Abstract
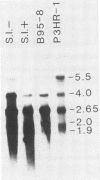
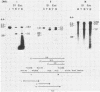
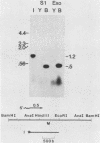
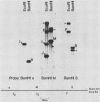
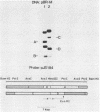
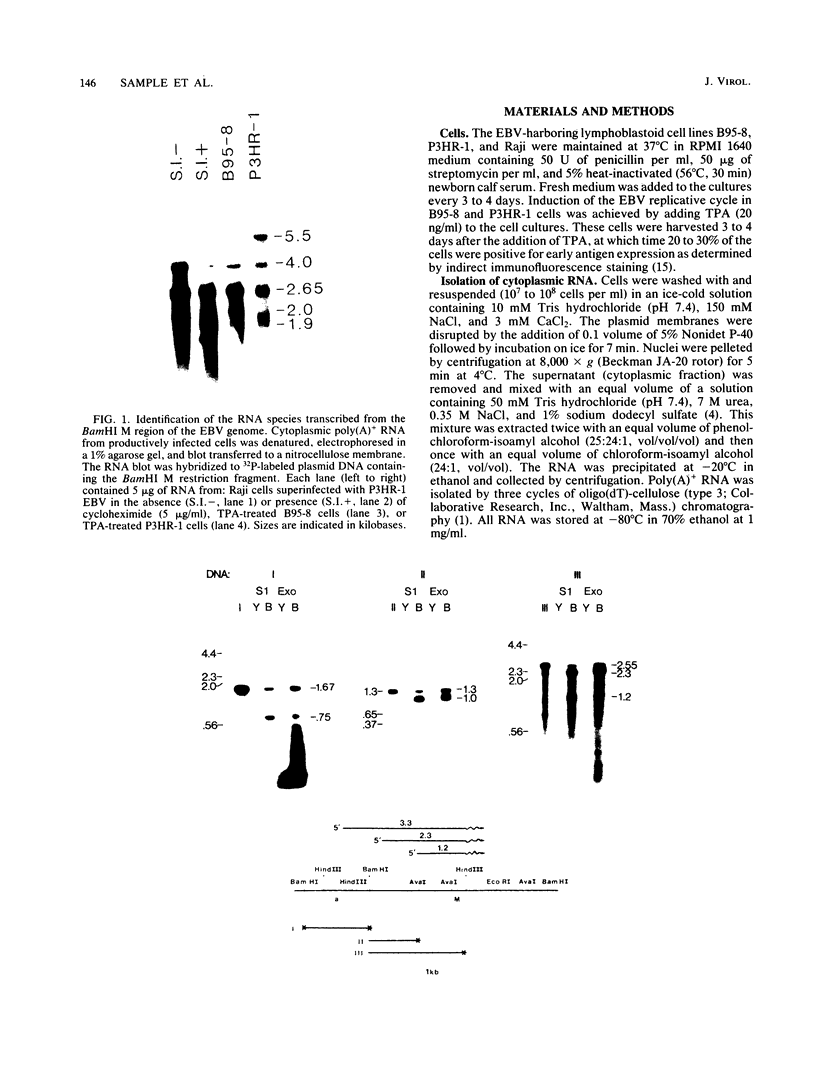
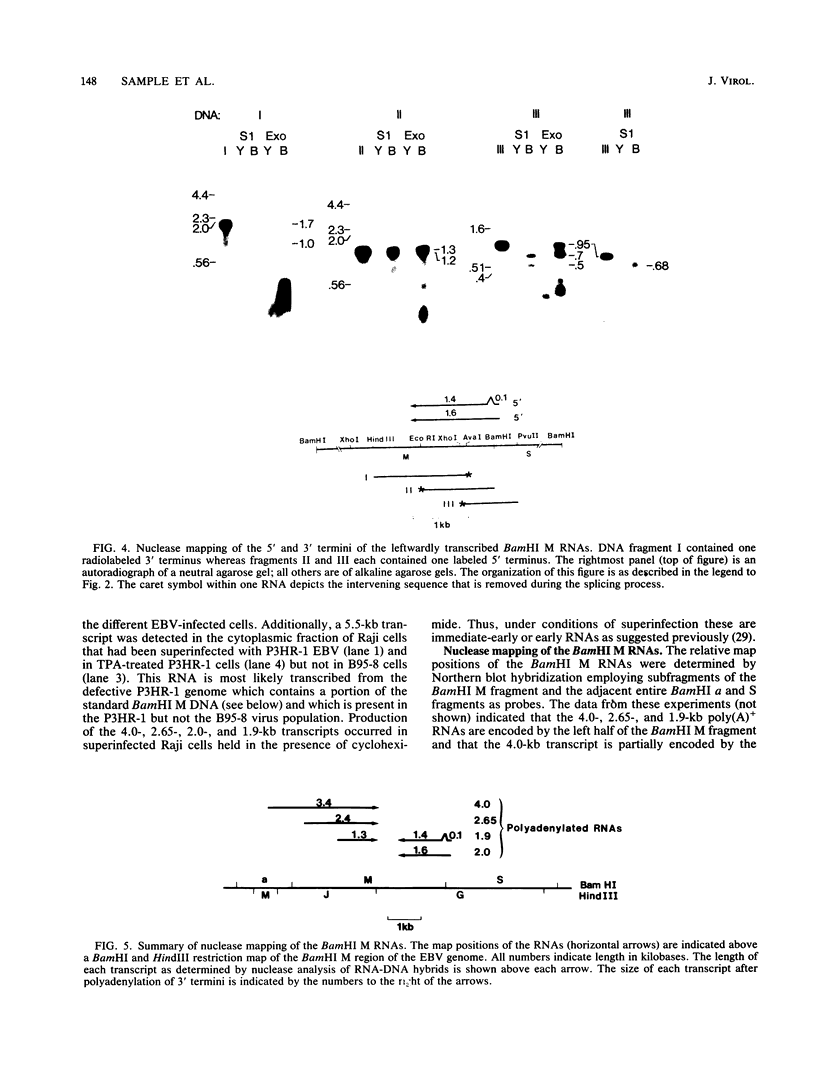
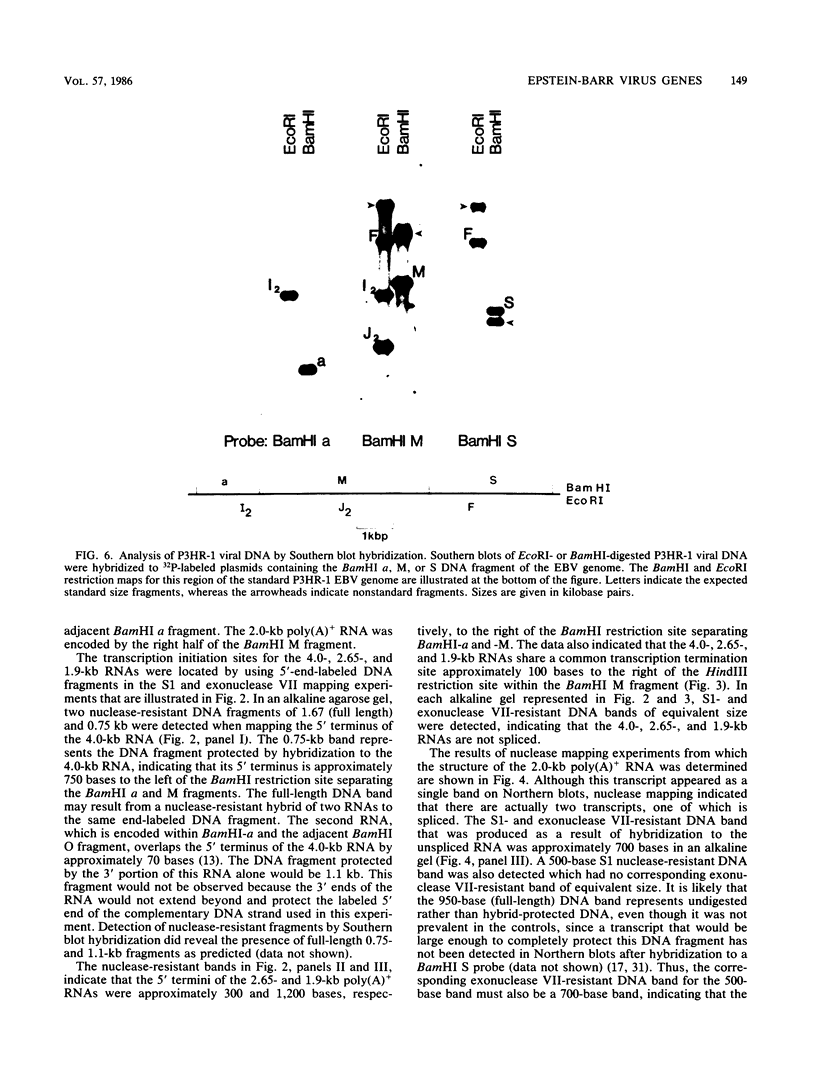
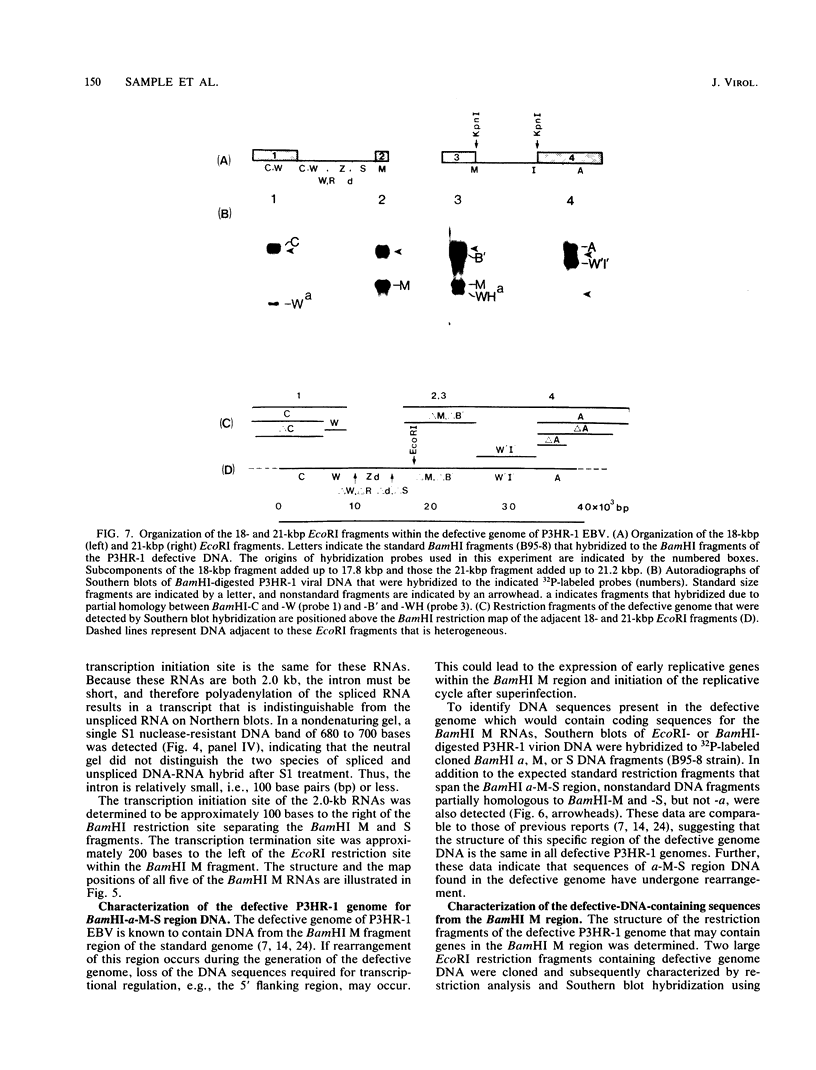
We used nuclease digestion to map RNA transcripts encoded in the BamHI M fragment of the Epstein-Barr virus (EBV) genome (strain B95-8). Of the five RNAs, three are rightwardly transcribed, have different cap sites but common 3' termini, and are unspliced. The two remaining RNAs are leftwardly transcribed and are 5' and 3' coterminal. One of these transcripts is spliced, resulting in the removal of a small intron from the 5' region of this RNA. We have previously published data which indicated that the BamHI M region is the first actively transcribed region of the viral genome during the replicative cycle, suggesting that one or more genes in this region is important in the initiation of EBV replication. We have now mapped two large EcoRI restriction fragments which span approximately 75% of the P3HR-1 defective genome and which contain DNA from the BamHI M region of the standard genome. The data indicate that only the coding and 5' flanking sequences for the leftwardly transcribed RNAs are intact within the defective genome. Fewer than 500 bases coding for the 3'-most regions of the rightwardly transcribed RNAs are intact, and it is unlikely that these encode functional native polypeptides. Therefore, it seems that transcriptional activation of the BamHI M-region genes is not mediated directly by the rearrangement of M genes in defective P3HR-1 EBV.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Zimber U., Hudewentz J., Epstein M. A. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980 Sep;35(3):603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Hudewentz J., Freese U. K., Zimber U. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J Virol. 1982 Sep;43(3):952–968. doi: 10.1128/jvi.43.3.952-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. S., Bornkamm G. W., zur Hausen H. Structure of defective DNA molecules in Epstein-Barr virus preparations from P3HR-1 cells. J Virol. 1984 Jul;51(1):199–207. doi: 10.1128/jvi.51.1.199-207.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. S., Gissmann L., Hayward S. D. Epstein-Barr virus (P3HR-1) defective DNA codes for components of both the early antigen and viral capsid antigen complexes. Virology. 1984 Aug;137(1):9–19. doi: 10.1016/0042-6822(84)90003-5. [DOI] [PubMed] [Google Scholar]
- Cohen L. K., Speck S. H., Roberts B. E., Strominger J. L. Identification and mapping of polypeptides encoded by the P3HR-1 strain of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4183–4187. doi: 10.1073/pnas.81.13.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Bornkamm G. W. Heterogeneity of Epstein-Barr virus. III. Comparison of a transforming and a nontransforming virus by partial denaturation mapping of their DNAs. J Virol. 1978 Jul;27(1):81–89. doi: 10.1128/jvi.27.1.81-89.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolyniuk M., Pritchett R., Kieff E. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J Virol. 1976 Mar;17(3):935–949. doi: 10.1128/jvi.17.3.935-949.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T., Stockwell P., Ginsburg M., Barrell B. Homology between two EBV early genes and HSV ribonucleotide reductase and 38K genes. Nucleic Acids Res. 1984 Jun 25;12(12):5087–5099. doi: 10.1093/nar/12.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M., Dambaugh T., Kieff E. Epstein-Barr virus DNA. IX. Variation among viral DNAs from producer and nonproducer infected cells. J Virol. 1981 May;38(2):632–648. doi: 10.1128/jvi.38.2.632-648.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle G., Henle W. Immunofluorescence in cells derived from Burkitt's lymphoma. J Bacteriol. 1966 Mar;91(3):1248–1256. doi: 10.1128/jb.91.3.1248-1256.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W., Henle G., Zajac B. A., Pearson G., Waubke R., Scriba M. Differential reactivity of human serums with early antigens induced by Epstein-Barr virus. Science. 1970 Jul 10;169(3941):188–190. doi: 10.1126/science.169.3941.188. [DOI] [PubMed] [Google Scholar]
- Hummel M., Kieff E. Epstein-Barr virus RNA. VIII. Viral RNA in permissively infected B95-8 cells. J Virol. 1982 Jul;43(1):262–272. doi: 10.1128/jvi.43.1.262-272.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes J., Leibold W., Klein G. Biological differences between Epstein-Barr virus (EBV) strains with regard to lymphocyte transforming ability, superinfection and antigen induction. Exp Cell Res. 1975 May;92(2):478–484. doi: 10.1016/0014-4827(75)90404-8. [DOI] [PubMed] [Google Scholar]
- Miller G., Rabson M., Heston L. Epstein-Barr virus with heterogeneous DNA disrupts latency. J Virol. 1984 Apr;50(1):174–182. doi: 10.1128/jvi.50.1.174-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Robinson J., Heston L., Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rabson M., Heston L., Miller G. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci U S A. 1983 May;80(9):2762–2766. doi: 10.1073/pnas.80.9.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sample J., Tanaka A., Lancz G., Nonoyama M. Identification of Epstein-Barr virus genes expressed during the early phase of virus replication and during lymphocyte immortalization. Virology. 1984 Nov;139(1):1–10. doi: 10.1016/0042-6822(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Seigneurin J. M., Vuillaume M., Lenoir G., De-Thé G. Replication of Epstein-Barr virus: ultrastructural and immunofluorescent studies of P3HR1-superinfected Raji cells. J Virol. 1977 Dec;24(3):836–845. doi: 10.1128/jvi.24.3.836-845.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Donovan J., Nonoyama M. Phosphonoacetic acid-resistant RNA of Epstein-Barr virus in productively infected cells. Virology. 1983 Jan 15;124(1):196–200. doi: 10.1016/0042-6822(83)90306-9. [DOI] [PubMed] [Google Scholar]
- Stoerker J., Glaser R. Rescue of transforming Epstein-Barr virus (EBV) from EBV-genome-positive epithelial hybrid cells transfected with subgenomic fragments of EBV DNA. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1726–1729. doi: 10.1073/pnas.80.6.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoerker J., Holliday J. E., Glaser R. Identification of a region of the Epstein-Barr virus (B95-8) genome required for transformation. Virology. 1983 Aug;129(1):199–206. doi: 10.1016/0042-6822(83)90406-3. [DOI] [PubMed] [Google Scholar]
- Yajima Y., Nonoyama M. Mechanisms of infection with Epstein-Barr virus. I. Viral DNA replication and formation of noninfectious virus particles in superinfected Raji cells. J Virol. 1976 Jul;19(1):187–194. doi: 10.1128/jvi.19.1.187-194.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H., Bornkamm G. W., Schmidt R., Hecker E. Tumor initiators and promoters in the induction of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1979 Feb;76(2):782–785. doi: 10.1073/pnas.76.2.782. [DOI] [PMC free article] [PubMed] [Google Scholar]