Abstract
β-O-Linked N-acetylglucosamine is a dynamic post-translational modification involved in protein regulation in a manner similar to phosphorylation. Removal of N-acetylglucosamine is regulated by β-N-acetylglucosaminidase (O-GlcNAcase), which was previously shown to be a substrate of caspase-3 in vitro. Here we show that O-GlcNAcase is cleaved by caspase-3 into two fragments during apoptosis, an N-terminal fragment containing the O-GlcNAcase active site and a C-terminal fragment containing a region with homology to GCN5 histone acetyl-transferases. The caspase-3 cleavage site of O-GlcNAcase, mapped by Edman sequencing, is a noncanonical recognition site that occurs after Asp-413 of the SVVD sequence in human O-GlcNAcase. A point mutation, D413A, abrogates cleavage by caspase-3 both in vitro and in vivo. Finally, we show that O-GlcNAcase activity is not affected by caspase-3 cleavage because the N- and C-terminal O-GlcNAcase fragments remain associated after the cleavage. Furthermore, when co-expressed simultaneously in the same cell, the N-terminal and C-terminal caspase fragments associate to reconstitute O-GlcNAcase enzymatic activity. These studies support the identification of O-GlcNAcase as a caspase-3 substrate with a novel caspase-3 cleavage site and provide insight about O-GlcNAcase regulation during apoptosis.
O-Linked β-N-acetylglucosamine (O-GlcNAc)2 is a dynamic, inducible post-translational modification, in which a monosaccharide N-acetylglucosamine is attached to serine or threonine residues of a protein (1–3). O-GlcNAc has been found on a number of nucleocytosolic proteins, ranging from transcription factors to cytoskeletal proteins (2, 4). Effects of O-GlcNAc appear to be protein-specific because O-GlcNAc can modulate protein function by regulating protein activity, protein-protein interaction, localization, or protein degradation (5, 6). O-GlcNAc modification is involved in signal transduction in a manner analogous to phosphorylation, in which O-GlcNAc and O-phosphate compete for sites on many proteins (5–10). O-GlcNAc plays important roles in various cell signaling pathways such as cellular stress, cell cycle, insulin signaling, and ubiquitin-proteasome pathways (2, 11–15).
Although phosphorylation is regulated by many kinases and phosphatases, O-GlcNAc modification is regulated by only two catalytic subunits: β-N-acetylglucosaminyl-transferase (OGT) and O-β-N-acetylglucosaminidase (O-GlcNAcase) (16–24). OGT is responsible for the addition of O-GlcNAc (5). OGT is essential for life because it is required for embryonic stem cell viability (21). O-GlcNAcase, which catalyzes the O-GlcNAc removal, is a neutral, nucleocytoplasmic β-glucosaminidase that is distinct from lysosomal hexosaminidases (17, 18). O-GlcNAcase is expressed in two splice variants: an active form of a 130-kDa protein, which is predominantly localized in the cytosol and cytoskeleton, and an inactive form of a 75-kDa protein, which is localized in the nucleus (25, 26). O-GlcNAcase is reported to be bifunctional, in which the N terminus possesses O-GlcNAcase activity, whereas the C terminus has histone acetyltransferase activity in vitro, suggesting that O-GlcNAcase may play a role in transcription activation (25, 27, 28). In addition to biochemical evidence, O-GlcNAcase has been shown to possess O-GlcNAcase activity in vivo as well (18, 26, 29).
There is growing evidence that O-GlcNAc modification is essential for cell survival (30–32). Loss of O-GlcNAc by OGT knock-out results in T-cell apoptosis and fibroblast growth arrest (21). Decreased O-GlcNAc levels through inhibition of production of UDP-GlcNAc, an OGT substrate, also cause impaired proliferation (33). In response to various forms of cellular stress, O-GlcNAc levels are increased, resulting in cells that are more thermotolerant (31). Increased O-GlcNAc levels also protects cardiomyocytes during ischemia/reperfusion (32). These findings suggest that O-GlcNAc may promote cell survival, and loss of O-GlcNAc could result in defective proliferation and cell death.
Apoptosis, or programmed cell death, is a naturally occurring process important in normal development and the maintenance of cellular homeostasis (34, 35). There are two major apoptotic pathways: the mitochondrial and the death receptor pathways (34–36). The mitochondrial pathway is triggered by activation of pro-apoptotic proteins at the mitochondria, whereas the death receptor pathway is activated by the death family receptors such as tumor necrosis factor receptor I and CD95 (Fas). Apoptotic stimulation by any of these pathways leads to activation of a cascade of a family of cysteine proteases called caspases (37). One of the most crucial effector caspases is caspase-3. Activation of caspase-3 leads to proteolysis of several protein substrates. This proteolytic cleavage usually results in either inactivation of proteins that protect living cells or activation of proteins that promote cell death (37–42).
Interestingly, O-GlcNAcase is an in vitro substrate of caspase-3 (26). However, it is unclear whether O-GlcNAcase is also a caspase-3 substrate in vivo and how caspase-3 cleavage affects the behavior of O-GlcNAcase. O-GlcNAcase cleavage may alter the properties of O-GlcNAcase, which may in turn have effects that could further alter O-GlcNAc levels and/or further influence apoptotic events. Hence, altered O-GlcNAc modification may be another hallmark of apoptotic pathways. In this study, we examined how apoptosis affects the behavior of O-GlcNAcase in vivo to better understand the role of O-GlcNAc in apoptosis.
EXPERIMENTAL PROCEDURES
Materials and Antibodies—Rabbit anti-O-GlcNAcase antibody was kindly provided by Dr. Wally Whiteheart (University of Kentucky). Anti-O-GlcNAc 110.6 was made by our laboratory. Rabbit anti-caspase-3 antibody was obtained from Stressgen. Anti-Fas mouse IgM CH-11 and rabbit anti-PARP antibodies were from Millipore. Anti-HA and anti-c-Myc (9E10) antibodies were from Santa Cruz Biotechnology, Inc. Anti-mouse IgM and secondary horseradish peroxidase-conjugated anti-mouse IgM were purchased from Sigma. Anti-rabbit and anti-chicken horseradish peroxidase-conjugated antibodies were from GE Healthcare and Promega, respectively. PUGNAc was purchased from Carbogen (Switzerland). Pan caspase inhibitor Z-VAD(OMe)-fmk and caspase-3 inhibitor (Z-D(OMe)E(OMe)VD(OMe)-fmk were purchased from Calbiochem. Recombinant caspase-3 was purchased from Biomol Research Laboratories, Inc.
Generation of O-GlcNAcase Antibodies—Through Sigma, we created a rabbit polyclonal antibody and a chicken polyclonal antibody specific to the N and C termini of O-GlcNAcase, respectively, using peptides purified from bacteria expressing amino acids 1–413 (N-terminally specific) and amino acids 414–916 (C-terminally specific) of the human O-GlcNAcase protein sequence. Prior to use, rabbit serum (for anti-N-terminal O-GlcNAcase antibody) or chicken IgY extracts (for anti-C-terminal O-GlcNAcase) were passed through antigen-coupled columns (coupled with either the N-terminal or the C-terminal O-GlcNAcase peptides) three times. The columns were washed with at least 20 column volumes of 20 mm Tris, pH 8.0, followed by 20 column volumes of 20 mm Tris, pH 8.0, and 500 mm NaCl. The antibodies were eluted by 100 mm glycine, pH 2.5, neutralized with 1 m Tris, pH 8.0, and concentrated into 40% glycerol in PBS.
Plasmid Constructions—Full-length O-GlcNAcase was subcloned into pET32 vector or pRK5-c-Myc vector with c-Myc tag located at the N terminus. O-GlcNAcase D413A clone was generated using QuikChange site-directed mutagenesis XL II (Stratagene) according to the manufacturer's protocol. Briefly, the O-GlcNAcase D413A mutant was generated by PCR amplification using DNA from either pRK5 or pET32 plasmids containing the full-length O-GlcNAcase as a template. The forward and reverse primers for PCR amplication from 5′ to 3′ ends were GCTAAAGCAAGTGTAGTTGCTGGTACCCCTTTAGTTGCAGC and GCTGCAACTAAAGGGGTACCAGCAACTACACTTGCTTTAGC, respectively. The N- and C-terminal O-GlcNAcase mutants were created by PCR amplification. The forward and reverse primers to amplify the N-terminal O-GlcNAcase were ACGCGTCGACCATGGTGCAGAAGGAGAGTCA and ATAAGAATGCGGCCGCTCAATCAACTACACTTGCTTTAG. The forward and reverse primers to amplify the C-terminal O-GlcNAcase were ACGCGTCGACCGGGACTCCTTTAGTTGCAGC and ATAAGAATGCGGCCGCTCACAGGCTCCGACCAAGTA, respectively. The PCR fragments were then subcloned into the pRK5-cMyc or pRK5-HA vectors with c-Myc tag or HA tag located at the N terminus of each construct. The amino acid sequences of c-Myc tag and HA tag are MEQKLISEEDLN and MGYPYDVPDYADLN, respectively. O-GlcNAcase and O-GlcNAcase mutant sequences were verified by sequencing.
Cell Culture, Transfection, and Apoptosis Induction—Jurkat cells were grown in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum (Gemini Bio-Products) and penicillin-streptomycin (Invitrogen). HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum and penicillin-streptomycin. The cells were transfected with expression vectors using Lipofectamine LTX (Invitrogen) according to the manufacturer's recommendation. To induce apoptosis, Jurkat cells were treated with 100 ng/ml of anti-Fas CH11 Ab. The same amount of anti-mouse IgM was used in control cells. The cells were harvested at the indicated times. To inhibit apoptosis in Jurkat cells, all of the caspase inhibitors were incubated with Jurkat cells 1 h prior to Fas stimulation. HeLa cells were treated with anti-Fas CH11 Ab and harvested 4 h after apoptotic induction.
Western Blot Analysis—After treatment, the cells were washed with PBS and lysed in either 10 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 25 mm KCl, and 0.5% Nonidet P-40 (for Jurkat cells) or 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 1% Nonidet P-40 (for HeLa cells) containing 1 mm phenylmethylsulfonyl fluoride, protease inhibitor cocktails, 10 mm β-glycerophosphate, and 5 mm sodium fluoride. The cells were incubated with lysis buffer on ice with occasional vortexing for 40 min and centrifuged at 13,000 rpm for 5 min. The supernatants were collected and separated using Tris-HCl gel electrophoresis (Bio-Rad). The separated proteins were transferred to nitrocellulose membrane and blocked with either 3% BSA in TBST (for anti-110.6, anti-c-Myc, and anti-HA antibodies) or 5% milk in TBST (for anti-O-GlcNAcase, anti-PARP, and anti-caspase-3 antibodies) for 1 h. The membranes were then probed with antibodies to O-GlcNAc (1:5,000), O-GlcNAcase (anti-full-length O-GlcNAcase; 1:5000; anti-N-terminal O-GlcNAcase; 1:1,000; anti-C-terminal O-GlcNAcase; 1:1,000), PARP (1:5,000), HA (1:5,000), c-Myc (1:5,000), and caspase-3 (1:2,500) overnight at 4 °C. Next, the membranes were washed with TBST five times for 10 min, probed with horseradish peroxidase-conjugated secondary antibodies for 1 h, and washed with TBST five times for 10 min followed by TBS for 10 min. Detection was carried out by ECL (GE Healthcare).
Immunoprecipitation—Cell extracts were incubated with an antibody specific to the C terminus of O-GlcNAcase at 4 °C overnight. The next day, the mixtures were incubated with IgY-agarose beads (Gallus Immunotech, Inc.) for 2 h at 4 °C. the beads were washed with cell extraction buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 1% Nonidet P-40). Laemmli buffer was added to the washed beads, boiled for 5 min, and subjected to SDS-PAGE followed by Western blot analysis.
Purification of Recombinant O-GlcNAcase—For expression of active O-GlcNAcase, the human O-GlcNAcase cDNA was subcloned into pET32 vector and transformed into BL21 bacteria. A freshly transformed colony was picked to inoculate 50 ml of LB and grown at 37 °C overnight. The next day, the inoculated LB were added to 1 liter of LB and grown at 37 °C to A600 = 0.6 before induction with 1 mm isopropyl-1-thio-β-d-galactopyranoside. The bacteria were next grown for 4 h at room temperature, harvested by centrifugation (3,000 × g, 15 min), and washed with cold PBS. The bacterial pellet was frozen overnight, slowly thawed and lysed by PBS supplemented with protease inhibitor cocktails and 1 mm phenylmethylsulfonyl fluoride. The bacterial DNA was sheared by sonication, and the lysate was cleared by centrifugation (30,000 × g, 30 min). The supernatant containing active O-GlcNAcase was precipitated with ammonium sulfate (45%), stirred for 1 h at 4 °C, and centrifuged at 30,000 × g for 30 min. The bacterial pellet was resuspended in nickel-nitrilotriacetic acid column equilibration buffer (20 mm Tris, pH 8.0, 0.5 m NaCl) with protease inhibitor cocktails and 1 mm phenylmethylsulfonyl fluoride. The resuspension was passed through a nickel-nitrilotriacetic acid column (Qiagen). The column was washed with 5 column volumes of equilibration buffer, 5 column volumes of wash buffer 1 (5 mm imidazole, pH 8.0, 20 mm Tris, pH 8.0, 0.5 m NaCl), and 5 column volumes of wash buffer 2 (30 mm imidazole, pH 8.0, 20 mm Tris, pH 8.0, 0.5 m NaCl). Recombinant O-GlcNAcase was eluted with 2 column volumes of elution buffer (500 mm imidazole, pH 8.0, 20 mm Tris, pH 8.0, 0.5 m NaCl). Active O-GlcNAcase was dialyzed in PBS at 4 °C overnight, concentrated using Centricon concentrator (Millipore), and stored in 40% glycerol. Recombinant O-GlcNAcase D413A mutant was also purified using the same protocol.
O-GlcNAcase Activity Assay—O-GlcNAcase activity assays were performed as previously described (18). O-GlcNAcase activity was measured directly using the synthetic substrate pNP-GlcNAc (Sigma).
Caspase-3 in Vitro Cleavage Assay—Caspase-3 cleavage assays were performed as previously described with some modifications (43). Briefly, wild-type O-GlcNAcase or O-GlcNAcase D413A mutant was incubated with recombinant caspase-3 in caspase-3 reaction buffer (50 mm Hepes, pH 7.4, 10 mm dithiothreitol, 1% sucrose, 0.1% CHAPS) for 2.5 h at 37 °C. The reaction was stopped by adding equal volume of 2× Laemmli buffer. Cleavage products were resolved onto a 7.5% SDS-PAGE and either stained with Coomassie Brilliant Blue G-250 or transferred to a nitrocellulose membrane (Bio-Rad) and immunoblotted with anti-O-GlcNAcase antibody.
Cleavage Site Mapping—Cleavage site mapping by Edman sequencing was performed as previously described (43). Briefly, 120 μg of purified recombinant pET32 O-GlcNAcase was incubated with 5,000 units of purified caspase-3 in caspase-3 reaction buffer for 2.5 h at 37 °C. Cleavage products were separated on a 7.5% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was stained with Coomassie Brilliant Blue R-250 for 1 min and destained in 50% methanol. Segments of polyvinylidene difluoride membrane containing O-GlcNAcase fragments were subjected to automated sequencing on an Applied Biosystems Procise® 484 cLC protein sequencing system according to the manufacturer's instructions. Note that one unit of caspase-3 was equal to 1 pmol/min at 30 °C using Ac-DEVD-pNA as substrate.
RESULTS
Cleavage of Recombinant O-GlcNAcase by Caspase-3 in Vitro—Our previous report showed that O-GlcNAcase could be cleaved by caspase-3 in vitro (26). To confirm that recombinant O-GlcNAcase was indeed a caspase-3 substrate, we cloned human O-GlcNAcase cDNA into pET32 vector, purified, and performed in vitro cleavage assay with recombinant caspase-3. Incubation of recombinant O-GlcNAcase with caspase-3 resulted in cleavage of O-GlcNAcase that yielded fragments distinguishable by gel electrophoresis from that found in the untreated O-GlcNAcase in a dose-dependent manner (Fig. 1A). We found that O-GlcNAcase was cleaved by caspase-3 into four different fragments with molecular masses of 150, 72, 70, and 60 kDa. This cleavage could be blocked by the caspase-3 inhibitor Ac-DEVD-CHO (Fig. 1B). In agreement with our previous report, cleavage of O-GlcNAcase by caspase-3 did not result in any changes in O-GlcNAcase enzyme activity (Fig. 1C).
FIGURE 1.
Cleavage of recombinant O-GlcNAcase by caspase-3 in vitro. A, 5 μg of human recombinant O-GlcNAcase was incubated with indicated amounts of recombinant caspase-3 in caspase-3 assay buffer at 37 °C for 2.5 h. The reaction was stopped by adding Laemmli buffer for Coomassie G-250 staining and Western analysis or 100 μm of caspase-3 inhibitor Ac-DEVD-CHO for O-GlcNAcase activity assay. Cleavage mixtures from in vitro caspase-3 cleavage assay were subjected to 7.5% SDS-PAGE followed by Coomassie G-250 staining. B, recombinant O-GlcNAcase was incubated with 200 units of caspase-3 in the absence or presence of various concentrations (as indicated) of peptide aldehyde inhibitor Ac-DEVD-CHO and subjected to 7.5% SDS-PAGE followed by immunoblotting (IB) against O-GlcNAcase antibody. C, cleavage mixtures from caspase-3 cleavage assay using various amounts of caspase-3 as indicated was analyzed for O-GlcNAcase activity in triplicate.
O-GlcNAcase Cleavage Occurs during Apoptosis in Vivo—Because O-GlcNAcase is a caspase-3 substrate in vitro, we next examined whether caspase-3 might also be responsible for cleavage of O-GlcNAcase in vivo. Apoptosis was induced in Jurkat cells using anti-Fas CH11 Ab, because Fas-mediated apoptosis in Jurkat cells is a well characterized model for apoptosis (44, 45). As seen by immunoblotting with the O-GlcNAcase-specific polyclonal antibody, O-GlcNAcase cleavage is observed after 2 h of apoptotic induction, suggesting that O-GlcNAcase was cleaved by an apoptotic enzyme in vivo (Fig. 2A). In addition, O-GlcNAcase cleavage occurred at the same time as that of PARP, which is a well known substrate of caspase-3 (40). O-GlcNAc levels on proteins in Jurkat cells were also altered during apoptosis, as seen by immunoblotting with the O-GlcNAc-specific monoclonal antibody, CTD 110.6 (Fig. 2B). This suggests that alterations in O-GlcNAc-modified proteins occurred specifically in response to Fas-mediated apoptosis. In addition, the patterns of changes in O-GlcNAc levels appeared to be specific depending on the proteins, suggesting that some proteins are more susceptible to decreased or increased O-GlcNAc levels, and these changes may be associated with their functions during apoptosis. Even though we could not rule out the possibility that changes in O-GlcNAc levels are not due to changes in protein levels upon apoptosis, we did not detect apparent changes of protein levels from G-250 staining of lysate from cells undergoing apoptotic compared with those from uninduced cells (supplemental Fig. S1). The addition of PUGNAc to cells prior to apoptosis induction also increased O-GlcNAc levels on proteins that showed decreased O-GlcNAc during apoptosis, suggesting that changes in O-GlcNAc levels were not solely due to protein degradation (supplemental Fig. S1).
FIGURE 2.
O-GlcNAcase is cleaved in cells undergoing apoptosis. A, Jurkat cells were treated with 100 ng/ml of either anti-mouse IgM (control) or anti-Fas CH11 mAb for different time periods. The cell lysates were subjected to 7.5% SDS-PAGE followed by Western blot analysis for O-GlcNAcase. Immunoblotting (IB) with PARP and actin antibodies were used as a control for apoptosis and loading respectively. B, O-GlcNAc levels in Jurkat cells undergoing Fas-mediated apoptosis. C, Jurkat cells were treated with 150 μm H2O2 or PBS (as control) for 8 h. The cell lysates were subjected to Western blot analysis with C-terminally specific O-GlcNAcase, PARP, and actin antibodies.
In addition to Jurkat cells, we also examined O-GlcNAcase cleavage during apoptosis in other cell lines. We found that O-GlcNAcase cleavage also occurs in HeLa cells during Fas-mediated apoptosis, indicating that O-GlcNAcase is a substrate of a protease that is activated in the Fas signaling pathway during apoptosis (data not shown). Next, we examined whether other types of apoptosis could induce O-GlcNAcase cleavage. We induced apoptosis in Jurkat cells using hydrogen peroxide, which works through the mitochondrial pathway instead of the receptor pathway (46). We found that O-GlcNAcase was also cleaved during hydrogen peroxide-induced apoptosis (Fig. 2C). These findings suggested that O-GlcNAcase is a substrate of a common protease that is activated during apoptosis in both apoptotic pathways.
O-GlcNAcase Is Cleaved by Caspase-3 during Fas-mediated Apoptosis in Jurkat Cells—To determine which protease is responsible for O-GlcNAcase cleavage during apoptosis in vivo, apoptosis was induced in Jurkat cells by anti-Fas CH11 Ab in the presence or absence of caspase inhibitors. We found that O-GlcNAcase cleavage during Fas-mediated apoptosis could be blocked by Z-VAD-fmk, a peptide inhibitor of caspases, and Z-DEVD-fmk, a peptide inhibitor of caspase-3 and caspase-3 like activities (Fig. 3A). These results indicated that caspase-3 or a protease with caspase-3 like activity is responsible for cleavage of O-GlcNAcase during Fas-mediated apoptosis in vivo.
FIGURE 3.
O-GlcNAcase is cleaved by caspase-3 during Fas-mediated apoptosis, and caspase-3 cleavage during apoptosis does not affect O-GlcNAcase enzymatic activity. A, Jurkat cells were treated with 100 ng/ml of either anti-mouse IgM (control) or anti-Fas CH11 mAb in the presence of dimethyl sulfoxide (DMSO, control) or 50 μm Z-VAD-fmk or 100 μm Z-DEVD-fmk for 6 h, harvested, and subjected to SDS-PAGE, and Western blot analysis for O-GlcNAcase cleavage and apoptosis using O-GlcNAcase, PARP, and actin antibodies. B, 20 μg of lysate from each sample were analyzed for O-GlcNAcase activity in triplicate.
Enzymatic Activity of O-GlcNAcase Is Not altered after Caspase-3 Cleavage—We next investigated how caspase-3 cleavage affected the enzymatic activity of O-GlcNAcase during apoptosis. Cell lysate from apoptotic Jurkat cells was used in the O-GlcNAcase activity assay using pNP-GlcNAc as a substrate. We found that even though O-GlcNAcase was almost completely cleaved in Jurkat cells during apoptosis (Fig. 3A), the activity of O-GlcNAcase remained the same (Fig. 3B). This was consistent with what was previously observed in this study, that nearly complete cleavage of recombinant O-GlcNAcase by caspase-3 in vitro had no effect on the activity of O-GlcNAcase.
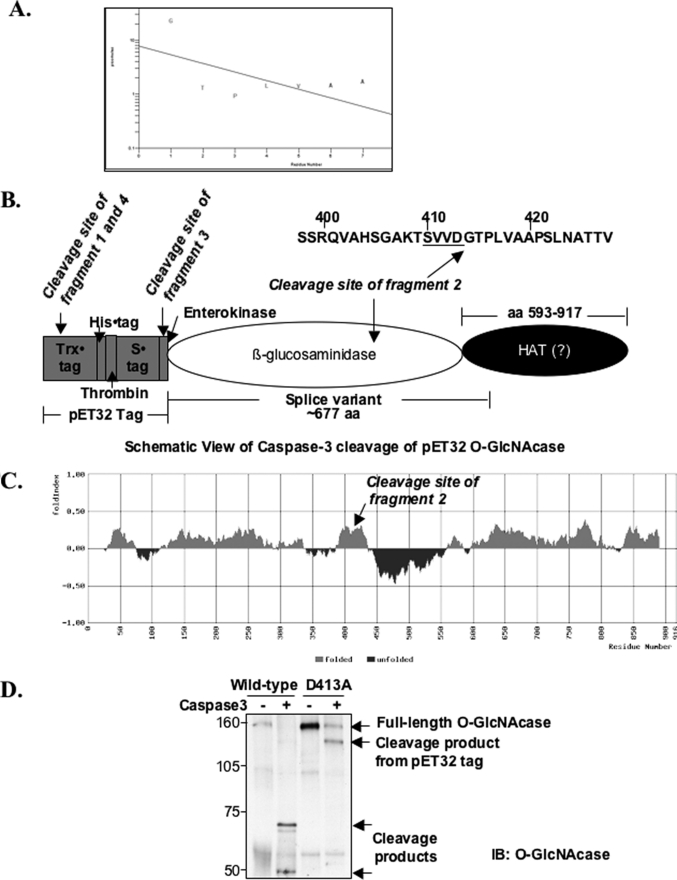
O-GlcNAcase Cleavage by Caspase-3 Occurs at a Noncanonical Site—Because O-GlcNAcase was shown to be a substrate of caspase-3, both in vitro and in vivo, we next determined the caspase-3 cleavage site of O-GlcNAcase. To map the cleavage site of O-GlcNAcase by caspase-3, recombinant O-GlcNAcase was incubated with recombinant caspase-3, and the proteolytic products were analyzed by Edman sequencing. Four cleavage products were chosen for sequencing (Fig. 1A). Amino acid sequencing of the cleavage products after transfer to a polyvinylidene difluoride membrane yielded the N-terminal sequences of GTPLVAA (Fig. 4, A and B). These results identified the caspase-3 cleavage site in O-GlcNAcase to Asp-413 of the tetrapeptide sequence SVVD, whereas other cleavage products resulted from cleavage within the tag from the pET32 vector. The cleavage site in O-GlcNAcase is not a canonical caspase-3 cleavage DXXD consensus and has not been shown to be a caspase-3 cleavage site (47, 48). Interestingly, using the FoldIndex program, which predicts whether a protein is disordered according to the average residue hydrophobicity and the net charge of the protein sequence (49), the caspase-3 cleavage site on O-GlcNAcase lies in between the folded N-terminal region and the intrinsically unfolded region that spans ∼200 amino acids and is connected to the folded C-terminal domain that has been shown to possess histone acetyltransferase activity (Fig. 4C) (27).
FIGURE 4.
Recombinant O-GlcNAcase is cleaved by caspase-3 after Asp-413 of the tetrapeptide sequence SVVD in human O-GlcNAcase amino acid sequence. 120 μg of recombinant O-GlcNAcase was incubated with 2500 units of caspase-3 at 37 °C for 2.5 h. The cleavage mixtures were subjected to 7.5% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, which was later stained with Coomassie R-250. Four bands that appeared in the cleavage mixtures but not in the untreated O-GlcNAcase were cut out and sequenced in an automated amino acid sequencer (see Fig. 1A for the four fragments used in this analysis). A, repetitive yield from N-terminal sequencing indicating the first seven amino acids in the N-terminal sequence of fragment 2. B, schematic view of caspase-3 cleavage of recombinant O-GlcNAcase, where the splice variant is also shown (25, 54). Trx·tag, thioredoxin tag. C, diagram showing the predicted folded and unfolded regions of human O-GlcNAcase using the FoldIndex program (49). D, wild-type O-GlcNAcase and D413A mutant were subjected to an in vitro caspase-3 cleavage assay. Cleavage products were analyzed by immunoblotting (IB) with O-GlcNAcase antibody.
To confirm that Asp-413 is indeed the caspase-3 cleavage site in O-GlcNAcase in vitro, we introduced a point mutation into O-GlcNAcase. We constructed a clone encoding full-length O-GlcNAcase with the single amino acid substitution D413A. Both the wild-type and the mutant O-GlcNAcase were subjected to caspase-3 cleavage assay. We found that in vitro cleavage of O-GlcNAcase by caspase-3 was completely abrogated in the alanine-containing mutant protein but not in the wild-type O-GlcNAcase, indicating that our identified caspase-3 cleavage site on O-GlcNAcase is correct (Fig. 4D). Antibodies specific to the N and C terminus of O-GlcNAcase according to this mapped cleavage site were also generated and tested (supplemental Fig. S2).
The identified site of caspase-3 cleavage in O-GlcNAcase suggests that upon caspase-3 cleavage O-GlcNAcase separates into two domains, the N-terminal domain, which possesses hexosaminidase activity, and the C-terminal domain, which has been shown to have histone acetyltransferase activity (27, 28). However, despite publications to the contrary, after many repeated attempts, we were unable to show that O-GlcNAcase possesses histone acetyltransferase activity using pET32 O-GlcNAcase plasmid generated by our lab (supplemental Fig. S3).
The Point Mutation D413A Abrogates Cleavage of O-GlcNAcase during Apoptosis in Vivo—To confirm that the caspase-3 cleavage site of O-GlcNAcase in vivo was the same as the mapped site in vitro, we generated and transfected an O-GlcNAcase D413A mutant into HeLa cells followed by apoptotic induction using anti-Fas CH11 Ab. Here, we found that the mutated alanine residue indeed prevented the mutant O-GlcNAcase from being cleaved during apoptosis (Fig. 5). In addition, the mutant itself was enzymatically active, and its activity was not altered after apoptosis (supplemental Fig. S4).
FIGURE 5.
The point mutation D413A abrogates cleavage of O-GlcNAcase during apoptosis in vivo. HeLa cells transfected with empty vector, wild-type O-GlcNAcase, or point-mutated D413A O-GlcNAcase were treated with 1 μg/ml of either anti-mouse IgM or anti-Fas mAb for 4 h. The cell lysates were subjected to SDS-PAGE and Western blot analysis for O-GlcNAcase. IB, immunoblotting.
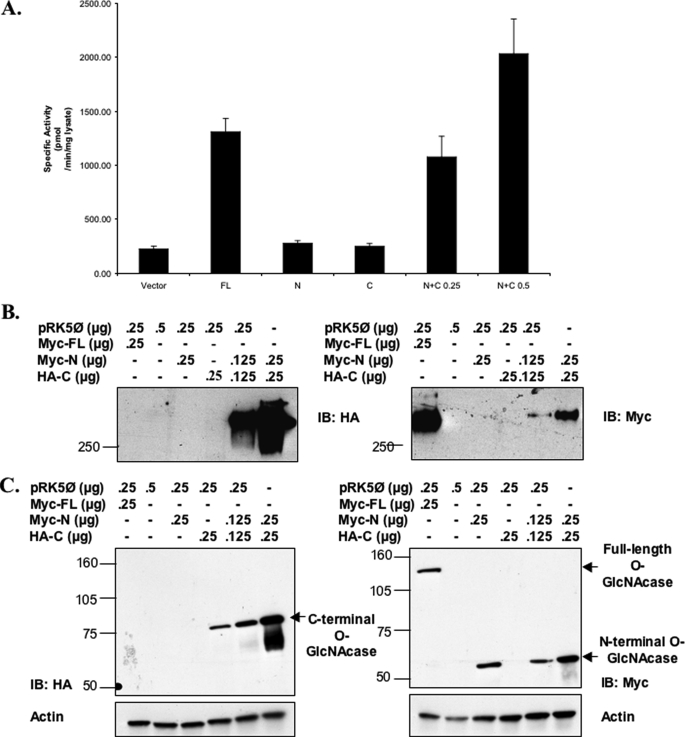
The N-terminal and C-terminal Fragments of O-GlcNAcase Remain Associated after Caspase-3 Cleavage—To gain a further insight into how O-GlcNAcase activity could be maintained even after being cleaved by caspase-3 during apoptosis, we generated truncated O-GlcNAcase mutants according to the mapped caspase-3 cleavage site that contained either the N terminus of O-GlcNAcase (amino acids 1–413) or the C terminus of O-GlcNAcase (amino acids 414–916). Each mutant was separately transfected into HeLa cells, and the O-GlcNAcase activity was measured and compared with that of cells transfected with the full-length O-GlcNAcase. Both mutants when expressed alone, however, did not show significant O-GlcNAcase activity compared with the full-length O-GlcNAcase, suggesting that both truncated O-GlcNAcase mutants lacked O-GlcNAcase enzymatic activity (Fig. 6A). Studies with cycloheximide showed that the lack of activity of the N- and C-truncated mutants were not due to protein degradation (data not shown). However, because O-GlcNAcase activity remained the same after the enzyme was cleaved, it was possible that both fragments of O-GlcNAcase might have remained associated after caspase-3 cleavage to maintain O-GlcNAcase enzymatic activity. To explore this possibility, we co-transfected the N-terminal O-GlcNAcase construct with a construct containing the C terminus of O-GlcNAcase. When both O-GlcNAcase fragments were co-expressed, O-GlcNAcase activity comparable with that of the full-length enzyme was detected, suggesting that O-GlcNAcase activity can be restored when both fragments of the enzyme are present, even when they are expressed as separate polypeptides (Fig. 6A and supplemental Fig. S5). According to this result, it appears likely that the maintained activity of O-GlcNAcase after caspase-3 cleavage during apoptosis is due to reassociation or retained association of the two fragments of the enzyme. To investigate the association between the N-terminal and C-terminal fragments of O-GlcNAcase, we performed Western blot analyses on lysates from cells expressing both fragments or either fragment of O-GlcNAcase alone, compared with that from cells expressing the full-length O-GlcNAcase in both native and denaturing conditions (Fig. 6, B and C). A complex with the molecular mass greater than 250 kDa was observed when both fragments of O-GlcNAcase were co-expressed and when the full-length O-GlcNAcase was expressed alone. Neither of the truncated O-GlcNAcase was detected from immunoblotting of a native gel but could be detected when eluted from the G-250-stained gel and ran in a denaturing condition (supplemental Fig. S6). To confirm that the N-terminal fragment of O-GlcNAcase could associate with the C-terminal fragment, immunoprecipitation was performed (Fig. 7) HeLa cells were transfected, and the cell lysates were immunoprecipitated with antibodies specific to the C-terminal O-GlcNAcase and analyzed by Western blotting. The N-terminal fragment was detected in the immunoprecipitates when both fragments of O-GlcNAcase were co-expressed in HeLa cells, suggesting that the N-terminal fragment indeed was associated with the C-terminal fragment of O-GlcNAcase. Furthermore, in HeLa cells in which apoptosis had been induced, more N terminus was found associated compared with control. Thus, the N-terminal portion of O-GlcNAcase associates with its C-terminal portion during apoptosis in vivo.
FIGURE 6.
Both the N and C termini of O-GlcNAcase are required for O-GlcNAcase activity. HeLa cells were transfected with DNA from empty pRK5 vector, full-length (FL) O-GlcNAcase, N-terminal O-GlcNAcase (N, amino acids 1–413), C-terminal O-GlcNAcase (C, amino acids 414–916), or both N- and C-terminal O-GlcNAcase plasmids (N+C). pRK5 vector, full-length O-GlcNAcase, and the N-terminal O-GlcNAcase plasmids contained c-Myc tag, whereas the C-terminal O-GlcNAcase plasmids contained the HA tag. The cells were harvested after 1 day of transfection, extracted, and analyzed for O-GlcNAcase expression and O-GlcNAcase activity. A, O-GlcNAcase activity from transfected HeLa cells. Plasmids and amount of DNA used (for co-transfection of O-GlcNAcase N and C termini, in μg) are indicated on the x axis. B and C, cell extracts were subjected to either native PAGE (B) or SDS-PAGE (C) and Western blot analysis for O-GlcNAcase using anti-HA and anti-c-Myc antibodies (left and right panels, respectively). Actin detection was used as loading controls. The plasmids and amounts of DNA used were as indicated. IB, immunoblotting.
FIGURE 7.
The N and C termini of O-GlcNAcase remain associated after caspase-3 cleavage. After a 1-day transfection, HeLa cells were treated with 1 μg/ml of either anti-mouse IgM (control) or anti-Fas CH11 mAb for 4 h. The cell extracts were subjected to immunoprecipitation using antibodies specific to the C terminus of O-GlcNAcase. Immunoprecipitated products were subjected to Western blot analysis using antibodies specific to the C-terminal O-GlcNAcase (upper panel) and the N-terminal O-GlcNAcase (lower panel). 1°, control immunoprecipitation of anti-C-terminal O-GlcNAcase antibody in blank extraction buffer. IgY, control immunoprecipitation using anti-chicken IgY antibody instead of anti-C-terminal O-GlcNAcase antibody. FL, full length. N+C, N and C termini. IB, immunoblotting.
DISCUSSION
In this present study, we report that O-GlcNAcase is cleaved by caspase-3 in cells undergoing apoptosis. We identify the caspase-3 cleavage site in O-GlcNAcase and demonstrate that after caspase-3 cleavage during apoptosis, both fragments of O-GlcNAcase remain associated or reassociate to maintain O-GlcNAcase enzymatic activity.
Our previous report has shown that O-GlcNAcase is an in vitro substrate of caspase-3, which is a key enzyme in apoptotic execution (26). Here, we show that O-GlcNAcase is cleaved into two fragments in cells undergoing Fas-mediated apoptosis, indicating that O-GlcNAcase is cleaved by a protease that is activated by apoptotic signals in vivo. These cleavage products appear at the same time as the cleavage of PARP, which is a known caspase-3 substrate (40). This indicates that O-GlcNAcase cleavage is an apoptotic event that occurs as early as PARP cleavage. The fact that O-GlcNAcase is a caspase-3 substrate is confirmed by O-GlcNAcase cleavage was abolished during Fas-mediated apoptosis in the presence of caspase-3 inhibitors, and in vitro cleavage of O-GlcNAcase by a recombinant caspase-3 enzyme yielded cleavage products with the same sizes as those in Jurkat and HeLa cells. From these results, we show that O-GlcNAcase is also a caspase-3 substrate in vivo and that O-GlcNAcase cleavage is another hallmark of apoptosis.
To better understand how cleavage by caspase-3 regulated O-GlcNAcase, we mapped the caspase-3 cleavage site in O-GlcNAcase by Edman sequencing using cleavage products derived in vitro. Interestingly, O-GlcNAcase is cleaved by caspase-3 at a noncanonical site, SVVD, because it has been well established that the preferred substrate recognition site of caspase-3 is DXXD (47, 48). An SVVD tetrapeptide is found in the BIR2 linker of an inhibitor of apoptosis protein DIAP-2. This tetrapeptide sequence is thought to be a caspase-3-binding site but not optimal for caspase-3 cleavage (50). Our results, however, indicated otherwise, because caspase-3 fails to cleave an O-GlcNAcase mutant where the aspartate residue in the SVVD sequence is replaced with alanine. In addition, another noncanonical caspase-3 cleavage site has been shown by the work of Kipp et al. (43), in which scaffold attachment factor A is cleaved by caspase-3 after an aspartate in the SALD sequence. It is suggested by Kipp et al. (43) that the requirement for aspartate in the P-4 position is not absolute because cleavage site recognition in native proteins may be more flexible, because of protein folding and cleavage site accessibility, as compared with peptide substrates.
Using pNP-GlcNAc (in the presence of large amount of GalNAc) as an in vitro substrate, we do not detect significant changes in O-GlcNAcase activity from either cleaved recombinant O-GlcNAcase or lysate from cells undergoing apoptosis. This is not surprising because some other caspase-3 substrates maintain complete activity upon cleavage, such as protein kinase Cδ and the phosphatase calcineurin (51, 52). We also observe by Western blot analysis that O-GlcNAc levels on proteins and/or O-GlcNAc-modified proteins are altered during apoptosis. Changes in O-GlcNAc levels are not global but instead seem to be specific to certain proteins, suggesting that there might be some changes in substrate recognition of either O-GlcNAcase or OGT.
Because O-GlcNAcase retains its enzymatic activity after caspase-3 cleavage, we generated truncated O-GlcNAcase mutants containing either the N-terminal or the C-terminal O-GlcNAcase. The N terminus of O-GlcNAcase has been shown to contain the key catalytic residue of O-GlcNAcase (28). Both mutants, when transfected into HeLa cells, however, failed to show significant O-GlcNAcase activity, indicating that the observed O-GlcNAcase activity after caspase-3 cleavage was not a result of the separation of both fragments of O-GlcNAcase. Cetinbas et al. (28) have previously reported that even though the N terminus possesses the N-acetylglucosaminidase activity, the activity of a recombinant O-GlcNAcase mutant containing only the N-terminal O-GlcNAcase is ∼1000-fold lower than that of the full-length O-GlcNAcase. Furthermore, a splice variant of O-GlcNAcase that lacks the C-terminal one-third of the full-length protein did not have any enzymatic activity (25, 26). This finding indicates that the C-terminal O-GlcNAcase is required for O-GlcNAcase to be fully active and may be important for catalytic function. Interestingly, when both halves of O-GlcNAcase were co-expressed in HeLa cells, the O-GlcNAcase activity was restored, suggesting that the O-GlcNAcase activity observed after caspase-3 cleavage might be a result from retained association or reassociation of the N- and C-terminal O-GlcNAcase after cleavage. This result was supported by Western analysis, in which a complex of O-GlcNAcase from cells expressing both fragments of O-GlcNAcase was detected at the molecular mass greater than 250 kDa by both antibodies specific to the tags of transfected N- and C-terminal O-GlcNAcase. Immunoprecipitation from cells expressing either the full-length O-GlcNAcase or both caspase-3 cleavage fragments of the enzyme in the presence/absence of Fas-mediated apoptosis also confirmed that the N and C termini of O-GlcNAcase interact with each other.
Most caspase-3 substrates when cleaved become unregulated or inactivated because caspase-3 targets separation of the regulatory and catalytic domain of its substrates (37). It is unclear why O-GlcNAcase is cleaved into two fragments by caspase-3 during apoptosis and then retains association or reunites with each other. We have attempted to determine the effects of O-GlcNAcase cleavage during apoptosis by introducing the uncleavable form of O-GlcNAcase (the D413A O-GlcNAcase mutant) to HeLa cells and examining apoptotic phenotypes such as caspase-3 activation, cleavage of PARP, and release of histones into the cytosol during Fas-mediated apoptosis. However, we could not detect any significant changes in apoptotic phenotypes compared with cells overexpressing the wild-type O-GlcNAcase (data not shown). We speculate that the process of recombining O-GlcNAcase fragments after caspase-3 cleavage occurs to alter substrate specificity of O-GlcNAcase, perhaps by affecting targeting proteins. Although O-phosphate removal can be regulated by several phosphatases, O-GlcNAc removal is regulated by only O-GlcNAcase catalytic subunit (2, 24). Separating and recombining both cleaved fragments is a possible mechanism for the enzyme to reconstruct its substrate recognition site or altering substrate targeting. This is a possibility because, using the FoldIndex program to predict disorder of O-GlcNAcase, the caspase-3 cleavage site on O-GlcNAcase appears to flank between the folded N-terminal domain and the intrinsically disordered region that is connected to the folded C-terminal domain (Fig. 4C). It has been reported that, in many proteins, disordered regions are involved in molecular recognition (53). Also, the disordered region may act as a spacer or linker that provides flexibility in substrate binding to the N- and C-terminal domains (53). Cleaving the two domains by caspase-3 may allow the complex to become more tightened and may alter its substrate recognition. Further studies on proteins that interact with O-GlcNAcase during apoptosis and how the interaction affects behaviors of those proteins will help elucidate the role of O-GlcNAcase in apoptosis.
Supplementary Material
Acknowledgments
We thank Zihao Wang for technical assistance with mass spectrometry and Dr. Chad Slawson for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HD13563 and CA42486 (to G. W. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
Footnotes
The abbreviations used are: O-GlcNAc, O-linkedβ-N-acetylglucosamine; O-GlcNAcase, O-linked β-N-acetylglucosaminidase; OGT, N-acetylglucosaminyl-transferase; PARP, poly(ADP)-ribose polymerase; HA, hemagglutinin; PBS, phosphate-buffered saline; Ab, antibody; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone.
References
- 1.Torres, C. R., and Hart, G. W. (1984) J. Biol. Chem. 2593308 –3317 [PubMed] [Google Scholar]
- 2.Wells, L., Vosseller, K., and Hart, G. W. (2001) Science 2912376 –2378 [DOI] [PubMed] [Google Scholar]
- 3.Hart, G. W., Housley, M. P., and Slawson, C. (2007) Nature 4461017 –1022 [DOI] [PubMed] [Google Scholar]
- 4.Whelan, S. A., and Hart, G. W. (2003) Circ. Res. 931047 –1058 [DOI] [PubMed] [Google Scholar]
- 5.Zachara, N. E., and Hart, G. W. (2002) Chem. Rev. 102431 –438 [DOI] [PubMed] [Google Scholar]
- 6.Kamemura, K., and Hart, G. W. (2003) Prog. Nucleic Acids Res. Mol. Biol. 73107 –136 [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre, T., Alonso, C., Mahboub, S., Dupire, M. J., Zanetta, J. P., Caillet-Boudin, M. L., and Michalski, J. C. (1999) Biochim. Biophys. Acta 147271 –81 [DOI] [PubMed] [Google Scholar]
- 8.Comer, F. I., and Hart, G. W. (2001) Biochemistry 407845 –7852 [DOI] [PubMed] [Google Scholar]
- 9.Chou, T. Y., Hart, G. W., and Dang, C. V. (1995) J. Biol. Chem. 27018961 –18965 [DOI] [PubMed] [Google Scholar]
- 10.Griffith, L. S., and Schmitz, B. (1999) Eur. J. Biochem. 262824 –831 [DOI] [PubMed] [Google Scholar]
- 11.Zachara, N. E., and Hart, G. W. (2004) Trends Cell Biol. 14218 –221 [DOI] [PubMed] [Google Scholar]
- 12.Zachara, N. E., O'Donnell, N., Cheung, W. D., Mercer, J. J., Marth, J. D., and Hart, G. W. (2004) J. Biol. Chem. 27930133 –30142 [DOI] [PubMed] [Google Scholar]
- 13.Slawson, C., Zachara, N. E., Vosseller, K., Cheung, W. D., Lane, M. D., and Hart, G. W. (2005) J. Biol. Chem. 28032944 –32956 [DOI] [PubMed] [Google Scholar]
- 14.Vosseller, K., Wells, L., Lane, M. D., and Hart, G. W. (2002) Proc. Natl. Acad. Sci. U. S. A. 995313 –5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, F., Su, K., Yang, X., Bowe, D. B., Paterson, A. J., and Kudlow, J. E. (2003) Cell 115715 –725 [DOI] [PubMed] [Google Scholar]
- 16.Iyer, S. P., and Hart, G. W. (2003) J. Biol. Chem. 27824608 –24616 [DOI] [PubMed] [Google Scholar]
- 17.Dong, D. L., and Hart, G. W. (1994) J. Biol. Chem. 26919321 –19330 [PubMed] [Google Scholar]
- 18.Gao, Y., Wells, L., Comer, F. I., Parker, G. J., and Hart, G. W. (2001) J. Biol. Chem. 2769838 –9845 [DOI] [PubMed] [Google Scholar]
- 19.Kreppel, L. K., and Hart, G. W. (1999) J. Biol. Chem. 27432015 –32022 [DOI] [PubMed] [Google Scholar]
- 20.Lubas, W. A., and Hanover, J. A. (2000) J. Biol. Chem. 27510983 –10988 [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell, N., Zachara, N. E., Hart, G. W., and Marth, J. D. (2004) Mol. Cell. Biol. 241680 –1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells, L., Kreppel, L. K., Comer, F. I., Wadzinski, B. E., and Hart, G. W. (2004) J. Biol. Chem. 27938466 –38470 [DOI] [PubMed] [Google Scholar]
- 23.Comer, F. I., and Hart, G. W. (2000) J. Biol. Chem. 27529179 –29182 [DOI] [PubMed] [Google Scholar]
- 24.Cohen, P. (2001) Eur. J. Biochem. 2685001 –5010 [DOI] [PubMed] [Google Scholar]
- 25.Comtesse, N., Maldener, E., and Meese, E. (2001) Biochem. Biophys. Res. Commun. 283634 –640 [DOI] [PubMed] [Google Scholar]
- 26.Wells, L., Gao, Y., Mahoney, J. A., Vosseller, K., Chen, C., Rosen, A., and Hart, G. W. (2002) J. Biol. Chem. 2771755 –1761 [DOI] [PubMed] [Google Scholar]
- 27.Toleman, C., Paterson, A. J., Whisenhunt, T. R., and Kudlow, J. E. (2004) J. Biol. Chem. 27953665 –53673 [DOI] [PubMed] [Google Scholar]
- 28.Cetinbas, N., Macauley, M. S., Stubbs, K. A., Drapala, R., and Vocadlo, D. J. (2006) Biochemistry 453835 –3844 [DOI] [PubMed] [Google Scholar]
- 29.Forsythe, M. E., Love, D. C., Lazarus, B. D., Kim, E. J., Prinz, W. A., Ashwell, G., Krause, M. W., and Hanover, J. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 10311952 –11957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulop, N., Marchase, R. B., and Chatham, J. C. (2007) Cardiovasc Res. 73288 –297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zachara, N. E., and Hart, G. W. (2004) Biochim. Biophys. Acta 167313 –28 [DOI] [PubMed] [Google Scholar]
- 32.Champattanachai, V., Marchase, R. B., and Chatham, J. C. (2007) Am. J. Physiol. 292C178 –C187 [DOI] [PubMed] [Google Scholar]
- 33.Boehmelt, G., Wakeham, A., Elia, A., Sasaki, T., Plyte, S., Potter, J., Yang, Y., Tsang, E., Ruland, J., Iscove, N. N., Dennis, J. W., and Mak, T. W. (2000) EMBO J. 195092 –5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengartner, M. O. (2000) Nature 407770 –776 [DOI] [PubMed] [Google Scholar]
- 35.Danial, N. N., and Korsmeyer, S. J. (2004) Cell 116205 –219 [DOI] [PubMed] [Google Scholar]
- 36.Kaufmann, S. H., and Hengartner, M. O. (2001) Trends Cell Biol. 11526 –534 [DOI] [PubMed] [Google Scholar]
- 37.Thornberry, N. A., and Lazebnik, Y. (1998) Science 2811312 –1316 [DOI] [PubMed] [Google Scholar]
- 38.Salvesen, G. S., and Dixit, V. M. (1997) Cell 91443 –446 [DOI] [PubMed] [Google Scholar]
- 39.Casciola-Rosen, L., Nicholson, D. W., Chong, T., Rowan, K. R., Thornberry, N. A., Miller, D. K., and Rosen, A. (1996) J. Exp. Med. 1831957 –1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholson, D. W., Ali, A., Thornberry, N. A., Vaillancourt, J. P., Ding, C. K., Gallant, M., Gareau, Y., Griffin, P. R., Labelle, M., Lazebnik, Y. A., Munday, N. A., Raju, S. M., Smulson, M. E., Yamin, T., Yu, V. L., and Miller, D. K. (1995) Nature 37637 –43 [DOI] [PubMed] [Google Scholar]
- 41.Ubeda, M., and Habener, J. F. (1997) J. Biol. Chem. 27219562 –19568 [DOI] [PubMed] [Google Scholar]
- 42.Schlegel, J., Peters, I., Orrenius, S., Miller, D. K., Thornberry, N. A., Yamin, T. T., and Nicholson, D. W. (1996) J. Biol. Chem. 2711841 –1844 [DOI] [PubMed] [Google Scholar]
- 43.Kipp, M., Schwab, B. L., Przybylski, M., Nicotera, P., and Fackelmayer, F. O. (2000) J. Biol. Chem. 2755031 –5036 [DOI] [PubMed] [Google Scholar]
- 44.Weis, M., Schlegel, J., Kass, G. E., Holmstrom, T. H., Peters, I., Eriksson, J., Orrenius, S., and Chow, S. C. (1995) Exp. Cell Res. 219699 –708 [DOI] [PubMed] [Google Scholar]
- 45.Algeciras-Schimnich, A., Shen, L., Barnhart, B. C., Murmann, A. E., Burkhardt, J. K., and Peter, M. E. (2002) Mol. Cell. Biol. 22207 –220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dumont, A., Hehner, S. P., Hofmann, T. G., Ueffing, M., Droge, W., and Schmitz, M. L. (1999) Oncogene 18747 –757 [DOI] [PubMed] [Google Scholar]
- 47.Talanian, R. V., Quinlan, C., Trautz, S., Hackett, M. C., Mankovich, J. A., Banach, D., Ghayur, T., Brady, K. D., and Wong, W. W. (1997) J. Biol. Chem. 2729677 –9682 [DOI] [PubMed] [Google Scholar]
- 48.Van Damme, P., Martens, L., Van Damme, J., Hugelier, K., Staes, A., Vandekerckhove, J., and Gevaert, K. (2005) Nat. Methods 2771 –777 [DOI] [PubMed] [Google Scholar]
- 49.Prilusky, J., Felder, C. E., Zeev-Ben-Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., Silman, I., and Sussman, J. L. (2005) Bioinformatics 213435 –3438 [DOI] [PubMed] [Google Scholar]
- 50.Silke, J., Ekert, P. G., Day, C. L., Hawkins, C. J., Baca, M., Chew, J., Pakusch, M., Verhagen, A. M., and Vaux, D. L. (2001) EMBO J. 203114 –3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb, P. R., Wang, K. Q., Scheel-Toellner, D., Pongracz, J., Salmon, M., and Lord, J. M. (2000) Apoptosis 5451 –458 [DOI] [PubMed] [Google Scholar]
- 52.Mukerjee, N., McGinnis, K. M., Gnegy, M. E., and Wang, K. K. (2001) Biochem. Biophys. Res. Commun. 2851192 –1199 [DOI] [PubMed] [Google Scholar]
- 53.Dunker, A. K., Brown, C. J., Lawson, J. D., Iakoucheva, L. M., and Obradovic, Z. (2002) Biochemistry 416573 –6582 [DOI] [PubMed] [Google Scholar]
- 54.Heckel, D., Comtesse, N., Brass, N., Blin, N., Zang, K. D., and Meese, E. (1998) Hum. Mol. Genet. 71859 –1872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.