of time to allow specialized studies, such cells were considered to be activated in vivo.
Animal Models of Liver Injury and Fibrosis
Liver injury and fibrosis were induced by intragastric gavage of carbon tetrachloride (CCl4) (1.0 ml per kg) every 4 days for four doses or by ligation of common bile duct as described by Kountouras and colleagues31 for 15 days.32 In some experiments, rats undergoing bile duct ligation (BDL) or CCl4received a soluble TGF-β receptor (5 mg/kg intraperitoneally) every 4 days. Animal protocols used to induce injury and fibrogenesis were approved by the institution’s animal care and guidelines committee.
Immunoprecipitation and Kinase Assays
Hepatic stellate cells were rinsed in phosphate-buffered saline (PBS), lysed in buffer (20 mmol/L Tris, 150 mmol/L NaCl, 5 mmol/L β-glycerolphosphate, 2 mmol/L dithiothreitol, 0.1 mmol/L Na3VO4, 10 mmol/L MgCl2, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton X-100) containing protease inhibitors (1 mg/ml leupeptin and 1 mmol/L phenylmethyl sulfonyl fluoride). Lysates were sonicated and centrifuged at 11,000 × g for 10 minutes at 4°C. Supernatants (200 μg of total protein) were incubated with 15 μl of specified immobilized antibodies to selectively immunoprecipitate the active MAPK from the lysate or with fusion protein bound to glutathione Sepharose beads to selectively pull down an active MAPK from the lysate, overnight at 4°C with gentle rotation. Pellets were suspended in kinase buffer (25 mmol/L Tris, 5 mmol/L β-glycerophosphate, 2 mmol/L dithiothreitol, 0.1 mmol/L Na3VO4, 10 mmol/L MgCl2) supplemented with 100 μmol/L ATP. Kinase reactions were performed at 30°C for 30 minutes followed by termination with 25 μl of sodium dodecyl sulfate sample buffer (62.5 mmol/L Tris-HCl, 2% w/v sodium dodecyl sulfate, 10% glycerol, 50 mmol/L dithiothreitol, 0.1% w/v bromophenol blue). After immunoprecipitation, samples were subjected to immunoblotting as below.
Immunoblotting
Stellate cells were lysed in sample buffer (containing 3% sodium dodecyl sulfate, 62.5 mmol/L Tris base, and 10% glycerol). Equal quantities of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions and transferred to nitrocellulose (Bio-Rad, Hercules, CA). Immunoblotting was performed using specified primary antibodies and horseradish peroxidase-conjugated secondary antibody. Specific signals were visualized using enhanced chemiluminescence (Pierce Biotechnology, Inc., Rockford, IL) and captured with a digital image system (Chemigenius2 photo-documentation system; Syngene, Cambridge, UK). Intensities of specific bands were quantified using standard software (Gene Tool, Syngene).
Immunocytochemistry
Cell cultures were washed with PBS and fixed with 4% paraformaldehyde for 10 minutes at room temperature. After quenching with 10% goat serum in PBS for 1 hour, specimens were incubated with anti-phospho-Smad3 and anti-Smad3 antibodies, respectively, and incubated overnight at 4°C. After washing with PBS, labeled proteins were detected with fluorescein-linked anti-rabbit IgG (1:250; Amersham Life Sciences, Arlington Heights, IL), washed, and mounted. Images were captured with a Zeiss LSM 510 META laser-scanning confocal microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY). Control samples were handled in a manner identical to experimental samples with the exception that they were not exposed to primary antibody or incubated with normal IgG.
Sense and Antisense Oligodeoxynucleotides (ODNs)
Egr-1 phosphorothioate-modified sense and antisense ODNs were synthesized based on published nucleotide sequences (Operon Technologies, Alameda, CA) Sequences corresponding to nucleotides −118 to −99 were used in this study and were as follows: rat Egr-1 antisense: 5′-AGTGTGCCCCTGCACCCCGC-3′ and rat Egr-1 sense: 5′-GCGGGGTGCAGGGGCACACT-3′, respectively. After sense or antisense ODNs (5 μmol/L) were added in the culture medium for 24 hours. Protein expression was examined by immunoblot.
Mobility Shift Assays
For preparation of nuclear extracts, cells were suspended in PBS and sonicated. After centrifugation at 1500 × g for 5 minutes. The cell pellet was resuspended in buffer containing 0.01 mol/L HEPES (pH 7.6), 1.5 mmol/L MgCl2, 0.5 mmol/L dithiothreitol, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, 1 mmol/L sodium vanadate, and 2 mmol/L KCl. After incubation on ice for 15 minutes, samples were resuspended in 10% Nonidet P-40 and centrifuged at 120,000 × g. The pellet was dissolved in high-salt buffer containing 2 mmol/L KCl, 0.42 mol/L NaCl, 0.01 mol/L HEPES (pH 7.6), 1. 5 mmol/L MgCl2, 0.5 mmol/L dithiothreitol, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, 1 mmol/L sodium vanadate, 25% glycerol, and 0.2 mmol/L EDTA. Finally, after centrifugation at 120,000 × g for 5 minutes, supernatants were collected and examined. Egr-1 consensus binding oligonucleotides (30 ng) (Santa Cruz Biotechnology) were end-labeled with [γ-32P] ATP by T4 polynucleotide kinase. The complementary oligonucleotides corresponding to the following region of the Smad3 promoter were used in the study; Egr-1 (−231 bp to −240 bp), 5-CGCCCCCGGC-3′, Egr-1 (−261 bp to −270 bp), 5′-GCCGGGGGCG-3′. After purification of the oligonucleotide probe with microspin G-50 column (Amersham Pharmacia Biotechnology, Piscataway, NJ), nuclear extract (10 μg) was incubated with radiolabeled probe (40,000 cpm) in the binding buffer containing 0.1 mol/L KCl, 25 mmol/L HEPES, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol, 20% glycerol, and 2 μg poly (dI-dC) at room temperature for 20 minutes. Binding reactions were resolved on 0.5× TBE, 6% acrylamide gel at 150 V for 2 hours. The gels were subsequently dried and autoradiography was performed. Egr-1 antibody (Santa Cruz Biotechnology) or smooth muscle α-actin antibody was added to the binding reaction 10 minutes before the addition of radiolabeled probe.
Statistics
Data are expressed as mean ± SE and n refers to the numbers of individual experiments performed. Individual cell preparations from unique isolations were used for all experiments. Differences among groups were determined using one-way analysis of variance followed by the Newman-Keuls procedure. The 0.05 level of probability was used as the criterion of significance.
Results
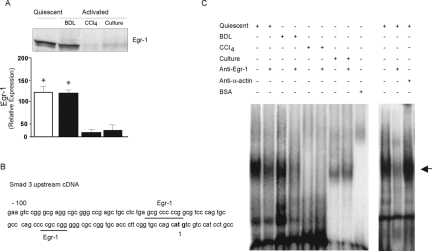
Smads Are Differentially Expressed in Stellate Cells after Liver Injury
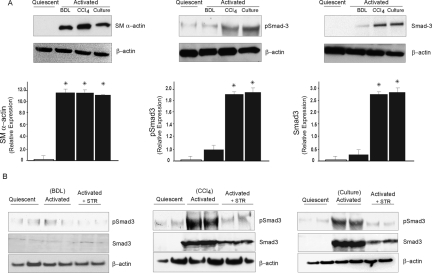
Smad and MAPK signaling families have been recognized as key components in TGF-β signaling.13 We and others have previously demonstrated that TGF-β regulates endogenous gene expression in stellate cells from injured livers.29 To determine whether Smads are involved in TGF-β signaling in stellate cell activation induced by different stimuli, we investigated phospho-Smad3 expression during stellate cell activation in vivo by using different models of injury (ie, BDL and CCl4), as well as a culture model of activation (the in vitro model is one in which cells are grown on plastic and exposed to high concentrations of serum—a process thought to mimic in vivo activation). We found that cells isolated from normal livers do not express smooth muscle α-actin, whereas those from either model of liver injury express abundant amounts of this protein, marking them as activated33 (Figure 1A, left). Minimal phospho-Smad3 and Smad3 was detectable in stellate cells from normal or BDL-injured livers (Figure 1A, middle and right, respectively). In contrast, phospho-Smad3 was abundant in stellate cells from CCl4-injured liver and after culture-induced activation. The antibodies used to detect phospho-Smad3 and total Smad3 did not show any cross-reactivity with recombinant Smad2 or to phospho-Smad1/5. This result suggested that Smads could be differentially involved in TGF-β signaling pathways after divergent types of injury. To further evaluate the effect of TGF-β on the regulation of Smad3 expression in vivo, we inhibited TGF-β signaling during liver injury with a soluble TGF-β receptor (Figure 1B). phospho-Smad3 expression in stellate cells from CCl4-injured liver or culture-induced activation was inhibited by ∼40 to 60% after TGF-β inhibition (Figure 1B).
Figure 1.
Smad3 expression and regulation by TGF-β in activated stellate cells in different liver injury models. Stellate cells were isolated from normal rat livers after BDL for 15 days and after four doses of CCl4 and immediately harvested as described in the Materials and Methods. In addition, cells from normal livers were isolated and allowed to undergo culture-induced activation as described in the Materials and Methods. A: Cell lysates (30 μg total protein) were subjected to immunoblotting using anti-phospho-Smad3, anti-Smad3 as described in the Materials and Methods (membranes for phospho-Smad3 were stripped and reprobed with anti-smooth muscle α-actin antibody and then stripped and reprobed with anti-β-actin antibody). Specific bands were scanned, quantified, normalized, and expressed graphically (n = 3, *P < 0.05 compared to quiescent stellate cells). Representative blots are shown in the figure. B: Liver injury was induced by BDL and CCl4, in addition, rats were exposed in vivo to a soluble TGF-β receptor type II (STR) construct, 5 mg/kg intraperitoneally as in the Materials and Methods. Cells were subsequently isolated and subjected to immunoblot to detect phospho-Smad3 and Smad3 as in the Materials and Methods. Culture-activated stellate cells were incubated in serum-free medium for 12 hours, after which medium containing STR (50 μg/ml) was added to the cells. Cells were harvested 24 hours later and subjected to immunoblotting as above. The blots shown are representative of three others.
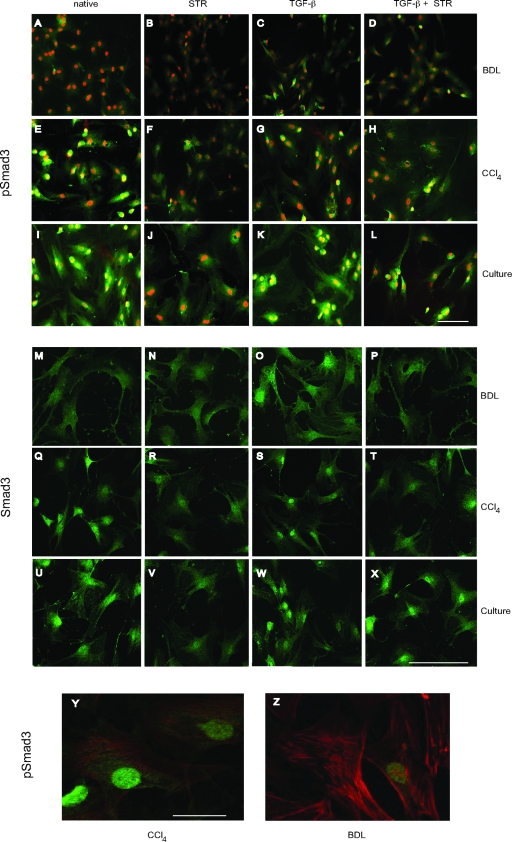
We next examined TGF-β mediated nuclear translocation of phospho-Smad3 and total Smad3 by immunocytochemical techniques (Figure 2). Phospho-Smad3 was absent in stellate cells isolated from BDL-injured liver (Figure 2, A–D); note that the signal shown in Figure 2A was identical to controls in which no primary antibody or a normal IgG was used. Cells isolated from CCl4-injured liver and culture-activated stellate cells expressed comparatively larger amounts of phospho-Smad3, distributed in the cytoplasm and prominent in the nucleus (Figure 2, E–L). In these latter models, inhibition of TGF-β binding to cell surface receptors using STR caused disappearance of phospho-Smad3 in both the cytoplasm and the nucleus of the cells (Figure 2, F–J). Stellate cells isolated from BDL-injured liver expressed small amounts of activated Smad3 on TGF-β stimulation (Figure 2C); STR had little observable effect (Figure 2D). After stimulation of stellate cells from CCl4-injured liver and culture-activated stellate cells with TGF-β, phospho-Smad3 was prominently translocated to the nucleus, an effect that was abrogated by STR (Figure 2, G, H, K, and L). Smad3 expression was identified in a pattern similar to phospho-Smad3 (Figure 2, M–X). To verify that the cells undergoing Smad3 signaling changes in BDL-injured livers were indeed in vivo-activated cells, we labeled cells after injury with anti-smooth muscle α-actin, a marker of the activated phenotype.33 We also labeled cells with anti-phospho-Smad3 antibody. We found that there was robust expression of smooth muscle α-actin, consistent with the activated phenotype (Figure 2, Y and Z). Notably, we found that phospho-Smad3 was not detected or was rare in stellate cells isolated from BDL-injured livers (Figure 2Z) as compared to the cells from CCl4-injured liver (Figure 2Y). These data provide further evidence that Smad3 is differentially activated in different models of liver injury, and that TGF-β causes Smad3 translocation.
Figure 2.
The effect of TGF-β on distribution of phospho-Smad3 and Smad3 in stellate cells. Stellate cells isolated from normal and injured livers (BDL and CCl4) were allowed to adhere for 12 hours in the presence of medium containing 20% serum. Cells were fixed and subjected to immunocytochemistry with anti-phospho-Smad3, Smad3, or in some cases, anti-smooth muscle α-actin antibodies. A, E, I, M, Q, and U: Representative examples of stellate cells from BDL- or CCl4-injured livers or after culture-induced activation. A–L: Stellate cells probed with anti-phospho-Smad3. M-X: Cells probed for Smad3. B, F, J, N, R, and V: Cells exposed to STR (50 μg/ml). C, G, K, O, S, and W: Cells exposed to TGF-β1 (5 ng/ml). D, H, L, P, T, and X: Stellate cells exposed to TGF-β plus STR (STR was added to cultured cells 30 minutes before addition of TGF-β for 12 hours). Y and Z: Cells isolated after CCl4 and BDL injury as in the Materials and Methods. Cells were allowed to adhere overnight, then fixed and dual-labeled with anti-phospho-Smad3 (FITC) and anti-smooth muscle α-actin (TRITC) antibodies as in the Materials and Methods. The images shown are representative of at least three others. Scale bars = 10 μm for A–L; = 70 μm for M–X, and 30 μm for Y and Z.
We further demonstrated that the cells isolated from different liver injury models were stellate cells. As seen in Supplemental Figure S1A at http://ajp.amjpathol.org; these cells clearly exhibit retinoid droplets and characteristic morphology of stellate cells. We have also documented the data for desmin expression (see Supplemental Figure S1B at http://ajp.amjpathol.org) to prove that these cells are stellate cells and not portal fibroblasts.33 In addition these cells have been shown to be highly fibrogenic because they express abundant amounts of collagen α1(1) (see Supplemental Figure S2 at http://ajp. amjpathol.org).
MAPKs Are Differentially Regulated in TGF-β Signaling
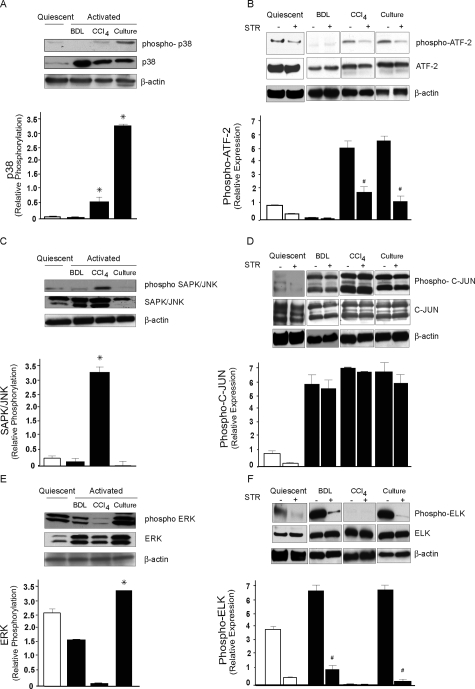
To analyze the involvement of MAPK in TGF-β-mediated liver wounding, expression and regulation of activated forms of p38, SAPK/JNK, and ERK1/2 subfamilies were examined by immunoblotting and detection of MAPK activity using specific assays. First phospho-p38 protein was up-regulated in stellate cells after culture-induced activation (Figure 3A) and p38 kinase activity was found to be significantly elevated in both culture-activated cells and in vivo in cells from CCl4-injured liver compared to quiescent stellate cells (Figure 3B). The increase in p38 kinase activity was inhibited by blocking TGF-β signaling. These data show that in stellate cells from culture-induced activation and CCl4-injured liver, p38 MAPK is differentially regulated. Although phospho-JNK/SAPK was stimulated to the greatest extent in cells from CCl4-injured livers, its kinase activity was high in cells after all forms of activation (Figure 3C) and TGF-β inhibition had minimal effect in abrogating JNK kinase activity in stellate cells from the various types of injury and activation (Figure 3, C and D), implying that JNK is also stimulated by a signaling network other than TGF. Notably, the immunoblotting using anti-SAPK/JNK antibody showed that JNK activation, although minimal in quiescent stellate cells (Figure 3C), was variable and restricted to JNK 1 isoform in activated stellate cells specifically in cells from CCl4-injured liver. Although the total JNK 2 (upper band isoform) was predominant, total JNK 1 was barely present in culture-activated cells. Phosphorylated ERK1/2 protein was notably down-regulated in cells from CCl4-injured liver compared to quiescent cells (Figure 3E). Interestingly, ERK1/2 kinase activity was found to be markedly reduced in CCl4-injured liver but elevated both in cells from culture-induced activation and BDL-injured liver and this effect was blocked by TGF-β inhibition (Figure 3F). These results demonstrate that TGF-β signaling proceeds through the ERK pathway in stellate cells after BDL and culture-induced activation. Apparently, TGF-β inhibition does not result in complete inhibition of p38 and ERK activities in cells from CCl4- and BDL-injured livers suggesting that these pathways may partially be regulated independently of TGF-β.
Figure 3.
MAPK expression and regulation by TGF-β in activated stellate cells in different liver injury models. Stellate cells were isolated from normal rat livers, BDL for 15 days and after four doses of CCl4 treatment as described in the Materials and Methods. A: Cell lysates (50 μg total protein) were subjected to immunoblotting using a specific phospho-p38 antibody. The membrane was stripped and reprobed with control p38 antibody. Data from different groups of cells were scanned, quantified, normalized, and presented graphically (n = 3, *P < 0.05 compared to quiescent stellate cells). B: Rats subjected to BDL- and CCl4-induced liver injury were exposed to STR as in the Materials and Methods or culture-activated stellate cells were incubated with STR as in Figure 1. Cellular proteins (200 μg of total protein) were used for immunoprecipitation with a specific threonine-tyrosine phospho-p38 MAP kinase antibody, followed by detection of phosphorylated p38 MAP kinase-specific substrate; ATF-2 by immunoblotting using phospho-ATF-2 antibody. As controls, cell lysates were subjected to immunoblotting using anti-ATF-2 antibody. Specific bands were scanned, quantified, normalized, and expressed graphically (n = 3, #P < 0.05 compared to controls without STR). C: Cell lysates (50 μg of total protein) were subjected to immunoblotting using a specific phospho-SAPK/JNK antibody. Membranes were stripped and reprobed with control SAPK/JNK antibody. For calculation of relative phosphorylation, bands corresponding to JNK 1 (46 kDa) were used and data from different group of cells were scanned, quantified, normalized, and presented graphically (n = 3, *P < 0.05 compared to quiescent stellate cells). D: Rats subjected to BDL- and CCl4-induced liver injury were exposed to STR as in the Materials and Methods or culture-activated stellate cells were incubated with STR as in Figure 1. An N-terminal C-JUN (1 to 89) fusion protein bound to glutathione Sepharose beads was used to selectively pull down SAPK/JNK from the cellular protein lysate (200 μg of total protein), followed by a kinase reaction. C-JUN phosphorylation was then measured by immunoblotting. As control, cellular proteins were immunoblotted with control C-JUN antibody. For relative quantification both bands for C-JUN phosphorylation (33 and 35 kDa) were used. E: Cell lysates (50 μg of total protein) were subjected to immunoblotting using a specific phospho-ERK antibody. Membranes were stripped and reprobed with control ERK antibody. For relative quantification both specific bands corresponding to phospho-ERK were used and data from different groups of cells were scanned, quantified, and presented graphically (n = 3, *P < 0.05 compared to quiescent stellate cells). F: Rats subjected to BDL- and CCl4-induced liver injury were exposed to STR as in the Materials and Methods or culture-activated stellate cells were incubated with STR as in Figure 1. Cellular proteins (200 μg of total protein) were used for immunoprecipitation with a specific monoclonal phospho-p44/42 antibody, followed by detection of phosphorylated p44/42 MAP kinase-specific substrate; ELK-1 by immunoblotting using phospho-ELK-1 antibody. As controls, cell lysates were subjected to immunoblotting using anti-ELK-1 antibody. Specific bands were scanned, quantified, normalized to the signal for β-actin, and presented graphically (n = 3, #P < 0.05 compared to controls without STR).
A Functional Role for TGF-β-Mediated MAPK Signal Transduction
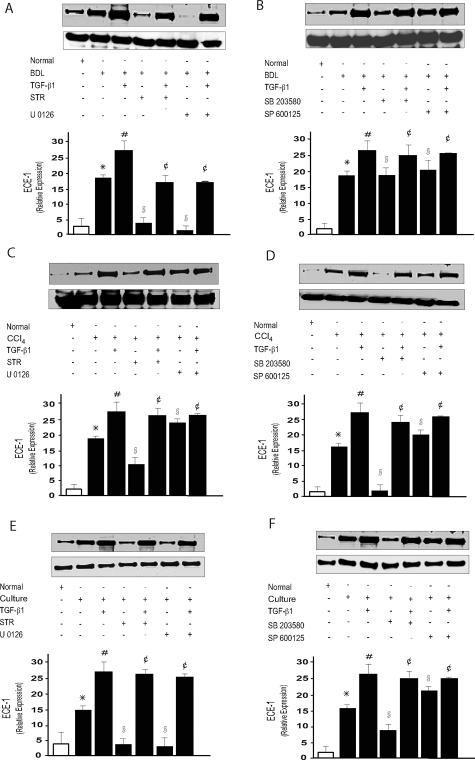
We next sought to explore functional roles for specific TGF-β signaling pathways. We chose the MAPK cascade as a prototypical pathway. Additionally, given that TGF-β plays an important role in controlling gene and protein expression,34 in particular, that of endothelin-converting enzyme-1 (ECE-1) expression (an enzyme that helps control ET-1 synthesis during wound healing), in stellate cells after liver injury,27 we asked whether a putative effect of TGF-β on ECE-1 expression could be mediated by the MAPK signaling pathway. ECE-1 protein levels in stellate cells from BDL-injured liver were increased twofold compared to those in normal stellate cells (Figure 4), consistent with previous data.27 Similarly, the levels of ECE-1 protein were also up-regulated in cells from CCl4-injured liver and culture-activated stellate cells. Exposure of stellate cells from injured livers and culture-activated cells to TGF-β further increased ECE-1 expression, whereas inhibition of MAPK signaling with U O126, a dual MEK1/MEK2 inhibitor, abrogated the effect of TGF-β on ECE-1 expression in stellate cells from BDL-injured livers and culture-activated cells but had no effect on cells from CCl4-injured liver (Figure 4, A, C, and E). In contrast, inhibition of MAPK signaling using SB 203580 inhibited ECE-1 expression in stellate cells from CCl4-injured liver and culture-activated cells but had no effect on cells from BDL-injured livers (Figure 4, B, D, and F). Inhibition of MAPK signaling using SP 600125, which is a specific JNK inhibitor, had minimal or no effect on ECE-1 expression in cells from either model of liver injury (Figure 4, B, D, and F). These data indicate that protein expression in stellate cells from BDL- and CCl4-injured livers is regulated by TGF-β and at least in part, follows two different MAPK pathways with ERK regulation being prominent in cells from BDL-injured livers and p38 MAPK being typical in stellate cells from CCl4-injured livers. Notably, the effects of U0126 in cells from BDL-injured livers to abrogate the expression of ECE-1 were more pronounced than those of STR, likewise, the effects of SB 203580 in cells from CCl4-injured liver and culture-activated cells than those of STR and thus supporting the finding that p38 and ERK in CCl4- and BDL-injured livers may be regulated independently of TGF-β.
Figure 4.
MAPK activation mediates the effect of TGF-β1 on ECE-1. After isolation of stellate cells from normal or BDL-injured (A, B) expression or CCl4-injured livers (C, D) (in the injury models, cells were cultured for 12 hours after isolation, then serum-free conditions were introduced), TGF-β (5 ng/ml), STR (50 μg/ml), PD98059 (10 μmol/L), SB 203580 (10 μmol/L), U 0126 (10 μmol/L), SP 600125 (50 μmol/L), or a combination of these compounds was added for 24 hours. In culture-activated cells (E, F), serum-free conditions were introduced after 5 days in activating condition, and cells were exposed to the compounds as above. Cell lysates (50 μg of total proteins) were prepared as described in the Materials and Methods and subjected to immunoblotting using anti-ECE-1 antibody. Data from different experimental conditions were scanned, specific ECE-1 bands were normalized to the signal for β-actin and presented graphically, the level of ECE-1 protein from BDL, CCl4 cells and culture-activated cells was set at 100 (For A to F, n = 3, *P < 0.05 compared to quiescent stellate cells; #P < 0.05 compared to BDL, CCl4, and culture-activated stellate cells; §P< 0.05 compared to BDL, CCl4, and culture-activated stellate cells; ¢P < 0.05 compared to BDL stellate cells, CCl4, and culture-activated stellate cells plus TGF-β).
To verify further the functionality of the signaling system highlighted above, we extended the model system to evaluate another readout known to be important in stellate cell activation. Thus, we examined, collagen α1(1) a prototypal TGF-β regulated gene in stellate cell activation.3 Stellate cells were isolated from BDL-injured livers, and were exposed to STR and MAP kinase inhibitors; collagen α1(1) mRNA was subsequently measured and found to be down-regulated by TGF-β inhibition and essentially completely by the MEK1/MEK2 inhibitor (U 0126), whereas p38 and SAPK/JNK inhibition had no effect (data not shown).
Differential Smad Expression after Liver Injury: Role of Egr-1
The discovery of divergent Smad3 expression in the BDL- and CCl4-injury models led us to ask whether Egr-1, a regulatory transcription factor, may regulate Smad transcriptional activity by binding to the Smad promoter. To test this hypothesis, we first investigated Egr-1 protein expression in stellate cells after liver injury; we found Egr-1 protein expression in stellate cells from normal and BDL-injured liver to be much higher than that from stellate cells after CCl4-injured liver or culture-induced activation (Figure 5A). Next, we identified two Egr-1 consensus-binding sites in the proximal Smad3 promoter, which might serve as regulatory elements (Figure 5B). Gel mobility shift assays were performed using Egr-1 oligonucleotides corresponding to Egr-1 consensus binding sequences and nuclear extracts were derived from stellate cells. We detected an Egr-1-DNA complex in stellate cells from normal and BDL-injured liver but not in cells from CCl4-injured liver (Figure 5C). This complex was inhibited by addition of specific Egr-1 antibody but not an unrelated (smooth muscle α-actin) antibody. An apparent smaller complex was detected in stellate cells after culture-induced activation, however, it was not altered by Egr-1 antibody, suggesting that it does not contain Egr-1. Finally, addition of anti-Egr-1 antibody to lysates abrogated the DNA-Egr-1 complex, but did not result in a typical supershift profile, consistent with previous Egr-1 supershift experiments.35
Figure 5.
Egr-1 regulated Smad3 expression after liver injury. Stellate cells were isolated from normal rat livers, 2 weeks after BDL and after four doses of CCl4 treatment as described in the Materials and Methods. A: Cellular Egr-1 was detected by immunoblotting using Egr-1-specific antibody. Specific bands were scanned, quantified, normalized and presented graphically (n = 3, *P < 0.05 compared to CC14 or culture). B: The Smad3 3′-5′ region is shown. Two underlined GC-rich Egr-1 binding sites are located within 100 bp of upstream cDNA. The start site is bolded. C: Cell nuclei from normal and injured liver stellate cells were isolated and prepared as described in the Materials and Methods were incubated with a γ-32P-labeled Egr-1 consensus oligonucleotides. Oligonucleotide-protein complex were analyzed by gel mobility shift assays. The arrow indicates the site of binding complex. A representative experiment is shown (n = 3).
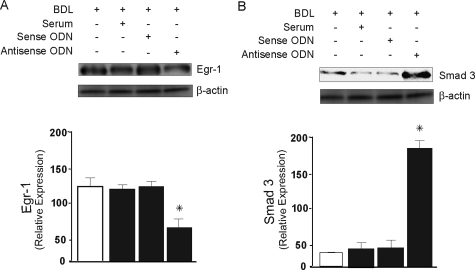
Finally, we examined the role of Egr-1 in Smad3 expression in stellate cells after BDL injury by inhibiting its expression with phosphorothioate-modified Egr-1 antisense ODNs. We selected Egr-1 ODNs based on reports of those previously described.36,37 We transduced cells from BDL-injured livers with 5′-fluorescein-labeled antisense ODNs; Egr-1 protein expression was decreased by 52% after inhibition of Egr-1 (Figure 6A) for 24 hours. When Egr-1 expression was inhibited by incubation of Egr-1 antisense ODNs for 24 hours, Smad3 was increased by 95% compared with control cells (Figure 6B). To verify the effect of Egr-1 inhibition on Smad3 expression in culture-activated cells, we examined the effect of Egr-1 inhibition using Egr-1 antisense oligonucleotides in culture-activated cells. This experiment revealed that there was no effect of Egr-1 inhibition on Smad3 expression (see Supplemental Figure S3 at http://ajp.amjpathol.org). These data indicate that Egr-1 inhibits Smad3 expression by regulation of Smad3 promoter activity in stellate cells activated by BDL-induced liver injury.
Figure 6.
Inhibition of Egr-1 results in increased expression of Smad3. Stellate cells from BDL-injured livers were isolated and allowed to adhere for 12 hours in the presence of medium containing 20% serum. Subsequently stellate cells were transduced with Egr-1 sense and antisense ODN as in the Materials and Methods. A: Cell lysates (30 μg of total protein) were subjected to immunoblotting using specific Egr-1 antibody as in the Materials and Methods. B: Cellular lysates were immunoblotted with anti-Smad3 antibody (membranes from A and B were striped and reprobed with anti-β-actin antibody; bottom). Specific signals from different experiments were scanned, normalized to the signal for β-actin, quantified, and expressed graphically (n = 3, *P < 0.05 compared to control cells).
Discussion
The major finding of this study was that different models of liver injury and stellate cell activation were associated with distinct TGF-β-mediated signaling. We demonstrated that TGF-β stimulated Smad3 signaling in stellate cells from CCl4-injured liver, but appeared to be inactive in a mechanistically distinct model of liver injury (ie, BDL). In contrast, TGF-β mediated MAPK signaling either partially or extensively in all of the models of injury. Further, the differential MAPK signaling in BDL-induced activation had a functional effect on ECE-1 expression in stellate cells. In the context of other data highlighting potential signaling specificity,18 the current data have broad implications for defining the roles of specific TGF-β-signaling pathways in wound healing.
We also found there to be functional consequences of TGF-β-MAPK signaling (Figure 4). Here, a MEK inhibition abrogated the effect of TGF-β on ECE-1 expression, whereas inhibition of p38 had no effect in cells from BDL-injured livers. In contrast, p38 inhibition in cells from CCl4-injured liver down-regulated expression of ECE-1, which was not affected by MEK inhibition in these cells. Previous work demonstrated that TGF-β stimulates endothelin-1 production in liver wound healing, an effect related to mRNA stabilization and increased ECE-1 protein expression in stellate cells.38 Increased ECE-1 protein expression is a result of de novo synthesis of mRNA binding proteins that are in turn regulated by TGF-β.38 A remarkable finding noted in our previous studies was a differential effect of TGF-β on sinusoidal endothelial cells and stellate cells38; TGF-β stimulated ET-1 production in normal liver endothelial cells but had no effect on endothelial cells from injured liver and the vice versa was true for stellate cells. Results of the current study raise a potential mechanism in which divergent cellular signaling pathways may explain differential functional effects.
In our study, activation of p38 and ERK 1/2 after CCl4- and BDL-induced injury, respectively, was evident in kinase assays established for p38 and ERK 1/2 but not by assessment of levels of phosphorylated proteins. Similarly JNK//2 activation in cells from all of the models of liver injury was only detectable in kinase assays (Figure 3, A and B, C and D, and E and F). This finding was somewhat unexpected, but is consistent with previous work on MAPK signaling molecules.39,40 For example, it has been shown that kinase activity and phosphorylation levels correlate poorly.39,40 It is also noteworthy that the absolute phosphorylation level detected by immunoblot is a rough estimate of actual enzymatic activity, and this could be attributable to the fact that protein phosphorylation is thought to be a transient initial step in the activation of kinase activity, often required for recruiting binding partners or facilitating a conformational change necessary for kinase activation.41,42 Notwithstanding, kinase activity is likely to be the most robust measure of signaling.
We demonstrated that phospho-Smad3 is differentially regulated in vivo in stellate cells in different injury models, consistent with data in a cholestatic liver injury model that showed Smad3 signaling in hepatocytes only.43,44 One question raised by our study is whether differential signaling may lead to differential functional effects in these two forms of liver injury and if so, what are the functional effects? Also, is it possible that there could be different cellular mechanisms in different forms of liver injury—a reflection of the apparent different Smad3 signaling pathways? The answers to these questions are currently open. An interesting point, although beyond the scope of the current study, is whether portal fibroblasts, which may be important effectors in biliary fibrogenesis,45,46 could also exhibit differential Smad3 activation.43
We have shown there to be apparent divergent signaling repertoires in stellate cells after BDL compared to other forms of activation. On one hand, this is surprising because many investigators believe that activation is a common response to injury, with a common phenotype. On the other hand, abundant evidence suggests that there is heterogeneity in stellate cell phenotypes.24 Thus, it may not be surprising that there could be substantial differences in signaling after different forms of injury and perhaps mechanisms underlying activation. Notwithstanding, we have considered that another possible explanation for the differences in signaling repertoire is that stellate cells isolated after BDL are fundamentally different from other populations of stellate cells, including perhaps that they are derived from a different cell pool than in other injury models.47 This is relevant given that other studies have found that portal fibroblasts are critical effectors in biliary fibrosis.46 However, we do not believe that this is the case in our system for the following reasons. We have gone to great length to characterize the cell types we report on. In each model, the cells we have isolated retain the characteristics of classic stellate cells.48 First, we have shown that these cells have classic stellate cell morphology, including the presence of retinoid droplets and expression of desmin (see Supplemental Figure S1B at http://ajp.amjpathol.org) that was remarkable also in cells isolated from the BDL-induced liver injury. These features are not characteristic of portal fibroblasts although in the early stages of biliary fibrosis, portal fibroblasts have been reported to express desmin49 that eventually disappears at advanced stages. Second, the activated stellate cells examined in this study uniformly express smooth muscle α-actin (Figure 1A) as previously reported.33 Although this feature may not be specific for stellate cells, electron microscopic studies conducted on experimental models of biliary fibrosis at different stages50 support our findings of smooth muscle α-actin being attributable to stellate cells in this particular model. It has been postulated that in the early stages of biliary fibrosis, hepatic stellate cells migrate to the areas adjacent to ductal proliferation where they become activated myofibroblasts.
Complete inhibition of ECE-1 expression by MEK1/MEK2 inhibitor in cells from BDL-induced liver injury suggests the presence of TGF-β-independent signaling pathways in different liver injury models. Consistent with previous data showing that collagen α(1) expression in hepatic stellate cells in culture may be TGF-β-independent,29 we have been able to show that collagen α(1) expression in hepatic stellate cells in BDL-induced liver injury may be regulated by ERK in a TGF-β-independent manner. Previous studies have demonstrated TGF-β-independent constitutive activation of Smad3 in activated stellate cells but not in quiescent cells.51 In contrast, others52 have shown TGF-β-responsive phosphorylation and nuclear localization of Smad3 in early stellate cell cultures but not in passaged stellate cells, while other studies have highlighted constitutive activation of Smad3 in response to TGF-β in a spontaneously immortalized line of activated rat hepatic stellate cells.53 Thus there appears to be a range of potential Smad3 responses, we speculate, dependent on the wide variety of conditions in which stellate cells and TGF-β signaling might be studied.
An important aspect of this study is that we have shown that Egr-1 appears to inhibit expression of Smad3. These data are highly consistent with the known function of Egr-1, because available data indicate that Egr-1 is an important regulator of gene transcription,54 ostensibly via Egr-1 binding to consensus GC-rich areas or Sp1 sites such as those identified in Figure 5B.55,56 Interestingly, previous studies have shown that TGF-β stimulates Egr-1 expression, and that this requires, as would be predicted, Smad3 and MAPKs.22,57 These two data sets therefore suggest that there is complex interplay between Egr-1 and TGF-β signaling partners (ie, Smad3 and MAPK). The stimulation of Egr-1 after BDL-induced injury but not after CCl4 administration was striking. The mechanism by which Egr-1 expression is differentially regulated in these two different wounding models remains unknown. Egr-1 is transiently induced by a variety of extracellular stimuli such as growth factors and cytokines.20Although there are clear similarities among the two different models of wound healing used in this study, an evolving theme is that there are important cell and molecular differences among different types of injury, which presumably may account for the stimulation of Egr-1 after BDL-induced but not after CCl4-induced injury. Our data supports this possibility.
The data presented here have important implications for not only the pathogenesis of wound healing, but also for the therapy of fibrogenesis. Although it is likely that inhibition of TGF-β signaling during ongoing wound healing would be of therapeutic benefit, it should be emphasized that interruption of TGF-β signaling is likely to have complex (and perhaps unpredictable) effects in the healing wound milieu.
Footnotes
Address reprint requests to Don C. Rockey, M.D., Division of Digestive and Liver Diseases, UT Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-8887. E-mail: don.rockey@utsouthwestern.edu.
Supported by the National Institutes of Health (grant R01 DK 50574) and the Burroughs Welcome Fund (translational scientist award to D.C.R.).
Supplemental material for this article can be found on http://ajp. amjpathol.org.
References
- Ashcroft GS, Roberts AB. Loss of Smad3 modulates wound healing. Cytokine Growth Factor Rev. 2000;11:125–131. doi: 10.1016/s1359-6101(99)00036-2. [DOI] [PubMed] [Google Scholar]
- Vaquero E, Molero X, Tian X, Salas A, Malagelada JR. Myofibroblast proliferation, fibrosis, and defective pancreatic repair induced by cyclosporin in rats. Gut. 1999;45:269–277. doi: 10.1136/gut.45.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Kushida M, Higashi K, Itoh J, Higashiyama R, Hong YY, Kawada N, Namikawa K, Kiyama H, Bou-Gharios G, Watanabe T, Okazaki I, Ikeda K. Cell type-specific intervention of transforming growth factor beta/Smad signaling suppresses collagen gene expression and hepatic fibrosis in mice. Gastroenterology. 2005;129:259–268. doi: 10.1053/j.gastro.2005.03.088. [DOI] [PubMed] [Google Scholar]
- Friedman SL, Rockey DC, Bissell DM. Hepatic fibrosis 2006: report of the Third AASLD Single Topic Conference. Hepatology. 2007;45:242–249. doi: 10.1002/hep.21459. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massague J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Wiercinska E, Wickert L, Denecke B, Said HM, Hamzavi J, Gressner AM, Thorikay M, ten Dijke P, Mertens PR, Breitkopf K, Dooley S. Id1 is a critical mediator in TGF-beta-induced transdifferentiation of rat hepatic stellate cells. Hepatology. 2006;43:1032–1041. doi: 10.1002/hep.21135. [DOI] [PubMed] [Google Scholar]
- Goumans MJ, Valdimarsdottir G, Itoh S, Lebrin F, Larsson J, Mummery C, Karlsson S, ten Dijke P. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFbeta/ALK5 signaling. Mol Cell. 2003;12:817–828. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Mol Biol Cell. 2005;16:4214–4224. doi: 10.1091/mbc.E05-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl B, Kweon YO, Frederick JP, Wang XF, Rippe RA, Brenner DA. The role of Smad3 in mediating mouse hepatic stellate cell activation. Hepatology. 2001;34:89–100. doi: 10.1053/jhep.2001.25349. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhou G, Hu MC, Yao Z, Tan TH. Activation of the hematopoietic progenitor kinase-1 (HPK1)-dependent, stress-activated c-Jun N-terminal kinase (JNK) pathway by transforming growth factor beta (TGF-beta)-activated kinase (TAK1), a kinase mediator of TGF beta signal transduction. J Biol Chem. 1997;272:22771–22775. doi: 10.1074/jbc.272.36.22771. [DOI] [PubMed] [Google Scholar]
- Yue J, Frey RS, Mulder KM. Cross-talk between the Smad1 and Ras/MEK signaling pathways for TGFbeta. Oncogene. 1999;18:2033–2037. doi: 10.1038/sj.onc.1202521. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Ninomiya-Tsuji J, Masuyama N, Nishita M, Fujisawa J, Shibuya H, Matsumoto K, Nishida E. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-beta-induced gene expression. J Biol Chem. 1999;274:27161–27167. doi: 10.1074/jbc.274.38.27161. [DOI] [PubMed] [Google Scholar]
- Schnabl B, Bradham CA, Bennett BL, Manning AM, Stefanovic B, Brenner DA. TAK1/JNK and p38 have opposite effects on rat hepatic stellate cells. Hepatology. 2001;34:953–963. doi: 10.1053/jhep.2001.28790. [DOI] [PubMed] [Google Scholar]
- Masamune A, Satoh M, Kikuta K, Sakai Y, Satoh A, Shimosegawa T. Inhibition of p38 mitogen-activated protein kinase blocks activation of rat pancreatic stellate cells. J Pharmacol Exp Ther. 2003;304:8–14. doi: 10.1124/jpet.102.040287. [DOI] [PubMed] [Google Scholar]
- Tsukada S, Westwick JK, Ikejima K, Sato N, Rippe RA. SMAD and p38 MAPK signaling pathways independently regulate alpha1(I) collagen gene expression in unstimulated and transforming growth factor-beta-stimulated hepatic stellate cells. J Biol Chem. 2005;280:10055–10064. doi: 10.1074/jbc.M409381200. [DOI] [PubMed] [Google Scholar]
- Brady LM, Beno DW, Davis BH. Bile acid stimulation of early growth response gene and mitogen-activated protein kinase is protein kinase C-dependent. Biochem J. 1996;316:765–769. doi: 10.1042/bj3160765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy B, Nathans D. Functional serum response elements upstream of the growth factor-inducible gene zif268. Mol Cell Biol. 1989;9:4889–4895. doi: 10.1128/mcb.9.11.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey TA, Fu C, Du B, Eksinar S, Kent KC, Bush H, Jr, Kreiger K, Rosengart T, Cybulsky MI, Silverman ES, Collins T. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000;105:653–662. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Ning H, Ishida W, Sodin-Semrl S, Takagawa S, Mori Y, Varga J. The early-immediate gene EGR-1 is induced by transforming growth factor-beta and mediates stimulation of collagen gene expression. J Biol Chem. 2006;281:21183–21197. doi: 10.1074/jbc.M603270200. [DOI] [PubMed] [Google Scholar]
- Liu C, Yao J, de Belle I, Huang RP, Adamson E, Mercola D. The transcription factor EGR-1 suppresses transformation of human fibrosarcoma HT1080 cells by coordinated induction of transforming growth factor-beta1, fibronectin, and plasminogen activator inhibitor-1. J Biol Chem. 1999;274:4400–4411. doi: 10.1074/jbc.274.7.4400. [DOI] [PubMed] [Google Scholar]
- Rockey DC, Chung JJ. Endothelin antagonism in experimental hepatic fibrosis. Implications for endothelin in the pathogenesis of wound healing. J Clin Invest. 1996;98:1381–1388. doi: 10.1172/JCI118925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Murphy LJ. Molecular pharmacology of endothelin converting enzymes. Biochem Pharmacol. 1996;51:91–102. doi: 10.1016/0006-2952(95)02036-5. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M. The endothelin system. A new target for therapeutic intervention. Circulation. 1994;89:1320–1322. doi: 10.1161/01.cir.89.3.1320. [DOI] [PubMed] [Google Scholar]
- Shao R, Yan W, Rockey DC. Regulation of endothelin-1 synthesis by endothelin-converting enzyme-1 during wound healing. J Biol Chem. 1999;274:3228–3234. doi: 10.1074/jbc.274.5.3228. [DOI] [PubMed] [Google Scholar]
- Hellerbrand C, Stefanovic B, Giordano F, Burchardt ER, Brenner DA. The role of TGFbeta1 in initiating hepatic stellate cell activation in vivo. J Hepatol. 1999;30:77–87. doi: 10.1016/s0168-8278(99)80010-5. [DOI] [PubMed] [Google Scholar]
- George J, Roulot D, Koteliansky VE, Bissell DM. In vivo inhibition of rat stellate cell activation by soluble transforming growth factor beta type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci USA. 1999;96:12719–12724. doi: 10.1073/pnas.96.22.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Shao R, Qian HS, George SE, Rockey DC. Gene transfer of the neuronal NO synthase isoform to cirrhotic rat liver ameliorates portal hypertension. J Clin Invest. 2000;105:741–748. doi: 10.1172/JCI7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kountouras J, Billing BH, Scheuer PJ. Prolonged bile duct obstruction: a new experimental model for cirrhosis in the rat. Br J Exp Pathol. 1984;65:305–311. [PMC free article] [PubMed] [Google Scholar]
- Proctor E, Chatamra K. Standardized micronodular cirrhosis in the rat. Eur Surg Res. 1984;16:182–186. doi: 10.1159/000128407. [DOI] [PubMed] [Google Scholar]
- Rockey DC, Boyles JK, Gabbiani G, Friedman SL. Rat hepatic lipocytes express smooth muscle actin upon activation in vivo and in culture. J Submicrosc Cytol Pathol. 1992;24:193–203. [PubMed] [Google Scholar]
- Kahari VM, Olsen DR, Rhudy RW, Carrillo P, Chen YQ, Uitto J. Transforming growth factor-beta up-regulates elastin gene expression in human skin fibroblasts. Evidence for post-transcriptional modulation. Lab Invest. 1992;66:580–588. [PubMed] [Google Scholar]
- Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- Hofer G, Grimmer C, Sukhatme VP, Sterzel RB, Rupprecht HD. Transcription factor Egr-1 regulates glomerular mesangial cell proliferation. J Biol Chem. 1996;271:28306–28310. doi: 10.1074/jbc.271.45.28306. [DOI] [PubMed] [Google Scholar]
- Fisher TL, Terhorst T, Cao X, Wagner RW. Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res. 1993;21:3857–3865. doi: 10.1093/nar/21.16.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R, Shi Z, Gotwals PJ, Koteliansky VE, George J, Rockey DC. Cell and molecular regulation of endothelin-1 production during hepatic wound healing. Mol Biol Cell. 2003;14:2327–2341. doi: 10.1091/mbc.02-06-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svegliati-Baroni G, Ridolfi F, Caradonna Z, Alvaro D, Marzioni M, Saccomanno S, Candelaresi C, Trozzi L, Macarri G, Benedetti A, Folli F. Regulation of ERK/JNK/p70S6K in two rat models of liver injury and fibrosis. J Hepatol. 2003;39:528–537. doi: 10.1016/s0168-8278(03)00291-5. [DOI] [PubMed] [Google Scholar]
- Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- Antonelli V, Bernasconi F, Wong YH, Vallar L. Activation of B-Raf and regulation of the mitogen-activated protein kinase pathway by the G (o) alpha chain. Mol Biol Cell. 2000;11:1129–1142. doi: 10.1091/mbc.11.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowal L, Provitera P, Scarlata S. Stable association between G{alpha}q and phospholipase Cbeta1 in living cells. J Biol Chem. 2006;281:23999–24014. doi: 10.1074/jbc.M512330200. [DOI] [PubMed] [Google Scholar]
- Seyhan H, Hamzavi J, Wiercinska E, Gressner AM, Mertens PR, Kopp J, Horch RE, Breitkopf K, Dooley S. Liver fibrogenesis due to cholestasis is associated with increased Smad7 expression and Smad3 signaling. J Cell Mol Med. 2006;10:922–932. doi: 10.1111/j.1582-4934.2006.tb00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa Y, Seki E, Adachi M, Taura K, Kodama Y, Siegmund SV, Schwabe RF, Brenner DA. Systemic mediators induce fibrogenic effects in normal liver after partial bile duct ligation. Liver Int. 2006;26:1138–1147. doi: 10.1111/j.1478-3231.2006.01346.x. [DOI] [PubMed] [Google Scholar]
- Ramm GA, Carr SC, Bridle KR, Li L, Britton RS, Crawford DH, Vogler CA, Bacon BR, Tracy TF. Morphology of liver repair following cholestatic liver injury: resolution of ductal hyperplasia, matrix deposition and regression of myofibroblasts. Liver. 2000;20:387–396. doi: 10.1034/j.1600-0676.2000.020005387.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Dranoff JA, Chan EP, Uemura M, Sevigny J, Wells RG. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- Guyot C, Combe C, Clouzeau-Girard H, Moronvalle-Halley V, Desmouliere A. Specific activation of the different fibrogenic cells in rat cultured liver slices mimicking in vivo situations. Virchows Arch. 2007;450:503–512. doi: 10.1007/s00428-007-0390-y. [DOI] [PubMed] [Google Scholar]
- Rockey DC, Chung JJ. Interferon gamma inhibits lipocyte activation and extracellular matrix mRNA expression during experimental liver injury: implications for treatment of hepatic fibrosis. J Invest Med. 1994;42:660–670. [PubMed] [Google Scholar]
- Beaussier M, Wendum D, Schiffer E, Dumont S, Rey C, Lienhart A, Housset C. Prominent contribution of portal mesenchymal cells to liver fibrosis in ischemic and obstructive cholestatic injuries. Lab Invest. 2007;87:292–303. doi: 10.1038/labinvest.3700513. [DOI] [PubMed] [Google Scholar]
- Tao LH, Enzan H, Hayashi Y, Miyazaki E, Saibara T, Hiroi M, Toi M, Kuroda N, Naruse K, Jin YL, Guo LM. Appearance of denuded hepatic stellate cells and their subsequent myofibroblast-like transformation during the early stage of biliary fibrosis in the rat. Med Electron Microsc. 2000;33:217–230. doi: 10.1007/s007950000022. [DOI] [PubMed] [Google Scholar]
- Liu C, Gaca MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem. 2003;278:11721–11728. doi: 10.1074/jbc.M207728200. [DOI] [PubMed] [Google Scholar]
- Dooley S, Delvoux B, Streckert M, Bonzel L, Stopa M, ten Dijke P, Gressner AM. Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 2001;502:4–10. doi: 10.1016/s0014-5793(01)02656-4. [DOI] [PubMed] [Google Scholar]
- Svegliati-Baroni G, Inagaki Y, Rincon-Sanchez AR, Else C, Sacco-manno S, Benedetti A, Ramirez F, Rojkind M. Early response of alpha2(I) collagen to acetaldehyde in human hepatic stellate cells is TGF-beta independent. Hepatology. 2005;42:343–352. doi: 10.1002/hep.20798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiada E, Razandi M, Levin ER. Egr-1 activates basic fibroblast growth factor transcription. Mechanistic implications for astrocyte proliferation. J Biol Chem. 1996;271:18576–18581. doi: 10.1074/jbc.271.31.18576. [DOI] [PubMed] [Google Scholar]
- Madden SL, Cook DM, Morris JF, Gashler A, Sukhatme VP, Rauscher FJ., III Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991;253:1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a re-pressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Zhang J, Lin Y, Zhu X, Zhao L, Ahmad M, Ehrengruber MU, Chen YE. Early stimulation and late inhibition of peroxisome proliferator-activated receptor gamma (PPAR gamma) gene expression by transforming growth factor beta in human aortic smooth muscle cells: role of early growth-response factor-1 (Egr-1), activator protein 1 (AP1) and Smads. Biochem J. 2003;370:1019–1025. doi: 10.1042/BJ20021503. [DOI] [PMC free article] [PubMed] [Google Scholar]