Abstract
The majority of mutations that cause isolated growth hormone deficiency type II are the result of aberrant splicing of transcripts encoding human growth hormone. Such mutations increase skipping of exon 3 and encode a 17.5-kDa protein that acts as a dominant negative to block secretion of full-length protein produced from unaffected alleles. Previously, we identified a splicing regulatory element in exon 3 (exonic splicing enhancer 2 (ESE2)), but we had not determined the molecular mechanism by which this element prevents exon skipping. Here, we show that two members of the serine/arginine-rich (SR) protein superfamily (ASF/SF2 and SC35) act antagonistically to regulate exon 3 splicing. ASF/SF2 activates exon 3 inclusion, but SC35, acting through a region just downstream of ESE2, can block such activation. These findings explain the disease-causing mechanism of a patient mutation in ESE2 that creates a functional SC35-binding site that then acts synergistically with the downstream SC35 site to produce pathological levels of exon 3 skipping. Although the precedent for SR proteins acting as repressors is established, this is the first example of a patient mutation that creates a site through which an SR protein represses splicing.
Pre-mRNA splicing is the process of intron removal and exon joining to form mature, protein-coding transcripts. This requires both cis-acting RNA elements and trans-acting factors. The main cis-acting elements are the 5′ splice site, 3′ splice site, polypyrimidine tract, and branch point sequence, each of which are defined by short, degenerate consensus sequences in higher eukaryotes (1, 2). Recognition of these elements by trans-acting factors, including five small nuclear ribonucleoproteins, results in assembly of the catalytically active spliceosome in which the two steps of splicing occur. Because of the lack of sequence conservation, the pairing of U1 snRNA with the 5′ splice site and U2 with the branch point results in a range of splice site strengths. This allows for regulation of splice site selection in cases of alternative splicing but at the same time creates a potential fidelity problem in cases where weak splice sites need to be properly recognized. These weak splice sites often require additional sequences known as enhancer elements that are recognized by regulatory splicing factors, such as SR3 proteins, which guide the spliceosome to the correct splice sites. Splicing enhancers are typically purine-rich and can be found in both exons and introns and are accordingly named exonic (ESEs) and intronic splicing enhancers. SR proteins are a large family of conserved proteins that have N-terminal RNA recognition motifs and C-terminal domains rich in serine-arginine dipeptides (RS domains). The RNA recognition motifs tether the proteins to enhancer elements, and the RS domains assist spliceosome assembly through protein-protein interactions (3).
SR proteins define exon boundaries by two mechanisms that are not mutually exclusive. First, SR proteins enhance splicing in an RS domain-independent manner by antagonizing adjacent silencer elements (4, 5). Second, they recruit the splicing machinery to splice sites in an RS domain-dependent manner. This results in the formation of bridge complexes across exons (6). For example, ASF/SF2 and SC35 can interact with the U1–70K subunit and with a protein that binds to the branch point region (U2AF35), thus physically bridging 5′ and 3′ splice sites (7–10).
The human growth hormone gene 1 is composed of five exons and four introns. Splicing of the wild-type pre-mRNA produces five isoforms (11). The protein product produced after removal of all four introns is the full-length, 22-kDa isoform. Aberrant splicing to an in-frame cryptic splice site in exon 3, which occurs 5–10% of the time, produces a transcript lacking the first 45 nucleotides of exon 3 and encodes a 20-kDa protein. The 20-kDa isoform apparently retains full biological activity (11–13). Complete skipping of exon 3, which occurs 0.1–5% of the time, results in production of a 17.5-kDa protein that acts as a dominant negative by blocking secretion of the full-length protein (14). Trace amounts of GH1 transcripts that skip exons 3 and 4 or exons 2–4 have also been identified and produce 11.3- and 7.4-kDa proteins, respectively (15).
Isolated growth hormone deficiency type II is an autosomal dominant growth hormone deficiency where the majority of mutations affect GH1 splicing (see Fig. 1A). These mutations result in increased production of exon 3 skipped transcripts encoding the 17.5-kDa protein. Mechanistically, the easiest mutations to understand are found at the splice sites bordering exon 3 that cause almost complete exon skipping. A second class of mutations alter purine-rich sequences in and around exon 3 and result in variable levels of exon 3 skipping depending on the specific mutation (16–21). These purine-rich sequences constitute splicing enhancer elements that help define the weak splice sites flanking exon 3 (21, 22). Exon 3 contains two such enhancers (ESE1 and ESE2) with an additional enhancer found in intron 3 (intronic splicing enhancer) (16, 19–23). ESE2 is a 15-nucleotide sequence upstream of the cryptic splice site in exon 3 (E3+19–33) (20). The molecular mechanism by which ESE2 prevents exon skipping has not been determined, and elucidation of such would be expected to explain the disease causing effects of a patient mutation within ESE2 (E3+29 A→G; A1338G) (18). We hypothesized that this patient mutation, as well as other ESE2 mutations, alter splicing by disrupting SR protein-binding sites, consistent with the behavior of other enhancer elements. Here, we show that two SR proteins, ASF/SF2 and SC35, bind to ESE2. However, when we analyzed the effects of these proteins on GH1 splicing in vivo, ASF/SF2 prevented exon 3 skipping, but SC35 actually caused increased exon skipping, opposite to the effect predicted. Further, we show that the increased exon 3 skipping seen with the patient mutation is due to the creation of a functional SC35-binding site in ESE2, which, in conjunction with a second SC35-binding site just downstream, synergistically increases exon 3 skipping.
FIGURE 1.
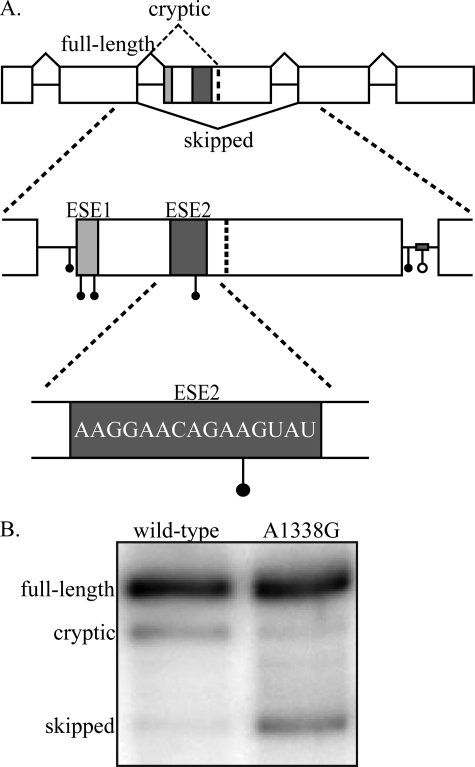
GH1 gene structure and disease causing mutations. A, the GH1 gene is composed of five exons and four introns. Inclusion of all five exons (top, solid black lines) produces a mature transcript that encodes the 22-kDa, biologically active, full-length protein product. Splicing to a cryptic splice site in exon 3 (top, dashed black line) results in production of a 20-kDa, biologically active protein. Complete skipping of exon 3 (bottom, black line) produces a 17.5-kDa protein that acts as a dominant negative to the full-length, 22-kDa protein. Splicing of primary transcripts is regulated by two exonic splicing enhancers (ESE1, light gray box; ESE2, dark gray box) and one intronic splicing enhancer in intron 3 (gray bar). Point mutations (solid circles) and a deletion mutation (open circle) identified in patients suffering from isolated growth hormone deficiency type II fall into the following regions: intron 2 3′ splice site, exon 3 5′ splice site/ESE1, intron 3 5′ splice site, and intronic splicing enhancer. One patient mutation has been identified in ESE2 (E3+29 A→G; A1338G). The accession number for the complete sequence of GH1 is NM_000515.3. B, splicing analysis of wild-type and A1338G transcripts. GH3 rat somatotroph cells were transfected with either wild-type GH1 or GH1 containing the A1338G patient mutation. Total RNA was analyzed by RT-PCR, and the resulting products were separated on denaturing, polyacrylamide gels.
EXPERIMENTAL PROCEDURES
Plasmid Construction—Wild-type GH1, A1338G, and exon 3 deletion mutants (Δ1–Δ10) were cloned into pXGH5 as previously described (20). The A1338G-Δ6 mutant was cloned using the QuikChange protocol (Stratagene) with the exon 3 Δ6 mutant as the template and the following primers: 5′-GGAACAGAGGTATTCATTCCTGC-3′ and 5′-GCAGGAATGATACCTCTGTTCC-3′. DSX-ΔE was a generous gift from Dr. Brenton R. Graveley. Annealed and phosphorylated DNA primers corresponding to wild-type (5′-AAAGGAACAGAAGTA-3′) and mutant (5′-ATAGTAATAGTAGTA-3′) ESE2 were inserted into DSX-ΔE at the BstBI site in exon 2 to generate the DSX-ESE2 and DSX-ESE2-QM constructs, respectively.
Protein Expression and Purification—Baculovirus-expressed ASF/SF2, SC35, and SRp20 were purified from infected Sf9 cells as previously described (24). Purified SR proteins from calf thymus were prepared as previously described (25) except proteins were resuspended in buffer D (20 mm Tris-HCl, pH 7.9, 100 mm KCl, 0.5 mm dithiothreitol, 0.2 mm EDTA, 5% glycerol).
In Vivo GH Splicing Analysis—GH3 cells (rat somatotrophs) were transfected with 500 ng of wild-type or mutant human growth hormone constructs in combination with the indicated amounts of pcDNA vector (empty) or vectors expressing the indicated SR proteins using TransIT LT1 (Mirus). Total RNA was isolated 48 h post-transfection using TRI reagent (Molecular Research Center), and RT-PCR was performed as described (21) with the following primers: RT, 5′-GGACAAGGCTGGTGGGCACTGG-3′; exon 2 forward, 5′-CCATCGTCTGCACCAGCTGGC-3′; and exon 5 reverse, 5′-CACAGCTGCCCTCCACAGAGC-3′.
RNA Affinity Chromatography—RNA affinity chromatography columns coupled with either ESE2 wild type (5′-AAAGGAACAGAAGUA-3′) or ESE2-QM (5′-AUAGUAAUAGUAGUA-3′) were prepared as previously described (26). Calf thymus SR proteins (70 μl) were first adjusted to 60% buffer D and then diluted with 60% buffer D to a final volume of 750 μl. Half of the calf thymus SR protein dilution was applied to each column and allowed to incubate at room temperature for 20 min. The flow-throughs were reapplied to the columns and incubated at room temperature for 10 min. The columns were then washed twice with 500 μl of 60% buffer D. Bound proteins were eluted in buffer D with increasing KCl concentrations (100–500 mm) and a final elution in 8 m urea/20 mm Tris-HCl, pH 8.0. Fractions were separated on 10% SDS gels and stained with Coomassie Blue using the Colloidal Coomassie Blue G-250 staining solutions kit (Invitrogen).
Mass Spectrometry and Data Base Analysis—Individual proteins were excised and subjected to in-gel trypsin digestion, and peptide mixtures were analyzed by matrix-assisted laser desorption time of flight and time of flight/time of flight tandem mass spectrometry using a Voyager 4700 mass spectrometer (Applied Biosystems, Framingham MA) in the Vanderbilt Proteomics Laboratory. Mass spectral data, in the form of peptide mass maps/fingerprints of the intact molecular peptide ions (M+H), as well as fragmentation data derived from individual peptide ions, were used to interrogate the Swiss-Prot and NCBI nr protein databases for statistically significant protein matches using GPS Explorer software (Applied Biosystems) running the MASCOT search engine (Matrix Science).
In Vitro Transcription and Splicing—Transcription and splicing of substrate RNAs was performed as previously described (20, 27). Briefly, the dsx constructs were linearized with MluI and transcribed with T7 RNA polymerase (Promega). For splicing reactions, 5 ng of substrate was spliced at 30 °C for 2 h in either 15 μl of HeLa nuclear extract or 10 μl of HeLa nuclear extract supplemented with the indicated SR proteins. The products were extracted and resolved on 8 m urea, 8% polyacrylamide gels and visualized by PhosphorImager analysis.
UV Cross-linking—UV cross-linking reactions were carried out in a final volume of 15 μl in the presence of 0.5 mm dithiothreitol, 2.5 mm MgCl2, 16.67 mm creatine phosphate, 100 nm radiolabeled ESE2 wild type transcript, and the indicated proteins and cold competitors. The reactions were set up and left on ice for 20 min, incubated at 30 °C for 20 min, and subjected to UV irradiation (254 nm at a distance of 6 cm) for 6 min. Then 30 μg of RNase A was added, followed by incubation at 37 °C for 30 min, after which SDS buffer was added, and the samples were loaded onto 10% SDS-PAGE gels. Cross-linking was visualized by PhosphorImager analysis.
RESULTS
The A1338G Patient Mutation in ESE2 Causes Increased Exon 3 Skipping—We previously identified a splicing regulatory element within exon 3 (ESE2) whose deletion causes exon skipping, but the mechanism of action for this sequence remained uncharacterized (20). Here, we sought to understand how splicing is activated by ESE2, an understanding that should also elucidate the mechanism underlying a previously reported isolated growth hormone deficiency type II-causing point mutation within ESE2 (E3+29 A→G; A1338G) (18). To first confirm that the A1338G mutation causes increased skipping of exon 3, we transfected a rat somatotroph cell line (GH3 cells) with either mutant or wild-type GH1 constructs and determined splicing patterns using RT-PCR. As shown, the A1338G mutation caused increased exon 3 skipping relative to wild type (Fig. 1B). As is typical of splicing enhancer sequences, ESE2 is purine-rich. Because ESE2 is known to play a role in exon 3 definition, mutations that result in increased exon skipping would logically disrupt enhancer function. However, this mutation is a transitional base substitution that maintains the purine content of ESE2, so it is not completely clear whether disruption of ESE2 is responsible for the splicing defect observed with the A1338G mutation. To understand the disease-causing mechanism of the A1338G mutation, we first needed to understand how ESE2 functions to activate splicing.
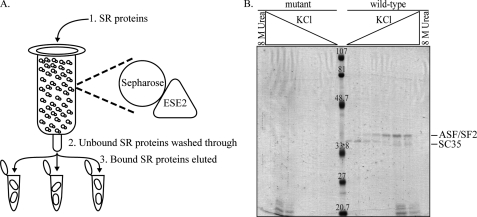
SC35 and ASF/SF2 Bind ESE2 in Vitro—Splicing enhancers typically bind one or more members of the SR protein superfamily to activate splicing. To determine which SR proteins bind to the wild-type ESE2 sequence, RNA affinity chromatography was performed using either the wild-type ESE2 sequence or ESE2 containing four point mutations that should completely block enhancer function (Fig. 2). RNA molecules were covalently linked to modified Sepharose resin, and purified calf thymus SR proteins were incubated with the columns. After washing, bound proteins were eluted with increasing salt and a final urea wash. Bound proteins were separated on SDS-PAGE gels and stained with Coomassie Blue (Fig. 2B). Two abundant proteins of ∼35 kDa bound to the wild-type RNA sequence, whereas no similar proteins bound to the mutant RNA sequence. These two bands were individually excised, analyzed by mass spectrometry, and identified as ASF/SF2 and SC35, two core SR protein family members.
FIGURE 2.
ASF/SF2 and SC35 bind to ESE2 in vitro. A, RNA affinity chromatography. RNA molecules were covalently linked to Sepharose beads, and SR proteins purified from calf thymus were passed over the column followed by extensive washing. Bound proteins were eluted with increasing salt concentrations (100, 150, 200, 250, 300, 400, 500, and 500 mm) and a final elution with 8 m urea. Elution fractions were run on SDS-PAGE gels and stained with Coomassie Blue. B, RNA affinity chromatography was performed using either wild-type ESE2 (5′-AAAGGAACAGAAGUA-3′) or mutant ESE2 (5′-AUAGUAAUAGUAGUA-3′) RNAs. Individual bands were cut out of the gel and identified by mass spectrometry as ASF/SF2 and SC35.
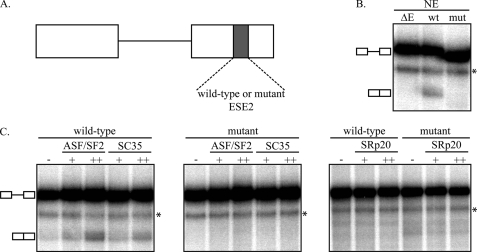
SC35 and ASF/SF2 Activate Splicing through ESE2 in a Heterologous, in Vitro Setting—The SR protein family contains eight core family members and a large number of related proteins (28). SC35 and ASF/SF2 are two of the core, conserved proteins, and their binding to ESE2 is consistent with enhancer function. To determine whether these proteins activate splicing through ESE2, we conducted in vitro splicing assays using an enhancer-dependent splicing construct derived from the Drosophila melanogaster gene doublesex (dsx). Splicing of this mini-gene requires the presence of an enhancer in the downstream exon (Fig. 3A). We created constructs derived from the dsx mini-gene in which the wild-type ESE2 sequence or the same mutant sequence used in Fig. 2 was cloned into the enhancer position. A construct lacking an enhancer sequence (ΔE) was used as a negative control. Splicing of these constructs was first tested in HeLa nuclear extract. As shown, constructs lacking an enhancer (ΔE) or containing the mutant ESE2 sequence (mut) were incapable of splicing, whereas spliced product formation was detectable when the wild-type ESE2 sequence was inserted (Fig. 3B). This shows that the wild-type ESE2 sequence functions as an enhancer in a heterologous setting and is consistent with previous work showing that disruption of ESE2 in GH1 transcripts leads to exon skipping (20). To test which SR proteins were responsible for splicing activation, we purified ASF/SF2, SC35, and an unrelated SR protein (SRp20) that did not bind to the RNA affinity column in Fig. 2. Splicing assays were then carried out in limiting amounts of HeLa nuclear extract supplemented with these SR proteins using the dsx constructs above. As shown in Fig. 3C, both ASF/SF2 and SC35 activated splicing in a dose- and enhancer-dependent manner. Mutation of ESE2 or supplementation with SRp20 did not result in detectable levels of splicing. Thus, SC35 and ASF/SF2 can functionally and specifically activate in vitro splicing through ESE2 in a heterologous setting.
FIGURE 3.
ASF/SF2 and SC35 activate in vitro splicing through ESE2 in a heterologous substrate. A, the dsx splicing reporter is a two exon, one intron substrate that requires the presence of an enhancer (gray box) in exon 2 for splicing. dsx constructs containing either wild-type or mutant ESE2 (same mutant sequence as in Fig. 2) sequences were used to analyze the ability of ASF/SF2 and SC35 to activate splicing through ESE2. B, wild-type (wt) and mutant (mut) constructs, as well as a construct lacking an enhancer (ΔE), were analyzed for splicing activity in HeLa nuclear extract. C, ASF/SF2, SC35, and SRp20 (50 or 200 ng) were added to limiting amounts of HeLa nuclear extract, and spliced product formation was determined. The asterisks denote nonspecific bands.
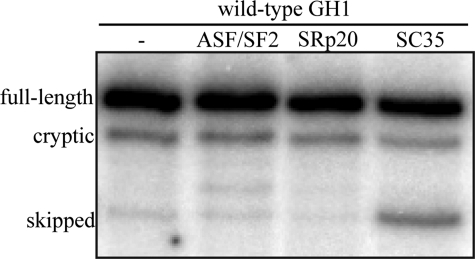
SC35 Causes Skipping of GH Exon 3 in Vivo—SR protein-binding sites are small and not highly conserved (29), so it is possible that activation of splicing in vitro and with heterologous substrates may not mimic in vivo function. As a first attempt to determine whether SC35 and ASF/SF2 activate GH1 exon 3 inclusion in vivo, we transfected GH3 cells with an expression vector containing the entire wild-type GH1 sequence either alone or in combination with SC35, ASF/SF2, or SRp20 expression vectors. With the wild-type sequence, only small amounts of exon 3 skipping should be observed, but the question was whether excess SR proteins might further reduce aberrant splicing. The cells were transfected, total RNA was isolated, and splicing patterns were analyzed by RT-PCR. As expected, the splicing pattern of GH1 was predominately full-length (80–90%) with ∼10–15% cryptic splice site usage and only barely detectable levels of exon 3 skipping (Fig. 4). This ratio remained essentially unchanged upon overexpression of ASF/SF2 and SRp20. However, we surprisingly detected significantly increased levels of exon 3 skipping upon overexpression of SC35. This was unexpected and inconsistent with activation by SC35 in the dsx setting but implies that the exact sequence context may be required to evaluate the ability of individual SR proteins to activate exon 3 inclusion. It also raises the possibility that SC35 and ASF/SF2 may antagonize one another.
FIGURE 4.
Analysis of the effects of SC35 and ASF/SF2 on GH1 splicing in vivo. GH3 cells were transiently transfected with wild-type GH1 and individual SR protein expression vectors. GH1 splicing was analyzed by RT-PCR, and the products were analyzed on denaturing, polyacrylamide gels.
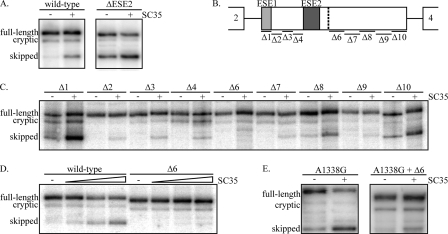
Sequence-specific Repression of Exon 3 by SC35—A simple model to explain regulation of exon 3 inclusion would be that ASF/SF2 and SC35 compete for binding to ESE2, as suggested by the RNA affinity purification experiments. However, given the differences between the in vitro and in vivo splicing assays above, we next sought to determine whether SC35-mediated exon 3 skipping requires the presence of ESE2 in the context of the full-length GH1 gene. GH3 cells were transfected with wild-type GH1 or a mutant GH1 construct containing a deletion of ESE2 (ΔESE2) in the presence or absence of co-transfection with SC35. As above, overexpression of SC35 with the wild-type construct led to increased exon 3 skipping (Fig. 5A). Interestingly, deletion of ESE2 did not abolish the ability of SC35 to promote exon skipping. The loss of ESE2 led to increased exon 3 skipping because of the loss of enhancer function, and co-transfection of SC35 further increased the level of skipping. This suggests that SC35 mediated this effect through a region other than ESE2 even though SC35 was found to bind to ESE2 by chromatography (Fig. 2B). To identify the sequence responsible for this skipping, a panel of deletions across exon 3 was used, and splicing patterns were determined in the presence and absence of SC35 (Fig. 5, B and C). Only one deletion, Δ6, did not show increased exon 3 skipping upon overexpression of SC35 even across a range of concentrations (Fig. 5, C and D). These results suggest that SC35 promotes skipping through a sequence just downstream of ESE2. Under wild-type conditions, with an intact ESE2 sequence, exon 3 is predominately included, but loss of ESE2 enables SC35 to promote skipping.
FIGURE 5.
Exon 3 deletion analysis and A1338G patient mutation analysis. A, GH3 cells were transiently transfected with either wild-type GH1 or ESE2-deleted GH1 (ΔESE2) in the presence or absence of SC35 overexpression. Spliced product formation was determined as above. B, GH1 exon 3 illustrating the deletion constructs. ESE1 is illustrated by a light gray box, and ESE2 is illustrated by a dark gray box. Note that Δ5 is the same as ΔESE2. C, GH3 cells were transiently transfected with the deletion mutants above in the presence or absence of SC35 overexpression and spliced products were analyzed as above. D, GH3 cells were transiently transfected with either wild-type or the Δ6 GH1 construct in the presence of increasing amounts of SC35 (100, 500, and 1500 ng). E, GH3 cells were transiently transfected with either the A1338G patient mutation or a GH1 construct containing both the A1338G mutation and the Δ6 mutation in the presence or absence of SC35 overexpression.
SC35-mediated Skipping and the A1338G Patient Mutation—The above results suggested a mechanism to explain the increased exon 3 skipping observed with the A1338G patient mutation (Fig. 1). As noted above, this mutation maintains the purine-rich ESE2 sequence yet blocks enhancer function. A possible explanation to account for these results is that the mutation creates an SC35-binding site within ESE2, at least as predicted by ESE Finder (30). This suggests that the mechanism of action of the A1338G mutation is to enable SC35 to mediate exon skipping through ESE2. If true, the expectation would be that the Δ6 mutation should still result in exon 3 skipping when combined with the A1338G mutation. Thus, we analyzed the effect of SC35 overexpression on exon 3 skipping in the A1338G background and when combined with the Δ6 mutation. As shown, co-transfection of SC35 resulted in increased exon 3 skipping with the A1338G mutation (Fig. 5E). However, when the A1338G and Δ6 mutations were combined, the splicing pattern resembled the wild-type pattern in the absence of excess SC35 and increased skipping in the presence of SC35 (Fig. 5E). Because no skipping was observed with the Δ6 mutation alone (Fig. 5C), this suggests that the A1338G mutation creates a site through which SC35 can mediate exon 3 skipping. Thus, the A1338G mutation and region 6 are each sufficient to produce SC35-mediated exon 3 skipping and apparently act in a synergistic manner to prevent exon 3 inclusion.
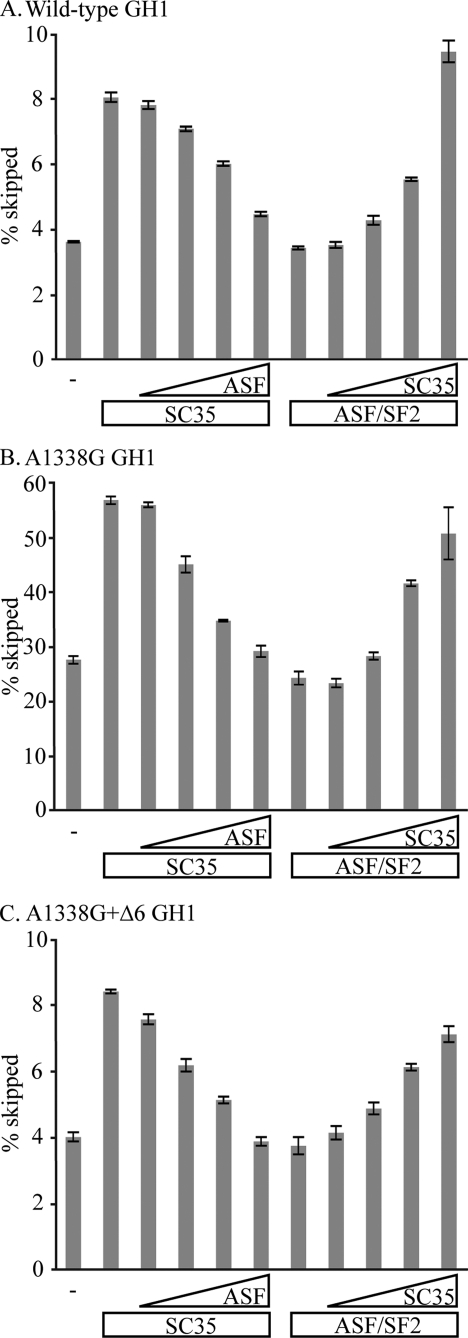
Antagonism between SC35 and ASF/SF2 in Exon 3 Splicing—In the wild-type setting, we were unable to observe activation of exon 3 splicing by ASF/SF2 mostly because the level of exon 3 skipping is too low to begin with (Fig. 4). However, if our model about how the A1338G and Δ6 mutations mediate skipping is correct, the prediction is that ASF/SF2 should be able to counteract SC35-mediated exon 3 skipping with these constructs. Thus, titration experiments were performed in which GH3 cells were transfected with variable combinations of SC35 and ASF/SF2 along with vectors expressing either wild-type, A1338G, or A1338G+Δ6 GH1 constructs. Splicing patterns and the amount of exon 3 skipping were then determined (Fig. 6). As shown, ASF/SF2 was able to counteract SC35-mediated exon 3 skipping in the wild-type construct (Fig. 6A). Likewise, SC35 could induce exon 3 skipping in the presence of ASF/SF2, although higher amounts of SC35 were needed to reach the same level of exon 3 skipping seen when SC35 was overexpressed alone. Similar analyses were performed with both the A1338G and A1338+Δ6 constructs (Fig. 6, B and C). For all three experiments, it is clear that the two proteins act antagonistically, with SC35 mediating skipping and ASF/SF2 promoting inclusion.
FIGURE 6.
Antagonistic properties of ASF/SF2 and SC35. GH3 cells were transfected with 500 ng of either SC35 or ASF/SF2 expression vector and increasing amounts (100, 250, 500, and 1000 ng) of the other expression vector. Empty vectors were used to balance each transfection to an equal mass of DNA. GH1 splicing was analyzed by RT-PCR and polyacrylamide gel electrophoresis. The amounts of exon 3 skipped transcripts in each lane were calculated as a percentage of all other transcripts within a single lane. The data are presented as the averages of at least n = 3. The error bars are S.E. A, wild-type GH1. B, A1338G mutant GH1. C, A1338G+Δ6 mutant GH1.
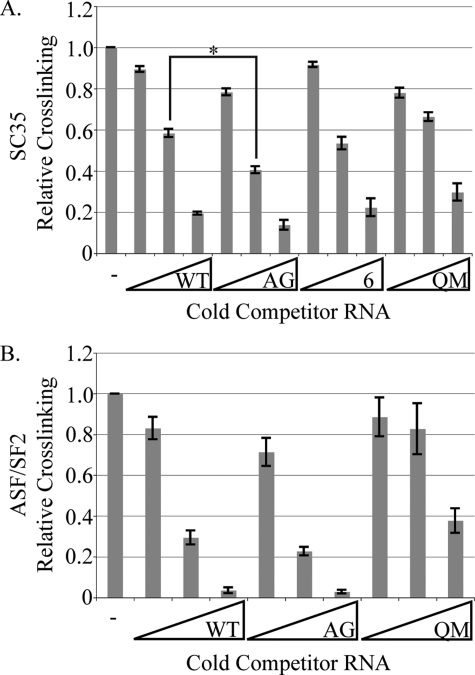
SC35 and ASF/SF2 Bind Sequences in Exon 3—We performed in vitro UV cross-linking experiments to examine differential binding of ASF/SF2 to ESE2 and SC35 to both ESE2 and region 6. Purified SC35 or ASF/SF2 were cross-linked to radiolabeled ESE2 RNA in the presence of increasing molar amounts of cold RNA competitors. The competitor RNAs included self-competitor (unlabeled ESE2), the A1338G patient mutation, region 6, and as a negative control, the QM sequence from the RNA affinity experiments. For SC35, the patient mutation was a better competitor than the wild-type sequence (Fig. 7A), unless a 50-fold molar excess was used. This is consistent with the effects of SC35 in the splicing assays. Also, consistent with earlier results, RNAs encompassing region 6 could also effectively displace SC35 from the wild-type ESE2 RNA, indicating that SC35 binds to region 6. For ASF/SF2, RNAs encompassing ESE2, both wild type and the A1338G patient mutation, were effective competitors, again consistent with ASF/SF2 binding to ESE2 to activate exon 3 inclusion. Thus, using both functional splicing assays and UV cross-linking assays, we have shown that SC35 and ASF/SF2 have differential effects on GH1 splicing.
FIGURE 7.
UV cross-linking of ASF/SF2 and SC35 to exon 3 sequences. In vitro transcribed, radiolabeled ESE2 wild type (WT, 5′-GGAAGGAACAGAAGUAU-3′) was cross-linked to either purified SC35 (A) or ASF/SF2 (B). Cold competition was performed with 1×, 10×, and 50× excess in vitro transcribed ESE2 wild type, ESE2 A1338G (5′-GGAAGGAACAGAGGUAU-3′), region 6 (5′-GGAACCCCCAGACCUCCCUC-3′), or ESE2 QM (5′-GGUAGUAAUAGUAGUAU-3′). The data are presented as the averages of n = 3. The error bars are S.E. The asterisk indicates statistical significance with a p value of 0.036.
DISCUSSION
Here, we sought to understand the mechanism by which ESE2 activates GH1 exon 3 inclusion, and in doing so, we have determined the disease-causing mechanism of the A1338G patient mutation in the GH1 gene. Two canonical SR proteins, ASF/SF2 and SC35, were identified as ESE2-binding proteins that could activate splicing of an enhancer-dependent splicing reporter through ESE2 in vitro. However, when the effects of overexpression of these proteins on GH1 splicing were analyzed, we discovered that SC35 did not activate exon inclusion but actually promoted exon 3 skipping. Although this finding was unexpected, it laid out a possible explanation for the mechanism behind the A1338G patient mutation. According to ESE2 finder, this mutation creates a binding site for SC35 in ESE2 leading to antagonism of ASF/SF2-mediated activation and skipping of exon 3. This prediction was supported by our UV cross-linking data where SC35 bound better to the patient mutation-containing ESE2 sequence. Such skipping requires the downstream SC35-binding site, suggesting that the two sites act synergistically to cause pathological levels of exon 3 skipping. This is the first time a disease-causing point mutation has been characterized as creating a SR protein-binding site that results in exon repression.
SC35 Synergistically Mediates Exon 3 Skipping through Region 6 and the A1338G Patient Mutation—The levels of exon 3 skipping caused by SC35 through region 6 (Fig. 6A) or the A1338G patient mutation (Fig. 6C) are approximately equivalent. Both of these constructs have a basal level of exon 3 skipping of about 4%, which doubles upon SC35 overexpression. If each of these sites was acting independently, then a construct containing both would be expected to skip exon 3 at additive levels. However, with the A1338G mutation and an intact region 6, the percentage of exon 3 skipping was 28% and doubled to 56% upon SC35 overexpression (Fig. 6B). This much higher level of exon 3 skipping shows that the effect of these two sites is synergistic, not additive. Synergism in this manner explains why patients with the A1338G mutation are growth hormone-deficient because the levels of exon 3 skipping increase dramatically.
SR Proteins and Repression of Splice Site Selection—SR proteins are generally characterized as activators of splicing. However, we show that SC35 can repress GH1 exon 3 inclusion. Although not common, there are reports showing that SR proteins can repress splicing (31, 32). Likewise, previous work has shown that the action of an individual SR protein depends on sequence context, similar to the differences we observed comparing activity in heterologous constructs versus GH1 splicing (31). Cystic fibrosis transmembrane conductance regulator exon 9 splicing is repressed by ASF/SF2 and SRp40 binding to an intronic splicing silencer, but these SR proteins activate splicing through the intronic splicing silencer in heterologous settings (31). Similarly, ASF/SF2 represses adenovirus 3a splicing through the 3RE repressor element but activates splicing through this element in a heterologous setting (32). These examples highlight the context-dependent nature of splicing regulatory elements.
In addition to the above examples, there is additional precedent for ASF/SF2 and SC35 acting as both activators and repressors of splicing. In splicing of the caspase-2 gene, ASF/SF2 and SC35 both cause exon skipping, and hnRNP A1, a well studied splicing repressor, activates exon inclusion (33). β-Tropomyosin exon 6A and 6B splicing is regulated by two intronic splicing enhancers, S3 and S4. ASF/SF2 recognizes S4 and activates exon 6A inclusion, whereas SC35 directly antagonizes ASF/SF2 resulting in exon 6A repression (34). However, both ASF/SF2 and SC35 activate exon 6B splicing through the S3 element (35).
Overall, our results and those just cited demonstrate the combinatorial nature of splicing regulation that enables very precise control of splice site recognition and selection. Combinations of splicing regulatory proteins binding to mostly short regulatory elements allows a somewhat limited number of factors to control a much larger number of splicing decisions. For GH1, mutations in one or more of these regulatory elements leads to aberrant skipping of exon 3. The current view is that skipping of exon 3 is a splicing fidelity issue rather than a regulated alternative splicing event, but it remains possible that one or more of the smaller human growth hormone isoforms may have development or tissue-specific functions.
Acknowledgments
We thank David Friedman and the Vanderbilt Proteomics Laboratory. We also thank Robin C. C. Ryther for valuable input.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 62487m (to J. G. P.) and DK 35592 (to J. A. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SR, serine/arginine-rich; ESE, exonic splicing enhancer; RT, reverse transcription.
References
- 1.Burge, C. B., Tuschl, T., and Sharp, P. A. (1999) in The RNA World (Gesteland, R. F., Cech, T. R., and Atkins, J. F., eds) 2nd Ed., pp. 525–560, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 2.Cartegni, L., Chew, S., and Krainer, A. (2002) Nat. Rev. Genet. 3 285–298 [DOI] [PubMed] [Google Scholar]
- 3.Birney, E., Kumar, S., and Krainer, A. R. (1993) Nucleic Acids Res. 21 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kan, J., and Green, M. (1999) Genes Dev. 13 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu, J., and Krainer, A. R. (2000) Genes Dev. 14 3166–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robberson, B. L., Cote, G., and Berget, S. M. (1990) Mol. Cell. Biol. 10 84094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamison, S. F., Pasman, Z., Wang, J., Will, C., Lührmann, R., Manley, J. L., and Garcia-Blanco, M. A. (1995) Nucleic Acids Res. 23 3260–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohtz, J. D., Jamison, S. F., Will, C. L., Zuo, P., Lührmann, R., Garcia-Blanco, M. A., and Manley, J. (1994) Nature 368 119–124 [DOI] [PubMed] [Google Scholar]
- 9.Wang, Z., Hoffmann, H. M., and Grabowski, P. J. (1995) RNA 1 21–35 [PMC free article] [PubMed] [Google Scholar]
- 10.Wu, J. Y., and Maniatis, T. (1993) Cell 75 1061–1070 [DOI] [PubMed] [Google Scholar]
- 11.DeNoto, F., Moore, D., and Goodman, H. (1981) Nucleic Acids Res. 9 3719–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asada, N., Takahashi, Y., Wada, M., Naito, N., Uchida, H., Ikeda, M., and Honjo, M. (2000) Mol. Cell. Endorinol. 162 121–129 [DOI] [PubMed] [Google Scholar]
- 13.Masuda, N., Watahiki, M., Tanaka, M., Yamakawa, M., Shimizu, K., Nagai, J., and Nakashima, K. (1988) Biochim. Biophys. Acta 949 125–131 [DOI] [PubMed] [Google Scholar]
- 14.Lecomte, C., Renard, A., and Martial, J. (1987) Nucleic Acids Res. 15 6331–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmetshofer, A., Zechner, D., Luger, T., and Barta, A. (1995) Mol. Cell. Endocrinol. 113 225–234 [DOI] [PubMed] [Google Scholar]
- 16.Cogan, J., Prince, M., Lekhakula, S., Bundey, S., Futrakul, A., McCarthy, E., and Phillips, J. A., III (1997) Hum. Mol. Genet. 6 909–912 [DOI] [PubMed] [Google Scholar]
- 17.Cogan, J., Ramel, B., Lehto, M., Phillips, J., III, Prince, M., Blizzard, R., de Ravel, T., Brammert, M., and Groop, L. (1995) J. Clin. Endocrinol. Metab. 80 3591–3595 [DOI] [PubMed] [Google Scholar]
- 18.Millar, D., Lewis, M., Horan, M., Newsway, V., Easter, T., Gregory, J., Fryklund, L., Norin, M., Crowne, E., Davies, S., Edwards, P., Kirk, J., Waldron, K., Smith, P., Phillips, J., III, Scanlon, M., Krawczak, M., Cooper, D., and Procter, A. (2003) Hum. Mut. 21 424–440 [DOI] [PubMed] [Google Scholar]
- 19.Moseley, C., Mullis, P., Prince, M., and Phillips, J., III (2002) J. Clin. Endocrinol. Metab. 87 847–852 [DOI] [PubMed] [Google Scholar]
- 20.Ryther, R. C. C., Flynt, A. S., Harris, B. D., Phillips, J. A., III, and Patton, J. G. (2004) Endocrinology 145 2988–2996 [DOI] [PubMed] [Google Scholar]
- 21.Ryther, R. C. C., McGuinness, L., Phillips, J. A., III, Moseley, C. T., Magoulas, C. B., Robinson, I. C. A. F., and Patton, J. G. (2003) Hum. Genet. 113 140–148 [DOI] [PubMed] [Google Scholar]
- 22.Procter, A., Phillips, J., III, and Cooper, D. (1998) Hum. Genet. 103 255–272 [DOI] [PubMed] [Google Scholar]
- 23.McCarthy, E. M. S., and Phillips, J. A., III (1998) Hum. Mol. Genet. 7 1491–1496 [DOI] [PubMed] [Google Scholar]
- 24.Barnard, D. C., and Patton, J. G. (2000) Mol. Cell. Biol. 20 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahler, A. M., Lane, W. S., Stolk, J. A., and Roth, M. B. (1992) Genes Dev. 6 837–847 [DOI] [PubMed] [Google Scholar]
- 26.Crawford, J. B., and Patton, J. G. (2006) Mol. Cell. Biol. 26 8791–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian, M., and Maniatis, T. (1994) Genes Dev. 8 1703–1712 [DOI] [PubMed] [Google Scholar]
- 28.Fu, X.-D. (1995) RNA 1 663–680 [PMC free article] [PubMed] [Google Scholar]
- 29.Blencowe, B. J. (2000) Trends Biochem. Sci. 25 106–110 [DOI] [PubMed] [Google Scholar]
- 30.Cartegni, L., Wang, J., Zhu, Z., Zhang, M., and Krainer, A. (2003) Nucleic Acids Res. 31 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buratti, E., Stuani, C., De Prato, G., and Baralle, F. E. (2007) Nucleic Acids Res. 35 4359–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanopka, A., Muhlemann, O., and Akusjarvi, G. (1996) Nature 381 535–538 [DOI] [PubMed] [Google Scholar]
- 33.Jiang, Z.-H., Zhang, W.-J., Rao, Y., and Wu, J. Y. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9155–9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallego, M. E., Gattoni, R., Stevenin, J., Marie, J., and Expert-Bezancon, A. (1997) EMBO J. 16 1772–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Expert-Bezancon, A., Sureau, A., Durosay, P., Salesse, R., Groeneveld, H., Lecaer, J. P., and Marie, J. (2004) J. Biol. Chem. 279 38249–38259 [DOI] [PubMed] [Google Scholar]