Abstract
Forkhead transcription factor Foxc2 is an essential regulator of the cardiovascular system in development and disease. However, the cellular and molecular functions of Foxc2 in vascular endothelial cells are still not fully understood. Here, through gene expression profiling in endothelial cells, we identified molecules associated with cell-extracellular matrix interactions, integrin β3 (Itgb3), integrin β5 (Itgb5), and fibronectin, as downstream targets of Foxc2. We found that Itgb3 expression is directly regulated by Foxc2 through multiple Forkhead-binding elements within two high homology regions in the Itgb3 promoter. Because Itgb3 is known to regulate angiogenesis, we further tested whether Foxc2 is directly involved in angiogenesis by regulating Itgb3 expression by in vitro experiments. Overexpression of Foxc2 significantly enhanced endothelial cell migration and adhesion, whereas this effect was strongly inhibited by Itgb3 neutralization antibody. In accordance with these results, pulmonary microvascular endothelial cells isolated from Foxc2 heterozygous mutant mice showed a marked reduction in Itgb3 expression and cell migration. Finally, ex vivo aortic ring assay to test the sprouting and microvessel formation revealed enhanced microvessel outgrowth by Foxc2 overexpression. Conversely, microvessel outgrowth from aortas of Foxc2 heterozygous mutant mice was reduced. Taken together, these results suggest that Foxc2 directly induces Itgb3 expression and regulates angiogenesis by Itgb3-mediated endothelial cell adhesion and migration.
Pathological and physiological angiogenesis is a complex event involving endothelial cell proliferation and migration and vascular tube formation. In this process, the penetration of new endothelial cells occurs in avascular zones by sprouting from existing vascular vessels (1). Dynamic conformation changes in extracellular matrix adhesion are an essential part of sprouting angiogenesis because the extracellular matrix provides a mechanical substrate in the cellular environment and regulates pathophysiological signals to neighboring cells. The role of integrins has been extensively studied in a variety of molecules participating in interactions between vascular endothelial cells and the extracellular matrix (2, 3).
Integrins represent a large family of heterodimeric adhesion receptors that interact with extracellular matrix components such as fibronectin and vitronectin. At present, there are 16α- and 8β-subunits identified, thereby forming 24 different integrins. Although accumulating evidence shows that αvβ3 and αvβ5 integrins play a pivotal role in the formation of blood vessels (2), Itgb3 and Itgb5 are dispensable for embryonic angiogenesis (4). However, given that Itgb3 and Itgb5 are highly expressed in vascular vessels in pathological angiogenesis, blockade of these integrins with specific antagonists inhibits angiogenesis in cancer as well as arthritis and ischemic retinopathy (2, 5, 6). Therefore, as a therapeutic approach to prevent pathological angiogenesis and tumor growth, Abegrin, a humanized monoclonal antibody that targets αvβ3 integrin, is being tested in the phase II clinical trial for anti-tumor therapy of melanoma patients (7).
The Foxc2 gene encodes a protein belonging to the Forkhead/Fox transcription factor family that shares an evolutionarily conserved DNA-binding domain. Our laboratory and others have shown that Foxc2 is an essential regulator of vascular/lymphatic vessel formation in cardiovascular development and disease. Disruption of Foxc2 in mouse causes perinatal lethality with defects in aortic arch patterning, arterial specification, and vessel remodeling during embryonic development (8–12). Furthermore, mutations in mouse and human result in lymphedema associated with abnormal lymphatic patterning, failure of lymphatic and venous valves, and lymphatic and venous dysfunction (13–16). However, the precise function of Foxc2 in the process of angiogenesis remains largely unknown. The goal of this study is therefore to elucidate the mechanisms by which Foxc2 functions in vascular endothelial cells. Here, we present the first evidence that Itgb3 is a novel downstream target of Foxc2 and that Foxc2 regulates angiogenesis by contributing primarily to endothelial cell migration and adhesion.
MATERIALS AND METHODS
Cell Culture—Immortalized mouse embryonic endothelial cells (MEECs)2 were obtained from Dr. Marie-Jose Goumans (The Netherlands Cancer Institute) and cultured on gelatin-coated tissue culture plates at 37 °C in a 5% CO2 incubator in Dulbecco's modified Eagle's medium containing 10% serum as described (17). Mouse pulmonary microvascular endothelial cells (PMVECs) were isolated and cultured as described (18).
Generation and Infection of Recombinant Adenovirus for Foxc2—To generate an adenovirus expression vector containing Foxc2, the complete coding sequence of mouse Foxc2 with XbaI sites at the 5′- and 3′-ends was first generated using Pfu DNA polymerase (Stratagene) and primers including XbaI sites (5′-TATCTAGAGGACGCATGCAGGCG-3′ and 5′-CTCTAGACTCAGTATTTGGTGCAGTGG-3′). The XbaI sites and the Kozak sequence upstream of the start codon for efficient translation are underlined and double-underlined, respectively. The initiation and stop codons are shown in boldface. The PCR product was subcloned into the EcoRV site of pBluescript II KS+ (Stratagene) and then sequenced for confirmation. The Foxc2 XbaI fragment was cloned into the XbaI site of the shuttle vector pAdTrack-CMV. To generate the adenovirus expression vector for Foxc2 (Ad-Foxc2) by homologous recombination, the resultant plasmid linearized with PmeI was cotransformed into Escherichia coli BJ5183 cells with the adenoviral backbone pAdEasy-1 vector containing green fluorescent protein to monitor the efficiency of transfection and infection. Adenovirus vectors (Ad-control and Ad-Foxc2) linearized with PacI were transfected into 293T cells to produce adenoviruses. Following three rounds of amplification in 293T cells, the adenoviruses were purified and titered based on the efficiency of infection by counting the number of green fluorescent protein-expressing cells as described previously (19). Infection of MEECs with the adenoviruses was performed as described previously (17).
Identification of Putative Fox-binding Elements (FBEs)—The alignment of mouse and human genomic sequences containing up to 30 kb upstream of the transcription start site of Itgb3 was visualized using mVISTA (genome.lbl.gov/vista/index.shtml). Among highly conserved regions (>50% identity), FBEs were subsequently selected according to the consensus sequence WAARYAAAYW (where W = Ala or Thr, R = Ala or Gly, and Y = Cys or Thr).
Real-time Reverse Transcription (RT)-PCR Analysis—Isolation of total RNA from endothelial cells and cDNA synthesis were performed using an RNeasy mini kit (Qiagen Inc.) and iScript (Bio-Rad), respectively. Real-time PCRs were carried out using the SYBR Green PCR Master Mix (Applied Biosystems) and iCycler (Bio-Rad) according to the manufacturers' instructions. The PCR primers used are shown in supplemental Table 1. The data were normalized by the expression levels of the ppi housekeeping gene.
Fluorescence-activated Cell Sorter Analysis—Dissociated PMVECs or MEECs were washed with phosphate-buffered saline (PBS) supplemented with 1% fetal calf serum and stained with phycoerythrin-conjugated anti-mouse CD61 (Itgb3) antibody (eBioscience) for 30 min on ice. 7-Aminoactinomycin D (Invitrogen) was also used to identify and remove a nonviable cell population. Cells were processed for cytometric analyses by LSR II (BD Biosciences) or for sorting by FACSAria (BD Biosciences) at the Vanderbilt University Flow Cytometry Core Facility.
Isolation of Endothelial Cells Stably Overexpressing Foxc2—MEECs were seeded at a density of 6 × 104 cells/well on 24-well tissue culture plates. The next day, these cells were transfected with 400 ng of the Foxc2 expression vector or pcDNA3.0 (Invitrogen) as a mock control using Lipofectamine and PLUS reagents (Invitrogen). After 3 days of transfection, the cells were continuously treated with Geneticin (5 mg/ml; Invitrogen) to collect a pool of Geneticin-resistant cells. The expression levels of Foxc2 were subsequently confirmed by semiquantitative RT-PCR (data not shown). The PCR primers for Foxc2 are shown in supplemental Table 1.
Cell Proliferation Assay—Cell proliferation was assessed by the incorporation of 5-bromo-2′-deoxyuridine (BrdUrd) into cellular DNA. MEECs were plated on gelatin-coated Lab-Tek chamber slides (Nalgene) at a density of 3.5 × 103 cells/chamber. Attached cells were cultured for 24 h in serum-reduced medium (0.5%) with a pulse of BrdUrd (25 mm) for the last 3 h. The cells were fixed with 4% paraformaldehyde immediately after the pulse and incubated with anti-BrdUrd monoclonal antibody (Sigma) for 1 h. After washing with Tris-buffered saline and 0.1% Tween 20, the cells were incubated with Alexa 568-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories). To visualize all cell nuclei, the slides were also stained with 4′,6-diamidino-2-phenylindole. Three random non-overlapping fields of view (total magnification ×200) were photographed in duplicate. The percentage of the uptake of BrdUrd into the cells was calculated based on the ratio of 4′,6-diamidino-2-phenylindole-positive and BrdUrd-positive cells to 4′,6-diamidino-2-phenylindole-positive and BrdUrd-negative cells. The experiment was repeated three times.
Construction of Luciferase Reporters—For construction of distal HHR-Itgb3-LUC, proximal Itgb3-LUC, and –793Itgb3-LUC, PCR fragments were amplified using a mouse bacterial artificial chromosome clone (RP24–94A3; BACPAC Resource Center) as template DNA. PCR-based mutagenesis of the FBEs in 1Mut-LUC (TCTATTT to TCTAGGT), 2Mut-LUC (AAAATAAA to AAGGTGGA), and 3Mut-LUC (AAAATAAA to AAGGTGGA) was performed using the distal HHR-Itgb3-LUC reporter as template DNA. All fragments were subsequently purified and ligated into the KpnI and NheI sites of the pGL3-basic vector or pGL3-promoter vector (Promega), followed by sequence confirmation. The primers used for plasmid construction are shown in supplemental Table 1.
Transfection and Luciferase Assay—MEECs were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For luciferase assays, the pRL-CMV reporter plasmid (Promega) containing Renilla luciferase as an internal control was cotransfected with the firefly luciferase reporter constructs described above. All transfections were carried out in triplicate on gelatin-coated 24-well plates. The transfected cells were harvested at 48 h after transfection, and luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega).
Chromatin Immunoprecipitation Assay—Cultured MEECs were washed with PBS and treated with formaldehyde to cross-link protein to DNA. Cellular lysates were obtained by scraping, followed by pulse ultrasonication to shear cellular DNA. After centrifugation, supernatants containing sheared chromatin were incubated with anti-Foxc2 antibody or control IgG (Abcam), followed by addition of protein A/G-Sepharose (Santa Cruz Biotechnology) overnight. After elution, immune complexes were subsequently treated with proteinase K at 55 °C for 1.5 h and extracted with phenol/chloroform and chloroform. Immunoprecipitated DNA was analyzed by PCR. The PCR primers used are shown in supplemental Table 1.
Adhesion Assay—Confluent MEECs stably transfected with the mock or Foxc2 expression vector were detached and preincubated with antibody against Itgb3 at 5 mg/ml or with control IgG for 30 min at 37 °C. After washing three times with Dulbecco's modified Eagle's medium containing 0.1% bovine serum albumin (BSA), 5 × 104 cells were plated on 96-well cell culture plates precoated with 10 μg/ml vitronectin for 1 h at 37 °C, blocked with 1% BSA for 1 h, and incubated for 30 min at 37 °C. The attached cells were stained and fixed with 0.1% crystal violet aqueous solution in 20% methanol for 10 min. After washing with water, the stained cells were dissolved with 1% SDS. The absorbance at 595 nm was measured using a microplate reader.
Wound Closure Assay—Endothelial cell migration was determined by wound closure assay. MEECs were grown to confluence on gelatin-coated 6-well plates in growth culture medium. Monolayer cells were mechanically wounded by scratching with the tip of a sterile plastic 10-ml pipette. After washing with PBS, the cells were cultured in serum-reduced medium (0.5%) for 24 h. The cells were treated with or without 10 ng/ml mouse recombinant vascular endothelial growth factor (VEGF) (R&D Systems) in the presence or absence of 5 mg/ml R-phycoerythrin-conjugated hamster anti-mouse β3 integrin monoclonal antibody (clone 2C9.G2; Armenian hamster IgG1κ; BD Biosciences) or IgG isotype control (clone A19-3; BD Biosciences). Residual wound areas were measured at 0 and 24 h by comparing phase-contrast micrographs of three fields of view (total magnification ×40) along the originally scraped areas in triplicate. The recovered areas were calculated using ImageJ software. To visualize the cell migration patterns after 24 h, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet.
Transwell Migration Assay—Transwells with 8-mm pores (Costar) were precoated with collagen I (20 mg/ml) overnight at 4 °C. The filters were subsequently blocked with 3% BSA in PBS to inhibit nonspecific migration. The lower wells of the chamber were filled with serum-free medium containing 0.1% BSA in the presence or absence of VEGF at 25 ng/ml. PMVECs from Foxc2+/– or control wild-type mouse suspensions were prepared from subconfluent cultures in serum-free medium containing 0.1% BSA. A total of 5 × 104 cells were added to the upper chamber and then incubated for 4 h at 37°C. Non-migrating cells on top of the filter were wiped out by a cotton swab, and the filters were fixed in 4% formaldehyde in PBS. Migrating cells were stained with 1% crystal violet and counted under a microscope.
Aortic Ring Assay—C57BL/6 mice (6–8 weeks old) were killed, and thoracic aortas were excised and immediately transferred to a culture dish with cold serum-free Dulbecco's modified Eagle's medium. The periaortic fibroadipose tissue was carefully removed, and aortic tubes were sagittally cut in two equal pieces to make two equivalent half-tubes. In the experiment shown in Fig. 7A, one of the half-tubes was immediately exposed to Ad-Foxc2 and the other to Ad-control each at 5 × 109 plaque-forming units in 0.1 ml of Dulbecco's modified Eagle's medium in 1.5-ml centrifugation tubes at 37 °C in 5% CO2 for 24 h. (This process was skipped for the experiment shown in Fig. 7B.) Infected tubes were then each cut into cross-sectional pieces of 0.5 mm in length using a razorblade. Pieces of aorta were placed on 48-well culture plates and overlaid with 150 ml of fibrinogen (Sigma) at 3 mg/ml. Polymerization was performed by the addition of 15 ml of thrombin (Sigma) at 1 unit/ml for 30 min, followed by the addition of 0.5 ml of EBM-2 (Lonza). The culture of aortic rings and analysis of microvessel outgrowth were as described previously (20).
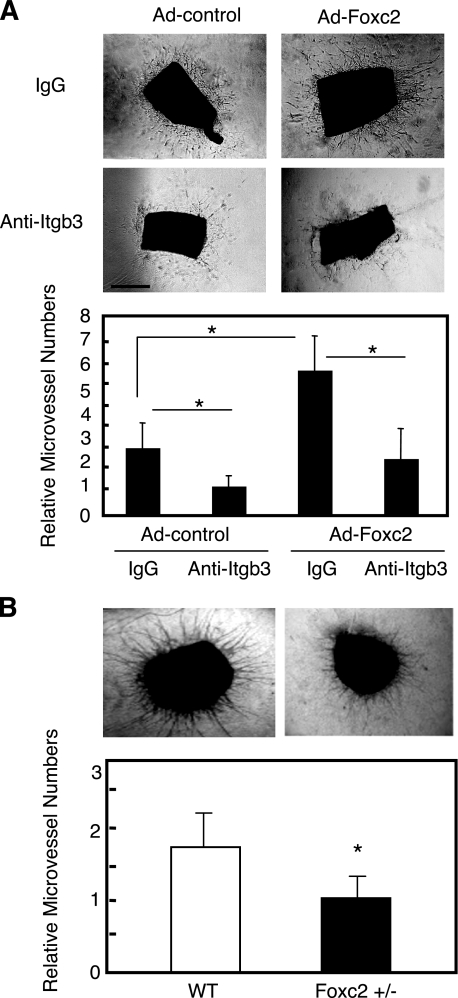
FIGURE 7.
Foxc2 regulates microvessel outgrowth through Itgb3 function from mouse aortic rings. A, aortic ring assay using wild-type aortas infected with Ad-Foxc2 or Ad-control. Thoracic aortas excised from wild-type (WT) mice were sagittally cut in two pieces, and a pair of equivalent tubes was infected with either Ad-Foxc2 or Ad-control. They were treated with either nonspecific control murine IgG or monoclonal antibody against Itgb3 at 20 mg/ml and then analyzed as described under “Materials and Methods”. B, aortic ring assay using Foxc2+/– mice. Thoracic aortas excised from Foxc2+/– mice were subjected to aortic ring assay as described above. Data are presented as the relative number of microvessels sprouting from aortic rings. Results are presented as the means ± S.D. (n = 9 or more). p values were determined by the corresponding sample indicated using Student's t test. *, p < 0.05 versus the corresponding control.
RESULTS
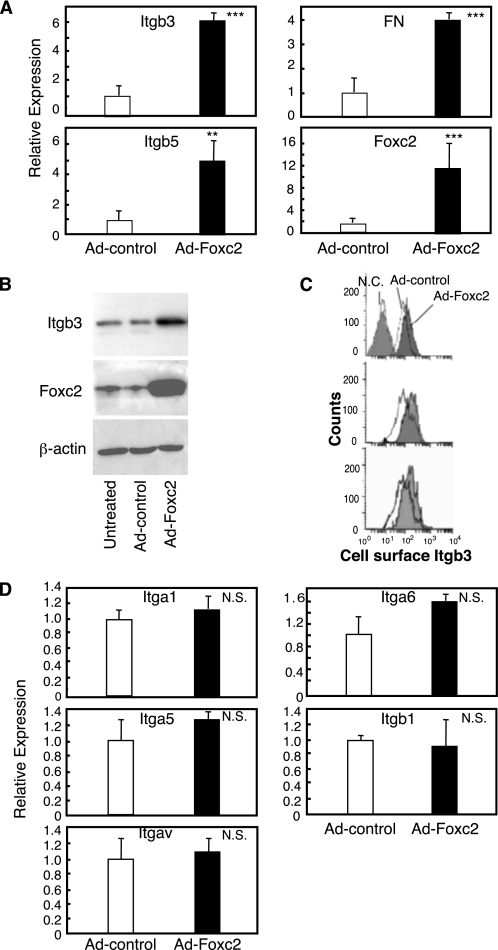
Foxc2 Directly Regulates Integrin β3 Expression in Endothelial Cells—To identify genes regulated by Foxc2 in endothelial cells, we performed gene expression profiling using immortalized MEECs by infection of recombinant adenovirus for Foxc2. On the basis of the 39,000 oligoprobe sets of Mouse Genome 430 2.0 Array (Affymetrix) we used, overexpression of Foxc2 in MEECs up-regulated 448 probe sets and down-regulated 156 probe sets. The details of the microarray results will be published elsewhere.3 Interestingly, we found that the expression of Itgb3 and Itgb5, known as components of αvβ3 and αvβ5 integrin heterodimers, and fibronectin (FN), a ligand for αvβ3 integrin, was significantly up-regulated. By real-time RT-PCR, we validated the induction of these genes by Foxc2 (Fig. 1A). We next confirmed that Itgb3 protein was indeed up-regulated in endothelial cells that overexpress Foxc2 by Western blot analysis (Fig. 1B). Consistently, the surface levels of Itgb3 expression were increased by Foxc2 as evaluated by flow cytometry (Fig. 1C). In contrast, Foxc2 did not regulate the expression of other integrins, including αv integrin (Itgav) (Fig. 1D). We therefore hypothesized that overexpression of Foxc2 could induce pro-angiogenic factors in endothelial cells because αvβ3 and αvβ5 integrins as well as FN were all up-regulated on certain tumor vasculature (2, 6, 21). Among these molecules, αvβ3 integrin is recognized as a promising therapeutic target for pathological angiogenesis.
FIGURE 1.
Foxc2 induces Itgb3 expression in endothelial cells. A, Foxc2 increases mRNA levels of Itgb3, Itgb5, and FN as detected by real-time RT-PCR. MEECs were infected with either Ad-control or Ad-Foxc2. Sixteen hours after infection, the cells were washed and cultured for 30 h. Total RNA was then isolated and subjected to real-time RT-PCR to detect expression levels of Itgb3, Itgb5, FN, and Foxc2. The experiment was independently repeated three times, and statistical significance was determined by Student's t tests. **, p < 0.01; ***, p < 0.005 versus Ad-control-infected cells. B, Foxc2 up-regulates Itgb3 protein in endothelial cells. Total protein lysates were prepared from MEECs infected with either Ad-Foxc2 or Ad-control and from untreated MEECs and subjected to Western blotting using antibodies against Foxc2 and Itgb3. Expression of β-actin is shown as a loading control. C, surface levels of Itgb3 expression in MEECs infected with either Ad-control or Ad-Foxc2 as measured by flow cytometry. Replicate analyses of three independent experiments are shown. The negative control (N.C.) indicates unstained MEECs. D, Foxc2 does not induce expression of Itga1, Itga5, Itga6, Itgav, and Itgb1. MEECs were infected with either Ad-control or Ad-Foxc2, and real-time RT-PCR was subsequently performed. The experiment was independently repeated three times, and statistical significance was determined by Student's t tests. N.S., non-significant.
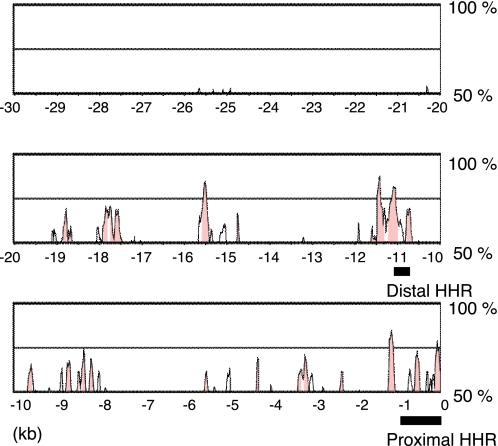
Because the molecular mechanisms for Itgb3 gene expression in angiogenesis are not fully understood, we further analyzed Foxc2-mediated Itgb3 gene expression in endothelial cells. A recent study showed that another Fox transcription factor, Foxf1, activates the mouse Itgb3 promoter by directly binding to the tandem repeats of FBEs located at region–871 to –815 of the Itgb3 locus in lung mesenchymal cells and induces mesenchymal cell migration (22). Notably, the reported FBEs are not found in the proximal HHR of the human Itgb3 promoter, as shown in Figs. 2 and 3A. Because we found that overexpression of Foxc2 induced Itgb3 mRNA expression in human umbilical vein endothelial cells (data not shown), we searched for additional FBEs farther upstream (up to 30 kb) of the Itgb3 locus (Fig. 2). In fact, we found a cluster of three FBEs located in the distal HHR between –11.1 and –10.8 kb upstream of Itgb3 (Figs. 2 and 3C). This finding led us hypothesize that the distal HHR is also responsible for Foxc2-induced Itgb3 expression.
FIGURE 2.
Alignment of 30-kb upstream regions from the transcription initiation site of Itgb3 between human and mouse using mVISTA. HHRs are shown in pink.
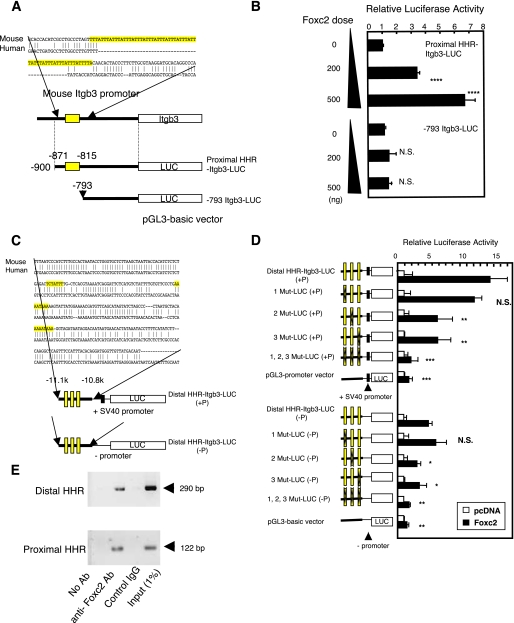
FIGURE 3.
Foxc2 regulates the proximal and distal promoters of Itgb3 in endothelial cells. A, schematic diagram of luciferase constructs for the proximal Itgb3 promoter (proximal HHR-Itgb3-LUC). Shown above are the mouse and human sequences for comparison. Note that the reported tandem repeats of the FBEs (highlighted) are located only in the mouse Itgb3 locus. B, Foxc2 activates the proximal Itgb3 promoter in a dose-dependent manner. MEECs were transfected with the Foxc2 expression vector together with either proximal HHR-Itgb3-LUC or its deletion construct, –793Itgb3-LUC. The -fold increase in normalized luciferase activity is shown. Statistical significance was determined by Student's t tests. ****, p < 0.001 versus the control. N.S., non-significant. C, schematic diagram of luciferase constructs for the distal Itgb3 promoter that consists of the three conserved FBEs (highlighted). Distal HHR-Itgb3-LUC (+P) and distal HHR-Itgb3-LUC (–P) indicate the presence and absence of the SV40 promoter in the constructs, respectively. Shown above are the mouse and human sequences for comparison. D, Foxc2 activates the distal Itgb3 promoter through two of the three FBEs. Luciferase assays were performed after transfection of MEECs with distal HHR-Itgb3-LUC or a series of mutant reporters (1Mut-LUC, 2Mut-LUC, 3Mut-LUC, and 1,2,3Mut-LUC) along with the Foxc2 expression vector. The -fold increase in normalized luciferase activity is shown. Data are presented as the means ± S.D. (n = 6) from two independent experiments. *, p < 0.05; **, p < 0.01 versus cells cotransfected with Foxc2 and pGL3-basic. E, Foxc2 binds to the FBEs in the distal or proximal HHRs in chromatin structure of endothelial cells. MEECs were subjected to chromatin immunoprecipitation assays using anti-Foxc2 antibody (Ab) or control IgG. Immunoprecipitated DNA was analyzed by PCR using primers specific to the proximal or distal HHR of mouse Itgb3.
To test the direct involvement of Foxc2 in the induction of Itgb3 expression, we next investigated whether Foxc2 could activate the Itgb3 promoter by regulating the FBEs in the distal and proximal HHRs of the mouse Itgb3 promoter. For analysis of the proximal HHR, we constructed luciferase reporters using the pGL3-basic vector that does not contain a basic promoter (Fig. 3A). On the other hand, for analysis of the distal HHR, we generated luciferase reporters using the pGL3-basic vector or the pGL3-promoter vector (Fig. 3C). The pGL3-promoter vector carries the SV40 promoter sequence and is designed to analyze enhancer sequences upstream of a gene of interest. In Fig. 3B, Foxc2 significantly activated the proximal HHR in a dose-dependent manner, whereas the deletion construct lacking the FBEs showed no induction of luciferase activity by Foxc2. These data suggest that similar to Foxf1, Foxc2 can activate the mouse Itgb3 proximal promoter. More important, Foxc2 was also able to activate the distal HHR through the FBEs except for the most upstream one (Fig. 3D). It should also be noted that essentially similar results were obtained in experiments with luciferase constructs in the presence or absence of the SV40 promoter. Next, we performed chromatin immunoprecipitation assays to test whether Foxc2 could directly bind to the FBEs in the distal or proximal HHRs in chromatin structure of endothelial cells. We found that Foxc2 specifically bound to both HHRs in vivo (Fig. 3E). Taken together, these findings indicate that Foxc2 directly induces Itgb3 expression through the multiple FBEs on the Itgb3 distal and proximal HHRs in endothelial cells.
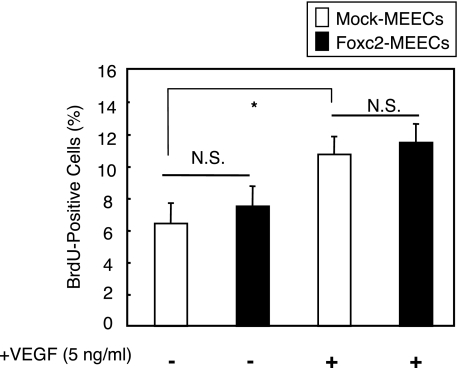
Foxc2 Regulates Itgb3-mediated Endothelial Cell Migration and Adhesion, but Not Proliferation—Angiogenesis is a multi-step process that includes endothelial cell proliferation, migration, and adhesion. Considering the direct induction of Itgb3 expression by Foxc2, we sought to determine whether Foxc2 regulates the angiogenic process in vitro. Using MEECs stably overexpressing Foxc2, the rate of BrdUrd incorporation in endothelial cells was determined in serum-reduced medium (0.5%) for 24 h. We observed no significant difference in the number of BrdUrd-positive cells between mock- and Foxc2-transfected MEECs (Fig. 4). In addition, although VEGF augmented cell division, Foxc2 overexpression along with VEGF did not show further increase in proliferation compared with the control cells treated with VEGF. These findings indicate that Foxc2 itself does not have the ability to stimulate the proliferation of endothelial cells.
FIGURE 4.
Proliferation assay for Foxc2-overexpressing endothelial cells. MEECs stably transfected with the mock or Foxc2 expression vector were cultured with or without VEGF (5 ng/ml) for 24 h in serum-reduced medium (0.5%) with a pulse of BrdUrd (BrdU) for the last 3 h. The cells were subsequently fixed and stained for BrdUrd and 4′,6-diamidino-2-phenylindole. The number of BrdUrd-positive cells and total cells was counted. Values are presented as the means ± S.D. from 10 microscopic fields from two independent experiments. Statistical significance was determined by Student's t tests. *, p < 0.05. N.S., non-significant.
We next analyzed the effect of Foxc2 on endothelial cell adhesion that is regulated by integrins during angiogenesis (23). Adhesion assays were performed using 96-well culture plates precoated by vitronectin. As shown in Fig. 5A, Foxc2 significantly increased adhesiveness compared with the control cells. Notably, consistent with our finding that Foxc2 directly regulated Itgb3 expression, Foxc2-enhanced cell adhesion was impaired by anti-Itgb3 blocking antibody.
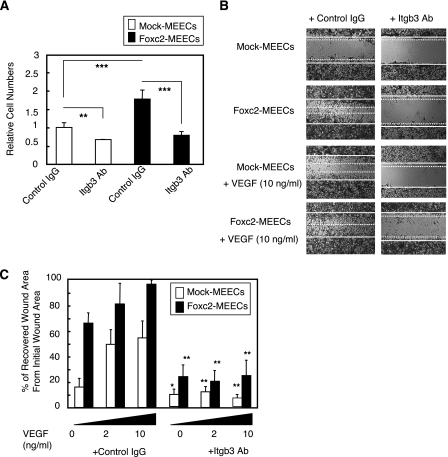
FIGURE 5.
Increased adhesion and migration in Foxc2-overexpressing endothelial cells. A, Foxc2-induced cell adhesion is inhibited by blocking Itgb3 function. MEECs stably transfected with the mock or Foxc2 expression vector were preincubated with anti-Itgb3 antibody (Ab) or control IgG and plated on vitronectin-coated 96-well plates for 30 min. The relative numbers of the adherent cells were measured by absorbance after crystal violet staining. Data are presented as the means ± S.D. from three independent experiments. Statistical significance was determined by Student's t tests. **, p < 0.01; ***, p < 0.005. B, Foxc2-induced cell migration is inhibited by blocking Itgb3 function. Confluent MEECs stably transfected with the mock or Foxc2 expression vector were scraped to induce a wound and cultured with anti-mouse Itgb3 antibody or control IgG in the presence or absence of VEGF (10 ng/ml). At 24 h after wounding, the cells were stained with crystal violet and photographed. Unbroken and dotted lines define the wounded areas by the initial scraping and the recovered areas after 24 h, respectively. Cell migration was evaluated as described under “Materials and Methods.” Representative experiments are shown. C, quantitative measurement of cell migration during wound closure of B. Values are presented as the means ± S.D. of nine microscopic fields. The statistical significance of treatment with anti-Itgb3 antibody was determined by Student's t tests. *, p < 0.05; **, p < 0.01 versus corresponding mock- or Foxc2-overexpressing cells treated with control IgG.
Finally, we performed wound closure assays to test whether Foxc2 is involved in endothelial cell migration. In agreement with the effect of Foxc2 on endothelial cell adhesion, wound closure assays further revealed that Foxc2 dramatically enhanced endothelial cell migration (Fig. 5, B and C). Of note, although Foxc2 did not induce endothelial cell proliferation under the same serum-reduced conditions (Fig. 4), the rate of Foxc2-enhanced wound closure was almost equivalent to that of VEGF treatment in the control cells. On the other hand, the addition of VEGF did not synergistically increase the rate of would closure by Foxc2. Furthermore, treatment with anti-Itgb3 blocking antibody significantly inhibited Foxc2-induced cell migration. Therefore, these data demonstrate that Foxc2 plays an important role in endothelial cell adhesion and migration, at least in part through Itgb3 function.
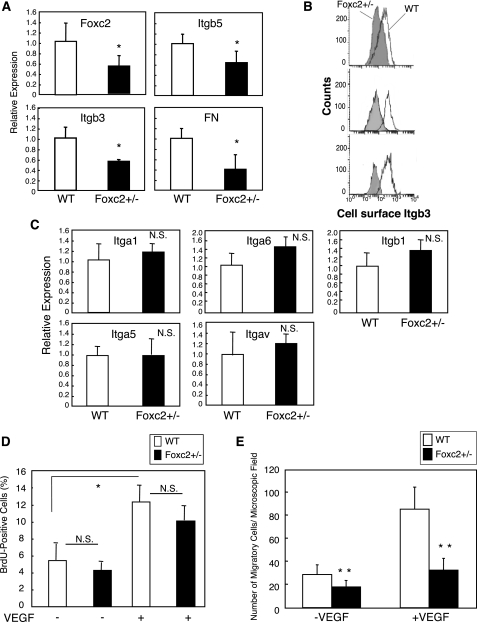
Loss of Foxc2 Function Diminishes Endothelial Cell Migration—To further test whether loss of Foxc2 affects the migration of endothelial cells, PMVECs were isolated form wild-type and Foxc2 heterozygous mutant (Foxc2+/–) mice. As shown in Fig. 6A, real-time RT-PCR analysis demonstrated a marked reduction in expression of Itgb3, Itgb5, and FN in endothelial cells isolated from Foxc2+/– mice. In accordance with this observation, the surface levels of Itgb3 expression measured by flow cytometry were markedly reduced in Foxc2+/– endothelial cells (Fig. 6B). On the other hand, no change in expression of other integrins was observed in Foxc2+/– endothelial cells compared with wild-type cells (Fig. 6C).
FIGURE 6.
Foxc2 deficiency results in a reduction in endothelial cell migration and Itgb3 expression. A, reduced mRNA expression of Itgb3, Itgb5, and FN in PMVECs isolated from Foxc2+/– mice. Total RNA was prepared from PMVECs isolated from wild-type (WT) and Foxc2+/– mice, and real-time RT-PCR was performed. Results are presented as the means ± S.D. from triplicate experiments. Statistical significance was determined by Student's t tests. *, p < 0.05 versus the wild type. B, surface levels of Itgb3 expression in PMVECs isolated from wild-type and Foxc2+/– mice measured by flow cytometry. Replicate analyses of independently isolated PMVECs for each genotype are shown. Note the reduced expression of Itgb3 in Foxc2+/– cells compared with wild-type cells. C, no change in expression of Itga1, Itga5, Itga6, Itgav, and Itgb1 in Foxc2+/– PMVECs measured by real-time RT-PCR. Results are presented as the means ± S.D. from triplicate experiments. Statistical significance was determined by Student's t tests. N.S., non-significant. D, loss of Foxc2 does not affect endothelial cell proliferation. PMVECs isolated from wild-type and Foxc2+/– mice were cultured with or without VEGF (5 ng/ml) for 24 h in serum-reduced medium (0.5%) with a pulse of BrdUrd (BrdU) for the last 3 h. The number of BrdUrd-positive cells and total cells was counted. Values are presented as the means ± S.D. from 10 microscopic fields from two independent experiments. Statistical significance was determined by Student's t tests. *, p < 0.05. E, migration assay using PMVECs isolated from wild-type and Foxc2+/– mice. Primary PMVECs were added to Transwells coated with collagen I and allowed to migrate toward serum-free medium with or without VEGF (25 ng/ml). The number of migrating cells from randomly selected microscopic fields/well was counted at 5 h after cell plating. Data are the means ± S.D. from three experiments with eight data points for each. **, p < 0.01 versus the corresponding control.
As in the case of overexpression of Foxc2, we found no significant difference in proliferation between wild-type and Foxc2+/– endothelial cells in the presence or absence of VEGF (Fig. 6D). In contrast, Transwell migration assay revealed that both basal and VEGF-induced migration of Foxc2+/– endothelial cells were significantly impaired compared with wild-type cells (Fig. 6E). Consistent with the results obtained from wound closure assay (Fig. 5, B and C), it should be noted that we tested the direct effects of Foxc2 on the migration of endothelial cells cultured in serum-free medium for 4 h. Therefore, these data confirm that Foxc2 plays a critical role in endothelial cell migration.
Foxc2 Regulates ex Vivo Angiogenesis—To address whether Foxc2 is really involved in angiogenesis, we used an aortic ring culture model that recapitulates the process of angiogenesis ex vivo by generating microvessels from isolated aortas on a fibrin/thrombin gel. First, segments of aortas from wild-type mice were infected with Ad-Foxc2 or Ad-control. Significantly, we observed abundant formation of vascular sprouts from aortic rings infected with Ad-Foxc2 compared with those infected with Ad-control (Fig. 7A). Consistent with the data obtained from wound healing assays and cell adhesion assays, anti-Itgb3 blocking antibody significantly inhibited Foxc2-induced ex vivo angiogenesis. On the other hand, fewer vascular sprouts were detected from aortic rings from Foxc2+/– mice compared with control aortic rings (Fig. 7B). These data suggest that Foxc2 in the endothelium controls angiogenesis ex vivo, partially through the direct regulation of Itgb3 expression.
DISCUSSION
Foxc2 is an essential regulator of the cardiovascular system in development and disease. However, the molecular mechanisms for a role of Foxc2 in angiogenesis remain to be elucidated. In this study, overexpression of Foxc2 in endothelial cells showed that Foxc2 regulates the expression of Itgb3, Itgb5, and FN, known pro-angiogenic factors. In addition, Foxc2 induces endothelial cell adhesion and migration partially through Itgb3 expression. Luciferase reporter assay and chromatin immunoprecipitation assay demonstrated that Foxc2 directly regulates the transcription of Itgb3 through the FBEs in the proximal and distal HHRs. On the other hand, loss of Foxc2 function in endothelial cells results in reduced expression of Itgb3, Itgb5, and FN as well as impaired cell migration. Aortic ring assay further demonstrated that Foxc2 directly regulates angiogenesis. Thus, Foxc2 regulates endothelial cell adhesion and migration by inducing expression of pro-angiogenic factors such as Itgb3 and thereby contributes to angiogenesis.
Foxc2-induced Expression of Itgb3 in Endothelial Cells—In addition to the multiple repeat FBEs in the proximal HHR whose sequences are only found in the mouse Itgb3 locus, we found novel FBEs in the distal HHR upstream of the Itgb3 locus (22). The distal HHRs are likely to be more critical for the induction of Itgb3 transcription by Foxc2 than the proximal HHRs because the FBEs in the distal HHR are highly conserved between human and mouse. When the distal HHR (–11.1 to –10.8 kb) was constructed in the pGL3-basic vector that does not carry the basic (SV40) promoter, it indeed had luciferase activity in response to Foxc2. This result suggests that the distal HHR itself has weak but significant promoter activity. Interestingly, we found that the second and third FBEs, but not the first FBE, are required for Foxc2-induced luciferase activity. We realized that the sequence (AAAATAAA) of the second and third FBEs in the distal HHR is similar to TATAAAAG, the p2 TATA box on the c-myc promoter. Given evidence that another Fox protein, FoxM1c, transactivates the c-myc promoter by directly binding to the p2 TATA box (24), it will be interesting to study whether Foxc2 binds to TATA box sequences.
Consistent with evidence that Foxc1 and Foxc2 transcription factors play dose-dependent redundant roles in cardiovascular development (10–12), like Foxc2, Foxc1 activates the Itgb3 promoter and regulates Itgb3 expression in endothelial cells (supplemental Figs. 1–3). In contrast to reduced expression of Itgb3 in endothelial cells isolated from adult lungs of Foxc1–/– and Foxc2+/– mice (Fig. 6B and supplemental Fig. 2), we found that Itgb3 expression was not obviously altered in CD31+ Flk1(VEGFR-2)+ endothelial cells isolated from Foxc2+/– and Foxc2–/– embryos compared with wild-type cells (supplemental Fig. 4). Because CD31+ Flk1+ endothelial cells were sorted from whole embryos, further analysis of Itgb3 expression during the angiogenic process in Foxc2 mutant embryos needs to be performed. Alternatively, embryonic expression of Itgb3 in endothelial cells may be tightly regulated by additional transcription factors (e.g. other Fox proteins) along with Foxc.
VEGF Signaling and Foxc2 Do Not Functionally Interact with Each Other in Cell Proliferation, Adhesion, and Migration—VEGF signaling enhances the expression and activation of several integrins. Moreover, the VEGF and integrin pathways often act synergistically to mediate the migration, proliferation, differentiation, and survival of endothelial cells (2). Recent studies emphasize the role of αvβ3 integrin as a “gatekeeper” of the VEGF-mediated process because VEGFR-2 binds to Itgb3 through its extracellular domain and phosphorylates Itgb3 via activation of c-Src, which in turn is crucial for VEGF-induced tyrosine phosphorylation of VEGFR-2 (25, 26). In this work, we have shown that there are no cooperative effects of Foxc2 and the VEGF signal on the control of endothelial cell proliferation and migration. However, we have found recently that VEGF-stimulated PI3K (phosphatidylinositide 3-kinase) and ERK (extracellular signal-regulated kinase) pathways modulate the transcriptional activity of Foxc2 for arterial gene expression in endothelial cells (27). Consequently, functional interaction between VEGF signaling and Foxc2 may take place in some aspects of blood vessel formation.
Foxc2 Regulates Angiogenesis—We demonstrated that Foxc2 mediates angiogenesis by aortic ring assay. Although there are several paradoxical reports (28–30), αvβ3 integrin is thought to induce tumor development and angiogenesis. A humanized monoclonal antibody against αvβ3 integrin is now being tested in the phase II clinical trial (7). Our results in this study support the idea that αvβ3 integrin is an inducer of angiogenesis because blockade of Itgb3 by the monoclonal antibody inhibited Foxc2-induced cell migration and angiogenesis. Notably, we have shown recently that Foxc transcription factors directly induce expression of the chemokine CXCR4 in endothelial cells and regulate endothelial cell migration toward its ligand, CXCL12 (32). Another gene regulated by Foxc2 is Dll4 (Delta-like 4) (12). Dll4 is implicated in vascular development (33–35) and is a strong candidate for clinical prevention of tumor angiogenesis because VEGF dynamically regulates Dll4 expression in tumor endothelial cells and inhibition of Dll4 blocks functional angiogenesis (36–39). Taken together, it is conceivable that Foxc2 regulates angiogenesis via multiple signaling pathways, including Itgb3, CXCR4, and Dll4.
Aberrant control of epithelial cell proliferation and angiogenesis underlies the initiation and growth of primary carcinomas. Recently, Mani et al. (40) showed that Foxc2 is expressed in several metastatic and non-metastatic tumor cells lines and that Foxc2 regulates epithelial-to-mesenchymal transition in basal-like breast cancer. However, a possible involvement of Foxc2 in tumor angiogenesis, as reviewed recently (41), has not yet been addressed. Because αvβ3 integrin plays a role in metastasis of tumors as well (31), it is possible that Foxc2 not only mediates metastasis through the regulation of αvβ3 integrin in breast cancer cells, but also regulates tumor angiogenesis.
Supplementary Material
Acknowledgments
We thank Drs. Ambra Pozzi and Jin Chen for helpful discussions and Drs. Marie-Jose Goumans and Peter ten Dijke for providing MEECs.
This work was supported, in whole or in part, by National Institutes of Health Grants HL067105, DK068547, and HL074121 (to T. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. 1–4, Table 1, and an additional reference.
Footnotes
The abbreviations used are: MEECs, mouse embryonic endothelial cells; PMVECs, pulmonary microvascular endothelial cells; Ad, adenovirus; FBEs, Fox-binding elements; RT, reverse transcription; PBS, phosphate-buffered saline; BrdUrd, 5-bromo-2′-deoxyuridine; HHR, high homology region; LUC, luciferase; BSA, bovine serum albumin; VEGF, vascular endothelial growth factor; FN, fibronectin; VEGFR-2, VEGF receptor type 2.
H. Hayashi and T. Kume, manuscript in preparation.
References
- 1.Carmeliet, P. (2005) Nature 438 932–936 [DOI] [PubMed] [Google Scholar]
- 2.Serini, G., Valdembri, D., and Bussolino, F. (2006) Exp. Cell Res. 312 651–658 [DOI] [PubMed] [Google Scholar]
- 3.Davis, G. E., and Senger, D. R. (2005) Circ. Res. 97 1093–1107 [DOI] [PubMed] [Google Scholar]
- 4.Rupp, P. A., and Little, C. D. (2001) Circ. Res. 89 566–572 [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson-Berka, J. L., Jones, D., Taylor, G., Jaworski, K., Kelly, D. J., Ludbrook, S. B., Willette, R. N., Kumar, S., and Gilbert, R. E. (2006) Investig. Ophthalmol. Vis. Sci. 47 1600–1605 [DOI] [PubMed] [Google Scholar]
- 6.Mahabeleshwar, G. H., Feng, W., Phillips, D. R., and Byzova, T. V. (2006) J. Exp. Med. 203 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulgrew, K., Kinneer, K., Yao, X. T., Ward, B. K., Damschroder, M. M., Walsh, B., Mao, S. Y., Gao, C., Kiener, P. A., Coats, S., Kinch, M. S., and Tice, D. A. (2006) Mol. Cancer Ther. 5 3122–3129 [DOI] [PubMed] [Google Scholar]
- 8.Winnier, G. E., Kume, T., Deng, K., Rogers, R., Bundy, J., Raines, C., Walter, M. A., Hogan, B. L., and Conway, S. J. (1999) Dev. Biol. 213 418–431 [DOI] [PubMed] [Google Scholar]
- 9.Iida, K., Koseki, H., Kakinuma, H., Kato, N., Mizutani-Koseki, Y., Ohuchi, H., Yoshioka, H., Noji, S., Kawamura, K., Kataoka, Y., Ueno, F., Taniguchi, M., Yoshida, N., Sugiyama, T., and Miura, N. (1997) Development (Camb.) 124 4627–4638 [DOI] [PubMed] [Google Scholar]
- 10.Kume, T., Jiang, H., Topczewska, J. M., and Hogan, B. L. (2001) Genes Dev. 15 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo, S., and Kume, T. (2006) Dev. Biol. 296 421–436 [DOI] [PubMed] [Google Scholar]
- 12.Seo, S., Fujita, H., Nakano, A., Kang, M., Duarte, A., and Kume, T. (2006) Dev. Biol. 294 458–470 [DOI] [PubMed] [Google Scholar]
- 13.Dagenais, S. L., Hartsough, R. L., Erickson, R. P., Witte, M. H., Butler, M. G., and Glover, T. W. (2004) Gene Expr. Patterns 4 611–619 [DOI] [PubMed] [Google Scholar]
- 14.Kriederman, B. M., Myloyde, T. L., Witte, M. H., Dagenais, S. L., Witte, C. L., Rennels, M., Bernas, M. J., Lynch, M. T., Erickson, R. P., Caulder, M. S., Miura, N., Jackson, D., Brooks, B. P., and Glover, T. W. (2003) Hum. Mol. Genet 12 1179–1185 [DOI] [PubMed] [Google Scholar]
- 15.Mellor, R. H., Brice, G., Stanton, A. W., French, J., Smith, A., Jeffery, S., Levick, J. R., Burnand, K. G., and Mortimer, P. S. (2007) Circulation 115 1912–1920 [DOI] [PubMed] [Google Scholar]
- 16.Petrova, T. V., Karpanen, T., Norrmen, C., Mellor, R., Tamakoshi, T., Finegold, D., Ferrell, R., Kerjaschki, D., Mortimer, P., Yla-Herttuala, S., Miura, N., and Alitalo, K. (2004) Nat. Med. 10 974–981 [DOI] [PubMed] [Google Scholar]
- 17.Goumans, M.-J., Valdimarsdottir, G., Itoh, S., Rosendahl, A., Sideras, P., and ten Dijke, P. (2002) EMBO J. 21 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, H., Lawson, W. E., Polosukhin, V. V., Pozzi, A., Blackwell, T. S., Litingtung, Y., and Chiang, C. (2007) Am. J. Pathol. 171 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamiyama, M., Pozzi, A., Yang, L., DeBusk, L. M., Breyer, R. M., and Lin, P. C. (2006) Oncogene 25 7019–7028 [DOI] [PubMed] [Google Scholar]
- 21.Hynes, R. O. (2007) J. Thromb. Haemostasis 5 Suppl. 1, 32–40 [DOI] [PubMed] [Google Scholar]
- 22.Malin, D., Kim, I. M., Boetticher, E., Kalin, T. V., Ramakrishna, S., Meliton, L., Ustiyan, V., Zhu, X., and Kalinichenko, V. V. (2007) Mol. Cell. Biol. 27 2486–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamalice, L., Le Boeuf, F., and Huot, J. (2007) Circ. Res. 100 782–794 [DOI] [PubMed] [Google Scholar]
- 24.Wierstra, I., and Alves, J. (2007) Biochem. Biophys. Res. Commun. 352 61–68 [DOI] [PubMed] [Google Scholar]
- 25.Borges, E., Jan, Y., and Ruoslahti, E. (2000) J. Biol. Chem. 275 39867–39873 [DOI] [PubMed] [Google Scholar]
- 26.Mahabeleshwar, G. H., Feng, W., Reddy, K., Plow, E. F., and Byzova, T. V. (2007) Circ. Res. 101 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi, H., and Kume, T. (2008) PLoS ONE 3 e2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bader, B. L., Rayburn, H., Crowley, D., and Hynes, R. O. (1998) Cell 95 507–519 [DOI] [PubMed] [Google Scholar]
- 29.Hynes, R. O. (2002) Nat. Med. 8 918–921 [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, L. E., Wyder, L., Lively, J. C., Taverna, D., Robinson, S. D., Huang, X., Sheppard, D., Hynes, R. O., and Hodivala-Dilke, K. M. (2002) Nat. Med. 8 27–34 [DOI] [PubMed] [Google Scholar]
- 31.Rolli, M., Fransvea, E., Pilch, J., Saven, A., and Felding-Habermann, B. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi, H., and Kume, T. (2008) Biochem. Biophys. Res. Commun. 367 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krebs, L. T., Shutter, J. R., Tanigaki, K., Honjo, T., Stark, K. L., and Gridley, T. (2004) Genes Dev. 18 2469–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duarte, A., Hirashima, M., Benedito, R., Trindade, A., Diniz, P., Bekman, E., Costa, L., Henrique, D., and Rossant, J. (2004) Genes Dev. 18 2474–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gale, N. W., Dominguez, M. G., Noguera, I., Pan, L., Hughes, V., Valenzuela, D. M., Murphy, A. J., Adams, N. C., Lin, H. C., Holash, J., Thurston, G., and Yancopoulos, G. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15949–15954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrington, L. S., Sainson, R. C., Williams, C. K., Taylor, J. M., Shi, W., Li, J. L., and Harris, A. L. (2008) Microvasc. Res. 75 144–154 [DOI] [PubMed] [Google Scholar]
- 37.Noguera-Troise, I., Daly, C., Papadopoulos, N. J., Coetzee, S., Boland, P., Gale, N. W., Lin, H. C., Yancopoulos, G. D., and Thurston, G. (2006) Nature 444 1032–1037 [DOI] [PubMed] [Google Scholar]
- 38.Ridgway, J., Zhang, G., Wu, Y., Stawicki, S., Liang, W. C., Chanthery, Y., Kowalski, J., Watts, R. J., Callahan, C., Kasman, I., Singh, M., Chien, M., Tan, C., Hongo, J. A., de Sauvage, F., Plowman, G., and Yan, M. (2006) Nature 444 1083–1087 [DOI] [PubMed] [Google Scholar]
- 39.Scehnet, J. S., Jiang, W., Kumar, S. R., Krasnoperov, V., Trindade, A., Benedito, R., Djokovic, D., Borges, C., Ley, E. J., Duarte, A., and Gill, P. S. (2007) Blood 109 4753–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mani, S. A., Yang, J., Brooks, M., Schwaninger, G., Zhou, A., Miura, N., Kutok, J. L., Hartwell, K., Richardson, A. L., and Weinberg, R. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myatt, S. S., and Lam, E. W. (2007) Nat. Rev. Cancer 7 847–859 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.