Abstract
Rap1 is a member of the Ras superfamily of small GTP-binding proteins and is localized on pancreatic zymogen granules. The current study was designed to determine whether GTP-Rap1 is involved in the regulation of amylase secretion. Rap1A/B and the two Rap1 guanine nucleotide exchange factors, Epac1 and CalDAG-GEF III, were identified in mouse pancreatic acini. A fraction of both Rap1 and Epac1 colocalized with amylase in zymogen granules, but only Rap1 was integral to the zymogen granule membranes. Stimulation with cholecystokinin (CCK), carbachol, and vasoactive intestinal peptide all induced Rap1 activation, as did calcium ionophore A23187, phorbol ester, forskolin, 8-bromo-cyclic AMP, and the Epac-specific cAMP analog 8-pCPT-2′-O-Me-cAMP. The phospholipase C inhibitor U-73122 abolished carbachol- but not forskolin-induced Rap1 activation. Co-stimulation with carbachol and 8-pCPT-2′-O-Me-cAMP led to an additive effect on Rap1 activation, whereas a synergistic effect was seen on amylase release. Although the protein kinase A inhibitor H-89 abolished forskolin-stimulated CREB phosphorylation, it did not modify forskolin-induced GTP-Rap1 levels, excluding PKA participation. Overexpression of Rap1 GTPase-activating protein, which blocked Rap1 activation, reduced the effect of 8-bromo-cyclic AMP, 8-pCPT-2′-O-Me-cAMP, and vasoactive intestinal peptide on amylase release by 60% and reduced CCK- as well as carbachol-stimulated pancreatic amylase release by 40%. These findings indicate that GTP-Rap1 is required for pancreatic amylase release. Rap1 activation not only mediates the cAMP-evoked response via Epac1 but is also involved in CCK- and carbachol-induced amylase release, with their action most likely mediated by CalDAG-GEF III.
Rap1 is a member of the Ras superfamily of small GTP-binding proteins and is known to be involved in cell adhesion, proliferation, and differentiation (1). Two isoforms of Rap1 exist, Rap1A and Rap1B, which are 95% identical at the amino acid sequence and appear to mediate similar actions (1). Like other members of this superfamily of proteins, Rap1 cycles between an inactive GDP-bound and an active GTP-bound form. A variety of intracellular signals regulate the cycle through unique guanine-nucleotide exchange factors (GEFs),2 which promote dissociation of GDP from Rap1 followed by binding of GTP, and GTPase-activating proteins (GAPs), which induce hydrolysis of GTP on Rap1 (1). Increases in intracellular levels of Ca2+ and diacylglycerol (DAG) activate Ca2+- and DAG-binding GEFs (CalDAG-GEF I and CalDAG-GEF III) (2), whereas increases in intracellular levels of cAMP activate the exchange proteins activated by cAMP (Epac1 and Epac2) (3, 4). Protein kinase A (PKA), another mediator of cAMP action, can phosphorylate Rap1, which is necessary for Rap1 activation in certain cell types, including neutrophils, fibroblasts, thyroid and enteroendocrine cells (5–8).
Evidence indicates that different endogenous Rap1GEFs exert physiological functions in various cells. For example, in neuronal PC12D cells, CalDAG-GEF I is involved in Ca2+- and DAG-induced Rap1 activation, resulting in the activation of ERK1/2 (9). In addition, CalDAG-GEF I is important for signal integration as well as granule secretion in platelets (10). Epac1 participates in the formation of endothelial cell tight junctions (11), whereas both Epac1 and Epac2 facilitate cAMP-induced exocytosis in pancreatic β-cells through an increase in Ca2+ release mediated by ryanodine-sensitive Ca2+ channels (12, 13). Recently, Chaudhuri et al. (14) presented evidence that Epac is a mediator of cAMP signaling in exocrine pancreas. Using cAMP analogs which activate Epac but not PKA, they showed effects of cAMP on carbachol-stimulated amylase secretion and zymogen activation. However, the downstream target of Epac was not identified.
A number of small GTP-binding proteins have been identified on the external surface of zymogen granules, and some, particularly Rab3D and Rab27B, have been implicated in the regulation of pancreatic exocytosis (15–20). Although Rap1 is also present on zymogen granules, as shown by both mass spectrometry and immunocytochemistry (20), there have been no studies on Rap1-dependent signaling and function in pancreatic acinar cells. Previous studies suggest that Rap1 may play a regulatory role in exocrine as well as endocrine cells. In parotid acinar cells, Rap1 is not only located on secretory granule membranes (21), but may also be involved in regulation of cAMP-stimulated amylase secretion (22). In a human endocrine BON cell line, Rap1 participates in neurotensin release (8). Recently, Shibasaki et al. (23) showed that Rap1 has a role in the regulation of insulin secretion stimulated by cAMP.
Since Rap1 is located on zymogen granules and Epac is a likely cAMP mediator in pancreatic acinar cells, the aim of the present study was to determine whether Rap1 activation is involved in the regulation of pancreatic amylase secretion. The present results show for the first time that Rap1 is required not only for cAMP-evoked response via Epac1 but also for cholecystokinin (CCK) and carbachol-stimulated amylase secretion, most likely mediated by CalDAG-GEF III. Thus, Rap1 plays an essential role as a common effector protein for secretion.
EXPERIMENTAL PROCEDURES
Materials—Collagenase was purchased from Crescent Chemical Company (Islandia, NY), bovine serum albumin (BSA) and soybean trypsin inhibitor (STI) were from Sigma, and Dulbecco's modified Eagle's medium was from Invitrogen. The following inhibitors and stimuli were used. Sulfated CCK octapeptide was from Research Plus (Bayonne, NJ); vasoactive intestinal peptide (VIP) was from American Peptide Company (Sunnyvale, CA); A23187 and GF-109203X were from Calbiochem; carbamylcholine chloride (carbachol), forskolin, phorbol 12-myristate 13-acetate (PMA), 8-bromo-cyclic AMP (8-Br-cAMP), dithiothreitol, EGTA, H-89, isopropyl β-d-thiogalactopyranoside, and U-73122 were from Sigma; 8-(p-chlorophenylthio)-2′-O-methyladenosine-3′,5′-cyclic monophosphate (8-pCPT-2′-O-Me-cAMP) was from Biolog Life Science Institute (Bremen, Germany), phenylmethylsulfonyl fluoride (PMSF) was from Pierce; and aprotinin and leupeptin were from Roche Applied Science. All other chemicals were of reagent grade.
Polyclonal antibodies against the following proteins were used. Phospho-Ser133-CREB was from Cell Signaling Technology (Beverly, MA); CREB-2, Rap1, Epac2, Rap1GAP, and RhoA were from Santa Cruz Biotechnology (Santa Cruz, CA); Epac1 and Epac2 were from Abcam Inc. (Cambridge, MA), and amylase was from U. S. Biological (Swampscott, MA).
Preparation of Isolated Pancreatic Acini—Acini were prepared by methods previously described (17, 18). In brief, pancreata were excised from fed adult male ICR mice weighing 22–27 g. Acini were isolated by collagenase digestion, followed by mechanical shearing and then filtered through 150-μm Nitex mesh, purified by sedimentation through 4% BSA in buffer, and suspended in Dulbecco's modified Eagle's medium containing 5 mg/ml BSA and 0.1 mg/ml STI.
Expression of Rap1, Epac, and CalDAG-GEF in Mouse Pancreatic Acini—The expression of Rap1A, Rap1B, Epac1, Epac2, CalDAG-GEF I, and CalDAG-GEF III in mouse pancreatic acini was assessed by reverse transcription (RT)-PCR. The following primers were used: mouse Rap1A, 5′-ATGCTGGAGATCCTGGACAC-3′ (sense) and 5′-TCTTTGCCAACTACCCGTTC-3′ (antisense); mouse Rap1B, 5′-ACTCCATCACAGCACAGTCG-3′ (sense) and 5′-AGTCGCATTTATTGCCAACC-3′ (antisense); mouse Epac1, 5′-GTTGTCGACCCACAGGAAGT-3′ (sense) and 5′-AGGAACAGGGCAGAGACAGA-3′ (antisense); mouse Epac2, 5′-ATTAATGGACGCCTGTTTGC-3′ (sense) and 5′-CCTCCTCAGGAACAAATCCA-3′ (antisense); mouse CalDAG-GEF I, 5′-CCAAGATGAGGCAGCTTTTC-3′ (sense) and 5′-CTTAGGCTTGGCAACTGAGG-3′ (antisense); mouse CalDAG-GEF III, 5′-GAACACTGTGCGGGATTTCT-3′ (sense) and 5′-CTTTGCTGCTTTGTCATGGA-3′ (antisense) (Invitrogen). The primers were designed with Invitrogen Oligoperfect Designer based on gene sequences obtained from the GenBank™ NCBI Sequence Viewer (available on the World Wide Web).
Subcellular Localization of Rap1 and Epac1—Subcellular fractionation of mouse pancreas was carried out by differential centrifugation, as initially described by Jamieson and Palade (24). Pancreas was homogenized in 0.3 m sucrose medium supplemented with 50 mm MOPS, pH 7.0, 2 mm EGTA, and 0.1 mm PMSF at 4 °C using a Teflon-glass homogenizer. After a 10-min spin at 300 × g to remove cell debris, crude particulate fraction enriched in zymogen granules was obtained by centrifugation at 800 × g for 10 min, whereas microsomal as well as cytosolic fractions were obtained by centrifugation at 100,000 × g for 45 min. Zymogen granule purification was carried out using Percoll gradients (20). Zymogen granule membranes were isolated by osmotically lysing granules in 150 mm sodium acetate, 10 mm MOPS, pH 7.0, 0.1 mm MgSO4, 0.1 mm PMSF, and 27 μg/ml nigericin for 15 min at 37 °C and then collecting the membranes by centrifugation at 38,000 rpm in a Ti 70.1 rotor for 1 h at 4 °C, followed by washing with 250 mm KBr.
Immunolocalization of Rap1 and Epac1—Freshly isolated pancreatic acini were stimulated with either 10 μm carbachol or 10 nm VIP for 10 min at 37 °C. Then acini were sedimented in test tubes and fixed for 30 min at room temperature with 4% paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4, rinsed with PBS, cryoprotected, and frozen with isopentane cooled with liquid nitrogen. Cryostat sections (5-μm thick) were mounted on SuperFrost Plus slides (Fischer) and processed for immunofluorescence localization as previously described (17). Briefly, the sections were incubated with the following primary antibodies: polyclonal rabbit anti-Epac1 (diluted 1:200), polyclonal rabbit anti-Rap1 (1:200), or polyclonal sheep anti-human salivary amylase (1:100 to 1:200) for 2 h at room temperature. Then they were washed three times with PBS. The sections were then incubated with secondary antibodies Cy3-conjugated donkey anti-rabbit IgG (1:200) or fluorescein-conjugated anti-sheep IgG (1:200) (Jackson Immunoresearch Laboratories, Inc.) for another 1 h and washed with PBS. Prolong Gold with 4,6-diamino-2-phenylindole was added to mounting medium to counterstain nuclei. Digitized images were collected with an Olympus Fluoview 500 confocal microscope as a Z-series (0.5-μm steps). A single image from the Z-stack was then chosen based on the most informative and sharpest fluorescence and processed using Photoshop CS software (Adobe Systems Inc., Mountain View, CA).
Pull-down Assay for the Determination of both Rap1 and RhoA Activation—Activation of Rap1 was measured using the glutathione S-transferase-RalGDS-Rap1-binding domain (RBD) as a pull-down reagent, which selectively binds the active GTP-bound form of Rap1 (25). RalGDS-RBD was expressed in Escherichia coli by induction with 0.1 mm isopropyl β-d-thiogalactopyranoside for 2 h at 37 °C and purified from bacterial lysates with glutathione-Sepharose. Acini were prepared for the assay as indicated above. Immediately, after incubation with agonist for the specified time, acini were washed once with ice-cold Tris-buffered saline, and then cold lysis buffer containing 50 mm Tris-HCl, pH 7.5, 1% Triton X-100, 500 mm NaCl, 10 mm MgCl2, 10% (v/v) glycerol, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm PMSF, 1 mm sodium orthovanadate, and 10 mm sodium fluoride was added. Acini were rapidly sonicated and cleared at 13,000 × g for 10 min at 4 °C. The resulting supernatants (1500–2000 μg of protein) were quickly transferred into tubes with glutathione S-transferase-Ral-GDS-RBD (80 μg), followed by a 2-h rotation at 4 °C. Samples were washed and subjected to Western blotting for immunodetection of GTP-Rap1 as described below. Fifteen μg from each pancreatic acini lysate was used to immunodetect total Rap1.
RhoA activation was determined as previously described (26). The pull-down assay was carried out in a similar manner to Rap1 using Rhotekin-RBD-agarose beads (Cell Biolabs, Inc., San Diego, CA).
cAMP-response Element-binding Protein (CREB) Phosphorylation—After acini were stimulated for 10 min at 37 °C, they were washed once using ice-cold PBS containing 1 mm sodium orthovanadate and 10 mm sodium fluoride and lysed in the lysis buffer (50 mm Tris-HCl, pH 7.4, 0.32 m sucrose, 1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm sodium orthovanadate, 10 mm sodium fluoride, 1 mm dithiothreitol), sonicated, and clarified at 13,000 × g for 10 min. Samples (10 μg from pancreatic acini lysates) were subjected to Western blotting for immunodetection of phospho-CREB and CREB-2 as described below.
Western Blotting—Immunodetection of proteins was carried out using SDS-polyacrylamide gels, which were transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked in 5% BSA dissolved in Tris-buffered saline containing 0.1% (v/v) Tween 20 for 2 h at room temperature. Corresponding rabbit polyclonal antibodies were diluted 1:1000 in 5% BSA, and the membrane was incubated with the antibody overnight at 4 °C, followed by treatment with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000 in 5% nonfat milk) for 1 h. After washing with Tris-buffered saline containing 0.1% (v/v) Tween 20, peroxidase activity was visualized using the SuperSignal West Femto sensitivity substrate kit (Pierce).
Amylase Secretion Studies—Freshly digested or overnight cultured acini were stimulated with CCK, carbachol, 8-Br-cAMP, 8-pCPT-2′-O-Me-cAMP, or VIP in 1-ml aliquots in plastic blood dilution vials for 30 min. Samples were then centrifuged for 30 s in a microcentrifuge, and the supernatant was assayed for amylase activity with Phadebas reagents (Magle Life Sciences, Lund, Sweden). Results were expressed as a percentage of initial acinar amylase content.
45Ca2+ Efflux—Cellular 45Ca2+ mobilization in isolated pancreatic acini was measured using a previously described procedure (27). Acini were suspended in Krebs-Henseleit bicarbonate buffer (KHB) (118 mm NaCl, 4.7 mm KCl, 1 mm NaH2PO4, 1.1 mm MgCl2, 2.5 mm CaCl2, 25 mm NaHCO3, 2.5 mg/ml d-glucose, and 0.1 mg/ml STI) and preincubated with 2 μCi/ml 45CaCl2 (New England Nuclear) for 1 h at 37 °C. Then labeled acini were washed twice with nonradioactive KHB, suspended in fresh KHB, and incubated with or without different secretagogues for specified times at 37 °C. 45Ca2+ remaining was calculated by difference between total 45Ca2+ and 45Ca2+ released from the acini into the extracellular medium.
Intracellular cAMP Content— cAMP generation was determined as previously described (28). Acini were preincubated for 30 min in KHB and then for 3 min in fresh KHB containing 1 mm IBMX. Acini were stimulated with different secretagogues for 12 min. cAMP was extracted in ethanol and measured using a cAMP colorimetric enzyme immunoassay kit according to the instructions provided by the manufacturer (Cayman Chemical Co., Ann Arbor, MI). Results were expressed as pmol/mg of protein.
Adenoviral Infection—To investigate the role of Rap1 activation in amylase release, we blocked the activation of endogenous Rap1 by expressing the Rap1-specific GTPase-activating protein, Rap1-GAP, in isolated pancreatic acini. Rap1GAP promotes the conversion of active GTP-bound Rap1 to the inactive GDP-bound form while having no effect on the activation of other GTPases (29). Recombinant adenovirus engineered for expression of Rap1GAP-green fluorescence protein (30) was provided by Dr. Patrick J. Casey (Duke University Medical Center, Durham, NC). The virus was amplified and purified using Adeno-X™ virus purification kits (BD Biosciences), and adenovirus expressing β-galactosidase was used as a control. Acini were infected with either β-galactosidase or Rap1GAP adenovirus (107 pfu/ml) during overnight culture at low density in 100-mm Petri dishes in Dulbecco's modified Eagle's medium enriched with 0.5% fetal bovine serum, 0.01% STI, and antibiotics at 37 °C with 5% CO2. The overexpression of Rap1GAP-green fluorescence protein in isolated pancreatic acini was analyzed by immunofluorescence and by immunoblotting using polyclonal Rap1GAP antibody as described above. Infection efficiencies were routinely over 90%, as reported previously (18). Infected acini were then used for studies of Rap1 or RhoA activation, amylase secretion, 45Ca2+ mobilization, or cAMP generation.
Statistical Analysis—Results were expressed as means ± S.E. of 3–6 separate experiments. The statistical analysis was performed by analysis of variance following by the Student-Newman-Keuls test. p ≤ 0.05 was considered statistically significant.
RESULTS
Both Rap1A and Rap1B Are Expressed in Pancreatic Acini— In our previous study, we showed that Rap1 protein is present on rat zymogen granules (20). Using RT-PCR, we have now found that both Rap1A and Rap1B mRNAs are expressed not only in the whole mouse pancreas but also in isolated pancreatic acini (data not shown). However, since the Rap1 antibody does not distinguish between the isoforms, we will refer to the protein as Rap1.
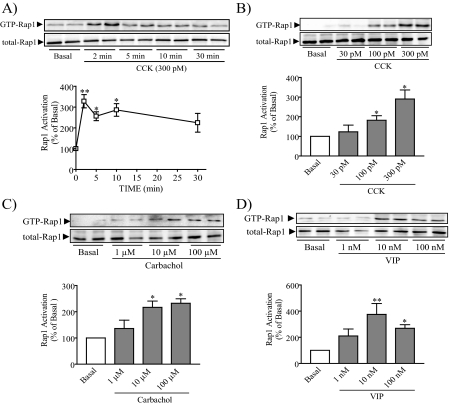
CCK and Other Secretagogues Induce a Rapid Increase in Rap1 Activation—We studied whether CCK, carbachol, and VIP activate Rap1 in pancreatic acini using a Rap1 pull-down assay. CCK provoked a rapid increase in Rap1 activation with the maximum effect between 2 and 10 min, after which GTP-Rap1 levels declined but remained elevated at 30 min (Fig. 1A). A similar time course was observed following stimulation with carbachol and VIP (data not shown). To allow more samples to be collected simultaneously, Rap1 activation was thereafter studied after a 10-min stimulation. CCK, carbachol, and VIP stimulation all led to a concentration-dependent increase with comparable maximal increases seen at 300 pm CCK, 10 μm carbachol, and 10 nm VIP (Fig. 1, B–D). VIP-stimulated Rap1 activation showed a lower response at the highest concentration. These results indicate that the most relevant secretagogues for mouse pancreatic acini all activate Rap1.
FIGURE 1.
Multiple secretagogues activate Rap1. A, isolated mouse pancreatic acini were treated with CCK for the specified time, after which a pull-down assay was used to detect the active form of Rap1. Activation of Rap1 was detected after 2 min, remained elevated at 5 and 10 min, and declined at 30 min. B–D, acini were stimulated with specific concentrations of CCK (B), carbachol (C), and VIP (D) for 10 min and then assayed for activation of Rap1. The upper panels show representative immunoblots for both GTP-Rap1 and total Rap1. The lower panels show quantitative analysis of Rap1 activation. Data shown are means ± S.E. (4–5 experiments) for activation of Rap1 expressed as a percentage of basal. *, p < 0.05; **, p < 0.01 versus basal.
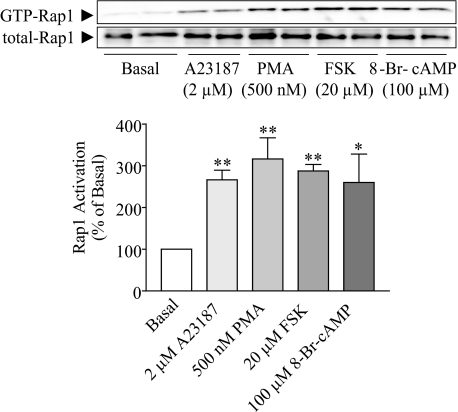
Ca2+, DAG, and cAMP Induce an Increase in Active Rap1 Levels— Both CCK and carbachol activate phospholipase Cβ (PLCβ), which generates the intracellular second messengers DAG and inositol 1,4,5-triphosphate (IP3). DAG activates protein kinase C (PKC), whereas IP3 stimulates the release of Ca2+ from the endoplasmic reticulum required for pancreatic amylase release (31). On the other hand, VIP increases intracellular cAMP levels in pancreatic acini (32). Since several intracellular signals are able to induce Rap1 activation (1), different stimulators and inhibitors were used to determine which intracellular messengers participated in secretagogue-induced Rap1 activation in acini. Both Ca2+ ionophore A23187 and phorbol ester PMA, which has a structure analogous to DAG, increased the amount of GTP-Rap1 by 140 and 170%, respectively (Fig. 2). Since PMA is a PKC activator, we studied whether the effect of PMA was direct or through PKC activation. Pretreatment with GF-109203X (1 μm), a PKC inhibitor (33), did not modify carbachol-induced GTP-Rap1 levels (data not shown). In accord with previous studies (34, 35), these results show that DAG, but not PKC, is involved in Rap1 activation. In addition, the specific adenylyl cyclase (AC) activator forskolin (36) and the permeable cAMP analog 8-Br-cAMP augmented Rap1 activation by 200 and 150%, respectively (Fig. 2). These findings indicate that the most important pathways involved in pancreatic amylase secretion, PLC/IP3/Ca2+, PLC/DAG, and AC/cAMP, are all able to activate Rap1.
FIGURE 2.
Rap1 is activated by Ca2+, DAG, and cAMP. Isolated pancreatic acini were stimulated by Ca2+ ionophore A23187, phorbol ester PMA, forskolin (FSK), and 8-Br-cAMP and then assayed for activation of Rap1. The upper panel shows a representative immunoblot for both GTP-Rap1 and total Rap1. The lower panel shows quantitative analysis for Rap1 activation. Data shown are means ± S.E. (four experiments) for activation of Rap1 expressed as a percentage of basal. *, p < 0.05; **, p < 0.01 versus basal.
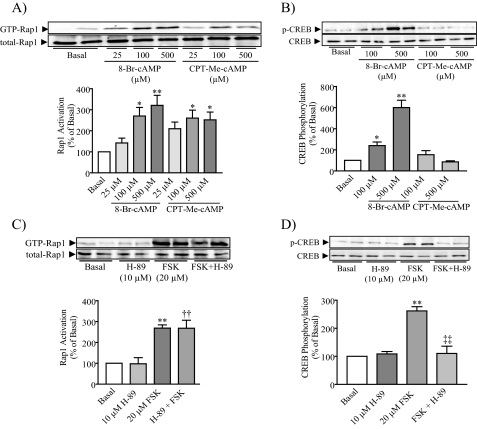
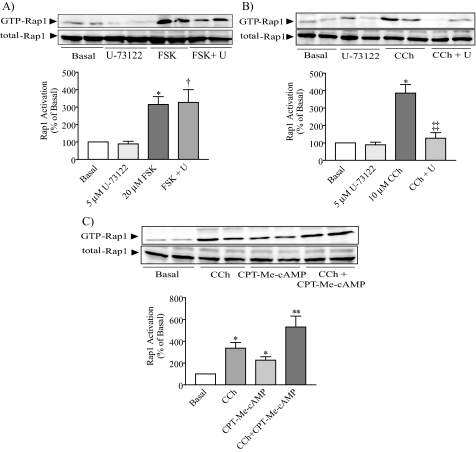
Rap1 Is Activated by the Epac-specific cAMP Analog 8-pCPT-2′-O-Me-cAMP but Not by PKA—Previous studies have showed that Rap1 may be activated by either Epac (3, 4) or both Epac and PKA (5, 6, 8). Based on the involvement of both PKA and Epac in cAMP-mediated rat pancreatic amylase secretion (14) and the participation of cAMP in Rap1 activation as shown in the current study, we sought first to determine whether Epac activates Rap1. Isolated pancreatic acini were incubated with the Epac-selective cAMP analog 8-pCPT-2′-O-Me-cAMP, which preferentially activates Epac without affecting PKA activity (37, 38), and 8-Br-cAMP, which can activate both Epac and PKA pathways (38). Both cAMP analogs increased Rap1 activation in a concentration-dependent manner (Fig. 3A). To confirm the selectively of the 8-pCPT-2′-O-Me-cAMP, its effect on the PKA-dependent phosphorylation of CREB was studied. 8-Br-cAMP increased CREB phosphorylation in a concentration-dependent manner, whereas 8-pCPT-2′-O-Me-cAMP did not, confirming that the 2′-O-Me-cAMP derivative acted via the Epac pathway without affecting the PKA pathway (Fig. 3B). These findings indicate that the Epac pathway is involved in cAMP-stimulated Rap1 activation.
FIGURE 3.
Epac, but not PKA, is involved in Rap1 activation. Either 8-Br-cAMP or the Epac-selective cAMP analog 8-pCPT-2′-O-Me-cAMP (CPT-Me-cAMP) was added to isolated pancreatic acini, and then both Rap1 activation (A) and CREB phosphorylation (B) were determined. 8-Br-cAMP stimulated both Rap1 activation and CREB phosphorylation, whereas CPT-Me-cAMP only stimulated Rap1 activation. PKA inhibition abolished the stimulated effect of forskolin on CREB phosphorylation (D) but did not modify forskolin (FSK)-stimulated Rap1 activation (C). The upper panels show representative immunoblots for GTP-Rap1, total Rap1, phospho-Ser133-CREB, and CREB-2. The lower panels show quantitative analysis of either Rap1 activation or CREB phosphorylation. Data shown are means ± S.E. (four experiments) for either Rap1 activation or CREB phosphorylation expressed as a percentage of basal. *, p < 0.05; **, p < 0.01 versus basal; ††, p < 0.01 versus H-89; ‡‡, p < 0.01 versus FSK.
Next we determined whether PKA might also participate in Rap1 activation using the potent and selective PKA inhibitor H-89 (10 μm) (39) added 30 min before forskolin stimulation. H-89 did not modify either basal or forskolin-evoked GTP-Rap1 levels (Fig. 3C). As expected forskolin-stimulated CREB phosphorylation was abolished in the presence of H-89, indicating that H-89 was able to inhibit PKA activity (Fig. 3D). These findings indicate that forskolin effect on Rap1 activation is most likely Epac-mediated, because it was not modified by the inhibitor of PKA.
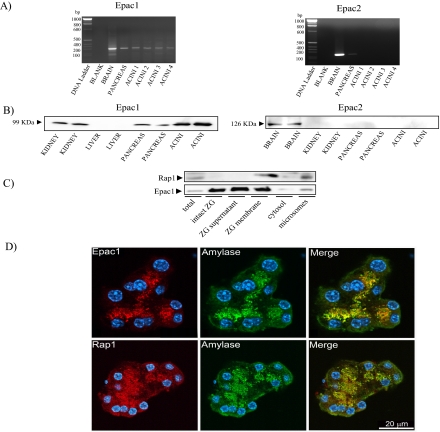
The Rap1GEF Epac1 Is Present in Pancreatic Acini—We next evaluated which Rap1GEFs are present in pancreatic acini. Because the Epac-specific cAMP analog, Ca2+, and DAG participated in Rap1 activation, Rap1 is most likely activated by two Rap1GEFs: Epacs and CalDAG-GEFs. RT-PCR showed that Epac1 mRNA expression was observed in pancreatic acini with a PCR product of the expected size (267 bp) (Fig. 4A). In addition, Epac1 protein was found in both intact pancreas and isolated pancreatic acini as well as in kidney by Western blotting analysis (Fig. 4B). Both Epac2 mRNA expression and protein were observed in the brain. A weak RT-PCR signal for Epac2 was observed in pancreas, consistent with its known presence in islets of Langerhans, but not in isolated pancreatic acini. Furthermore, Epac2 protein could not be detected in the pancreas using two different antibodies (Fig. 4B). Together, these findings indicate the existence of a functional cAMP/Epac1/Rap1 pathway in pancreatic acinar cells.
FIGURE 4.
Epac1 is present in pancreatic acini. Both Epac1 and Epac2 expression were assessed in pancreatic acini by RT-PCR (A) and Western blotting (B) as detailed under “Experimental Procedures.” Brain and kidney were used as positive control for Epac1, whereas only brain was used for Epac2. Epac1 mRNA expression was determined in pancreatic acini and yielded a product of the expected size (267 bp) (A), and Epac1 protein was present in mouse pancreatic acini, as indicated by Western blotting analysis (B). Epac2 protein was not present in acini; a second distinct antibody gave similar results. C, in zymogen granule (ZG) fractionation, Rap1 was only present in ZG membrane fraction, whereas Epac1 was present not only in the ZG supernatant but also in ZG membrane fractions. Note that 15 μg of protein from each fraction was applied to the gel, but most (>90%) of the ZG protein was present in the supernatant following ZG lysis. D, immunohistochemistry was used to localize Epac1 or Rap1 (red) and compare it to amylase (green); nuclei were stained with 4,6-diamino-2-phenylindole (blue). Epac1 as well as Rap1 were localized on ZG in close proximity to amylase.
Rap1 Is Integral to Zymogen Granule Membranes, whereas Epac1 Is Peripherally Associated with Them—Both subcellular fractionation and zymogen granule purification were carried out to localize Epac1 and Rap1 in pancreatic acini. Rap1 was enriched in zymogen granule membranes compared with total lysate, consistent with its integral association with the membranes. In contrast, Epac1 was present in both intact zymogen granule and the supernatant following zymogen granule lysis, which represents the proteins either present in the granule content or associated with the membrane prior to lysis. However, a small amount of Epac1 (<10% total) was also present in zymogen granule membrane fraction. Both Rap1 and Epac1 were also present in the microsomal fraction, but neither was present in the cytosol (Fig. 4C). In addition, the intracellular localization of both Epac1 and Rap1 was investigated by immunohistochemistry in both nonstimulated and stimulated conditions. Immunofluorescent staining of both Epac1 and Rap1 showed a punctuate localization in the area of the acini normally occupied by zymogen granules (Fig. 4D). Dual labeling of Epac1 or Rap1 and a granule content protein, amylase, showed them to be in close proximity. Thus, both Epac1 and Rap1 appear to be associated closely with zymogen granules (Fig. 4D). However, Rap1 was not present in all of zymogen granules, indicating that Rap1 might regulate the exocytosis of a specific zymogen granule pool. In addition, there were also regions where Epac1 or Rap1 and amylase did not overlap. These results are in accordance with the presence of both proteins in the microsomal fraction (Fig. 4C). Since this fraction is expected to contain plasma membranes, Golgi membranes, and lysosomal and endocytotic membranes as well as a fraction of zymogen granule membranes (15), it is likely that Epac1 and Rap1 are also present on some of these membranes. Neither 10 nm VIP nor 10 μm carbachol stimulation altered the immunostaining pattern or subcellular fractionation, indicating no release of either Epac1 or Rap1 into the cytosol (data not shown).
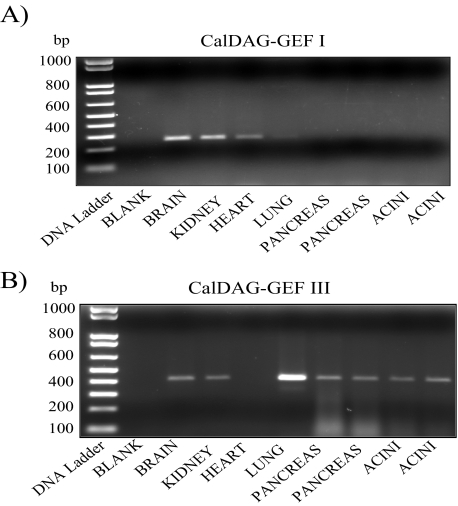
RT-PCR Shows That CalDAG-GEF III Is Expressed in Pancreatic Acini—Four isoforms of the CalDAG-GEFs exist: CalDAG-GEF I, CalDAG-GEF II, CalDAG-GEF III, and CalDAG-GEF IV (2, 39). The first three isoforms are widely distributed (2), whereas CalDAG-GEF IV, the most recently identified member, is present only in mast cells (40). Since CalDAG-GEF I and CalDAG-GEF III are able to activate Rap1 (2), we studied the expression of these two isoforms in mouse pancreatic acini. Only CalDAG-GEF III mRNA expression was observed not only in whole pancreas but also in isolated pancreatic acini with a PCR product of the expected size (414 bp) (Fig. 5). These findings indicate that CalDAG-GEF III is most likely the protein responsible to integrate signaling downstream of the rise in Ca2+ and DAG levels, thereby activating Rap1.
FIGURE 5.
CalDAG-GEF III is expressed in pancreatic acini. The expression of CalDAG-GEFs was assessed in pancreas as well as pancreatic acini by RT-PCR. The brain and kidney were used as positive control. CalDAG-GEF I (A) and CalDAG-GEF III (B) RT-PCR products yielded bands of the expected size (257 and 414 bp, respectively). Only CalDAG-GEF III was expressed not only in whole pancreas but also in pancreatic acini. Results shown are representative of multiple experiments.
Epac1- and Carbachol-evoked Pathways Act Independently to Induce Rap1 Activation—Because the PLC products, DAG and Ca2+, are known to activate CalDAG-GEFs (1, 41), we pretreated acini with the PLC inhibitor U-73122 (5 μm) for 10 min in the presence or absence of either carbachol or forskolin to determine the contribution of PLC activation to carbachol- and forskolin-evoked response. The PLC inhibitor abolished Rap1 activation induced by carbachol but did not modify forskolin-evoked GTP-Rap1 levels (Fig. 6, A and B). These findings indicate that carbachol-induced Rap1 activation is dependent on PLC activity. In addition, since carbachol and the Epac-selective cAMP analog increase GTP-Rap1 levels, in other experiments we tested whether the co-stimulation of both can increase the activation of Rap1 more than either alone. The co-stimulation with both stimulators induced an additive effect (Fig. 6C). Altogether, these data indicate that both Epac1 pathway and carbachol-evoked pathway act independently to induce Rap1 activation.
FIGURE 6.
The Epac1 pathway and the carbachol-induced pathway act independently to activate Rap1. Acini were pretreated with the PLC inhibitor U-73122 and then stimulated with either forskolin (A) or carbachol (B) for 10 min, and Rap1 activation was analyzed. The presence of the PLC inhibitor decreased carbachol-evoked GTP-Rap1 levels without affecting the response of forskolin. C, in other experiments, pancreatic acini were treated with either carbachol or 8-pCPT-2′-O-Me-cAMP (CPT-Me-cAMP) alone or with a combination of both for 10 min. The results showed that the co-addition of both stimulators induces an additive effect on Rap1 activation. The upper panels show representative immunoblots for GTP-Rap1 and total-Rap1. The lower panels show a quantitative analysis of Rap1 activation. Data shown are means ± S.E. (3–5 experiments) for Rap1 activation expressed as a percentage of basal. FSK, forskolin; CCh, carbachol. *, p < 0.05; **, p < 0.01 versus basal; †, p < 0.01 versus U-73122; ‡‡, p < 0.01 versus carbachol.
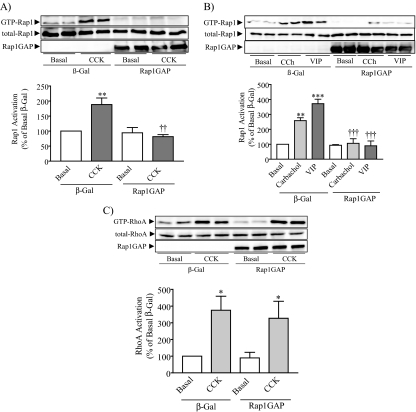
Rap1GAP Overexpression Abolishes Secretagogue-evoked Rap1 Activation and Inhibits Amylase Secretion—The expression of Rap1 on zymogen granules (20) and the activation of Rap1 by the principal secretagogues and intracellular signals which participate in amylase secretion suggested that Rap1 may be involved in the regulation of stimulated amylase secretion. To block the increase in active Rap1, we overexpressed Rap1GAP by means of an adenoviral vector (25). Rap1GAP overexpression blocked the ability of CCK, carbachol, and VIP to activate Rap1 in isolated pancreatic acini (Fig. 7, A and B). The specificity of the Rap1GAP was demonstrated by the observation that CCK-stimulated RhoA activation (26) was not affected by Rap1GAP overexpression (Fig. 7C).
FIGURE 7.
Rap1GAP overexpression blocks the activation of Rap1 by CCK, carbachol, and VIP. Acini were infected overnight with adenovirus expressing either β-galactosidase (vector control) or Rap1GAP and then stimulated with 300 pm CCK (A), 10 μm carbachol (CCh), and 10 nm VIP (B) for 10 min. An inhibition of Rap1 activation was observed when the Rap1GAP-overexpressing acini were stimulated with CCK, carbachol, and VIP. The upper panel shows a representative immunoblot for GTP-Rap1, total Rap1, or Rap1GAP. The lower panel shows the quantitative analysis of Rap1 activation. Data shown are means ± S.E. (four experiments) for activation of Rap1 expressed as a percentage of basal. **, p < 0.01; ***, p < 0.001 versus basal; ††, p < 0.01; †††, p < 0.001 versus CCK, carbachol, or VIP. C, to study the specificity of Rap1GAP, another set of acini were stimulated with 1 nm CCK, and then a RhoA pull-down assay was carried out; Rap1GAP overexpression did not affect CCK-induced RhoA activation. The upper panel shows a representative immunoblot for GTP-RhoA, total RhoA, or Rap1GAP. The lower panel shows the quantitative analysis of RhoA activation. Data shown are means ± S.E. (three experiments) for activation of RhoA expressed as a percentage of basal. *, p < 0.05 versus basal.
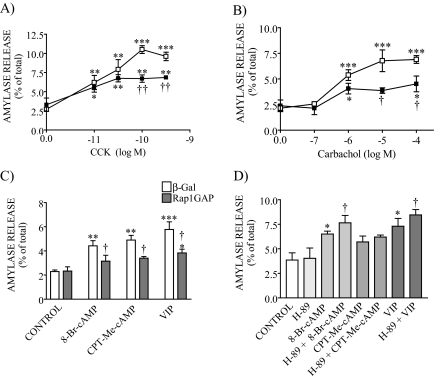
The overexpression of Rap1GAP had no effect on basal amylase release but decreased both CCK- and carbachol-evoked amylase secretion by 40% compared with the control adenovirus (Fig. 8, A and B). This effect was seen across the secretagogue concentration-response curve.
FIGURE 8.
GTP-Rap1 is involved in evoked pancreatic amylase release. β-Galactosidase (β-Gal; vector control) and Rap1GAP-overexpressing acini were stimulated with different concentrations of either CCK (A), carbachol (B), and cAMP-evoked secretagogues (C) for 30 min, and amylase release was measured. An inhibition of amylase secretion (40%) was observed when the Rap1GAP-overexpressing acini were stimulated with high concentrations of either CCK or carbachol. In addition, Rap1GAP overexpression decreased 8-Br-cAMP-, CPT-Me-cAMP-, and VIP-stimulated amylase secretion by 60%. Data shown are means ± S.E. (4–6 experiments) of amylase release expressed as a percentage of total. □, β-galactosidase-expressing cells; ▪, Rap1GAP-overexpressing cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control; †, p < 0.05; ††, p < 0.01 versus CCK, carbachol, 8-Br-cAMP, CPT-Me-cAMP, or VIP response in β-galactosidase-expressing cells. D, pancreatic acini were preincubated for 10 min with the PKA inhibitor H-89 and then stimulated with 8-Br-cAMP, CPT-Me-cAMP, or VIP. Secretagogue-stimulated amylase release was not modified in the presence of the inhibitor. Data shown are means ± S.E. (four experiments) of amylase release expressed as a percentage of the total. *, p < 0.05 versus control; †, p < 0.05 versus 8-Br-cAMP or VIP.
We also studied whether the activation of endogenous Epac mediates cAMP-stimulated amylase secretion response by use of 8-pCPT-2′-O-Me-cAMP. The Epac-specific cAMP analog weakly stimulated amylase secretion by 114%, producing an increase similar to that induced by 8-Br-cAMP (93%) and by VIP (153%) (Fig. 8C). We also studied the effect of the combination of 8-pCPT-2′-O-Me-cAMP and carbachol on amylase release. Although carbachol alone increased amylase release by 200%, the co-stimulation with both secretagogues potently stimulated it by 400% (supplemental Fig. 1), indicating that the combination of 8-pCPT-2′-O-Me-cAMP and carbachol induced a synergistic effect on amylase secretion. Such a synergistic effect has previously been reported for the combination of CCK and VIP in mouse pancreatic acini (32).
We next determined whether Rap1 mediates the cAMP-evoked amylase release. Rap1GAP overexpression reduced the response to 8-Br-cAMP, 8-pCPT-2′-O-Me-cAMP, and VIP by 60% compared with the control adenovirus (Fig. 8C). These results indicate that Epac1/Rap1 pathway is involved in cAMP-stimulated amylase secretion. Since VIP increases intracellular cAMP levels in pancreatic acini (29), both PKA and Epac1 pathways could mediate the effect of VIP on amylase release. However, the PKA inhibitor H-89 did not modify the responses to 8-Br-cAMP, 8-pCPT-2′-O-Me-cAMP, and VIP (Fig. 8D). These findings show that in mouse pancreatic acini, the cAMP effect is largely mediated by the Epac1/Rap1 pathway and not by PKA.
Neither Epac1 nor Active Rap1 Affects Ca2+ Mobilization in Pancreatic Acini—We also studied whether Epac1 induces Ca2+ mobilization in pancreatic acini, since the Epac-selective cAMP analog 8-pCPT-2′-O-Me-cAMP is a stimulator of Ca2+-induced Ca2+ release in pancreatic β-cells (12). Carbachol (data not shown) and CCK increased Ca2+ mobilization, whereas VIP and 8-pCPT-2′-O-Me-cAMP did not (supplemental Fig. 2A), indicating that Epac1 does not evoke Ca2+ release in pancreatic acini. We next evaluated whether Rap1 activation was an effect of Ca2+ mobilization and not a cause. Preventing Rap1 activation by overexpressing Rap1GAP did not affect either CCK- or carbachol-induced Ca2+ mobilization (supplemental Fig. 2B; CCK response only shown).
GTP-Rap1 Does Not Influence cAMP Generation—To confirm that Rap1 was activated by cAMP but did not influence cAMP production, we measured acinar cAMP levels. VIP, but not CCK and carbachol, increased cAMP generation in mouse pancreatic acini (data not shown), and the stimulating effect of VIP was not influenced by Rap1GAP (supplemental Fig. 2C).
DISCUSSION
The results of the current study demonstrate that numerous secretagogues and second messengers as well as the Epac-selective cAMP analog 8-pCPT-2′-O-Me-cAMP are able to activate Rap1. Additionally, we show that in mouse pancreatic acini, Rap1 activation is PKA- and PKC-independent, because the PKA inhibitor H-89 as well as the PKC inhibitor GF-109203X did not modify forskolin- and carbachol-stimulated Rap1 activation, respectively. Our data also demonstrate that the two Rap1GEFs, Epac1 and CalDAG-GEF III, are expressed in mouse pancreatic acini. We also demonstrate that the presence of the PLC inhibitor U-73122 did not modify Rap1 activation induced by forskolin but abolished carbachol-evoked GTP-Rap1 levels. In addition, the co-stimulation with carbachol and 8-pCPT-2′-O-Me-cAMP produces an additive effect on Rap1 activation. These findings are consistent with the existence of two independent mechanisms by which Rap1 may be activated in acini: the Epac1 pathway activated by cAMP and the CalDAG-GEF III pathway evoked by DAG and Ca2+ mobilization. Finally, the major finding of the current study is that Rap1 is required for stimulated amylase secretion, since inhibiting Rap1 activity by overexpression of Rap1GAP reduces not only the action of cAMP on amylase release but also CCK- and carbachol-stimulated amylase secretion. Furthermore, we demonstrate that cAMP-stimulated amylase release is unaffected by the PKA inhibitor H-89, suggesting that in mouse pancreatic acini, amylase release evoked by cAMP is PKA-independent and mediated by the Epac1/Rap1 pathway. Together, our findings show that different secretagogues, including CCK, acetylcholine, and VIP, mediated by stimulation of CCK, muscarinic, and VPAC receptors, respectively, generate second messengers, including DAG, Ca2+, and cAMP, which are able to activate Rap1 via either CalDAG-GEF III or Epac1 and thereby regulate amylase secretion in pancreatic acinar cells. In this way, Rap1 plays an important role as a common effector for the cAMP/Epac1, PLC/IP3/Ca2+, and PLC/DAG pathways (Fig. 9). A role for Rap1 as a common effector is also found in hematopoietic cells, such as platelets and neutrophils, as well as in fibroblasts (42).
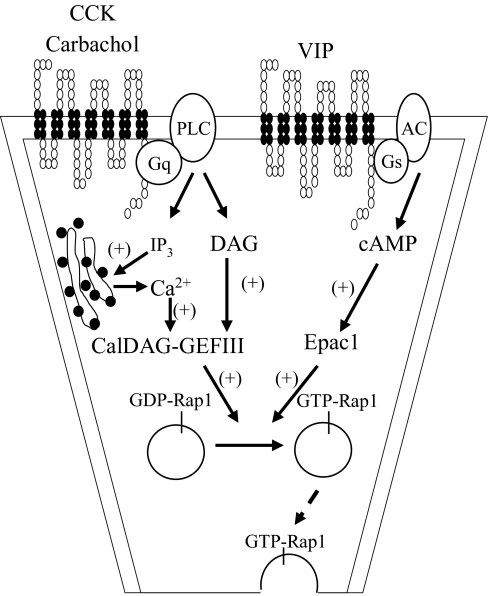
FIGURE 9.
Proposed model for Rap1-mediated response on stimulated amylase release. Activation of Rap1 occurs after the stimulation by secretagogues CCK, carbachol, and VIP in pancreatic acini. Our findings show that different second messengers, including DAG, Ca2+, and cAMP generated from activation of different receptors, CCK and muscarinic as well as VPAC receptors, are able to mediate the activation of Rap1 via either CalDAG-GEFIII or Epac1 and thereby regulate amylase secretion in pancreatic acinar cells. Note that the broken line indicates a less well defined pathway.
In the present study, we found that cAMP-induced amylase release was strongly reduced in Rap1GAP-expressing cells, whereas CCK- as well as carbachol-induced amylase secretion was reduced to a lesser extent (Fig. 8, A–C). The residual responses suggested that molecular signals other than Rap1 were also involved in CCK- and carbachol-evoked responses. Ca2+, the primary signaling molecule involved in pancreatic enzyme release, is believed to be the major mediator of both secretagogues (31).
Several lines of evidence indicate that in some cell types Rap1 is activated by both Epac and PKA. Rap1 participates in neurotensin secretion (8) and may be involved in parotid amylase release (21, 22) downstream of both PKA and Epac signaling pathways. However, our current study demonstrated that in acinar cells Rap1 is activated by Epac1, Ca2+, and DAG, but not by PKA (Figs. 2 and 3). One explanation for this difference appears to be whether or not PKA has a relevant role in exocytotic process. In those cells in which Rap1 is activated by both Epac and PKA, PKA is required for exocytosis. In neuroendocrine BON cells, PKA is involved in neurotensin secretion, since the PKA inhibitor H-89 reduced forskolin-induced neurotensin release (8). In parotid acinar cells, cAMP is the primary signal for amylase release, and PKA phosphorylation of the proteins associated with regulated exocytosis is one of the critical events in this process (38). By contrast, in mouse pancreatic acinar cells, PKA does not appear to participate in the process. Although PKA catalyzes the phosphorylation of regulatory proteins associated with pancreatic exocytotic process (43, 44), our current study showed that the Epac1/Rap1 pathway, but not the PKA pathway, has a relevant role in cAMP-stimulated amylase release (Fig. 8, C and D). Altogether, the data and earlier work indicate that in mouse pancreatic acinar cells, both cAMP/PKA and cAMP/Epac1/Rap1 pathways co-exist but appear to act in distinct process.
Despite considerable structural similarity, both Epac1 and Epac2 appear to be functionally different. Several lines of evidence show that Epac1 acts through pathways involving direct protein-protein interaction with Rap1, whereas Epac2 acts through other proteins, such as Rim2 (Rab3-interacting molecule 2) and Piccolo. In mouse epididymal spermatozoa heads, Epac1 interacts with Rap1, inducing its activation (45). In hematopoietic cells, the Epac1/Rap1 pathway regulates cellular adhesion and chemotaxis (46, 47). In pancreatic β-cells, Epac2 interacts with Rim2 (48) as well as Piccolo and then forms a Epac2-Rim2-Piccolo complex that regulates Ca2+-dependent exocytosis mediated by ryanodine-sensitive Ca2+ channels, an important event in cAMP-induced insulin secretion (12, 49). Although Epac1 has also been implicated in the regulation of insulin secretion (13), recently, Shibasaki et al. (23) showed that in pancreatic β-cells, Rap1 is downstream of Epac2 and is involved in the regulation of insulin secretion. Our observation that Epac2 is not present in pancreatic acini (Fig. 4B) is consistent with the lack of proteins on zymogen granules that commonly interact with Epac2, Rim2, and Piccolo (20). These results suggest that two different types of Epac-mediated exocytosis exist, those activated by the cAMP/Epac1 pathway and those activated by the cAMP/Epac2 pathway, and that the participation of one or another functional pathway depends on which subtype of Epac protein is present in the cell.
Several reports show that Epac has different localization, depending on the cell type. Epac1 is localized in the acrosomal region of the sperm head and is associated with Ca2+-induced acrosomal exocytosis (50). In addition, in parotid acinar cells, where Epac1 has also been linked to the exocytotic process, Epac1 is localized in the intracellular and plasma membrane fractions (22). By contrast, in neuroendocrine line PC12 and thyrocyte cells, Epac subcellular localization is at the nuclear membrane but not at the plasma membrane (51, 52). In pancreatic β-cells, Epac2 and Rap1 are co-localized with insulin (23, 53) and shown to mediate cAMP-dependent PKA-independent exocytosis. In the current study, using immunohistochemistry, we show that both Epac1 and Rap1 were co-localized with amylase present in zymogen granules in either nonstimulated or stimulated conditions (Fig. 4D). Using subcellular fractionation, we found that Rap1 is localized on zymogen granule membranes, which is consistent with its attachment to the membranes by a geranylgeranylated moiety (54). By contrast, Epac1 is more likely to be associated with zymogen granule membranes through protein-protein interaction (Fig. 4C). In addition, we showed that in stimulated conditions, both Epac1 and Rap1 do not translocate to the cytosol. These findings indicate that the localization of Rap1 and Epac1 in zymogen granules in nonstimulated and stimulated conditions may promote their interaction. Pancreatic acinar exocytosis involves several distinct stages, namely tethering, docking, or formation of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex (priming), culminating in the fusion of zymogen granules with the luminal plasma membrane of the acinar cells (28). Since the most known small GTP-binding proteins, Rab proteins, are implicated in tethering/docking steps (17, 55), both Rap1 and Epac1 may participate in some step of exocytosis.
A number of small GTP-binding proteins have been shown to be involved in the regulation of pancreatic exocytosis (15–20). However, there is limited understanding of the molecular mechanisms involved. A possibility that has been considered is that the activation of Rap1 causes a fusion between zymogen granules and plasma membrane mediated by cytoskeleton. Rap1 could facilitate the movement of zymogen granules during exocytotic process and/or regulate the exocytotic event through interactions with proteins located on cytoskeleton. The presence of Rap1 on zymogen granule membranes (Fig. 4C) and the participation of Rap1 in cytoskeletal assembly in human platelets (56, 57) support this possibility. In accordance with this hypothesis, both Rac and Rho, two other small GTP-binding proteins belonging to Rho superfamily, have been implicated in the regulation of CCK-induced amylase release through an actin cytoskeleton-dependent cellular process (26), suggesting the cytoskeleton as a possible target for the action of multiple GTP-binding proteins, including Rap1 in pancreatic acinar cells.
The current report not only helps elucidate the physiological significance of Rap1 but also sheds light on the mechanisms involved in the exocytotic process of zymogen granules. We conclude that Rap1 belongs to a group of diverse small GTP-binding proteins involved in the regulation of exocrine secretory process. Rap1 not only mediates the action of cAMP on amylase release with Epac1 as an intermediate but is also involved in CCK- and carbachol-stimulated amylase secretion, probably via CalDAG-GEF III. It can be activated by multiple secretagogues and second messengers, playing a critical role as a common effector protein for cAMP/Epac1, PLC/IP3/Ca2+, and PLC/DAG pathways.
Supplementary Material
Acknowledgments
We thank Nancy Vogel for technical support with Rap1GAP adenovirus amplification and Bradley B. Nelson for technical assistance with immunohistochemistry.
This work was supported, in whole or in part, by National Institutes of Health Grant DK-41128 (to J. A. W.). This work was also supported by Michigan Gastrointestinal Peptide Center Grant P30 DK-34933. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: GEF, guanine-nucleotide exchange factor; 8-Br-cAMP, 8-bromo-cyclic AMP; BSA, bovine serum albumin; CalDAG-GEF, Ca2+- and diacylglycerol guanine-nucleotide exchange factor; CCK, cholecystokinin; 8-pCPT-2′-O-Me-cAMP, 8-(4-chlorophenylthio)-2′-O-methyladenosine-3′,5′-cyclic monophosphate; CREB, cAMP-response element-binding protein; DAG, diacylglycerol; GAP, GTPase-activating protein; GF-109203X, 3-[1-(3-dimethylaminopropyl)-3-indolyl]-3(3-indolyl)maleimide; H-89, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide dihydrochloride; IP3, inositol 1,4,5-triphosphate; MOPS, (N-morpholino)propanesulfonic acid; PBS, phosphate-buffered saline; PKA, protein kinase A; PKC, protein kinase C; PLCβ, phospholipase Cβ; PMA, phorbol 12-myristate 13-acetate; PMSF, phenylmethylsulfonyl fluoride; RBD, Rap1-binding domain; RT, reverse transcription; STI, soybean trypsin inhibitor; VIP, vasoactive intestinal peptide.
References
- 1.Bos, J. L. (2005) Curr. Opin. Cell Biol. 17 123-128 [DOI] [PubMed] [Google Scholar]
- 2.Yamashita, S., Mochizuki, N., Ohba, Y., Tobiume, M., Okada, Y., Sawa, H., Nagashima, K., and Matsuda, M. (2000) J. Biol. Chem. 275 25488-25493 [DOI] [PubMed] [Google Scholar]
- 3.de Rooij, J., Zwartkruis, F. J., Verheijen, M. H., Cool, R. H., Nijman, S. M., Wittinghofer, A., and Bos, J. L. (1998) Nature 396 474-477 [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki, H., Springett, G. M., Mochizuki, N., Toki, S., Nakaya, M., Matsuda, M., Housman, D. E., and Graybiel, A. M. (1998) Science 282 2275-2279 [DOI] [PubMed] [Google Scholar]
- 5.Quilliam, L. A., Mueller, H., Bohl, B. P., Prossnitz, V., Sklar, L. A., Der, C. J., and Bokoch, G. M. (1991) J. Immunol. 147 1628-1635 [PubMed] [Google Scholar]
- 6.Lerosey, I., Pizon, V., Tavitian, A., and de Gunzburg, J. (1991) Biochem. Biophys. Res. Commun. 175 430-436 [DOI] [PubMed] [Google Scholar]
- 7.Tsygankova, O. M., Saavedra, A., Rebhun, J. F., Quilliam, L. A., and Meinkoth, J. L. (2001) Mol. Cell Biol. 21 1921-1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, J., O'Connor, K. L., Cheng, X., Mei, F. C., Uchida, T., Townsend, C. M., Jr., and Evers, B. M. (2007) Mol. Endocrinol. 21 159-171 [DOI] [PubMed] [Google Scholar]
- 9.Guo, F., Kumahara, E., and Saffen, D. (2001) J. Biol. Chem. 276 25568-25581 [DOI] [PubMed] [Google Scholar]
- 10.Crittenden, J. R., Bergmeier, W., Zhang, Y., Piffath, C. L., Liang, Y., Wagner, D. D., Housman, D. E., and Graybiel, A. M. (2004) Nat. Med. 10 982-986 [DOI] [PubMed] [Google Scholar]
- 11.Kooistra, M. R. H., Corada, M., Dejana, E., and Bos, J. L. (2005) FEBS Lett. 579 4966-4972 [DOI] [PubMed] [Google Scholar]
- 12.Kang, G., Joseph, J. W., Chepurny, O. G., Monaco, M., Wheeler, M. B., Bos, J. L., Schwede, F., Genieser, H. G., and Holz, G. G. (2003) J. Biol. Chem. 278 8279-8285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang, G., Chepurny, O. G., Rindler, M. J., Collis, L., Chepurny, Z., Li, W. H., Harbeck, M., Roe, M. W., and Holz, G. G. (2005) J. Physiol. 566 173-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri, A., Husain, S. Z., Kolodecik, T. R., Grant, W. M., and Gorelick, F. S. (2007) Am. J. Physiol. 292 G1403-G1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göke, B., Williams, J. A., Wishart, M. J., and De Lisle, R. C. (1992) Am. J. Physiol. 262 C493-C500 [DOI] [PubMed] [Google Scholar]
- 16.Pinxteren, J. A., O'Sullivan, A. J., Larbi, K. Y., Tatham, P. E. R., and Gomperts, B. D. (2000) Biochimie (Paris) 82 385-393 [DOI] [PubMed] [Google Scholar]
- 17.Chen, X., Edwards, J. A., Logsdon, C. D., Ernst, S. A., and Williams, J. A. (2002) J. Biol. Chem. 277 18002-18009 [DOI] [PubMed] [Google Scholar]
- 18.Chen, X., Ernst, S. A., and Williams, J. A. (2003) J. Biol. Chem. 278 50053-50060 [DOI] [PubMed] [Google Scholar]
- 19.Chen, X., Li, C., Izumi, T., Ernst, S. A., Andrews, P. C., and Williams, J. A. (2004) Biochem. Biophys. Res. Commun. 323 1157-1162 [DOI] [PubMed] [Google Scholar]
- 20.Chen, X., Walker, A. K., Strahler, J. R., Simon, E. S., Tomanicek-Volk, S. L., Nelson, B. B., Hurley, M. C., Ernst, S. A., Willams, J. A., and Andrews, P. C. (2006) Mol. Cell Proteomics 5 306-312 [DOI] [PubMed] [Google Scholar]
- 21.D'Silva, N. J., Jacobson, K. L., Ott, S. M., and Watson, E. L. (1998) Am. J. Physiol. 274 C1667-C1673 [DOI] [PubMed] [Google Scholar]
- 22.Shimomura, H., Imai, A., and Nashida, T. (2004) Arch. Biochem. Biophys. 431 124-128 [DOI] [PubMed] [Google Scholar]
- 23.Shibasaki, T., Takahashi, H., Miki, T., Sunaga, Y., Matsumura, K., Yamanaka, M., Zhang, C., Tamamoto, A., Satoh, T., Miyazaki, J.-I., and Seino, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19333-19338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamieson, J. D., and Palade, G. E. (1967) J. Cell Biol. 34 597-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke, B., Akkerman, J. W., and Bos, J. L. (1997) EMBO J. 16 252-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi, Y., and Williams, J. A. (2005) Am. J. Physiol. 289 C22-C32 [DOI] [PubMed] [Google Scholar]
- 27.Williams, J. A., Korc, M., and Dormer, R. L. (1978) Am. J. Physiol. 4 E517-E524 [DOI] [PubMed] [Google Scholar]
- 28.Sabbatini, M. E., Villagra, A., Davio, C. A., Vatta, M. S., Fernández, B. E., and Bianciotti, L. G. (2003) Am. J. Physiol. Gastrointest. Liver Physiol. 285 G929-G937 [DOI] [PubMed] [Google Scholar]
- 29.Mochizuki, N., Ohba, Y., Kiyokawa, E., Kurata, T., Murakami, T., Ozaki, T., Kitabatake, A., Nagashima, K., and Matsuda, M. (1999) Nature 400 891-894 [DOI] [PubMed] [Google Scholar]
- 30.Wittchen, E. S., Worthylake, R. A., Kelly, P., Casey, P. J., Qilliam, L. A., and Burridge, K. (2005) J. Biol. Chem. 280 11675-11682 [DOI] [PubMed] [Google Scholar]
- 31.Williams, J. A., and Yule, D. I. (2006) in Physiology of the Gastrointestinal Tract (Johnson L. R., ed) 4th Ed., pp. 1337-1369, Academic Press, Inc., New York
- 32.Burnham, D. B., McChesney, D. J., Thurston, K. C., and Williams, J. A. (1984) J. Physiol. 349 475-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toullec, D., Pianetti, P., Coste, H., Bellevergue, P., Grand-Perret, T., Ajakane, M., Baudet, V., Boissin, P., Boursier, E., Loriolle, F., Duhamel, L., Charon, D., and Kirilovsky, J. (1991) J. Biol. Chem. 266 15771-15781 [PubMed] [Google Scholar]
- 34.M'Rabet, L., Coffer, P., Zwartkruis, F., Franke, B., Segal, A. W., Koenderman, L., and Bos, J. L. (1998) Blood 92 2133-2140 [PubMed] [Google Scholar]
- 35.McLeod, S. J., Ingham, R. J., Bos, J. L., Kurosaki, T., and Gold, M. R. (1998) J. Biol. Chem. 273 29218-29223 [DOI] [PubMed] [Google Scholar]
- 36.Heisler, S. (1983) Can. J. Physiol. Pharmacol. 61 1168-1176 [DOI] [PubMed] [Google Scholar]
- 37.Enserink, J. M., Christensen, A. E., de Rooij, J., van Triest, M., Schwede, F., Genieser, H. G., Doskeland, S. O., Blank, J. L., and Bos, J. L. (2002) Nat. Cell Biol. 4 901-906 [DOI] [PubMed] [Google Scholar]
- 38.Seino, S., and Shibasaki, T. (2005) Physiol. Rev. 85 1303-1342 [DOI] [PubMed] [Google Scholar]
- 39.Chijiwa, T., Mishima, A., Hagiwara, M., Sano, M., Hayashi, K., Inoue, T., Naito, K., Toshioka, T., and Hidaka, H. (1990) J. Biol. Chem. 265 5267-5272 [PubMed] [Google Scholar]
- 40.Yang, Y., Li, L., Wong, G. W., Krislis, S. A., Madhusudhan, M. S., Sali, A., and Stevens, R. L. (2002) J. Biol. Chem. 277 25756-25774 [DOI] [PubMed] [Google Scholar]
- 41.Ebinu, J. O., Bottorff, D. A., Chan, E. Y., Stang, S. L., Dunn, R. J., and Stone, J. C. (1998) Science 280 1082-1086 [DOI] [PubMed] [Google Scholar]
- 42.Zwartkruis, F. J. T., and Bos, J. L. (1999) Exp. Cell Res. 253 157-165 [DOI] [PubMed] [Google Scholar]
- 43.Burnham, D. B., and Williams, J. A. (1984) Am. J. Physiol. 246 G500-G508 [DOI] [PubMed] [Google Scholar]
- 44.Burnham, D. B., Sung, C. K., Munowitz, P., and Williams, J. A. (1988) Biochim. Biophys. Acta 969 33-39 [DOI] [PubMed] [Google Scholar]
- 45.Amano, R., Lee, M. J., Goto, N., and Harayama, H. (2007) J. Reprod. Dev. 53 123-133 [DOI] [PubMed] [Google Scholar]
- 46.Enserink, J. M., Price, L. S., Methi, T., Mahic, M., Sonnenberg, A., Bos, J. L., and Taskén, K. (2004) J. Biol. Chem. 279 44889-44896 [DOI] [PubMed] [Google Scholar]
- 47.Gupta, M., and Yarwood, S. J. (2005) J. Biol. Chem. 280 8109-8116 [DOI] [PubMed] [Google Scholar]
- 48.Kashima, Y., Miki, T., Shibasaki, T., Ozaki, N., Miyazaki, M., Yano, H., and Seino, S. (2001) J. Biol. Chem. 276 46046-46053 [DOI] [PubMed] [Google Scholar]
- 49.Fujimoto, K., Shibasaki, T., Yokoi, N., Kashima, Y., Matsumoto, M., Sasaki, T., Tajima, N., Iwanaga, T., and Seino., S. (2002) J. Biol. Chem. 277 50497-50502 [DOI] [PubMed] [Google Scholar]
- 50.Branham, M. T., Mayorga, L. S., and Tomes, C. N. (2006) J. Biol. Chem. 281 8656-8666 [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z., Dillon, T. J., Pokala, V., Mishra, S., Labudda, K., Hunter, B., and Stork, P. J. (2006) Mol. Cell Biol. 26 2130-2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dremier, S., Milenkovic, M., Blancquaert, S., Dumont, J. E., Doskeland, S. O., Maenhaut, C., and Roger, P. P. (2007) Endocrinology 148 4612-4622 [DOI] [PubMed] [Google Scholar]
- 53.Eliasson, L., Ma, X., Renström, E., Barg, S., Berggren, P. O., Galvanovskis, J., Gromada, J., Jing, X., Lundquist, I., Salehi, A., Sewing, S., and Rorsman, P. (2003) J. Gen. Physiol. 121 181-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takai, Y., Sasaki, T., and Matozaki, T. (2001) Physiol. Rev. 81 153-208 [DOI] [PubMed] [Google Scholar]
- 55.Savina, A., Fader, C. M., Damiani, M. T., and Colombo, M. I. (2005) Traffic 6 131-143 [DOI] [PubMed] [Google Scholar]
- 56.Fischer, T. H., Gatling, M. N., Lacal, J. C., and White, G. C., II (1990) J. Biol. Chem. 265 19405-19408 [PubMed] [Google Scholar]
- 57.Nagata, K., and Nozawa, Y. (1995) Br. J. Haematol. 90 180-186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.