Abstract
The highly negatively charged membrane sialoglycoprotein leukosialin, CD43, is shed during neutrophil activation. This is generally thought to enhance cell adhesion. We here describe two novel consequences of this shedding, during neutrophil activation by phorbol esters or by chemoattractants after TNF-α priming. CD43 proteolysis was investigated by Western blotting, using a polyclonal antibody to CD43 intracellular domain. Our data emphasize the importance of a juxtamembranous cleavage of about 50% of membrane CD43 molecules by cathepsin G. Indeed, it is inhibited by α1-antichymotrypsin and cathepsin G inhibitor I and is reproduced by exogenous purified cathepsin G. The resulting membrane-anchored C-terminal fragment, CD43-CTF, becomes susceptible to presenilin/γ-secretase, which releases CD43 intracytoplasmic domain: preincubation with three different γ-secretase inhibitors, before PMN treatment by agonists or by purified cathepsin G, results in the accumulation of CD43-CTF. Because CD43 binds E-selectin, we also investigated the effect of the soluble extracellular domain CD43s, released by cathepsin G juxtamembranous cleavage, on neutrophil adhesion to endothelial cells. A recombinant CD43s-Fc fusion protein inhibited neutrophil E selectindependent adhesion to endothelial cells under flow conditions, while it had no effect on neutrophil static adhesion. We thus propose that, in addition to its potential pro-adhesive role, CD43 proteolysis results in: (i) the release, by cathepsin G, of CD43 extracellular domain, able to inhibit the adhesion of flowing neutrophils on endothelial cells and thus to participate to the natural control of inflammation; (ii) the release and/or the clearance, by presenilin/γ-secretase, of CD43 intracellular domain, thereby regulating CD43-mediated signaling.
The regulated proteolysis of transmembrane proteins represents an important mechanism of cell functions modulation. The inflammation resolution involves, for example, the shedding of cytokine receptors and adhesion molecules, which down-regulates leukocyte adhesion to endothelium. These regulations result from a decreased membrane expression of receptors and/or from the release of soluble fragments, competing with their membrane counterparts. The majority of shed proteins identified to date are cleaved by metalloproteinases or by neutrophil-derived serine proteases (1). Leukosialin, CD43, is the predominant cell surface sialoprotein of leukocytes (2) and has both anti-adhesive and adhesive properties. Its function has been mainly studied on lymphocytes, where CD43 behaves both as a negative regulator of T cell proliferation and adhesion and as a positive regulator of memory T cell trafficking (3, 4). Although its expression is normally restricted to leukocytes, CD43 is present on colon carcinomas and on several nonhematopoietic cell lines (5, 6). In these cell lines, CD43 is processed by a presenilin/γ-secretase-mediated regulated intramembrane proteolysis (RIP)3 (7). RIP refers to a sequential proteolysis of various type I membrane proteins, including the amyloid precursor protein of Alzheimer disease, the Notch receptor, CD44, and E-cadherin (8). RIP is initiated usually by a metalloproteinase, which induces the shedding of the receptor ectodomain, followed by an intramembrane processing of the cell-bound fragment by a PS/γ secretase. This releases the receptor intracellular domain in the cytoplasm. In the case of CD43 in cancer cell lines, this domain translocates in the nucleus and causes the up-regulation of various genes (9).
CD43 has been recently described as a ligand for E-selectin (10, 11). Its expression on polymorphonuclear neutrophils (PMN) is 10-fold higher than P-selectin glycoprotein ligand-1, PSGL-1,4 the main leukocyte ligand for P-selectin, which also binds E-selectin. Despite CD43 abundance, data on its precise role in PMN responses are scarce. CD43 is shed during PMN activation (12–15) and during adhesion and spreading (2, 16), together with other E-selectin ligands PSGL-1 and CD44 (17, 18). A soluble form of CD43, CD43s, representing the entire extracellular domain, has been described in plasma and identified to galactoglycoprotein (Galgp) (19). CD43 proteolysis on PMN was claimed to involve serine proteases and metalloproteinases. The precise analysis of the proteolysis mechanism was hampered by the fact that CD43 soluble fragments do not transfer on blotting membranes (15) and required an immunoprecipitation of radiolabeled neutrophil membranes. We produced a polyclonal antibody against a recombinant CD43 intracellular domain to re-assess the molecule proteolysis in the light of recent data on: (i) the γ-secretase processing of CD43 in cancer cells; (ii) the description of CD43 as a leukocyte ligand for endothelial E-selectin. We here describe for the first time, in human PMN activated by pro-inflammatory stimuli, a cathepsin G and γ-secretase mediated processing of CD43 with putative important functional consequences.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents—FITC-anti-CD43 mAb (1G10) and IgG1 were from BD Pharmingen (San Diego, CA), anti-CD18 clone IB-4 from Ancell (Bayport, MN), anti-CD62E, FITC-anti-CD62L mAbs, and PE-anti-CD146 from Beckman Coulter (Roissy, France). The rabbit anti-CD43cyto pAb was obtained as described (20). Human neutrophil elastase (NE), cathepsin G, and proteinase 3 were purified as described (21). Purified cathepsin G was also purchased from Calbiochem (Darmstadt, Germany) and Athens Technology (Athens, Georgia). Human IgGs were purified on protein A-Sepharose from an AB blood group normal human serum. The CD43s-Fc fusion protein was produced in COS cells, as described (22) and purified on protein A-Sepharose. Tumor necrosis factor α, TNF-α, was from PeproTech (Rocky Hill, NJ). Recombinant human VCAM-1-Fc was from R&D (Abingdon, UK). α1-anti-chymotrypsin (α1-CT), cathepsin G inhibitor I, and MG-132 were from Calbiochem, Compound E from Alexis (Lausanne, Switzerland), Tapi-0, and Tapi-1 from Peptides International (Louisville, KY), while Marimastat, CH46644 and CH6631 were a kind gift from Dr. John Bird (Celltech Chiroscience, Cambridge, UK). All other inhibitors and reagents were from Sigma-Aldrich.
Cells—Human neutrophils and endothelial cells HUVEC were isolated as described (20, 23) from platelet-depleted blood and umbilical vein, respectively, from healthy volunteers. Neutrophils were suspended in Hanks balanced solution with Ca2+ and Mg2+ containing bovine serum albumin (0.1%, HBSS2+-BSA). In our hands, this BSA concentration has no effect on CD43 proteolysis (2). HUVECs were cultured on gelatin-coated flasks (Costar Corporation, Cambrige, MA) in M199 supplemented with penicillin, streptomycin, l-glutamine, Hepes (10 mm), heparin (50 μg/ml), endothelial cell growth supplement ECGS (20 μg/ml), and 20% fetal calf serum. 3rd- and 4th passage cells were used exclusively. CHO cells were either transiently transfected with CD43 cDNA or stably transfected with cDNAs encoding both CD43 and core 2 β-1,6-N-acetylglucosaminyl transferase (C2GnT), as described (24), then further transiently transfected to enhance protein expression.
Neutrophil Activation, Flow Cytometry, and Western Blotting—Freshly isolated neutrophils were distributed in BSA-coated tubes and gently tumbled every 5 min during their activation at 37 °C either with phorbol myristate acetate (PMA) or with TNF-α and N-formyl-l-methionyl-l-leucyl-l-phenylalanine (fMLP), as described in the figure legends. The reaction was stopped by adding ice-cold phosphate-buffered saline containing phenylmethylsulfonyl fluoride (1 mm) and 1,10-phenanthroline (1 mm), to prevent any further CD43 cleavage. PMN were centrifuged at 4 °C for 30 s at 3,800 × g and submitted to Western blotting or flow cytometry as described (20).
Assays for PMN Adhesion to HUVEC—Static adhesion: confluent HUVECs, subcultured in 48-well 0.5% gelatin-coated plates, were pretreated for 4 h with 10 ng/ml TNF-α and rinsed with medium. PMN were then added (2 × 106 per ml of M199 with 0.1% BSA) and allowed to adhere for 30 min at 37 °C. Non-adherent cells were removed by rinsing and adherent PMN and HUVEC were detached with 0.5 mg/ml collagenase for 15 min at 37 °C. They were then labeled with PE-anti-CD146, to distinguish CD146-negative PMN from CD146-positive endothelial cells. PMN were counted by flow cytometry with a constant time setting.
Flow Assay—Cell adhesion was analyzed, as described (25), by real-time videomicroscopy with a ×10 objective, in a flow chamber containing a 0.5% gelatin-precoated glass coverslip with confluent HUVECs, which had been stimulated for 4 h with 0.2 ng/ml TNF-α and washed to remove the TNF-α. Controlled flow rates, generating a wall shear rate of 100 s–1, were applied alternatively to two syringes, resulting in sequential perfusion of 37 °C warmed HBSS2+-BSA and PMN suspension (2 × 106/ml), for 4 min each, over the endothelial cells layer. This was followed by a period of washout with warm buffer, and video microscopic recordings were made between minutes 1 and 2, as described (26). The Histolab software (Microvision Instruments, Evry, France) allowed quantifying the number of adherent cells visible in at least three fields.
Statistical Analysis—Data were compared using a paired Student's t test analysis. Statistical significance was defined as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
RESULTS
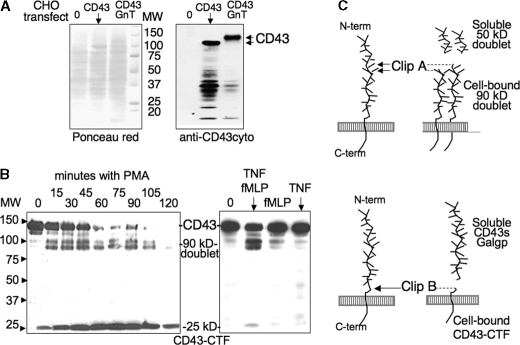
Cell-associated CD43 Fragments Analyzed by Western Blotting—Confirming a previous report (15), we observed that extracellular soluble CD43 fragments fail to transfer onto blotting membranes, whatever membrane (nitrocellulose, polyvinylidene difluoride, positively charged PALL membranes), blotting buffer (pH 4–11), or anti-CD43 mAb clones (L60, MEM59, 1G10) were used. We thus raised an anti-CD43cyto intrapolyclonal antibody to analyze by Western blotting the fragments containing CD43 intracellular domains and remaining membrane-anchored after proteolysis. The antibody specificity was analyzed by Western blot with CHO cell lines transfected with CD43 without and with Core2GnT, resulting in bands corresponding to the previously reported 115- and 130-kDa glycosylated forms of CD43 (24) (Fig. 1A). Analysis of PMA-activated PMN (Fig. 1B) confirmed previous data, because it revealed cell-associated fragments complementary in size to the reported soluble CD43 fragments observed by immunoprecipitation of 125I-labeled neutrophils (15, 27): (i) a cell-bound 90-kDa doublet, resulting from clip A (Fig. 1C) and complementary to the reported soluble 50-kDa doublet; (ii) a cell-bound CD43 C-terminal fragment, CD43-CTF, of 25 kDa, resulting from clip B and complementary to the reported soluble 120-kDa fragment.
FIGURE 1.
Analysis of CD43 proteolysis by Western blot with the anti-CD43cyto pAb, showing the remaining membrane-associated CD43 fragments. A, specificity of the anti-CD43cyto pAb tested by Western blotting of lysates from native CHO cells (lane 0) and CHO transfected with CD43 without and with the Core2 GnT. Blotted membranes were analyzed with the anti-CD43cyto (right), while total proteins were detected with Ponceau red (left). B, kinetic analysis of PMA-induced proteolysis of CD43, analyzed by Western blotting: freshly isolated neutrophils (2 × 106/ml in HBSS2+/0,1% BSA) were incubated at 37 °C with 3 ng/ml PMA and stopped at increasing incubation time points (left panel) or with TNF-α (5 ng/ml) for 30 min then with fMLP (10–6 m) for another 30-min incubation, or with TNF-α or fMLP alone (right panel) and compared with cells incubated with buffer (lane 0). Neutrophil lysates, submitted to gel electrophoresis on 7–15% gradient acrylamide, were analyzed by Western blotting with the anti-CD43cyto antibody and an horseradish peroxidase-labeled secondary anti-rabbit IgG antibody and revealed by chemiluminescence. C, schematic representation of soluble and membrane CD43 fragments.
The intensity of the 90-kDa doublet was variable and appeared to be related to the “spontaneous” activation of PMN during the isolation procedure: As we had observed with the soluble 50-kDa doublet (27), it was often observed on freshly isolated PMN, appeared upon incubation at cell concentrations above 106/ml without any stimuli, and was significantly prevented by avoiding the lysis of contaminating erythrocytes and by the presence of 1 mg/ml BSA in incubation medium. It is not known whether the doublet represents two very close split sites, or a single split resulting in fragments that differ by post-translational modifications such as glycosylation. Kinetic studies showed that the small 25-kDa CD43-CTF appeared later than the 90-kDa doublet and was the main fragment after 30 min of incubation (Fig. 1B). PMN activation by TNF-α (10 ng/ml) or fMLP (10–6 m) did not modify significantly CD43, as mentioned previously (14), while priming with 5 ng/ml TNF-α for 30 min, followed by 30 min of activation with fMLP (10–6 m) resulted in CD43 cleavage similar to PMA, but the 25-kDa CD43-CTF was either weakly present or not detected (Fig. 1B).
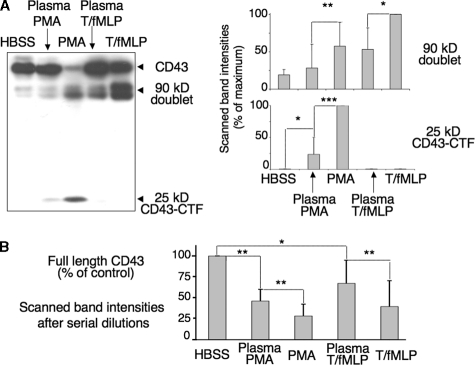
CD43 Proteolysis in Plasma—Western blot analysis confirmed that plasma inhibits CD43 proteolysis (Fig. 2A). It significantly prevented the decrease of the CD43 full-length band, and the appearance of CD43 fragments promoted by high PMA concentrations (100 ng/ml) or, to a lesser extent, by TNF-α (5 ng/ml) with fMLP (10–6 m). This inhibition, not modified by plasma heat-inactivation (30 min, 56 °C), was impressive but not complete. Although neutrophil activation in plasma resulted in little or no increase of the 90-kDa doublet, as compared with control cells in buffer, some 25-kDa CD43-CTF appeared with plasma/PMA-activated cells (Fig. 2A). This suggests that plasma efficiently protects the extended extracellular domain of CD43 from proteolysis, while the juxtamembranous cleavage site is less accessible to plasma inhibitors. Sample loads of Fig. 2A blots, aimed to detect the 25-kDa CD43-CTF fragment, resulted in saturating signals for the full-length CD43 band. However, serial dilutions of PMN lysates showed a significant decrease of full-length CD43 band intensities upon activation in plasma with PMA and TNF/fMLP, respectively (Fig. 2B).
FIGURE 2.
CD43 proteolysis in plasma. 2 × 106/ml PMN, suspended in HBSS2+-BSA or in undiluted autologous heparinated plasma, were incubated with PMA (100 ng/ml) for 30 min at 37 °C or with TNF-α (5 ng/ml) for 30 min, followed by the addition of fMLP (10–6 m) and another 30-min incubation. Cells lysates were analyzed by Western blotting, as in Fig. 1. A, Western blot of one representative experiment and mean ± S.D. of the intensities of the 90-kDa doublet and 25-kDa scanned bands from four similar experiments. Results are expressed as percent of the maximal level observed with TNF/fMLP (90-kDa doublet) or PMA (25 kDa). B, native CD43 bands shown in the blot A resulted in saturating signals. Quantitative evaluation of residual full-length CD43 was then calculated from Western blot analysis of serial dilutions of PMN lysates, after activation with or without plasma as above. Scanned band intensities, normalized with the scanned actin bands, are expressed as percent of the initial level of full-length CD43 of PMN incubated in HBSS (n = 4).
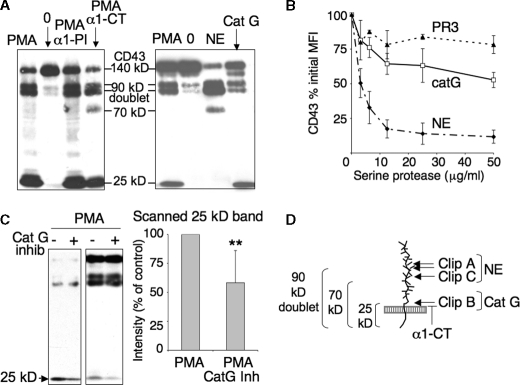
Role of Neutrophil Serine Proteases in CD43 Shedding—Inhibition of CD43 proteolysis may involve plasma protease inhibitors. Because the main plasma serpins are α1-proteinase inhibitor α1-PI and α1-anti-chymotrypsin α1-CT, we analyzed the effects of these inhibitors on CD43 proteolysis. Western blotting analysis revealed that whereas α1-PI does not seem to prevent any cleavage, α1-CT inhibits the degradation of a newly appearing 70-kDa fragment and decreases the intensity of the 25-kDa CD43-CTF (Fig. 3A). α1-PI is known to inhibit all the serine proteases released by PMN upon PMA-triggered degranulation, namely elastase, cathepsin G, and proteinase 3, PR3, with the highest affinity for elastase. By contrast, α1-CT inhibits cathepsin G but neither elastase nor PR3 (28). These results thus proposed cathepsin G as a candidate for the cleavage of the 70-kDa fragment into a 25-kDa cell-bound CTF. Flow cytometry analysis of CD43 membrane expression, after neutrophil incubation with exogenous purified serine proteases, showed no significant cleavage by PR3, while elastase and cathepsin G resulted in up to 80 and 40% CD43 down-regulation respectively (Fig. 3B). As shown in Fig. 3A and in agreement with previous data (29), Western blot analysis of PMN incubated with purified elastase resulted in a 90-kDa doublet, similar to that observed with PMA. Elastase also produced a 70-kDa fragment. Interestingly, cathepsin G promoted a juxtamembranous cleavage, leaving a 25-kDa cell-associated fragment analogous to the CD43-CTF observed after PMN activation by PMA (Fig. 3A). Data shown in Fig. 3A were obtained with our homemade cathepsin G preparation, but identical results were obtained with two commercial cathepsin G (data not shown). Finally, the specific cathepsin G inhibitor 1 (10 μm) partially inhibited the appearance of the 25-kDa CD43-CTF resulting from PMA activation Fig. 3C).
FIGURE 3.
Role of neutrophil serine proteases in CD43 shedding. A, left panel, CD43 fragments resulting from PMA activation in the presence of plasma serpins (left panel). 2 × 106/ml PMN were preincubated for 15 min with α1-PI (500 μg/ml) or α1-CT (100 μg/ml) and activated for 30 min with 3 ng/ml PMA in HBSS2+-BSA. Right panel, CD43 fragments resulting from the addition of exogenous purified serine proteases to resting PMN. 10 × 106/ml PMN in phosphate-buffered saline were treated with human purified elastase (NE) (15 μg/ml) or cathepsin G (Cat G) (25 μg/ml) for 20 min at 37 °C. Cell lysates were analyzed by Western blotting revealed by anti-CD43cyto pAb. B, dose response effect of purified elastase, cathepsin G, and PR3 on CD43 expression. PMN were incubated as described above with increasing concentrations of serine proteases, then labeled with FITC-anti-CD43 and analyzed by flow cytometry. C, inhibition, by cathepsin G inhibitor 1, of the cleavage resulting in 24-kDa CD43-CTF. PMN, preincubated or not for 15 min with cathepsin G inhibitor-1 10 μm, were activated with PMA and processed as in A. Two different experiments are shown, where PMA induced high and low levels of CD43 proteolysis, as well as a quantitative analysis of the scanned 25-kDa band (mean ± S.D. of four experiments). Data are expressed as percent of the control band observed upon PMA activation without inhibitor. D, schematic representation of serine protease-induced CD43 fragments.
We thus propose (Fig. 3D) that PMA-induced CD43 proteolysis includes: (i) clips A and C, mediated by elastase-like enzyme(s) and resulting in 90- and 70-kDa membrane-spanning fragments, respectively; (ii) a juxtamembranous clip B, presumably mediated by cathepsin G, producing the 25-kDa CD43-CTF. Our results also show that clip C appears to be followed immediately by clip B and thus only detected if clip B is prevented by α1-CT.
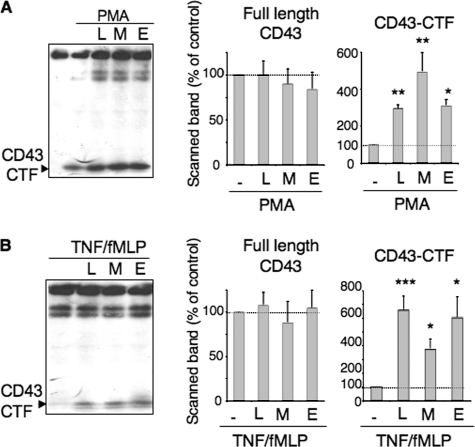
Role of Presenilin/γ-Secretase (PS/γ-Secretase)—In cancer cells, an intramembrane cleavage of CD43 by PS/γ-secretase has been described (7). We thus investigated a possible role of γ-secretase in CD43 processing in neutrophils. In most instances, RIP-induced intracytoplasmic fragments are difficult to individualize, either because they translocate to the nucleus or because they are degraded by the proteasome. PS/γ-secretase activity is thus indirectly measured by the accumulation of membrane CTF in the presence of γ-secretase inhibitors. The presence of three different inhibitors of γ-secretase, L685,458, MG132, or compound E, during neutrophil activation by PMA (Fig. 4A), significantly increased the amount of the 25-kDa CD43-CTF observed on Western blots. Moreover, in the presence of γ-secretase inhibitors, the 25-kDa CD43-CTF now appeared on Western blots from neutrophils activated by TNF/fMLP (Fig. 4B). Neutrophil incubation with γ-secretase inhibitors in the absence of agonist did not produce any CD43-CTF fragment (data not shown). γ-Secretase inhibition thus reveals the occurrence of a juxtamembranous cleavage, upon PMN activation by TNF/fMLP, which was not detected normally, because of CD43-CTF rapid degradation by γ-secretase. We did not detect smaller putative intracytoplasmic fragments resulting from this further degradation, even when cell pellets were immediately boiled in Laemmli buffer to solubilize nuclear proteins before Western blot analysis (data not shown).
FIGURE 4.
Accumulation of the 25-kDa CD43-CTF in the presence of γ-secretase inhibitors. PMN were preincubated with γ-secretase inhibitors (5 μm L685,458 (L), 15 μm MG132 (M), 20 nm compound E (E)) for 2 h at 37°C, then further incubated without or with either PMA (3 ng/ml) for 30 min (A) or with 5 ng/ml TNF-α for 30 min followed by fMLP (10–6 m) for another 30 min (B). The reaction was stopped, and cell lysates processed for Western blotting as described under “Experimental Procedures.” Left panels show one representative experiment. Right panels show the mean ± S.D. of the scanned band intensities of full-length CD43, expressed as percent of initial levels of PMN incubated in buffer and of CD43-CTF, expressed as percent of the levels observed with PMN activated with PMA or TNF/fMLP in the absence of inhibitors (n = 3 experiments).
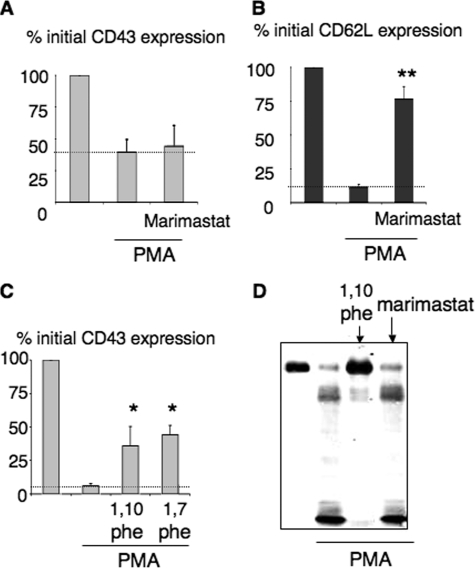
Absence of Metalloproteinase Involvement in CD43 Shedding—RIP intramembranous proteolysis is usually triggered by matrix metalloproteinases, MMPs. We thus tested the effects of various MMP inhibitors on CD43 shedding. PMA-induced CD43 cleavage was not inhibited by metalloproteinase inhibitors, such as TAPI-0, TAPI-1, and CH4644 and CH6631, that block various MMPs, including MMP8 and MMP9 (data not shown) and the broad MMP and TACE inhibitor marimastat (BB 2516) (Fig. 5, A and D). In parallel experiments, marimastat efficiently blocked the shedding of L-selectin (CD62L) on the same cells (Fig. 5B). CD43 down-regulation was inhibited by 1,10-phenanthroline (Fig. 5, C and D), but also by the inactive analogue 1,7-phenanthroline (Fig. 5C), confirming previous data (17) and suggesting an effect unrelated to metalloproteinase inhibition. Metalloproteinases thus do not seem to be involved in PMA-induced cleavage.
FIGURE 5.
Absence of involvement of metalloproteinases in CD43 shedding. Flow cytometry analysis of CD43 (A) and CD62L (B) membrane expression after PMN activation with limiting PMA concentration (3 ng/ml), without and with marimastat (6 μm)(n = 7). C, flow cytometry analysis of CD43 membrane expression after PMN activation by high PMA concentrations (10 ng/ml) in the presence of 1,10- or 1,7-phenanthroline (1 mm)(n = 3). D, Western blot analysis of PMN lysates after 30 min of incubation with 3 ng/ml PMA in the presence of 1,10-phenanthroline (1 mm) or marimastat (6 μm).
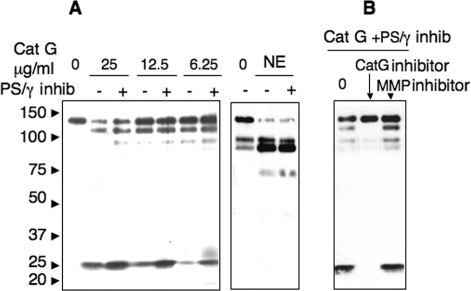
CD43 Is Processed by Presinilin/γ-Secretase as a Consequence of Cathepsin G Cleavage—We thus wondered if cathepsin G could fulfill the function of MMPs in the RIP processing and prepare CD43 for γ-secretase processing. As shown in Fig. 6A, exogenous purified cathepsin G resulted in a CD43-CTF fragment, which was further degraded byγ-secretase. Indeed, the intensity of the CD43-CTF fragment, resulting from increasing doses of cathepsin G, was further enhanced by PMN preincubation with γ-secretase inhibitor compound E (97.3 ± 64.6% increase, n = 7, p = 0.007). To exclude the effects of putative enzymes contaminating our cathepsin G preparations, we repeated these experiments with two commercial cathepsin G sources and obtained the same results (data not shown). By contrast, the intensities of CD43 fragments released by exogenous elastase were not modified by compound E (Fig. 6A). Finally, CD43-CTF fragments appearing upon PMN treatment by cathepsin G, in the presence of the γ-secretase inhibitor, disappeared completely in the presence of the specific cathepsin G inhibitor I, but were not affected by the MMP inhibitor marimastat (Fig. 6B). This result shows that the cleavage of CD43 by purified cathepsin G, in the absence of any PMN agonist, is sufficient to allow CD43 further processing by PS/γ-secretase.
FIGURE 6.
Cathepsin G triggers CD43-CTF further processing by γ-secretase. A, PMN (2.106/ml), preincubated (+) or not (–) with γ-secretase inhibitor compound E (20 nm) for 2 h at 37°C, were then treated for 20 min with buffer (lane 0), with decreasing concentrations of purified cathepsin G (left panel) or with purified elastase (15 μg/ml) (right panel). B, PMN 2 × 106/ml were preincubated at 37 °C with γ-secretase inhibitor compound E (20 nm) for 2 h and with cathepsin G inhibitor I (CatG) (10 μm) or metalloproteinase inhibitor (MMP) marimastat (6 μm) for 15 min, then treated for 20 min with purified cathepsin G (25 μg/ml). Cell lysates were analyzed by Western blot as described under “Experimental Procedures.”
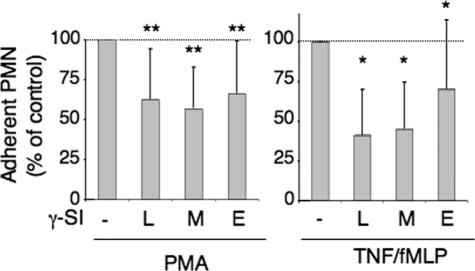
Role of Presinilin/γ-Secretase in the Regulation of Neutrophil Functions—γ-Secretase inhibitors had no effect on neutrophil degranulation, promoted by PMA or TNF/fMLP, reflected by CD11b up-regulation, as measured by flow cytometry (n = 5, data not shown). They had no effect either on the oxidative burst, measured with the fluorescent probe 2′-7′-dichlorofluorescein hydrodiacetate (n = 5, data not shown). By contrast, all threeγ-secretase inhibitors significantly inhibited neutrophil adhesion to gelatin, promoted either by TNF/fMLP or by PMA (Fig. 7).
FIGURE 7.
Effect of γ-secretase inhibitors on neutrophil adhesion. PMN (2 × 106/ml) were preincubated with γ-secretase inhibitors (5 μm L685,458 (L), 15 μm MG132 (M), 20 nm compound E (E)) in HBSS without Ca2+/Mg2+, for 2 h at 37°C. They were diluted (v/v) with HBSS containing 2 mm Ca2+/Mg2+ (to reach 1 mm final concentration) and added to gelatin-coated plates. After 15 min at 37 °C, cells were activated either with PMA (5 ng/ml) or with TNF (5 ng/ml) for 15 min and fMLP (10–6 m) for 30 min. After washing off non-adherentcells, the amount of adherent cells was measured by protein titration with a Micro BCA Protein Assay (Pierce). Results are expressed as percent of the levels reached upon PMN activation with PMA or TNF/fMLP without inhibitors.
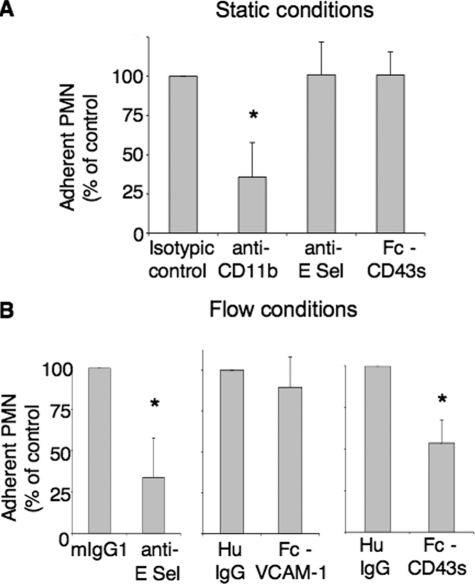
Modulation of PMN Adhesion to Endothelial Cells by CD43s, the Soluble Ectodomain of CD43 Released after Cell Activation—We tested the effect of a recombinant chimeric Fc-CD43s molecule on neutrophil adhesion to endothelial cells. To preserve the function of the natural molecule, CD43s was expressed in COS cells, which contain the enzymatic machinery required for a post-translational glycosylation similar to neutrophils (24). We used 10–30 μg/ml Fc-CD43s, based on normal plasma CD43s (Galgp) concentration, previously estimated at 10 μg/ml (19). Moreover, a homemade sandwich ELISA, using anti-CD43-coated plates and biotinylated wheat germ agglutinin and standardized with the recombinant Fc-CD43s protein, resulted in a mean of 7 ± 6.7 μg/ml of CD43s in normal plasma (n = 7, data not shown). Our results show that Fc-CD43s has no effect on PMN static adhesion to extracellular matrix (gelatin, fibronectin, data not shown) or to TNF-activated endothelial cells (Fig. 8A). We confirmed with blocking antibodies that this adhesion is β2 integrin-dependent but E-selectin-independent.
FIGURE 8.
Effect of CD43s on PMN adhesion to endothelial cells. A, lack of effect on static adhesion: PMN were preincubated with control IgG2a or anti-CD18 mAb or TNF-α-activated HUVECs preincubated with IgG1, anti-E-selectin mAb (anti-CD62E) or recombinant Fc-CD43s (10 μg/ml), for 20 min. PMN were then allowed to adhere to TNF-α-activated HUVECs under static conditions (n = 6). B, inhibition of PMN adhesion under shear stress: TNF-α-activated HUVECs were preincubated for 10 min with either control IgG1 or anti-E-selectin mAb (left), recombinant Fc-CD43s (10 μg/ml) or control human IgGs (middle), recombinant Fc-VCAM-1 (10 μg/ml) or control human IgGs (right). Neutrophils (106/ml) were then perfused together with IgGs or recombinant molecules and tested in a flow-based adhesion assay, as described under “Experimental Procedures.” Data give the numbers of adherent neutrophils, as percent of control adhesion obtained in the presence of control IgGs (mean ± S.D., n = 4).
By contrast, PMN adhesion to TNF-activated HUVECs under flow conditions, partially prevented by anti-E-selectin antibodies, was also significantly inhibited by 10 μg/ml Fc-CD43s, when compared with human IgGs purified from an AB normal serum, as a control for the Fc portion (Fig. 8B). The inhibition level was the same with 10 μg/ml and 30 μg/ml CD43s (data not shown). In similar conditions, 10 μg/ml recombinant Fc-VCAM-1 had no effect on PMN adhesion (Fig. 8B). These results suggest that CD43s interferes with the E-selectin-dependent adhesion of PMN to endothelial cells under flow.
DISCUSSION
Our analysis of CD43 proteolysis, resulting from neutrophil activation, confirms several points proposed previously (15). Indeed, the size of membrane-anchored C-terminal fragments, detected with our anti-CD43cyto antibody, are mostly complementary of those previously described for soluble N-terminal fragments. We demonstrate that CD43 proteolysis includes three simultaneous or sequential proteolytic steps, schematized in Figs. 1C and 3D: clips A and C, distant from the membrane and a juxtamembranous clip B.
The double clip A, previously described as a spontaneous cleavage (15) indeed corresponds to a protein domain very sensitive to proteolysis. This cleavage was observed upon PMN incubation without stimuli, particularly in buffers devoid of BSA, and it increased with the cell concentration. This suggests that it is mediated by proteases either constitutively present on neutrophils, but normally blocked by plasma inhibitors or albumin, or induced on PMN membrane by the isolation procedure. This cleavage was enhanced by PMA or by TNF/fMLP and we confirm that it results in CD43 fragments similar to those observed with exogenous elastase (27, 29). In addition, purified elastase results in a minor cleavage C, closer to the membrane and leaving a 70-kDa membrane-spanning fragment. A similar 70-kDa fragment was produced during PMN activation, but was only detected in the presence of α1-antichymotrypsin α1-CT, which prevents its further degradation. These results strongly suggest that, during PMN activation, (an) elastase-like enzyme(s) cleave(s) CD43 at clip A and C sites, distant from the cell membrane and resulting in cell-bound 90-kDa doublet and 70-kDa fragments. The clip A site is more exposed and preferentially cleaved. The minor 70-kDa fragment seems to be immediately further processed by the α1CT-sensitive clip B enzyme (as schematized in Fig. 3C).
The identification of plasma Galgp as the entire extracellular CD43 domain released in the fluid phase (19) makes it likely that the juxtamembranous clip B is involved in the physiologic ectodomain shedding of CD43. This clip had been suggested by the down-regulation of a T305 epitope and claimed to be exclusively induced by PMA (15). Our anti-CD43cyto pAb allowed to clearly demonstrate, for the first time, the occurrence of such juxtamembranous clip, which results in a 25-kDa membrane inserted C-terminal fragment CD43-CTF fragment. We also show that this cleavage is not only triggered by PMA but also by pro-inflammatory stimuli, such as TNF-α priming followed by chemoattractant activation. In the latter condition, the resulting CD43-CTF is only detected in the presence of γ-secretase inhibitors, as discussed below. Clip B also occurs during PMN apoptosis, because we previously described a 25-kDa membrane-anchored CD43 fragment on apoptosis-induced membrane-derived microparticles (20).
The juxtamembranous Clip B had been proposed to result from an unknown “CD43-ase” insensitive to serine protease or metalloproteinase inhibitors (15). We here show that, unlike clip A, which occurs “spontaneously” upon incubation of isolated PMN, clip B requires a strong activation induced by PMA or by TNF together with fMLP, while it is not observed with TNF-α or fMLP alone. This is reminiscent of the mobilization of azurophilic granules, which requires PMN priming together with chemoattractants. Azurophilic granule serine proteases, which are then released, are elastase, cathepsin G and PR3. All are strongly cationic enzymes, likely to spontaneously interact with such a negatively-charged preeminent membrane protein as CD43. Our flow cytometry data confirm that elastase and cathepsin G result in CD43 shedding, while PR3 has no significant effect (29).
Western blot analysis of CD43 proteolysis by cathepsin G, which had not been investigated until now, reveals that cathepsin G could be a good candidate for the juxtamembranous clip B. Indeed: (i) both α1-CT, which inhibits cathepsin G but neither elastase nor PR3, and a specific cathepsin G inhibitor peptide partially inhibited PMA-induced CD43 proteolysis and prevented the appearance of the 25-kDa CD43-CTF fragment; (ii) PMN treatment with purified cathepsin G from three different sources resulted in a juxtamembranous cleavage, producing a 25-kDa CD43-CTF similar to that observed after PMN activation by PMA. Moreover, the analysis of CD43 sequence (30) reveals a putative cathepsin G site, Val-Pro-Phe, P3-P1 (31), 10 amino acids away from the transmembrane sequence. Such a cleavage would release an extracellular soluble fragment with the same C-terminal Phe-226 as galactoglycoprotein (Galgp), the soluble CD43 form described in normal plasma (19). The clip B cleavage of CD43, resulting from neutrophil activation, is thus likely to be mediated by cathepsin G.
The lack of inhibition of PMA-induced CD43 proteolysis by DFP, aprotinin or α1-PI, known inhibitors of elastase and cathepsin G, remains puzzling and different hypothesis have been discussed previously (2, 15). The large quantities of proteases released by PMA-activated neutrophils may overwhelm and inactivate protease inhibitors. α1PI is known to be inactivated by PMN membrane-bound MMPs, MMP8 and MT6-MMP/MMP25 (32, 33), and may not be functional at sites close to PMN membrane, particularly during cell adhesion. Moreover, a competition between neutrophil elastase and cathepsin G for binding α1-PI has been described, resulting in a lack of control of cathepsin G by this inhibitor when elastase is present (34). Finally, the lack of effect of some inhibitors may be related to their accessibility to the immediate membrane surrounding. Indeed, degranulation releases endogenous proteases at the immediate proximity of CD43 juxtamembranous cleavage site. The charge or size of some but not other protease inhibitors may limit their access, through the cell glycocalyx, to this cleavage site. This would also explain the partial inhibition promoted by the specific cathepsin G inhibitor I.
Following the description of a γ-secretase-mediated RIP of CD43 in cancer cell lines (7), we investigated a similar cleavage of CD43 during neutrophil activation. The γ-secretase component presenilin-1 has been described in neutrophil azurophilic granules (35). A functional presenilin/γ-secretase complex could then result from azurophilic granule fusion with the plasma membrane. Interestingly, γ-secretase inhibitors have recently been shown to accelerate neutrophil apoptosis (36), suggesting that some of the γ-secretase products may be antiapoptotic. Upon PMN activation by PMA or by TNF-α/fMLP, we indeed observed a significant accumulation of the 25-kDa CD43-CTF, when cells were preincubated with three different γ-secretase inhibitors. This effect of γ-secretase inhibitors was also observed on the 25-kDa CD43-CTF, resulting from PMN treatment by exogenous cathepsin G. The juxtamembranous clip promoted by cathepsin G could thus be sufficient to render CD43 susceptible to γ-secretase cleavage. This role of cathepsin G is unexpected, because in most cases it is performed by metalloproteinases.
A role for MMPs in CD43 shedding had been proposed, since PMA-induced CD43 shedding was inhibited by 1,10 orthophenanthroline. We tested here various broad metalloproteinase inhibitors, able to inhibit MMPs and/or membrane sheddases such as TACE: None of them had significant effect on CD43 down-regulation by PMA, except for 1,10-orthophenanthroline, but a similar inhibition was observed with the inactive analogue 1,7-orthophenanthroline, as previously mentioned (15). Metalloproteinases thus do not appear to be involved in CD43 proteolysis on PMN.
We addressed the question of the role of the γ-secretase-mediated release of CD43 intracellular fragments, by preventing this release with PS/γ-secretase inhibitors. Our results show that γ-secretase inhibitors have no effect on neutrophil degranulation or oxidative burst, but significantly inhibit neutrophil adhesion. Although this inhibition may extend beyond CD43, to other putative membrane receptors processed by γ-secretase, these results suggest that neutrophil adhesion requires the release of intracellular domains from shed transmembrane molecules.
PMN activation by inflammatory stimuli results in the shedding of half of membrane exposed CD43. The full-length unshed CD43 molecules interact with cytoskeletal linker proteins ezrin, radixin and moesin (ERM), are redistributed in the uropod and participate to cell motility (37, 38). The proteolysis by PS/γ-secretase of CD43-CTF fragments, remaining in the membrane after CD43 shedding, may be seen as the simple clearing of useless fragments by a “membrane proteasome” (39). By analogy with the E-cadherin cleavage by γ-secretase, that dissociates catenins from the cytoskeleton (40), γ-secretase effect on CD43 could be the release of CD43CTF-bound ERM, so that actin bundles would only interact with full-length CD43 molecules. Similarly, the removal of CD43-CTF cytoplasmic domains, endowed with signaling functions (4, 41) may be crucial to concentrate this signaling on CD43 molecules with entire functional ectodomains. Indeed, lymphocyte migration has been shown to require both the ectodomain and the intracellular domain of CD43 (4). Intramembrane proteolysis, such as γ-secretase-mediated RIP, may represent a general way of switching off all signals transmitted by transmembrane molecules after the shedding of their extracellular region (39). Finally, PS/γ-secretase processing may also release CD43 intracellular fragments endowed with nuclear localization and transcription properties. Indeed, a nuclear localization sequence has been described in CD43 cytoplasmic tail and, in carcinoma cell lines, CD43 intracellular domain was shown to translocate in the nucleus and cause an up-regulation of target genes (9). Gene transcription is known to be triggered in neutrophils upon stimulation by pro-inflammatory agonists, which also induce CD43 shedding. CD43 intracytoplasmic fragments could regulate these transcriptions, if they localize in the nucleus. Genes that would be differentially expressed in neutrophils in the absence or presence of γ-secretase inhibitors deserve investigation.
We here show that clip A is mostly prevented by plasma, presumably due to high levels of plasma serpins. However, CD43 shedding has been shown to occur in vivo on circulating PMN during hemodialysis (42) and during PMN migration to inflammation sites (43). Our data suggest that the juxtamembranous clip B partially escapes plasma inhibition. This cleavage was only weakly detected by Western blotting, presumably because of the immediate degradation of CD43-CTF by PS/γ-secretase. Interestingly, the decrease of native CD43, induced either by PMA or TNF/fMLP in plasma or by increasing concentrations of exogenous cathepsin G, reached a plateau of about 50% of the initial PMN CD43 expression. This level is reminiscent of the 50% down-regulation of CD43 observed in vitro during PMN adhesion and migration across activated endothelium (16) or in vivo during PMN migration to inflamed rheumatoid synovial fluids (43). This could reflect an heterogeneity among CD43 molecules expressed on PMN, half of them being less exposed to cathepsin G cleavage, possibly due to steric hindrance by cis-interacting membrane molecules.
If PMN activation in whole blood only allows CD43 juxtamembranous cleavage by cathepsin G, while preventing cleavages distant from the membrane, this would release the entire extracellular domain of CD43, CD43s. CD43s has indeed been described in human plasma and identified to the Galgp protein (19). We addressed the question of a possible function for CD43s, by analogy with soluble PSGL-1, which has been shown to have anti-inflammatory functions (44). Interestingly, soluble leukosialin has been reported to inhibit the adhesion of colon carcinoma cells to endothelial cells (22). We postulated that CD43s, acting as an E-selectin ligand (10, 11), could compete with cell surface selectin ligands to reduce leukocyte rolling. Our data confirm this hypothesis, because we show that CD43s inhibits the E-selectin-dependent adhesion of neutrophils to endothelium under flow conditions, while it does not interfere with their integrin-mediated static adhesion. CD43s effects were observed at a concentration compatible with normal plasma concentrations (see “Results”). Moreover, CD43s levels may be enhanced locally in inflamed microvessels, where cytokine-activated neutrophils adhere to the endothelium, a situation known to result in CD43 shedding (2, 16). The effect of CD43s is reminiscent of the reported partial inhibition of leukocyte rolling by recombinant Fc-PSGL-1, a complete inhibition being only obtained with a mixture of PSGL-1 and L-selectin blocking antibodies (44).
Therefore, CD43 shedding during neutrophil activation results in fluid phase CD43s with adhesion regulatory functions. The inhibition of PMN adhesion to endothelium by local high concentrations of CD43s may participate to the decrease of PMN recruitment required for the resolution of inflammation, as was proposed for selectins ectodomain shedding (1). This has to be added to the numerous roles of O-glycosylated membrane receptors in immune responses (45).
Neutrophil cathepsin G participates, together with elastase, to the shedding of various adhesion molecules or cytokine receptors and has been shown to interfere with neutrophil migration by several ways (reviewed in Ref. 46). Our data demonstrate a new function for cathepsin G, CD43 processing, which may be important for leukocyte migration, in view of CD43 known participation to leukocyte motility and trafficking (4, 37, 38).
Acknowledgments
We thank John Bird and Martyn Robinson, from Celltech Chiroscience Ltd, Cambridge, UK, for providing metalloproteinase inhibitors.
This work was supported, in whole or in part, by National Institutes of Health NCI Grant CA 33000. This work was also supported in part by the Association pour l'Utilisation du Rein Artificiel AURA, Baxter, and Amgen. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RIP, regulated intramembrane proteolysis; α1PI, α1-proteinase inhibitor (α1-antitrypsin); a1-CT,α1 anti-chymotrypsin; BSA, bovine serum albumin; CD43-CTF, CD43 C-terminal fragment; CD43s, soluble CD43; fMLP, N-formyl-l-methionyl-l-leucyl-l-phenylalanine; Galgp, galactoglycoprotein; HBSS, Hanks balanced solution; HUVEC, human umbilical vein endothelial cells; MMP, matrix metalloproteinase; NE, neutrophil elastase; PMA, phorbol myristate acetate; PMN, polymorphonuclear neutrophil; PR3, proteinase 3; TNF, tumor necrosis factor; FITC, fluorescein isothiocyanate; PSGL, P-selectin glycoprotein ligand; mAb, monoclonal antibody; PS, presenilin.
L. Halbwachs-Mecarelli, unpublished observation.
References
- 1.Garton, K. J., Gough, P. J., and Raines, E. W. (2006) J. Leuk. Biol. 79 1105–1116 [DOI] [PubMed] [Google Scholar]
- 2.Nathan, C., Xie, Q. W., Halbwachs-Mecarelli, L., and Jin, W. W. (1993) J. Cell Biol. 122 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manjunath, N., Correa, M., Ardman, M., and Ardman, B. (1995) Nature 377 535–538 [DOI] [PubMed] [Google Scholar]
- 4.Mody, P. D., Cannon, J. L., Bandukwala, H. S., Blaine, K. M., Schilling, A. B., Swier, K., and Sperling, A. I. (2007) Blood 110 2974–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sikut, R., Nilsson, O., Baeckstrom, D., and Hansson, G. C. (1997) Biochem. Biophys. Res. Commun. 238 612–616 [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Rodriguez, J., Andersson, C. X., Laos, S., Baeckstrom, D., Sikut, A., Sikut, R., and Hansson, G. C. (2002) Tumour Biol. 23 193–201 [DOI] [PubMed] [Google Scholar]
- 7.Andersson, C. X., Fernandez-Rodriguez, J., Laos, S., Baeckstrom, D., Haass, C., and Hansson, G. C. (2005) Biochem. J. 387 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selkoe, D. J., and Wolfe, M. S. (2007) Cell 131 215–221 [DOI] [PubMed] [Google Scholar]
- 9.Andersson, C. X., Fernandez-Rodriguez, J., Laos, S., Sikut, R., Sikut, A., Baeckstrom, D., and Hansson, G. C. (2004) Biochem. Biophys. Res. Commun. 316 12–17 [DOI] [PubMed] [Google Scholar]
- 10.Fuhlbrigge, R. C., King, S. L., Sackstein, R., and Kupper, T. S. (2006) Blood 107 1421–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto, M., Atarashi, K., Umemoto, E., Furukawa, Y., Shigeta, A., Miyasaka, M., and Hirata, T. (2005) J. Immunol. 175 8042–8050 [DOI] [PubMed] [Google Scholar]
- 12.Campanero, M. R., Pulido, R., Alonso, J. L., Pivel, J. P., Pimentel-Muinos, F. X., Fresno, M., and Sanchez-Madrid, F. (1991) Eur. J. Immunol. 21 3045–3048 [DOI] [PubMed] [Google Scholar]
- 13.Kuijpers, T. W., Hoogerwerf, M., Kuijpers, K. C., Schwartz, B. R., and Harlan, J. M. (1992) J. Immunol. 149 998–1003 [PubMed] [Google Scholar]
- 14.Rieu, P., Porteu, F., Bessou, G., Lesavre, P., and Halbwachs-Mecarelli, L. (1992) Eur. J. Immunol. 22 3021–3026 [DOI] [PubMed] [Google Scholar]
- 15.Remold-O'Donnell, E., and Parent, D. (1994) J. Immunol. 152 3595–3605 [PubMed] [Google Scholar]
- 16.Lopez, S., Seveau, S., Lesavre, P., Robinson, M. K., and Halbwachs-Mecarelli, L. (1998) Cell Adhes. Commun. 5 151–160 [DOI] [PubMed] [Google Scholar]
- 17.Bazil, V., and Strominger, J. L. (1994) J. Immunol. 152 1314–1322 [PubMed] [Google Scholar]
- 18.Gardiner, E. E., De Luca, M., McNally, T., Michelson, A. D., Andrews, R. K., and Berndt, M. C. (2001) Blood 98 1440–1447 [DOI] [PubMed] [Google Scholar]
- 19.Schmid, K., Hediger, M. A., Brossmer, R., Collins, J. H., Haupt, H., Marti, T., Offner, G. D., Schaller, J., Takagaki, K., Walsh, M. T., et al. (1992) Proc. Natl. Acad. Sci. U.S.A. 89 663–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nusbaum, P., Laine, C., Bouaouina, M., Seveau, S., Cramer, E. M., Masse, J. M., Lesavre, P., and Halbwachs-Mecarelli, L. (2005) J. Biol. Chem. 280 5843–5853 [DOI] [PubMed] [Google Scholar]
- 21.Renesto, P., Halbwachs-Mecarelli, L., Nusbaum, P., Lesavre, P., and Chignard, M. (1994) J. Immunol. 152 4612–4617 [PubMed] [Google Scholar]
- 22.Sawada, R., Tsuboi, S., and Fukuda, M. (1994) J. Biol. Chem. 269 1425–1431 [PubMed] [Google Scholar]
- 23.Jaffe, E. A., Nachman, R. L., Becker, C. G., and Minick, C. R. (1973) J. Clin. Investig. 52 2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierhuizen, M. F., Maemura, K., and Fukuda, M. (1994) J. Biol. Chem. 269 4473–4479 [PubMed] [Google Scholar]
- 25.Legendre, P., Salsmann, A., Rayes, J., Trassard, O., Kieffer, N., and Baruch, D. (2006) J. Thromb. Haemost. 4 236–246 [DOI] [PubMed] [Google Scholar]
- 26.Radford, D. J., Luu, N. T., Hewins, P., Nash, G. B., and Savage, C. O. (2001) Arth. Rheum. 44 2851–2861 [DOI] [PubMed] [Google Scholar]
- 27.Halbwachs-Mecarelli, L., Bessou, G., Lesavre, P., Renesto, P., and Chignard, M. (1996) Blood 87 1200–1202 [PubMed] [Google Scholar]
- 28.Travis, J., and Salvesen, G. S. (1983) Annu. Rev. Biochem. 52 655–709 [DOI] [PubMed] [Google Scholar]
- 29.Remold-O'Donnell, E., and Parent, D. (1995) Blood 86 2395–2402 [PubMed] [Google Scholar]
- 30.Shelley, C. S., Remold-O'Donnell, E., Davis, A. E., 3rd, Bruns, G. A., Rosen, F. S., Carroll, M. C., and Whitehead, A. S. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 2819–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima, K., Powers, J. C., Ashe, B. M., and Zimmerman, M. (1979) J. Biol. Chem. 254 4027–4032 [PubMed] [Google Scholar]
- 32.Owen, C. A., Hu, Z., Lopez-Otin, C., and Shapiro, S. D. (2004) J. Immunol. 172 7791–7803 [DOI] [PubMed] [Google Scholar]
- 33.Nie, J., and Pei, D. (2004) Exp. Cell Res. 296 145–150 [DOI] [PubMed] [Google Scholar]
- 34.Korkmaz, B., Poutrain, P., Hazouard, E., de Monte, M., Attucci, S., and Gauthier, F. L. (2005) Am. J. Resp. Cell Mol. Biol. 32 553–559 [DOI] [PubMed] [Google Scholar]
- 35.Mirinics, Z. K., Calafat, J., Udby, L., Lovelock, J., Kjeldsen, L., Rothermund, K., Sisodia, S. S., Borregaard, N., and Corey, S. J. (2002) Blood Cells, Mol. Dis. 28 28–38 [DOI] [PubMed] [Google Scholar]
- 36.Park, H. Y., Park, J. I., Baek, D. W., Lee, S. Y., Lee, M. J., Jin, J. O., Kim, J. W., Hong, Y. S., Lee, Y. H., and Kwak, J. Y. (2006) Int. Immunopharmacol. 6 1061–1069 [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Mateos, P., Campanero, M. R., del Pozo, M. A., and Sanchez-Madrid, F. (1995) Blood 86 2228–2239 [PubMed] [Google Scholar]
- 38.Seveau, S., Keller, H., Maxfield, F. R., Piller, F., and Halbwachs-Mecarelli, L. (2000) Blood 95 2462–2470 [PubMed] [Google Scholar]
- 39.Kopan, R., and Ilagan, M. X. (2004) Nat. Rev. 5 499–504 [DOI] [PubMed] [Google Scholar]
- 40.Marambaud, P., Shioi, J., Serban, G., Georgakopoulos, A., Sarner, S., Nagy, V., Baki, L., Wen, P., Efthimiopoulos, S., Shao, Z., Wisniewski, T., and Robakis, N. K. (2002) EMBO J. 21 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker, J., and Green, J. M. (1999) J. Immunol. 162 4109–4114 [PubMed] [Google Scholar]
- 42.Alvarez, V., Pulido, R., Campanero, M. R., Paraiso, V., de Landazuri, M. O., and Sanchez-Madrid, F. (1991) Kidney Int. 40 899–905 [DOI] [PubMed] [Google Scholar]
- 43.Lopez, S., Halbwachs-Mecarelli, L., Ravaud, P., Bessou, G., Dougados, M., and Porteu, F. (1995) Clin. Exp. Immunol. 101 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridger, V. C., Hellewell, P. G., and Norman, K. E. (2005) Am. J. Pathol. 166 945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuboi, S., and Fukuda, M. (2001) Bioessays 23 46–53 [DOI] [PubMed] [Google Scholar]
- 46.Pham, C. T. (2006) Nat. Rev. Immunol. 6 541–550 [DOI] [PubMed] [Google Scholar]