Abstract
The significance of Hedgehog (HH) signaling in the development of basal cell carcinoma (BCC) has been established. Although several target genes of HH signaling have been described previously, their precise role in tumorigenesis and cell proliferation is not yet known. To identify genes responsible for tumor formation in BCC, we screened a DNA microarray database of human BCC cases; the orphan G-protein-coupled receptor GPR49 was found to be up-regulated in all cases. GPR49 is a novel gene reported to be a marker of follicular and other tissue stem cells. Using real-time quantitative RT-PCR analysis, significant expression of GPR49 mRNA was observed in 19 of 20 BCC cases (95%) compared with controls. Up-regulation of GPR49 was confirmed by in situ hybridization. Moreover, knockdown of mouse Gpr49 showed suppression of cell proliferation in a mouse BCC cell line, and overexpression of GPR49 in human immortalized keratinocyte HaCaT cells induced proliferation. Furthermore, HaCaT cells overexpressing GPR49 showed tumor formation when transplanted into immunodeficient mice. In addition, inhibition of the HH signaling pathway in a mouse BCC cell line down-regulated endogenous Gpr49, whereas activation of HH signaling in mouse NIH3T3 cells up-regulated endogenous GPR49. These results suggest that GPR49 is expressed downstream of HH signaling and promotes cell proliferation and tumor formation in cases of BCC.
Basal cell carcinoma (BCC) is a common malignant tumor of the skin. Histopathologically, they usually arise from the lowermost layers of the epidermis and comprise various histological subtypes. Interestingly, BCC cells have many features in common with follicular epithelium.1
Recent studies have shown that BCC frequently has abnormalities of the Hedgehog (HH) signaling pathway. HH signaling plays a key role in vertebrate development as it is involved in multiple biological processes such as cell differentiation, proliferation, and growth.2 Also, several genes overexpressed in BCC have been reported to be related to HH signaling.3 However, the genes expressed as a target of HH signaling and their functions in tumorigenesis are still not precisely understood. Further understanding of the molecules expressed in BCC and their relation between HH signaling is therefore required.
G-Protein-coupled receptor GPR49, also known as LGR5/HG38/FEX, belongs to the leucine-rich repeat containing G-protein-coupled receptors (LGRs) structurally similar to glycoprotein hormone receptors including thyroid-stimulating hormone receptor, follicle-stimulating hormone receptor, and luteinizing hormone receptor.4 LGRs are grossly divided into three subgroups: glycoprotein hormone receptors; the subgroup of GPR48, GPR49, and LGR6; and the receptors of relaxin family ligands represented by LGR7 and LGR8.4,5 Unlike LGRs in the other two subgroups, the ligands of GPR48, GPR49, and LGR6 have not been identified. A recent report showed that GPR49 null mice exhibited neonatal lethality characterized by ankyloglossia and gastrointestinal distension.6 Interestingly, a recent report showed that GPR49 is expressed on stem cells in the small intestine and colon, and has also been suggested that stem cells in the hair follicle are GPR49-positive.7 Thus, GPR49 may play a significant role in the biology of stem cells. Although the function of GPR49 in cancer is poorly understood, overexpression of GPR49 has been reported in some studies.8,9 In particular, it is overexpressed in some hepatocellular carcinoma with β-catenin mutation and is thought to be a target of Wnt-β catenin signaling.8
Here we report that GPR49 is markedly up-regulated in almost all cases of BCC under the control of HH signaling, and that it plays an important role in cell proliferation and tumor formation.
Materials and Methods
Samples
Tumor samples were collected from patients at Keio University Hospital and other affiliated hospitals. Tumor and normal skin were snap-frozen after surgical removal and stored at −80°C until use. We also used paraffin-embedded sections of skin tumors. The experiment was approved by the ethics committee of Keio University School of Medicine and all samples were taken after written informed consent was obtained from the patients.
GeneChip Database and Gene Expression Analysis
The GeneChip was analyzed with the GeneExpress software system (Gene Logic Inc., Gaithersburg, MD). This database contains gene expression signatures from about 10,000 clinical samples from a variety of normal and disease conditions on the Affymetrix HG-U133 GeneChip.
DNA microarray analysis was independently performed in our laboratory with the Gene Spring Gx software system (Tomy Digital Biotechnology, Tokyo, Japan) containing gene expression signatures from four BCC and four normal skin samples on the Agilent whole human Genome oligo DNA MicroArray 4×44k (Agilene Technologies, Santa Clara, CA). For microarray hybridization, we followed the manufacturer’s protocol. Difference in gene expression was quantified as the fold change in gene expression between sets of BCC-derived and normal skin tissues. Genes were considered as being differentially expressed at a significance of P < 0.01. We then listed the genes expressed more than six fold in BCC and hypothetical genes were eliminated.
Quantitative Real Time Polymerase Chain Reaction
Total RNA were isolated from tissues and cell lines with RNeasy Mini Kit including DNAase treatment (Quiagen KK, Tokyo, Japan). cDNA was synthesized with a First-Strand cDNA Synthesis Kit (GE Health care, Piscataway, NJ). Quantitative real time PCR (QRT-PCR) analysis was performed on an ABI7700 using SYBR Green PCR Core Reagents (Applied Biosystems, Warrington, UK). Primer sequences for QRT-PCR studies are shown in Table 1. Human and mouse GAPDH was used as a reference. Fold-induction values were calculated using the 2−ΔΔCt method. All experiments were performed in triplicate and were repeated at least five times in separate experiments; representative data are shown.
Table 1.
Primer Sequences for Quantitative RT-PCR
| Gene | Primer orientation | Sequence |
|---|---|---|
| Human GAPDH | Forward | 5′-CCAGCCGAGCCACATCGCTC-3′ |
| Reverse | 5′-ATGAGCCCCAGCCTTCTCCAT-3′ | |
| Human GPR49 | Forward | 5′-GAGGATCTGGTGAGCCTGAGAA-3′ |
| Reverse | 5′-CATAAGTGATGCTGGAGCTGGTAA-3′ | |
| Human GLI1 | Forward | 5′-GAAGACCTCTCCAGCTTGGA-3′ |
| Reverse | 5′-GGCTGACAGTATAGGCAGAG-3′ | |
| Human GLI2 | Forward | 5′-TGGCCGCTTCAGATGACAGATGTTG-3′ |
| Reverse | 5′-CGTTAGCCGAATGTCAGCCGTGAAG-3′ | |
| Mouse Gapdh | Forward | 5′-TGCACCACCAACTGCTTAG-3′ |
| Reverse | 5′-GGATGCAGGGATGATGTTT-3′ | |
| Mouse Gpr49 | Forward | 5′-GAGTCAACCCAAGCCTTAGTATCC-3′ |
| Reverse | 5′-CATGGGACAAATGCAACTGAAG-3′ | |
| Mouse Gli1 | Forward | 5′-CATTCCACAGGACAGCTCAA-3′ |
| Reverse | 5′-TGGCAGGGCTCTGACTAACT-3′ | |
| Mouse Ptch1 | Forward | 5′-CAAACTTTGACCCCTTGGAA-3′ |
| Reverse | 5′-AAAACAAGGGGCACATCAAG-3′ |
In Situ Hybridization
Digoxigenin-labeled GPR49 sense and antisense probes were generated from the 120-bp fragment of GPR49 (corresponding to nucleotides 2548 to 2667, GenBank NM_003667). The protocol for in situ hybridization is described elsewhere.10 Briefly, de-paraffinized sections were treated with thermolysin 4 mg/ml and proteinase K 10 μg/ml, and hybridized with sense and anti-sense probes. After hybridization, the slides were washed and treated with RnaseA for 30 minutes and washed with NTE buffer (0.5M NaCl, 1 mmol/L EDTA, 10 mmol/L Tris-HCl pH 8.0) at 37°C. The sections were then washed and rinsed with NT buffer (150 mmol/L NaCl, 100 mmol/L Tris-HCl pH 7.5). Nonspecific staining was blocked with 5% normal goat serum and incubated with anti-digoxigenin-alkaline phosphatase (Roche Diagnostics, Basel, Switzerland) diluted 1:250 in NT buffer, followed by washing. Sections were visualized by BCIP/NBT liquid substrate (0.15 mg/ml 5-bromo-4-chloro-3-indolyl phosphate solution salt, 0.3 mg/ml Nitrotetrazolium Blue Chrolide, 1 mmol/L MgCl2, 100 mmol/L Tris pH 9.5).
Cell Culture and Reagents
Human immortalized keratinocyte HaCaT cells were a gift from Dr. Norbert Fusenig (German Cancer Research Center, Heidelberg, Germany). NIH3T3 cells and HaCaT cells were maintained as described elsewhere.11 The mouse BCC cell line ASZ001 was kindly provided by Dr. Ervin Epstein (Department of Dermatology, University of California, San Francisco, CA) and Dr. Matthew P. Scott (Department of Developmental Biology, Howard Hughes Medical Institute, Stanford University School of Medicine). It was maintained as reported previously.12 Cyclopamine (Biomol Int., Philadelphia, PA) was dissolved in 0.19% ethanol and added to ASZ001 culture at a concentration of 2 or 10 μmol/L. Purmorphamine (Calbiochem, San Diego, CA) was dissolved in dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO) and added to NIH3T3 culture at a concentration of 2 μmol/L. As a control, the same amount of 0.19% ethanol and dimethyl sulfoxide was added.
Plasmids and Transfection
Mouse Gli1 expression vector, Gli-consensus reporter gene (8×3′GBS-luc) and its mutant reporter gene (8×3′mutGBS-luc) were gifts from H. Sasaki (Riken, Kobe, Japan).13 Human GPR49 expression vector was constructed by inserting human GPR49 full coding cDNA (RZPD, Berlin, Germany) to pcDNA3 (Invitrogen, Carlsbad, CA). Transfection was performed using Fugene6 (Roche Diagnostics, Basel, Switzerland). Expression vector-transfected cells were treated with G418 (Invitrogen) and resistant colonies were collected without cloning.
RNA Interference
RNA interference was performed using shRNA.14 Briefly, 64- nucleotide hairpin-loop sequences (containing the 21-nucleotide shRNA targeted sequence) were generated (Sigma, Tokyo, Japan). GPR49 shRNA targeted sequences were as follows. GPR49-585: 5′-GAACAAAAUACACCACAUA-3′, GPR49-662: 5′-GAAUCCACUCCCUGGGAAA-3′. As a control, we used a random 21-nucleotide sequence that does not target any known genes (B-Bridge International Inc, Mountain View, CA) to exclude a non-specific effect of the hairpins. These genes were subsequently inserted upstream of the H1RNA polymerase III promoter in a pENTR221-H1R-stuffer vector. The pENTR221 rfA cassette “expressing shRNA under the control of the human H1 promoter” was changed to the pDEST-SI-CMSCVpuro vector (CMV-based vector) using the GATEWAY system (Invitrogen) for transient transfection. pENTR221-H1R-stuffer vector and pDEST-SI-CMSCVpuro vector were provided by Dr. T. Kiyono (National Cancer Center, Tokyo, Japan). Transfected cells were treated with puromycin (Invitrogen) for 2 weeks. Puromycin-resistant colonies were collected by trypsinization and used without cloning.
Cell Proliferation Assay
Antibiotic-resistant cells were seeded in 6-well plates at 5 × 104 cells/well and incubated as described above. After seeding, cells were counted every 24 hours in a Burker-Turk counting chamber (CIS, Japan). WST-1 assay (Roche Diagnostics) and BrdU incorporation assay (Roche Diagnostics) were performed according to the manufacturer’s protocol. An apotosis detection assay was performed using Annexin V-PE Apoptosis Detection Kit 1 (BD Pharmingen, San Diego, CA) and flow cytometry according to the manufacturer’s protocol. All experiments were performed in triplicate and repeated separately at least five times, and representative data are shown.
Tumor Formation Assay
GPR49-overexpressed HaCaT cells and pcDNA3-transfected HaCaT cells were transplanted into NOG mice (NOD/Shi-scid/IL-2γ−/− mouse) at 1 × 10 7 cells without cloning. GPR49-overexpressed HaCaT cells were transplanted onto the right side of the back, and pcDNA3-transfected HaCaT cells were transplanted onto the left side. Tumor size was measured every week. Twenty-four weeks after transplantation, transplanted sites with or without tumor were resected and samples were used for histological analysis and RNA extraction. Experiments were performed in ten mice. We used mouse anti-human Ki-67 antibody (Dako, Carpinteria, CA) for Immunohistochemistry to estimate cell proliferation.
Statistical Analysis
Statistical analysis was performed using Statcel2 software (OMS, Saitama, Japan). Statistically significant differences were determined by Student’s t-test using P = 0.05 as the level of significance.
Results
Overexpression of GPR49 in BCC
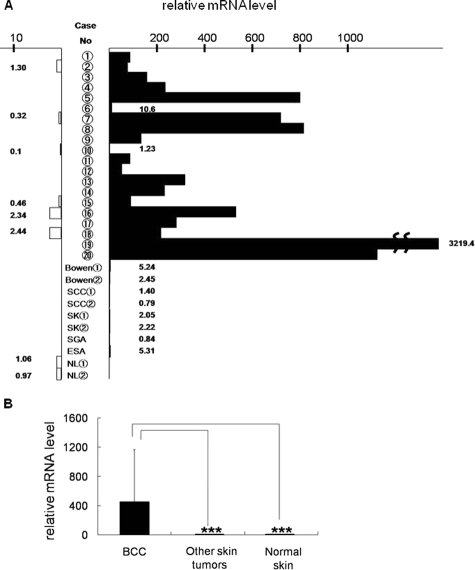
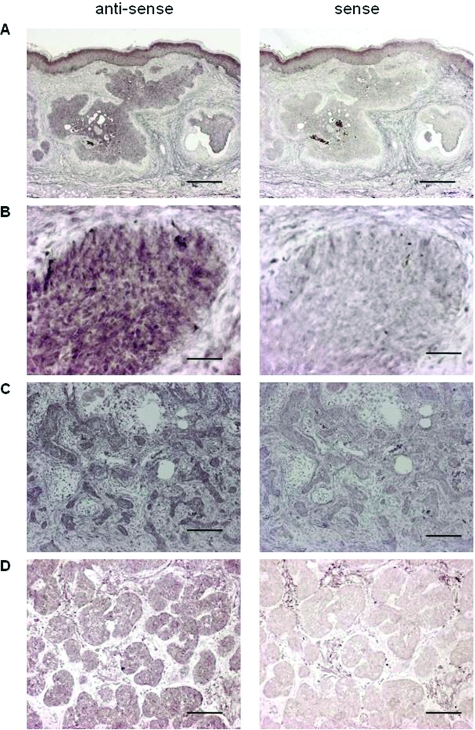
Among the genes found to be expressed in BCC from GeneChip analysis databases, GPR49 showed a significant increase compared to normal skin (Supplementary Figure S1, http://ajp.amjpathol.org). DNA microarray analysis in our samples also showed GPR49 expression to be about 8.3-fold higher than normal skin (Supplementary Table S1, http://ajp.amjpathol.org). Expression of GPR49 was confirmed by QRT-PCR using 20 cases of BCC (Table 2) together with GLI1 and GLI2, which were reported to be overexpressed in BCC3 and to be transcription factors involved in HH signaling. Of these 20 cases, 19 showed GPR49 expression levels more than three times higher than the control (Figure 1, A and B) with a mean increase of about 450-fold). As we could not obtain a specific antibody against GPR49 suitable for immunohistochemical analysis, we performed in situ hybridization to determine whether tumor cells express the gene. The anti-sense signal level was higher compared to the sense probe, and the signal expression site coincided with the tumor nest in various histological subtypes (Figure 2). On the other hand, expression by QRT-PCR was low in other epidermal skin tumors (Figure 1, A and B). These results suggest that the overexpression of GPR49 is a characteristic feature of BCC.
Table 2.
Expression Level of GPR49, GLI1, GLI2 in BCC*
| Case | Pattern | Lesion | GPR49 | GLI1 | GLI2 |
|---|---|---|---|---|---|
| Case 1 | Nodular | Face | 85.3 | 574.7 | 72 |
| Case 2 | Nodular | Face | 78.8 | 664 | 9.5 |
| Case 3 | Nodular | Face | 105.8 | 229.99 | 6.9 |
| Case 4 | Nodular | Face | 234.7 | 726 | 70.8 |
| Case 5 | Nodular | Face | 799.9 | 753.3 | 19.5 |
| Case 6 | Superficial | Trunk | 10.6 | 63.54 | 2.3 |
| Case 7 | Nodular | Face | 718.5 | 927 | 17.8 |
| Case 8 | Nodular | Face | 815.6 | 641 | 12.4 |
| Case 9 | Superficial | Face | 132.7 | 514.9 | 9.3 |
| Case 10 | Superficial | Trunk | 1.2 | 5.6 | 0.1 |
| Case 11 | Nodular | Face | 86.5 | 161.97 | 3.2 |
| Case 12 | Superficial | Trunk | 53.3 | 410.3 | 15.6 |
| Case 13 | Superficial | Trunk | 316.8 | 2362.1 | 96.7 |
| Case 14 | Superficial | Trunk | 231.6 | 359.3 | 184.7 |
| Case 15 | Nodular | Face | 90.2 | 512 | 26.6 |
| Case 16 | Nodular | Face | 530.17 | 3983 | 141.6 |
| Case 17 | Nodular | Face | 281.9 | 2101.8 | 30.7 |
| Case 18 | Nodular | Face | 217.9 | 1364.4 | 25.9 |
| Case 19 | Nodular | Face | 3219.4 | 7417.3 | 168.6 |
| Case 20 | Superficial | Trunk | 1126.4 | 6880.3 | 147.27 |
Each gene expression value represents the ratio of mRNA in tumor to that in normal skin mRNA.
Figure 1.
Expression of GPR49 in BCC. A: QRT-PCR of GPR49. The mRNA levels of GPR49 in 20 BCC, 8 other skin tumors, 6 normal skin in the vicinity of BCC, and 2 normal skin samples from non-cancerous patients (NL) are estimated by QRT-PCR. (Closed column: mRNA level in tumors. Open column: mRNA level in normal skin). In other types of skin tumors such as Bowen’s disease, squamous cell carcinoma (SCC), sweat gland adenocarcinoma (SGA), eccrine spiradenocarcinoma (ESA), and seborrheic keratosis (SK), expression of GPR49 is negligible compared to BCC. B: The mean value and SD of the mRNA levels of GPR49 in BCC, other skin tumors and normal skin. About 450-fold higher levels of GPR49 are shown in BCC as compared with normal skin.
Figure 2.
In situ hybridization of GPR49 in BCC. De-paraffinized sections were hybridized with anti-sense (left) or sense (right) probes for GPR49. A: Case 17. Low power view (scale bar = 1000 μm). Signal expression site hybridized with anti-sense probe coincides with the tumor nest. B: High magnification of case 17 (scale bar = 100 μm). Signal of anti-sense probe coincides with cytoplasm. C: Sclerosing type BCC (scale bar = 250 μm). D: Micronodular type BCC (scale bar = 250 μm).
Function of GPR49 in Cell Proliferation
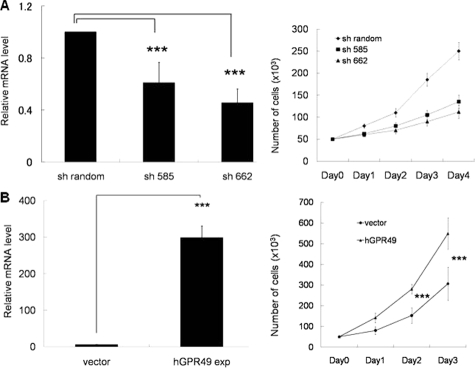
To identify the function of GPR49, we first performed RNA interference in BCC cell line. As no human BCC cell lines are available, we extensively tried to establish human BCC cell lines. However, we could neither obtain transplantable BCC nor establish BCC cell lines. So we used a mouse BCC cell line, ASZ001.15 As expected, this cell line expressed mouse Gpr49 and high HH signaling activity. We constructed two sequences of shGPR49RNAi vectors (sh585 and sh662) and transfected into ASZ001 cells, and puromycin-resistant cells were collected. Knockdown experiments using sh585 and sh662 decreased GPR49 mRNA levels by 40% and 50%, respectively, compared to controls (Figure 3A) and the knockdown effect of sh585 and sh662 persisted up to day 5 after cell selection was completed. Cell growth was markedly decreased by both shGPR49RNAi vectors in comparison with the control, and suppression of cell proliferation was dependent on the knock down effect (Figure 3A). Suppression of cell proliferation was also confirmed in WST-1 and BrdU incorporation assays (data not shown). To exclude any apoptotic effect and differentiation effect of knocking down mouse Gpr49 gene, we performed an apoptosis detection assay and analyzed the expression level of involucrin and loricrin, both of which are markers of keratinization. No significant difference was seen between control cells and mouse Gpr49 knockdown cells (data not shown).
Figure 3.
Function of Gpr49 in cell proliferation. A: Knockdown effect of shRNA against mGpr49 in ASZ001 cell proliferation. Left: Gene expression level of mouse Gpr49 in ASZ001 harboring short-hairpin RNAi against GPR49 was measured by QRT-PCR. ASZ001 cells were transfected with shRandom, sh585 or sh662, and puromycine-resistant cells were collected for RNA extraction and cell proliferation assay. Knockdown experiments using sh585 and sh662 decreased GPR49 mRNA levels by 40% and 50%, respectively, compared to controls. Right: Cell counting. ASZ001 cells harboring shRNAi were plated on 24-well dishes (5 × 104 cells/well). Trypsinized cells were counted every 24 hours for 4 consecutive days. When the expression level of mouse Gpr49 was knocked down, cell proliferation was down-regulated in comparison with the shRandom (sh585 and sh662 to shRandom: P < 0.01 at day 3 and day 4). Also, down-regulation of cell proliferation was dependent on the knock down effect. B: The effect of overexpressing of GPR49 on cell proliferation. Left: HaCaT cells were transfected with hGPR49 expression vector, and G-418–resistant colonies were collected for RNA extraction and cell proliferation assay. The gene expression level of Gpr49 was measured by QRT-PCR. HaCaT cells harboring GPR49- expression vector showed expression of GPR49, about 300 times higher than cells transfected with empty vector. Right: Cell counting. HaCaT cells harboring hGPR49 expression or empty vector were plated on 24-well dishes (5 × 104 cells/well). Trypsinized cells were counted every 24 hours for 3 consecutive days. When hGPR49 is overexpressed in HaCaT cells, upregulation of cell proliferation was observed in comparison with cells transfected with empty vector. ***P < 0.01, bars = SD.
In addition, the direct effect of GPR49 expression was evaluated by transfecting human GPR49 expression vector into HaCaT cells. As we could not find or create a specific antibody against GPR49 suitable for western blotting, we performed studies using QRT-PCR. Expression of GPR49 in neomycin-resistant GPR49-transfected cells (HaCaT-GPR49exp cells) was 300 times higher than empty vector transfectants (HaCaT-vector cells), and HaCaT-GPR49exp cells showed increased cell growth (Figure 3B). Upregulation of cell proliferation was also confirmed in WST-1 assay (data not shown). In the course of cell culture, no apoptosis or cell differentiation was observed (data not shown).
Function of GPR49 in Tumor Formation
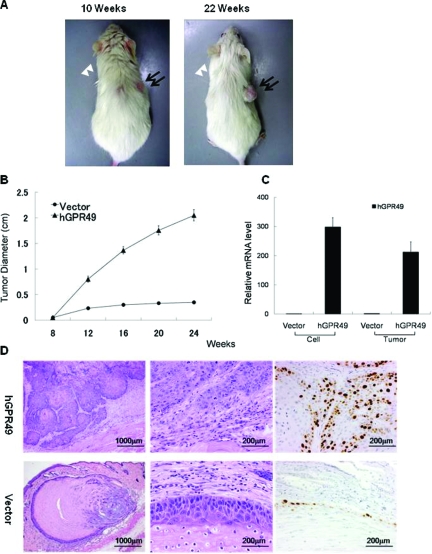
To evaluate further the function of GPR49 in vivo, we attempted to transplant shRNA-transfected ASZ001 cells into NOG mice, but we could not establish a stable mGpr49 knockdown ASZ001 cell culture, probably due to a growth-suppressive effect of shRNA. Then we transplanted HaCaT-GPR49exp cells into NOG mice. HaCaT cells are immortal in vitro, but they are also nontumorigenic and reform an orderly structured and differentiated epidermal tissue when transplanted into nude mice.16 So we thought its normal keratinocyte-like feature of HaCaT cells would be useful to evaluate whether overexpression of GPR49 in this cell line promotes tumorigenicity. As expected, tumor formation was observed about 10 weeks after transplantation. HaCaT-vector cells also showed a cystic mass formation in three of ten mice but it did not become larger. On the other hand, HaCaT-GPR49exp cells showed tumor formation and enlargement in all mice (Figure 4A and B). Overexpression of GPR49 in the tumor of HaCaT-GPR49exp cells was confirmed to be as much as 200 times higher than HaCaT-vector cells or nodules derived from HaCaT-vector cells (Figure 4C). In histological examination, nodules of empty vector transfected cells showed a small cystic structure containing keratinizing material and calcification. The cyst wall was composed of a thin and uniform epithelial structure (Figure 4D). On the other hand, the solid tumor made from HaCaT-GPR49exp cells displayed dyskeratosis, cancer pearls with a high tendency toward proliferation and interstitial invasion. Cell proliferation of the tumor made from HaCaT-GPR49exp cells was examined by Ki-67 immunohistochemical staining, and HaCaT-GPR49exp cells showed significantly increased expression of Ki-67 (Figure 4D). These findings indicate that GPR49 promotes cell proliferation and tumor formation.
Figure 4.
hGPR49-overexpressed HaCaT cells showed tumor formation in NOG mice. A: Tumor formation at 10 weeks (left panel) and 22 weeks (right panel). HaCaT cells overexpressing hGPR49 (1 × 107 cells) were transplanted subcutaneously on the back of NOG mice on the right side. Cells with empty vector were also transplanted onto the back of the same mouse on the left side. Cells with hGPR49 formed tumors (closed arrows) in all ten mice, while only a nodule (open arrowheads) was formed in three mice with cells harboring empty vector. B: Measurement of tumor size. The diameters of tumors were measured every other week after transplantation. Apparent tumor formation was observed 10 weeks after transplantation. All ten tumors derived from GPR49-overexpressed cells continued to grow until 24 weeks, while three nodules derived from cells with empty vector did not show enlargement. C: Gene expression level of GPR49 in innoculated cells and in formed tumors was measured by QRT-PCR. Tumors derived from GPR49-overexpressed HaCaT cells showed high levels of hGPR49, about 200 times higher than the nodule with empty vector (bars = SD). D: Histological examination. Tumors derived from GPR49-overexpressed HaCaT cells showed aberrant proliferation and invasion. Keratinization was observed in some tumor nests and had features resembling squamous cell carcinoma (upper panels). The nodule derived from HaCaT cells with empty vector showed a cystic structure with keratinization and calcification (lower panels). Left panels: low magnification (scale bar = 1000 μm), Middle panels: high magnification (scale bar = 200 μm). Right panels: immunohistochemical staining of Ki-67. Significantly increased expression of Ki-67 coincident with nucleus is observed in tumors derived from GPR49-overexpressed HaCaT cells (scale bar = 200 μm).
Relation between GPR49 and HH Signaling
Using 20 cases of BCC, expression of GPR49, GLI1 and GLI2 was confirmed by QRT-PCR as listed in Table 2. So we analyzed the correlation between the expression level of GPR49 and GLI1, GLI2 to evaluate the relation between HH signaling and GPR49. Statistical analysis using Pearson’s coefficient was calculated for all combinations. A high correlation was seen between the expression levels of GPR49 and GLI1 at r = 0.802. Also, a mild correlation was observed between GPR49 and GLI2 (r = 0.539), GLI1 and GLI2 (r = 0.707) (Table 3). As HH signaling is activated in BCC, we speculated here that expression of GPR49 is highly related to HH signaling.
Table 3.
Pearson Coefficient Analysis of Each Gene Expressed in BCC**
| GPR49 | GLI1 | GLI2 | |
|---|---|---|---|
| GPR49 | 0.804424 | 0.539395 | |
| GLI1 | 0.804424 | 0.707205 | |
| GLI2 | 0.539395 | 0.707205 |
Pearson correlation coefficient (two-tailed) was calculated pairwise using Statcel2 software for all combinations. (P < 0.01).
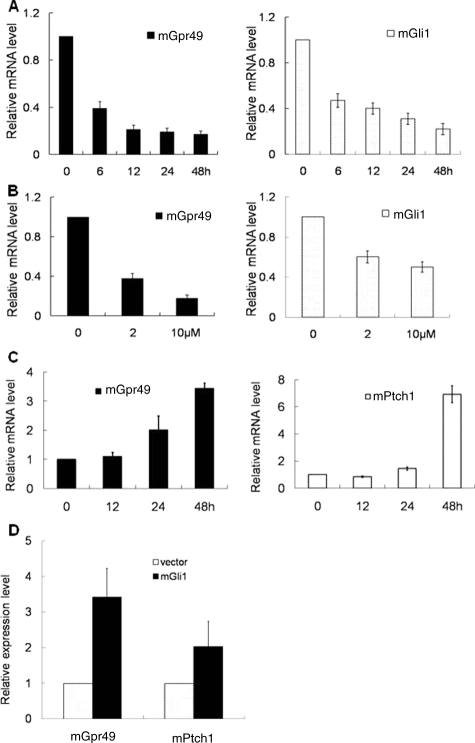
To further evaluate the relation between HH signaling and GPR49, we performed an in vitro assay using cell lines. Activation and suppression of HH signaling was confirmed by the reporter constructs of HH signaling.13 Luciferase activity of 8×3′GBS-luc is much higher than 8×3′mutGBS-luc in the condition of activated HH signaling. (Supplementary Figure S1, http://ajp.amjpathol.org). We treated ASZ001 with cyclopamine, a known inhibitor of HH signaling. Down-regulation of mouse Gpr49 expression was observed together with down-regulation of mouse Gli1, a target of HH signaling,3 and this down-regulation was dependant on the time after treatment and the concentration of cyclopamine (Figure 5A and B). Next, to see whether GPR49 expression is up-regulated by HH signaling, we treated mouse embryonic fibroblast NIH3T3 cells, which are known to respond to HH signaling, with the HH agonist purmorphamine. The expression of mouse Gpr49 was increased together with mouse Ptch1, another target of HH signaling, in a time-dependent manner (Figure 5C). Up-regulation of mouse GPR49 was also confirmed when we transfected mouse Gli1 expression vector13 to NIH3T3 (Figure 5D). These findings indicate that GPR49 is regulated by HH signaling.
Figure 5.
HH signaling regulates Gpr49 expression. A: Time course of mRNA expression of endogenous mGpr49 and mGli1 in ASZ001 cells treated with cyclopamine. Cells were treated with 10 μmol/L of cyclopamine, and expression of mGpr49 (left) and mGli1 (right) mRNA was measured by QRT-PCR at the times shown in the figure. The fold decrease of mRNA levels in cyclopamine-treated to non-treated cells at each sampling time was normalized setting the baseline value at 1. The figure is one of the five repeated experiments. Suppression of mGpr49 and mGli1 expression was dependent on the time course. The expression level of mGpr49 was persistently decreased by 20% of initial time in 48 hours after treatment. Statistical analysis using Pearson’s coefficient was calculated for each time’s combination and a high correlation was seen between the expression levels of mGpr49 and mGli1 at r = 0.985 (P < 0.01). B: Dose dependency of mRNA expression of mGpr49 and mGli1 on the concentration of cyclopamine. ASZ001 cells were treated with 0, 2, and 10 μmol/L of cyclopamine, and expression of mGpr49 (left) and mGli1 (right) was measured by QRT-PCR 48 hours after treatment. Values are shown as a ratio relative to cyclopamine 0 μmol/L. Suppression of mRNA levels of mGpr49 and mGli1 was dependent on the concentration of cyclopamine. C: Time course of mRNA expression of mGpr49 and mPtch1 in NIH3T3 cells treated with purmorphamine. Cells were treated with 2 μmol/L of purmorphamine for the time indicated in the figure. Ptch1 is a well-known target of HH signaling. Upregulation of mGpr49 (left) expression was dependent on the time course, as with mPtch1 (right). Statistical analysis using Pearson’s coefficient was calculated for each time’s combination and a high correlation was seen between the expression levels of mGpr49 and mPtch1 at r = 0.945 (P < 0.01). D: After transfection of mouse Gli1-expression vector to NIH3T3. mRNA levels of mGpr49 and mPtch1 were assayed at the time indicated in the figure. The ratios of mRNA level of mGli1-transfected to vector-transfected cells were estimated, and the values are shown as the ratio to the value at 0 hours. When Gli1 is expressed in NIH3T3 cells, the gene expression level of mGpr49 and mPtch1 were elevated.
Discussion
Our studies demonstrated that GPR49 is specifically overexpressed in BCC and plays a significant role in tumor formation and cell proliferation.
DNA microarray analysis in our samples showed GPR49 expression to be about 8.3-fold higher than normal skin. This result is reliable because our data includes almost all genes reported to be overexpressed in BCC.3,17,18 In QRT-PCR study, GPR49 was markedly overexpressed in 19 of 20 BCC samples of nodular and superficial types in comparison with normal tissue samples. On the other hand, other types of malignant epidermal tumors did not show significant upregulation of GPR49. Together with the result of in situ hybridization, our study suggests that the overexpression of GPR49 is a characteristic feature of BCC. Several reports show that BCC is a tumor of hair follicle origin. In particular, follicular bulge stem cells and their progeny with high self-renewal capacity are suggested to play the role in BCC formation.19 In support of this idea, the expression of keratin 6a, a marker of the follicular bulge stem cells20 or their progeny,21 was 6.8-fold higher in BCC than in normal skin in our DNA microarray analysis (data not shown). Recent report shows that GPR49 is the second most highly up-regulated gene as assessed by differential expression arraying on isolated hair follicle stem cells.22 Together with the report that GPR49 marks stem cells in intestine and hair follicle,7 our experiment suggest that BCC is similar to hair follicle stem cells.
Our data also showed that down-regulation of Gpr49 suppressed ASZ001 cell proliferation without inducing differentiation or apoptosis, and GPR49-overexpressing HaCaT cells showed cell proliferation in vitro. Furthermore, GPR49-overexpressing HaCaT cells showed tumor formation when transplanted into NOG mice. Even though the nodules of HaCaT-vector cells showed epidermal cyst-like structures on histological examination, the tumors of GPR49-overexpressing HaCaT cells showed not only nuclear atypia but also a complicated nest with cancer pearls and invasion into the stroma. Also, increased expression of Ki-67 in the tumors of GPR49-overexpressing HaCaT cells was observed. Though the tumor showed features of squamous cell carcinoma instead of BCC, this could be explained by differences in the character of the BCC progenitor cells and more highly differentiated HaCaT cells. These histological alterations by GPR49 overexpression revealed that GPR49 functions as an oncogene. A recent report showed that stem cells in the small intestine and colon overexpress GPR49, and also possibly in the hair follicle.7 But it was unclear whether GPR49 has a functional role or whether it is simply a stem cell marker. Our results show that GPR49 does have a role in cell proliferation, at least in some cell types. As discussed in the emerging concept of “cancer stem cells,” some subpopulations of tumor cells possessing somatic stem cell markers have a high proliferative characteristics.23 But this concept is somewhat controversial since this phenomenon goes against the original concept that somatic stem cells are defined as those with slow cell cycling. However, this phenomenon is now reported in various types of cancers, and the current idea is that the difference between somatic and cancer stem cells is explained by how strictly self-renewal is regulated.23 Therefore, considering these theories, we speculate that BCC cells possess the features somewhat similar to follicular stem cells, while dysregulated expression of the stem cell marker GPR49 led to abnormal cell proliferation and tumor formation.
Some studies have also suggested that G-protein-coupled receptors are involved in carcinogenesis.24 In particular GPR48, which is also a member of the LGRs, plays a significant role in invasion and metastasis of carcinoma cell.25 GPR49 has potential SH2- and SH3-interacting sequences in the C-terminal tail and it may be able to link to additional signal transduction cascades. Therefore, together with our observations about functional analysis, GPR49 may be closely involved in the tumorigenesis of BCC. These data also suggested that GPR49 will be a novel target of therapy in BCC. Because a numbers of drugs currently in use are agonists or antagonists of G protein-coupled receptors, we speculate that antagonists of GPR49 will also be a drug for treating BCC. So, future studies to identify the ligand of GPR49 would be useful.
Our results indicated that gene expression of GPR49 and GLI1 showed significant correlation in BCC tumor samples. Moreover, inhibition of the HH signaling pathway in a mouse BCC cell line down-regulated endogenous Gpr49, and activation of HH signaling in mouse NIH3T3 cells up-regulated endogenous GPR49. These findings indicate that GPR49 is regulated by HH signaling. Previous reports showed that GPR49 is regulated by Wnt-β -catenin signaling,7,8 but our data suggested that GPR49 is also regulated by HH signaling in BCC. However, we could not find the GLI binding site or related sequences13 in the promoter region of GPR49. Also, luciferase assay using reporter constructs of the GPR49 promoter region did not show significant activity (data not shown). However, the regulation of HH signaling target genes in mammals seems complicated. For instance, GLI binding sites of N-MYC, which is also thought to be a target gene of HH signaling, may exist in the second intron and about 50 kb upstream of the transcription start site.26 There may also some intermediary step exist to induce GPR49. One possibility is that GPR49 is expressed via the activation of Wnt-β catenin signaling, since past studies suggested that activation of HH signaling induces activation of Wnt-β catenin signaling.27 However, we could not successfully prove this hypothesis in our BCC cases because immunohistochemistry of β catenin in BCC cases did not show nuclear accumulation (data not shown). And reporter activity of TOP flash/FOP flash28 was negligible in ASZ001 in comparison with the high activity seen in hepatocellular carcinoma cells in which Wnt-β catenin signaling had been activated (data not shown). So at present, we do not know whether GPR49 is up-regulated by HH signaling directly or whether there are any intermediate steps. Further analysis is necessary to reveal the precise regulation of the gene expression.
In conclusion, stem cell marker GPR49 is expressed in BCC and plays a significant role in the pathogenesis of BCC. Because some cancer cells and tissue development share the HH signaling pathway, GPR49 might be a key molecule with roles common to both the carcinogenesis and normal tissue development pathways.
Acknowledgments
We thank Dr. Ervin Epstein (Department of Dermatology, University of California San Francisco) and Dr. Matthew P. Scott (Department of Developmental Biology, Howard Hughes Medical Institute, Stanford University School of Medicine) for providing ASZ001 cells; Norbert Fusenig (Division of Carcinogenesis and Differentiation in vitro, German Cancer Research Center) for providing the HaCaT cells; Dr. Hiroshi Sasaki (Laboratory for Embryonic Induction, RIKEN Center for Developmental Biology) for providing mouse Gli1 expression vector, 8×3′GBS-luc reporter and 8× mutant3′GBS-luc reporter; Dr. Toru Kiyono (Virology Division, National Cancer Center Research Institute) for providing pENTR221 vector and pDEST-SI-CMSCVpuro vector; and Drs. Syunichi Miyakawa (Department of Dermatology, Kawasaki Municipal Hospital), Shinichi Takahashi (Department of Dermatology, University of Tokyo Dental College, Ichikawa Hospital), Yasuki Hata (Department of Dermatology, Kanagawa Saiseikai Hospital), and Junki Ogawa (Department of Dermatology, Nihon Koukan Hospital) for providing samples of BCC cases.
Footnotes
Address reprint requests to Michiie Sakamoto. Department of Pathology, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan. E-mail: msakamot@sc.itc.keio.ac.jp.
Supported by Grant-in-Aid for the 21st Century Center of Excellence program and Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; The Third Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labor and Welfare of Japan; and research encouraging scholarship of Keio University School of Medicine.
Supplemental material for this article can be found on http:// ajp.amjpathol.org.
References
- Schirren CG, Rütten A, Kaudewitz P, Diaz C, McClain S, Burgdorf WH. Trichoblastoma and basal cell carcinoma are neoplasms with follicular differentiation sharing the same profile of cytokeratin intermediate filaments. Am J Dermatopathol. 1997;19:341–350. doi: 10.1097/00000372-199708000-00005. [DOI] [PubMed] [Google Scholar]
- Wicking C, McGlinn E. The role of hedgehog signalling in tumorigenesis. Cancer Lett. 2001;173:1–7. doi: 10.1016/s0304-3835(01)00676-0. [DOI] [PubMed] [Google Scholar]
- Bonifas JM, Pennypacker S, Chuang PT, McMahon AP, Williams M, Rosenthal A, De Sauvage FJ, Epstein EH., Jr Activation of expression of hedgehog target genes in basal cell carcinomas. J Invest Dermatol. 2001;116:739–742. doi: 10.1046/j.1523-1747.2001.01315.x. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, van Duin M, Hsueh AJ. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol. 2000;14:1257–1271. doi: 10.1210/mend.14.8.0510. [DOI] [PubMed] [Google Scholar]
- Hsu SY. New insights into the evolution of the relaxin-LGR signaling system. Trends Endocrinol Metab. 2003;14:303–309. doi: 10.1016/s1043-2760(03)00106-1. [DOI] [PubMed] [Google Scholar]
- Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, Kuwabara Y, Baribault H, Tian H, Hsueh AJ. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol. 2000;24:9736–9743. doi: 10.1128/MCB.24.22.9736-9743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sakamoto M, Fujii G, Tsuiji H, Kenetaka K, Asaka M, Hirohashi S. Overexpression of orphan G-protein-coupled receptor. Gpr49, in human hepatocellular carcinomas with βcatenin mutations. Hepatology. 2003;37:528–533. doi: 10.1053/jhep.2003.50029. [DOI] [PubMed] [Google Scholar]
- McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, Kirschmeier P, Samatar AA. Identification of Orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–426. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- Scharffetter K, Lankat-Buttgereit B, Kreig T. Localization of collagen mRNA in normal and scleroderma skin by in-situ hybridization. Eur J Clin Invest. 1998;18:9–17. doi: 10.1111/j.1365-2362.1988.tb01158.x. [DOI] [PubMed] [Google Scholar]
- Jin W, Kim BC, Tognon C, Lee HJ, Patel S, Lannon CL, Maris JM, Triche TJ, Sorensen PH, Kim SJ. The ETV6-NTRK3 chimeric tyrosine kinase suppresses TGF-beta signaling by inactivating the TGF-beta type II receptor. Proc Natl Acad Sci USA. 2005;102:16239–16244. doi: 10.1073/pnas.0503137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Aszterbaum M, Zhang X, Bonifas JM, Zachary C, Epstein E, McCormick F. A role of PDGFRα in basal cell carcinoma proliferation. Proc Natl Acad Sc. 2001;98:9255–9259. doi: 10.1073/pnas.151173398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit PE, Scott MP, Epstein EH., Jr Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat Med. 1999;5:1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichberger T, Regl G, Ikram MS, Neill GW, Philpott MP, Aberger F, Frischauf AM. FoxE1. A new transcriptional target of GLI2, is expressed in human epidermis and basal cell carcinoma. J Invest Dermatol. 2004;122:1180–1187. doi: 10.1111/j.0022-202X.2004.22505.x. [DOI] [PubMed] [Google Scholar]
- Cui C, Elsam T, Tian Q, Seykora JT, Grachtchouk M, Dlugosz A, Tseng H. Gli proteins up-regulate the expression of basonuclin in basal cell carcinoma. Cancer Res. 2004;64:5651–5658. doi: 10.1158/0008-5472.CAN-04-0801. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Leonardi S, Tanori M, Pasquali E, Pierdomenico M, Rebessi S, Di Majo V, Covelli V, Pazzaglia S, Saran A. Hair cycle-dependent basal cell carcinoma tumorigenesis in Ptc1neo67/+ mice exposed to radiation. Cancer Res. 2006;66:6606–6614. doi: 10.1158/0008-5472.CAN-05-3690. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;16:303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu LH, Coulombe PA. Keratin expression provides novel insight into the morphogenesis and function of the companion layer in hair follicles. J Invest Dermatol. 2007;127:1061–1073. doi: 10.1038/sj.jid.5700673. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nature Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Spillane JB, Henderson MA. Cancer stem cells: a review. ANZ J Surg. 2007;77:464–468. doi: 10.1111/j.1445-2197.2007.04096.x. [DOI] [PubMed] [Google Scholar]
- Li S, Huang S, Peng SB. Overexpression of G protein-coupled receptors in cancer cells: involvement in tumor progression. Int J Oncol. 2005;27:1329–1339. [PubMed] [Google Scholar]
- Gao Y, Kitagawa K, Hiramatsu Y, Kikuchi H, Isobe T, Shimada M, Uchida C, Hattori T, Oda T, Nakayama K, Nakayama KI, Tanaka T, Konno H, Kitagawa M. Up-regulation of GPR48 induced by down-regulation of p27Kip1 enhances carcinoma cell invasiveness and metastasis. Cancer Res. 2006;66:11623–11631. doi: 10.1158/0008-5472.CAN-06-2629. [DOI] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Boonchai W, Walsh M, Cummings M, Chenevix-Trench G. Expression of beta-catenin, a key mediator of the WNT signaling pathway, in basal cell carcinoma. Arch Dermatol. 2000;136:937–938. doi: 10.1001/archderm.136.7.937. [DOI] [PubMed] [Google Scholar]
- Coghlan MP, Culbert AA, Cross DA, Corcoran SL, Yates JW, Pearce NJ, Rausch OL, Murphy GJ, Carter PS, Roxbee Cox L, Mills D, Brown MJ, Haigh D, Ward RW, Smith DG, Murray KJ, Reith AD, Holder JC. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]