Abstract
Chronic gastritis induced by Helicobacter pylori is the strongest known risk factor for peptic ulceration and distal gastric cancer, and adherence of H. pylori to gastric epithelial cells is critical for induction of inflammation. One H. pylori constituent that increases disease risk is the cag pathogenicity island, which encodes a secretion system that translocates bacterial effector molecules into host cells. Decay-accelerating factor (DAF) is a cellular receptor for H. pylori and a mediator of the inflammatory response to this pathogen. H. pylori induces DAF expression in human gastric epithelial cells; therefore, we sought to define the mechanism by which H. pylori up-regulates DAF and to extend these findings into a murine model of H. pylori-induced injury. Co-culture of MKN28 gastric epithelial cells with the wild-type H. pylori cag+ strain J166 induced transcriptional expression of DAF, which was attenuated by disruption of a structural component of the cag secretion system (cagE). H. pylori-induced expression of DAF was dependent upon activation of the p38 mitogen-activated protein kinase pathway but not NF-κB. Hypergastrinemic INS-GAS mice infected with wild-type H. pylori demonstrated significantly increased DAF expression in gastric epithelium versus uninfected controls or mice infected with an H. pylori cagE- isogenic mutant strain. These results indicate that H. pylori cag+ strains induce up-regulation of a cognate cellular receptor in vitro and in vivo in a cag-dependent manner, representing the first evidence of regulation of an H. pylori host receptor by the cag pathogenicity island.
Helicobacter pylori induces an inflammatory response in the stomach that persists for decades and increases the risk not only for peptic ulceration but also for gastric adenocarcinoma and non-Hodgkin's lymphoma of the stomach (1, 2). Gastric adenocarcinoma is the second leading cause of cancer-related death in the world, and chronic gastritis induced by H. pylori is the strongest known risk factor for this malignancy (1, 3–7). However, only a fraction of infected persons ever develop cancer, underscoring the importance of defining mechanisms that regulate biological interactions between H. pylori and their hosts that promote transformation.
Although the vast majority of H. pylori in colonized hosts are free-living, ∼20% bind to gastric epithelial cells, and adherence is important in the induction of injury (8). BabA is an outer-membrane protein (OMP)2 encoded by the strain-specific gene babA2, which binds the Lewisb (Leb) histo-blood-group antigen on gastric epithelial cells (9, 10). BabA binding specificities reflect H. pylori strain adaptation to different glycosylation patterns that predominate in a particular host population, and BabA-mediated Leb binding can be altered by both bacterial phase variation and genetic recombination (11). Another H. pylori adhesin, SabA, binds the sialyl-Lewisx (sLex) antigen, which is an established tumor antigen and marker of gastric dysplasia (12). Gastric inflammation induced by H. pylori up-regulates the expression of sLex on epithelial cells, which amplifies interactions between this molecule and SabA.
We recently identified another H. pylori receptor, Decay-accelerating factor (DAF), that is up-regulated after bacterial contact (13). DAF is an intrinsic regulator of complement that is attached to the outer leaflet of the cell membrane by a glycophosphatidylinositol anchor (14). DAF protects cells from complement activation on their surfaces by dissociating membrane-bound C3 convertases that are required for cleaving C3 and further propagating the complement cascade. DAF can also be utilized as a cellular receptor by several pathogenic organisms associated with chronic inflammatory diseases, including uropathogenic diffusely adhering Escherichia coli, coxsackieviruses, echoviruses, and enteroviruses (14–18).
Expression of DAF is increased within H. pylori-infected human gastric tissue compared with uninfected mucosa, and the intensity of expression is directly related to the density of H. pylori colonization and severity of inflammation (19, 20). We recently demonstrated that DAF influences the inflammatory response to H. pylori as infected DAF-deficient mice developed significantly less severe inflammation compared with infected wild-type mice, suggesting that the interaction between H. pylori and DAF is important for pathogenesis (13).
In addition to host effectors that mediate injury, H. pylori constituents can also regulate pathogenic responses. After adherence, H. pylori strains that possess a type IV secretion system encoded by the cag pathogenicity island translocate CagA and components of peptidoglycan into host cells (21–24). CagA subsequently undergoes Src- and Abl-dependent tyrosine phosphorylation and activates a eukaryotic phosphatase (SHP-2), eventuating in dephosphorylation of host cell proteins and cellular morphological changes (22, 23, 25, 26). H. pylori peptidoglycan components delivered by the cag secretion system are recognized by the intracellular pattern recognition receptor NOD1, which initiates cell-signaling events including activation of NF-κB (24). In vivo, the presence of the cag island also influences the topography of colonization, as H. pylori cag- strains predominate within the mucus gel layer, whereas cag+ strains are found immediately adjacent to epithelial cells (27). Compared with cag- strains, H. pylori cag+ strains augment the risk for severe pathologic outcomes, such as peptic ulceration and gastric cancer (1). Because adherence likely plays a critical role in pathogenesis, we sought to delineate the host and bacterial factors that mediate H. pylori induction of DAF. We demonstrate that H. pylori cag+ strains up-regulate DAF expression in gastric epithelial cells in vitro and in vivo in a cag-dependent manner and that this induction is mediated by p38 MAP kinase activation.
EXPERIMENTAL PROCEDURES
Reagents and Constructs—Actinomycin D, cycloheximide, the p38 inhibitor SB203580, and the JNK1/2/3 inhibitor JNK inhibitor II were obtained from Calbiochem, and the MEK1/2 inhibitor PD98059 was obtained from Cayman Chemical. Anti-DAF IA10 (BD Pharmingen), anti-phospho-ERK1/2 (Thr-202/Tyr-204), anti-phospho-MAPKAPK-2 (Thr-334), and anti-phospho-c-Jun (Ser-73) antibodies (Cell Signaling) were used for Western analysis. The anti-DAF antibody MCA1614 (AbD Serotec) was used for immunohistochemistry. The pNF-κB luciferase vector (Clontech) and pRL Renilla luciferase vector (Promega) were used for NF-κB luciferase studies. Dominant-negative mutant IκBα S32A/S36A and dominant-negative IκB kinase β K44A constructs were used for NF-κB inhibition studies (generous gifts of Dr. Andrew Neish, Emory University School of Medicine) (28).
Cell Culture—MKN28 human gastric epithelial cells (kindly provided by Dr. Robert Coffey, Vanderbilt University) were grown in RPMI 1640 (Invitrogen) with 10% heat-inactivated fetal bovine serum and 20 μg/ml gentamicin in an atmosphere of 5% CO2 at 37 °C.
Bacterial Strains—Experiments were performed with the H. pylori cag+ strains J166 and 7.13 (13, 29). Isogenic cagA, cagE, and cagM null mutants were constructed by insertional mutagenesis using aphA (conferring kanamycin resistance) as previously described (30, 31) and were selected on Brucella agar with kanamycin (25 μg/ml). Heat-killed H. pylori were generated by heating the bacteria to 80 °C for 10 min. H. pylori lysates were generated by sonication as previously described (32). Lysates were then sterilized using a 0.2-μm pore size filter.
Western Analysis—MKN28 gastric epithelial cells were grown to confluence, then cultured in serum-free medium for 24 h and then co-cultured with H. pylori for specified times at a multiplicity of infection (m.o.i.) of 100. H. pylori-infected and uninfected MKN28 cells were lysed in radioimmune precipitation assay buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 1% Triton X-100, 0.1% SDS), and protein concentrations were quantified by the BCA assay (Pierce) (31). Proteins (30 μg) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Pall Corp., Ann Arbor, MI). DAF levels were examined in gastric cells by Western blotting using an anti-DAF (1:1000, IA10) antibody. Levels of the phosphorylated MAPK targets ERK1/2, MAPKAPK-2, and c-Jun were detected using the respective antibodies described above (1:1000). Primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies (Santa Cruz) and visualized by Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences) according to the manufacturer's instructions. Western blots were imaged, and band intensities were quantified using the ChemiGenius Gel Bio Imaging System (Syngene).
Real-time Quantitative RT-PCR—MKN28 gastric epithelial cells were grown to confluence, cultured in serum-free medium for 24 h, and then co-cultured with H. pylori for specified times (m.o.i. = 100). RNA was prepared from H. pylori:gastric cell co-cultures using the RNeasy RNA purification kit (Qiagen) following the manufacturer's instructions. Reverse transcriptase PCR was performed using TaqMan reverse transcription reagents (Applied Biosystems), which was followed by real-time quantitative PCR using the TaqMan Gene Expression Assay and a 7300 real-time PCR system (Applied Biosystems). Daf and gapdh cDNA were quantified using a TaqMan® Gene Expressions primer set purchased from Applied Biosystems. -Fold induction of daf mRNA was determined from the threshold cycle values normalized for gapdh mRNA expression, and this ratio was then normalized to the value derived from cells cultured with medium alone.
Transfections and Luciferase Assay—MKN28 cells were transiently transfected using FuGENE 6 reagent (Roche Applied Science) per the manufacturer's instructions. Cells were allowed to incubate with the transfection mixture for 24 h, cultured in serum-free medium for an additional 24 h, and then co-cultured with H. pylori strain J166 (m.o.i. = 100). Samples were assayed for luciferase activity on a TD-20/20 Luminometer (Turner Designs) using the Dual Luciferase® reporter kit (Promega) according to the manufacturer's instructions.
Experimental Animal Infections—All procedures were approved by the Institutional Animal Care Committee of Vanderbilt University. Male INS-GAS transgenic mice on the FVB/N background 6–8 weeks of age were challenged with either sterile Brucella broth, wild-type H. pylori strain 7.13, or a 7.13 cagE- mutant by oral gavage as previously described (33). Mice were euthanized at 4, 12, and 24 weeks post-challenge. At necropsy, linear strips extending from the squamocolumnar junction through proximal duodenum were fixed in 10% neutral-buffered formalin, paraffin-embedded, and cut at 5 μm increments. Sections were then deparaffinized, and DAF immunohistochemical (IHC) staining was carried out as previously described (29) using the anti-DAF antibody MCA1614 (Serotec). A single pathologist (E. Harris), experienced in murine pathology and blinded to treatment groups, scored DAF IHC staining on an ordinal scale from 0 to 4 by as previously described (34).
To assess colonization, gastric tissue was homogenized, plated, and incubated under microaerobic conditions at 37 °C for 5–6 days as previously described (33). Colonies were verified as H. pylori by Gram's stain, urease, catalase, and oxidase reactions as described (33). Successful colonization was confirmed by IHC staining using an anti-H. pylori antibody.
Gastric Epithelial Cell Isolation and Detection of DAF Protein by Flow Cytometry—Gastric epithelial cells were isolated from frozen stomach samples using a dissociation and dispersion technique as previously described (35). Briefly, epithelial cells were treated with 10 mm dithiothreitol at room temperature for 30 min and then with 1.0 mm EDTA for 60 min at 4 °C. Dispersed cells were filtered through a 0.4 μm filter to isolate single cells. Cell surface DAF protein was stained using hamster anti-mouse DAF antibody conjugated with phycoerythrin (BD Pharmingen) (1:50 dilution) at 4 °C for 45 min. After washing, cells were fixed and permeabilized with 0.1% paraformaldehyde and ice-cold methanol. To confirm epithelial lineage, cells were also stained with a mouse anti-pan cytokeratin antibody conjugated with fluorescein isothiocyanate (Abcam, Cambridge, MA) (1:50 dilution) for 30 min at 4 °C. Cells were acquired using a LSR II flow cytometer (BD Biosciences) and analyzed by Flowjo (Star tree, Ashland, OR).
Extraction of Total RNA from Murine Gastric Mucosa—Serial sectioning of frozen gastric tissue was performed using a cryostat. The thickness of the serial sections was 7 μm. Hematoxylin and eosin stains were performed on the first and last section in the series to determine cellular composition of the tissue and serial sections were selected in which the majority cell type was epithelial. RNA was isolated using the RNeasy RNA extraction kit according to the manufacturer's instructions. daf mRNA expression was measured using real-time RT-PCR as described above and normalized to levels of gapdh mRNA. Taqman® probes were used to detect daf and gapdh expression; Mm00438377_m1 and Rodent GAPDH control reagents, respectively (Applied Biosystems).
Statistical Analysis—An analysis of variance one-way analysis of variance and the Tukey-Kramer post test were used for analysis of in vitro data. The Mann-Whitney U test of intergroup comparisons was used for analysis of in vivo IHC and real-time PCR data. The Newman-Keuls test was used for analysis of flow cytometric data. Significance was defined as p ≤ 0.05. All calculations were performed with the GraphPad Prism 4 statistical analysis software package (GraphPad Software, Inc.).
RESULTS
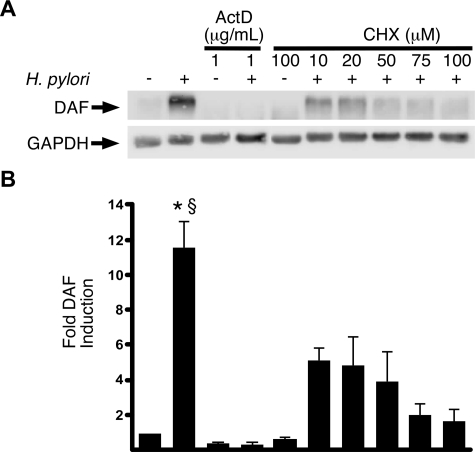
H. pylori Induction of DAF Is Regulated at the Transcriptional Level—We previously demonstrated that H. pylori up-regulates DAF in vitro (13). To determine whether DAF induction was transcriptionally or post-transcriptionally mediated, the H. pylori cag+ strain J166 was co-cultured with MKN28 gastric epithelial cells that had been pretreated with either actinomycin D (inhibitor of transcription) or cycloheximide (inhibitor of translation). DAF protein expression was assessed after 24 h of co-culture (Fig. 1). Actinomycin D completely blocked DAF induction in response to H. pylori (p < 0.001), and inhibition of translation by cycloheximide blocked DAF induction in a dose-dependent manner. Vehicle-treated, H. pylori-infected cells expressed significantly more DAF than H. pylori-infected cells that had been pretreated with cycloheximide (p < 0.01), and DAF expression in cycloheximide-treated, infected cells was not significantly higher than vehicle-treated, uninfected control cells. These results indicate that up-regulation of DAF by H. pylori in human gastric epithelial cells is mediated at a transcriptional level.
FIGURE 1.
H. pylori induces the transcriptional up-regulation of DAF in gastric epithelial cells. MKN28 gastric epithelial cells were pretreated with either actinomycin D (ActD) or cycloheximide (CHX) at the indicated concentrations and then co-cultured with the H. pylori cag+ strain J166 for 24 h at an m.o.i. = 100. A, Western blot analysis was performed using an anti-DAF antibody as described under “Experimental Procedures.” -, cells incubated with medium alone. A representative blot is shown. Anti-GAPDH blots served as normalization controls. B, densitometry represents data from three independent experiments. Error bars, S.E. *, p < 0.001 J166 versus control cells or ActD-treated cells. §, p < 0.01 J166 versus cycloheximide-treated cells.
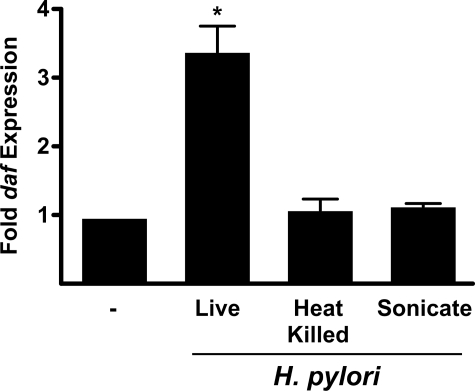
H. pylori Induction of daf Requires Viable Bacteria—Bacteria can activate epithelial signaling pathways via multiple mechanisms. To determine whether live H. pylori are necessary for daf induction or if inert bacterial components are sufficient, we incubated MKN28 cells with viable bacteria or with H. pylori that had either been heat-killed or lysed by sonication and assessed daf mRNA expression using real-time quantitative RT-PCR (Fig. 2). Co-culture of cells with live H. pylori, as expected, significantly induced daf mRNA. However, incubation with either heat-killed or sonicated H. pylori failed to induce expression of daf. These results indicate that induction of daf in gastric epithelial cells is dependent upon an active interplay with viable bacteria.
FIGURE 2.
H. pylori induction of daf requires viable bacteria. MKN28 cells were incubated with medium alone (-) or live, heat-killed, or sonicates of H. pylori strain J166 for 2 h. Levels of daf mRNA were determined by real-time qRT-PCR as described under “Experimental Procedures” and normalized to corresponding levels of gapdh mRNA. Results are expressed as -fold increase in daf mRNA in H. pylori-infected versus uninfected samples. Error bars, S.E. *, p < 0.001 versus uninfected cells.
DAF Induction Is Mediated by a Functional Type IV Secretion System—The requirement for viable H. pylori to induce daf raised the possibility that bacterial components intimately involved in epithelial contact may mediate daf expression. The cag pathogenicity island encodes a bacterial type IV secretion system that translocates effector molecules such as peptidoglycan and CagA into host cells after binding, thus affecting cell function. Therefore, we determined if H. pylori-mediated up-regulation of DAF is cag pathogenicity island-dependent.
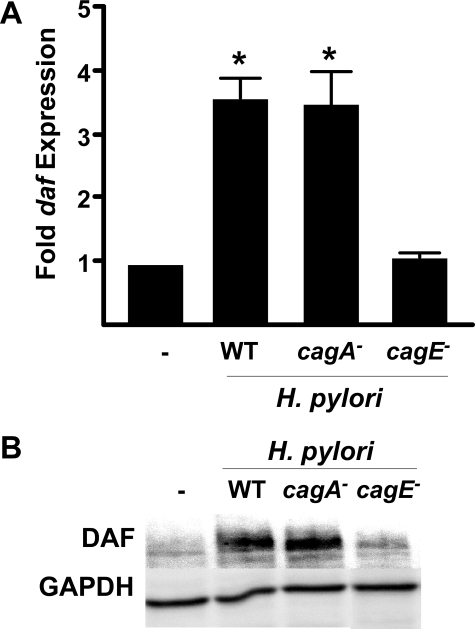
MKN28 cells were co-cultured with either wild-type H. pylori or isogenic cagA or cagE null mutant derivatives. Real-time qRT-PCR analysis demonstrated that co-culture with the cagA- mutant induced daf expression to levels similar to those induced by the wild-type strain (Fig. 3A). However, co-culture with the cagE- mutant failed to induce daf, and expression levels were no different from levels in uninfected cells. Western blot analysis confirmed that inactivation of cagE significantly attenuates the ability of H. pylori to induce DAF (Fig. 3B). Experiments were also performed with an independent H. pylori cag+ strain, 7.13, which readily infects animals and has been shown to induce gastric cancer in Mongolian gerbils and hypergastrinemic mice (29, 33, 36, 37). Similar to results obtained with strain J166, real-time qRT-PCR results showed that daf induction was dependent upon cagE but not cagA. The importance of the cag secretion system was more rigorously confirmed by demonstrating that inactivation of another cag gene encoding a structural component of the type IV secretion system (cagM) similarly attenuated daf expression (data not shown). These results indicate that H. pylori induction of DAF is dependent upon a functional cag secretion system but not CagA per se.
FIGURE 3.
DAF induction is mediated by a functional cag secretion system but not CagA. MKN28 cells were co-cultured with wild-type (WT) H. pylori strain J166 or isogenic cagA- or cagE- mutants at an m.o.i. = 100. A, levels of daf mRNA were determined by real-time qRT-PCR after 2 h of co-culture and were normalized to corresponding levels of gapdh mRNA. Results are expressed as -fold increase in daf mRNA in H. pylori-infected versus uninfected samples. Error bars, S.E. *, p < 0.05 versus uninfected cells. B, cell extracts were used for Western blot analysis using an anti-DAF antibody. A representative blot of multiple repetitions performed on three occasions is shown. Anti-GAPDH blots served as normalization controls.
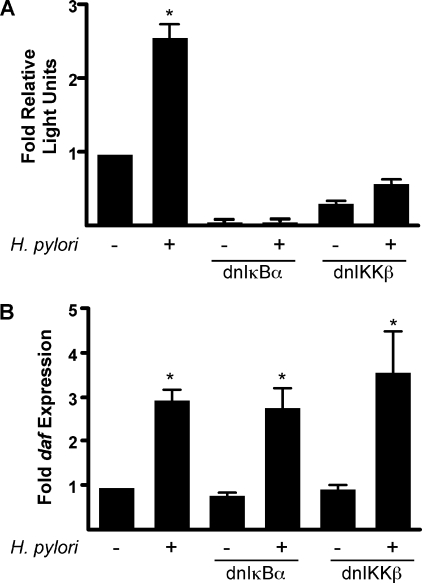
H. pylori Induction of daf Occurs via a NF-κB-independent Pathway—Activation of the transcription factor NF-κB by H. pylori is mediated by the cag secretion system (24, 38–40). The daf promoter contains a κB response element, and activation of NF-κB leads to the up-regulation of DAF in response to proinflammatory stimuli (41–44). To define the role of NF-κB in H. pylori-induced DAF expression, MKN28 cells were transiently transfected with constructs that express either a dominant-negative IκBα or dominant-negative IκB kinase β as well as a NF-κB-responsive luciferase reporter construct. As expected, H. pylori strain J166 significantly increased NF-κB-mediated luciferase activity, which was abolished by the dominant-negative IκBα and IκB kinase β constructs (Fig. 4A). However, inhibition of NF-κB had no effect on the ability of H. pylori to induce daf (Fig. 4B).
FIGURE 4.
NF-κB is not required for H. pylori induction of daf. MKN28 cells were transiently transfected with a NF-κB-responsive luciferase reporter construct and either a dominant negative-IκBα (dnIκBα) or a dominant negative IκB kinase β (dnIKKβ) expression construct. Cells were then co-cultured with H. pylori strain J166 at a m.o.i. = 100. A, NF-κB-driven firefly luciferase activity was assayed on a luminometer after 6 h of co-culture and normalized to Renilla luciferase activity. Error bars, S.E. *, p < 0.001 versus uninfected cells. B, levels of daf mRNA were determined by real-time qRT-PCR after 2 h of co-culture with the H. pylori strain J166 and were normalized to corresponding levels of gapdh mRNA. Results are expressed as -fold increase in daf mRNA in H. pylori-infected versus uninfected samples. Error bars, S.E. *, p < 0.05 versus uninfected cells.
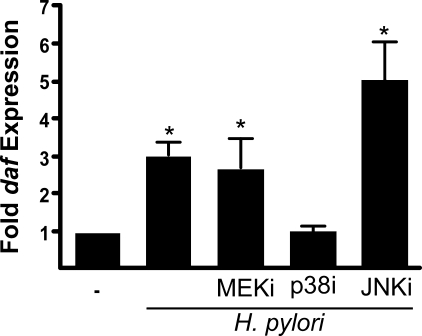
Activation of p38 Mediates daf Up-regulation by H. pylori— Our group and others have shown that H. pylori cag+ strains activate MAP kinases such as ERK, p38, and JNK in a cag-dependent manner (31, 45). As a prelude to defining the role of these signaling molecules in the transcriptional up-regulation of daf, we first confirmed the efficacy of ERK, p38, and JNK inhibitors used in subsequent experiments. Total MKN28 cell lysates were assessed for phospho-ERK, the p38 target phospho-MAPAP kinase-2 (46), and the JNK target phospho-c-Jun (47) by Western blot after stimulation with H. pylori. The specified concentration of each inhibitor was sufficient to attenuate H. pylori-induced activation of each respective MAPK (supplemental Fig. 1). MKN28 cells were then pretreated with inhibitors of MEK, p38, or JNK, and daf mRNA expression was quantified by real-time qRT-PCR. Inhibition of p38 blocked the induction of daf by H. pylori strains J166 (Fig. 5) and 7.13 (data not shown), whereas inhibition of ERK had no effect. Inhibition of JNK resulted in slightly higher levels of daf than observed in the H. pylori-infected vehicle-treated control; however, this difference was not statistically significant. These data indicate that H. pylori-induced daf expression is mediated in a p38 MAPK-dependent manner.
FIGURE 5.
Activation of p38 is required for daf up-regulation by H. pylori. MKN28 cells were pretreated with pharmacological inhibitors of MEK1/2 (MEKi, PD98095, 50 μm), p38 (SB203580, 10 μm), JNK (JNK inhibitor II (JNKi), 10 μm), or vehicle control (DMSO) (-) for 30 min and then co-cultured with the H. pylori strain J166 at a m.o.i. = 100. Levels of daf mRNA were determined by real-time qRT-PCR after 2 h of co-culture and were normalized to corresponding levels of gapdh mRNA. Results are expressed as -fold increase in daf mRNA in H. pylori-infected versus uninfected samples. Error bars, S.E. *, p < 0.05 versus uninfected cells.
Inactivation of a Component of the cag Secretion System Attenuates H. pylori Induction of DAF in Vivo—To determine whether the in vitro observations in MKN28 cells mirrored events within colonized gastric mucosa, DAF expression was assessed in transgenic hypergastrinemic INS-GAS mice. INS-GAS mice overexpress gastrin and spontaneously develop gastric cancer, but this requires the virtual lifetime of the animal (48). Infection with H. pylori accelerates this process and closely models lesions found in human disease (33, 48, 49). Therefore, we infected INS-GAS mice with the H. pylori cag+ strain 7.13, which readily infects rodents, and investigated DAF expression (29, 33, 36, 37).
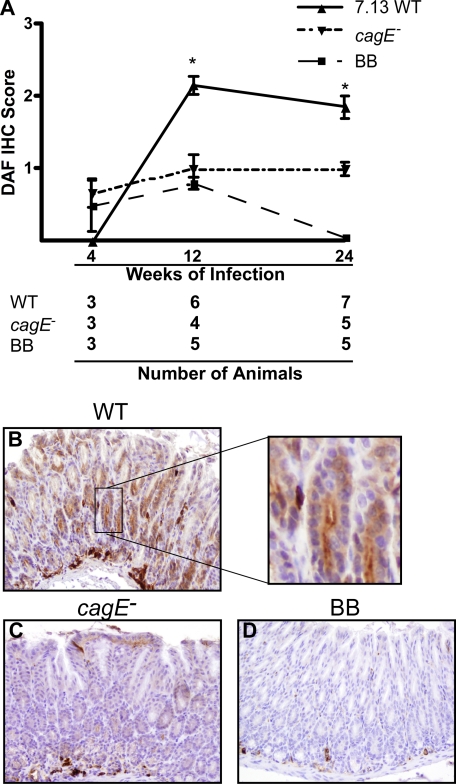
Mice were challenged with Brucella broth alone, wild-type strain 7.13, or a 7.13 cagE- isogenic mutant for 4, 12, and 24 weeks. DAF expression was detected using immunohistochemistry and scored on an ordinal scale from 0 to 4 as previously described (34). DAF staining in uninfected mice was localized to stromal plasma cells, lymphocytes, and endothelial cells, with focal weak staining of surface foveolar epithelial cells (Fig. 6D). There were no differences in DAF staining detected at 4 weeks post-challenge among the groups. However, mice infected with wild-type H. pylori strain 7.13 for 12 weeks and 24 weeks demonstrated significantly more abundant DAF staining versus uninfected mice (Fig. 6A). DAF staining was accentuated along the luminal surface in gastric epithelial cells that comprise the foveolar pits and was often accompanied by light diffuse cytoplasmic staining (Fig. 6B, inset). The intensity of DAF staining in mice infected with the cagE- mutant (Fig. 6C) was significantly attenuated compared with mice infected with wild-type H. pylori and was similar to uninfected mice.
FIGURE 6.
Inactivation of a component of the cag secretion system attenuates H. pylori induction of DAF in vivo. A, INS-GAS mice were challenged with Brucella broth (BB) control, wild-type (WT) H. pylori cag+ strain 7.13, or an isogenic 7.13 cagE- mutant for 4, 12, or 24 weeks. A, immunohistochemical staining of DAF was performed and scored on an ordinal scale from 0 to 4 by a single pathologist. Error bars, S.E. *, p < 0.05 WT 7.13 versus Brucella broth or cagE-. B–D, representative DAF IHC-stained sections from mice challenged for 12 weeks with wild-type H. pylori strain 7.13 (B), 7.13 cagE- isogenic mutant (C), or Brucella broth alone (D). Magnification, 20×.
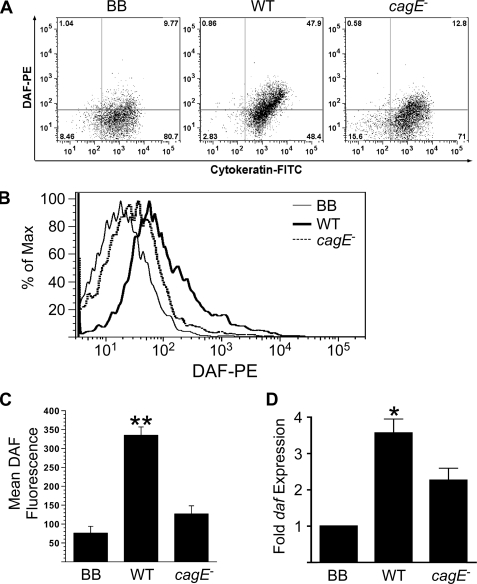
To confirm increased DAF expression on gastric epithelial cells, individual cells were isolated from frozen gastric mucosal tissue from INS-GAS mice that had been infected with H. pylori for 12 weeks as described under “Experimental Procedures.” Cells were fluorescently stained for DAF, and the epithelial marker cytokeratin and protein expression was quantified using flow cytometry. DAF staining was significantly increased on gastric epithelial cells of mice infected with wild-type H. pylori strain 7.13 compared with mice challenged with the cagE- isogenic mutant or broth alone (Fig. 7, A–C).
FIGURE 7.
H. pylori-induced expression of DAF is mediated by the cag secretion system in vivo. A–C, gastric epithelial cells were harvested from frozen tissue obtained 12 weeks post-challenge from INS-GAS mice challenged with Brucella broth (n = 5), wild-type H. pylori strain 7.13 (n = 6), or an isogenic 7.13 cagE- mutant (n = 4), stained with phycoerythrin (PE)-conjugated anti-DAF antibodies and fluorescein isothiocyanate (FITC)-conjugated anti-cytokeratin antibodies, and then used for flow cytometric analysis. A, representative dot plots of DAF and cytokeratin expression demonstrate increased DAF expression on gastric epithelial cells from mice challenged with wild-type (WT) H. pylori strain 7.13 compared with either Brucella broth (BB) alone or a cagE- isogenic mutant. B, representative histogram of DAF expression on gastric epithelial cells shows an increase in DAF fluorescence as a shift in fluorescence profile to the right in cells infected with wild type strain 7.13 versus uninfected cells. C, epithelial expression of DAF as determined by flow cytometry was significantly higher in mice infected with wild-type H. pylori (n = 6) when compared with mice challenged with either the cagE- isogenic mutant (n = 4) or Brucella broth control (n = 5). Error bars, S.E. **, p < 0.01 versus uninfected cells. D, total RNA was isolated from frozen gastric tissue, and daf mRNA expression was analyzed by real-time RT-PCR. daf mRNA expression was significantly increased in mice infected with wild-type H. pylori strain 7.13 (n = 5) versus mice challenged with the cagE- isogenic mutant (n = 4) or Brucella broth alone (n = 5). Error bars, S.E. *, p < 0.05 versus uninfected cells.
To further confirm the immunohistochemistry data, daf mRNA was quantified in gastric tissue using real-time RT-PCR. Total RNA was extracted from frozen tissue from INS-GAS mice 12 weeks post-challenge. Mice infected with wild-type H. pylori strain 7.13 demonstrated significantly increased expression of daf compared with uninfected mice or mice infected with the cagE- mutant (p < 0.05, Fig. 7D). Collectively, these in vivo findings recapitulate our in vitro data and confirm that a functional cag secretion system is required for H. pylori-mediated induction of DAF in gastric epithelial cells.
DISCUSSION
Our results have demonstrated that 1) H. pylori cag+ strains induce DAF expression in a cag pathogenicity island-dependent manner that does not require CagA, 2) H. pylori-induction of daf is abolished by inhibition of p38, and 3) an in vivo model of H. pylori-induced gastritis recapitulates our in vitro observations by demonstrating a requirement for a functional cag secretion system to induce DAF in epithelial cells. Collectively, these data indicate that H. pylori utilizes the cag island to affect the expression of DAF, potentially increasing adherence capacity, which may be important for initial and chronic colonization of its host.
Increased pathologic outcomes have been associated with infection by H. pylori cag+ strains, but the mechanism by which these strains increase disease risk is not completely understood. Several studies have highlighted the importance of the translocated effector protein CagA, which is responsible for aberrant activation of multiple signaling pathways. These include activation of β-catenin, SHP-2, and Grb-2, molecules that have been implicated in carcinogenesis. However, our results demonstrate that CagA is not required for increased expression of DAF.
Our finding that a functional cag secretion system is sufficient for H. pylori-mediated induction of DAF implicates additional bacterial factors that may be translocated into host cells leading to DAF induction. A candidate molecule for such induction is the bacterial cell wall component peptidoglycan. Peptidoglycan motifs that are recognized by NOD1 are delivered into host cells via the cag secretion system, and an important signaling event mediated by NOD1 is activation of NF-κB (24). However, although other investigators have shown that DAF regulation is responsive to NF-κB activation by pro-inflammatory stimuli (43, 50), our data demonstrate that NF-κB activation is not necessary for DAF induction by H. pylori. Another intriguing hypothesis based on a recent investigation (51) is that binding of CagL to cell surface α5β1 integrins can alter local membrane dynamics and eventuate in the assembly of focal adhesions that trigger integrin signaling cascades (52). Activation of integrin signaling may then subsequently induce expression of DAF.
Listeria monocytogenes induces IL-8 secretion via NOD1 activation in a NF-κB and p38-dependent manner (53), and H. pylori-induced secretion of IL-8 is dependent upon p38 activation and NOD1 activation of NF-κB (24, 45, 54, 55). Although the mechanism through which NOD1 induces activation of p38 remains unclear (56), there are several mechanisms by which p38 may promote increased DAF expression. Activation of p38 can transactivate the transcription factor CREB, which has previously been shown to transcriptionally up-regulate DAF in intestinal epithelial cells (41, 42, 44). Alternatively, daf mRNA transcript stability has been shown to be increased by activation of p38 in monocytic cell lines (57). Investigations into the mechanism by which p38 mediates DAF induction are currently ongoing in our laboratory.
An interesting question is whether up-regulation of DAF in H. pylori-infected cells is dependent on the DAF receptor per se. Previous studies have demonstrated that DAF can orchestrate epithelial pro-inflammatory responses in other cell systems. For example, co-culture of DAF-expressing T84 intestinal epithelial cells with Dr+ diffusely adhering E. coli leads to activation of ERK 1/2, p38, and JNK, which eventuates in IL-8 secretion (58). However, we have recently demonstrated that suppression of DAF in gastric epithelial cells via siRNA does not affect H. pylori-induced IL-8 production,3 a response that has previously been shown to be dependent on MAPK signaling (44). Therefore, these findings are not consistent with a model of DAF autoregulation in H. pylori-infected gastric epithelial cells.
Murine models provide valuable insights into host, bacterial, and environmental factors involved in H. pylori-induced gastric injury and inflammation. The INS-GAS model of gastritis has been used extensively for the study of H. pylori-induced inflammation and injury (33, 48, 49, 59–61). Utilizing this model, we have shown that the pattern of DAF up-regulation mirrors our in vitro studies; specifically, a functional cag secretion system plays an active role in the induction of epithelial DAF. Because H. pylori cag+ strains are found in closer juxtaposition to gastric epithelium than cag- strains (27), our current results suggest that DAF may represent one of several receptors that are up-regulated during chronic inflammation and which contribute to the persistence of more virulent H. pylori strains.
In addition to a role in maintaining chronic inflammation during infection, DAF has also been shown to play a role in tumorigenesis. Increased expression of DAF by transformed cells has been linked with resistance to immune clearance (62–64). Increased DAF expression is present in gastric cancer precursor lesions such as intestinal metaplasia, gastric adenomas, and gastric dysplasia, suggesting that aberrant expression of DAF precedes the development of gastric cancer (65). Our results implicating the cag pathogenicity island in DAF up-regulation may also help to explain why persons infected with H. pylori cag+ strains are at significantly increased risk for the development of gastric cancer versus those infected with cag- strains.
In conclusion, H. pylori induces the transcriptional up-regulation of the cellular receptor DAF. DAF induction is mediated by the cag secretion system but does not require the translocated effector protein CagA. DAF induction is also mediated by activation of p38 MAPK. In vivo, a functional cag secretion system is important for the induction of DAF by H. pylori in a murine model of gastritis. Collectively, these data have identified a novel mechanism by which H. pylori cag+ strains may tightly regulate their interactions with gastric epithelial cells and lower the threshold for more severe disease.
Supplementary Material
Acknowledgments
We greatly appreciate the generous gifts of the dominant-negative IκBα and dominant-negative IκB kinase β expression constructs provided by Dr. Andrew Neish (Emory University School of Medicine, Atlanta, GA). We also thank Lydia Wroblewski, Toni Nagy, Shannon Allen, and Karen Edelblum for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK73902, DK58587, and DK7795 (to R. M. P.) and DK053620 (to K. T. W.). This work was also supported by Vanderbilt Digestive Diseases Research Center Grant DK058404 and by the Office of Medical Research, Department of Veterans Affairs (to K. T. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: OMP, outer-membrane protein; DAF, decay-accelerating factor; NF-κB, nuclear factor-κB; NOD, nucleotide binding oligomerization domain; MAPK, mitogen-activated protein (MAP) kinase; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; m.o.i., multiplicity of infection; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; RT, reverse transcription; qRT, quantitative real-time; IHC, immunohistochemical; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
D. P. O'Brien, J. Romero-Gallo, B. G. Schneider, R. Chaturvedi, A. Delgado, E. J. Harris, U. Krishna, S. R. Ogden, D. A. Israel, K. T. Wilson, and R. M. Peek, Jr., unpublished data.
References
- 1.Peek, R. M., Jr., and Blaser, M. J. (2002) Nat. Rev. Cancer 2 28-37 [DOI] [PubMed] [Google Scholar]
- 2.Moss, S. F., and Sood, S. (2003) Curr. Opin. Infect. Dis. 16 445-451 [DOI] [PubMed] [Google Scholar]
- 3.Moss, S. F., and Blaser, M. J. (2005) Nat. Clin. Pract. Oncol. 2 90-97 [DOI] [PubMed] [Google Scholar]
- 4.Correa, P. (2004) Gut 53 1217-1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard, T. G., Drakes, M. L., and Czinn, S. J. (2004) Curr. Opin. Gastroenterol. 20 10-15 [DOI] [PubMed] [Google Scholar]
- 6.Ernst, P. B., Peura, D. A., and Crowe, S. E. (2006) Gastroenterology 130 188-206 [DOI] [PubMed] [Google Scholar]
- 7.Beswick, E. J., Suarez, G., and Reyes, V. E. (2006) World J. Gastroenterol. 12 5599-5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hessey, S. J., Spencer, J., Wyatt, J. I., Sobala, G., Rathbone, B. J., Axon, A. T., and Dixon, M. F. (1990) Gut 31 134-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhard, M., Lehn, N., Neumayer, N., Boren, T., Rad, R., Schepp, W., Miehlke, S., Classen, M., and Prinz, C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12778-12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilver, D., Arnqvist, A., Ogren, J., Frick, I. M., Kersulyte, D., Incecik, E. T., Berg, D. E., Covacci, A., Engstrand, L., and Boren, T. (1998) Science 279 373-377 [DOI] [PubMed] [Google Scholar]
- 11.Solnick, J. V., Hansen, L. M., Salama, N. R., Boonjakuakul, J. K., and Syvanen, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2106-2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahdavi, J., Sonden, B., Hurtig, M., Olfat, F. O., Forsberg, L., Roche, N., Angstrom, J., Larsson, T., Teneberg, S., Karlsson, K. A., Altraja, S., Wadstrom, T., Kersulyte, D., Berg, D. E., Dubois, A., Petersson, C., Magnusson, K. E., Norberg, T., Lindh, F., Lundskog, B. B., Arnqvist, A., Hammarstrom, L., and Boren, T. (2002) Science 297 573-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Brien, D. P., Israel, D. A., Krishna, U., Romero-Gallo, J., Nedrud, J., Medof, M. E., Lin, F., Redline, R., Lublin, D. M., Nowicki, B. J., Franco, A. T., Ogden, S., Williams, A. D., Polk, D. B., and Peek, R. M., Jr. (2006) J. Biol. Chem. 281 13317-13323 [DOI] [PubMed] [Google Scholar]
- 14.Servin, A. L. (2005) Clin. Microbiol. Rev. 18 264-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowicki, B., Hart, A., Coyne, K. E., Lublin, D. M., and Nowicki, S. (1993) J. Exp. Med. 178 2115-2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarkson, N. A., Kaufman, R., Lublin, D. M., Ward, T., Pipkin, P. A., Minor, P. D., Evans, D. J., and Almond, J. W. (1995) J. Virol. 69 5497-5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shafren, D. R. (1998) J. Virol. 72 9407-9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergelson, J. M., Mohanty, J. G., Crowell, R. L., St John, N. F., Lublin, D. M., and Finberg, R. W. (1995) J. Virol. 69 1903-1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berstad, A. E., and Brandtzaeg, P. (1998) Gut 42 522-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rautemaa, R., Rautelin, H., Puolakkainen, P., Kokkola, A., Karkkainen, P., and Meri, S. (2001) Gastroenterology 120 470-479 [DOI] [PubMed] [Google Scholar]
- 21.Odenbreit, S., Puls, J., Sedlmaier, B., Gerland, E., Fischer, W., and Haas, R. (2000) Science 287 1497-1500 [DOI] [PubMed] [Google Scholar]
- 22.Backert, S., Ziska, E., Brinkmann, V., Zimny-Arndt, U., Fauconnier, A., Jungblut, P. R., Naumann, M., and Meyer, T. F. (2000) Cell. Microbiol. 2 155-164 [DOI] [PubMed] [Google Scholar]
- 23.Selbach, M., Moese, S., Hauck, C. R., Meyer, T. F., and Backert, S. (2002) J. Biol. Chem. 277 6775-6778 [DOI] [PubMed] [Google Scholar]
- 24.Viala, J., Chaput, C., Boneca, I. G., Cardona, A., Girardin, S. E., Moran, A. P., Athman, R., Memet, S., Huerre, M. R., Coyle, A. J., DiStefano, P. S., Sansonetti, P. J., Labigne, A., Bertin, J., Philpott, D. J., and Ferrero, R. L. (2004) Nat. Immunol. 5 1166-1174 [DOI] [PubMed] [Google Scholar]
- 25.Higashi, H., Tsutsumi, R., Muto, S., Sugiyama, T., Azuma, T., Asaka, M., and Hatakeyama, M. (2002) Science 295 683-686 [DOI] [PubMed] [Google Scholar]
- 26.Tammer, I., Brandt, S., Hartig, R., Konig, W., and Backert, S. (2007) Gastroenterology 132 1309-1319 [DOI] [PubMed] [Google Scholar]
- 27.Camorlinga-Ponce, M., Romo, C., Gonzalez-Valencia, G., Munoz, O., and Torres, J. (2004) J. Clin. Pathol. 57 822-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng, H., Wu, H., Sloane, V., Jones, R., Yu, Y., Lin, P., Gewirtz, A. T., and Neish, A. S. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 290 96-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franco, A. T., Israel, D. A., Washington, M. K., Krishna, U., Fox, J. G., Rogers, A. B., Neish, A. S., Collier-Hyams, L., Perez-Perez, G. I., Hatakeyama, M., Whitehead, R., Gaus, K., O'Brien D, P., Romero-Gallo, J., and Peek, R. M., Jr. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10646-10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peek, R. M., Jr., Blaser, M. J., Mays, D. J., Forsyth, M. H., Cover, T. L., Song, S. Y., Krishna, U., and Pietenpol, J. A. (1999) Cancer Res. 59 6124-6131 [PubMed] [Google Scholar]
- 31.Crawford, H. C., Krishna, U. S., Israel, D. A., Matrisian, L. M., Washington, M. K., and Peek, R. M., Jr. (2003) Gastroenterology 125 1125-1136 [DOI] [PubMed] [Google Scholar]
- 32.Gewirtz, A. T., Yu, Y., Krishna, U. S., Israel, D. A., Lyons, S. L., and Peek, R. M., Jr. (2004) J. Infect. Dis. 189 1914-1920 [DOI] [PubMed] [Google Scholar]
- 33.Fox, J. G., Wang, T. C., Rogers, A. B., Poutahidis, T., Ge, Z., Taylor, N., Dangler, C. A., Israel, D. A., Krishna, U., Gaus, K., and Peek, R. M., Jr. (2003) Gastroenterology 124 1879-1890 [DOI] [PubMed] [Google Scholar]
- 34.Shattuck-Brandt, R. L., Lamps, L. W., Heppner Goss, K. J., DuBois, R. N., and Matrisian, L. M. (1999) Mol. Carcinog. 24 177-187 [PubMed] [Google Scholar]
- 35.Whitehead, R. H., VanEeden, P. E., Noble, M. D., Ataliotis, P., and Jat, P. S. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 587-591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero-Gallo, J., Harris, E. J., Krishna, U., Washington, M. K., Perez-Perez, G. I., and Peek, R. M., Jr. (2008) Lab. Investig. 88 328-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franco, A. T., Johnston, E., Krishna, U., Yamaoka, Y., Israel, D. A., Nagy, T. A., Wroblewski, L. E., Piazuelo, M. B., Correa, P., and Peek, R. M., Jr. (2008) Cancer Res. 68 379-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glocker, E., Lange, C., Covacci, A., Bereswill, S., Kist, M., and Pahl, H. L. (1998) Infect. Immun. 66 2346-2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kudo, T., Lu, H., Wu, J. Y., Ohno, T., Wu, M. J., Genta, R. M., Graham, D. Y., and Yamaoka, Y. (2007) Gastroenterology 132 1024-1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandt, S., Kwok, T., Hartig, R., Konig, W., and Backert, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9300-9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, D. J., and Lublin, D. M. (1993) J. Immunol. 150 151-160 [PubMed] [Google Scholar]
- 42.Ewulonu, U. K., Ravi, L., and Medof, M. E. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 4675-4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andoh, A., Kinoshita, K., Rosenberg, I., and Podolsky, D. K. (2001) J. Immunol. 167 3887-3893 [DOI] [PubMed] [Google Scholar]
- 44.Holla, V. R., Wang, D., Brown, J. R., Mann, J. R., Katkuri, S., and DuBois, R. N. (2005) J. Biol. Chem. 280 476-483 [DOI] [PubMed] [Google Scholar]
- 45.Keates, S., Keates, A. C., Warny, M., Peek, R. M., Jr., Murray, P. G., and Kelly, C. P. (1999) J. Immunol. 163 5552-5559 [PubMed] [Google Scholar]
- 46.Rouse, J., Cohen, P., Trigon, S., Morange, M., Alonso-Llamazares, A., Zamanillo, D., Hunt, T., and Nebreda, A. R. (1994) Cell 78 1027-1037 [DOI] [PubMed] [Google Scholar]
- 47.Derijard, B., Hibi, M., Wu, I. H., Barrett, T., Su, B., Deng, T., Karin, M., and Davis, R. J. (1994) Cell 76 1025-1037 [DOI] [PubMed] [Google Scholar]
- 48.Wang, T. C., Dangler, C. A., Chen, D., Goldenring, J. R., Koh, T., Raychowdhury, R., Coffey, R. J., Ito, S., Varro, A., Dockray, G. J., and Fox, J. G. (2000) Gastroenterology 118 36-47 [DOI] [PubMed] [Google Scholar]
- 49.Fox, J. G., Rogers, A. B., Ihrig, M., Taylor, N. S., Whary, M. T., Dockray, G., Varro, A., and Wang, T. C. (2003) Cancer Res. 63 942-950 [PubMed] [Google Scholar]
- 50.Lidington, E. A., Haskard, D. O., and Mason, J. C. (2000) Blood 96 2784-2792 [PubMed] [Google Scholar]
- 51.Kwok, T., Zabler, D., Urman, S., Rohde, M., Hartig, R., Wessler, S., Misselwitz, R., Berger, J., Sewald, N., Konig, W., and Backert, S. (2007) Nature 449 862-866 [DOI] [PubMed] [Google Scholar]
- 52.Backert, S., and Selbach, M. (2008) Cell Microbiol., in press [DOI] [PubMed]
- 53.Opitz, B., Puschel, A., Beermann, W., Hocke, A. C., Forster, S., Schmeck, B., van Laak, V., Chakraborty, T., Suttorp, N., and Hippenstiel, S. (2006) J. Immunol. 176 484-490 [DOI] [PubMed] [Google Scholar]
- 54.Aihara, M., Tsuchimoto, D., Takizawa, H., Azuma, A., Wakebe, H., Ohmoto, Y., Imagawa, K., Kikuchi, M., Mukaida, N., and Matsushima, K. (1997) Infect. Immun. 65 3218-3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma, S. A., Tummuru, M. K., Blaser, M. J., and Kerr, L. D. (1998) J. Immunol. 160 2401-2407 [PubMed] [Google Scholar]
- 56.Strober, W., Murray, P. J., Kitani, A., and Watanabe, T. (2006) Nat. Rev. Immunol. 6 9-20 [DOI] [PubMed] [Google Scholar]
- 57.Frevel, M. A., Bakheet, T., Silva, A. M., Hissong, J. G., Khabar, K. S., and Williams, B. R. (2003) Mol. Cell. Biol. 23 425-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Betis, F., Brest, P., Hofman, V., Guignot, J., Bernet-Camard, M. F., Rossi, B., Servin, A., and Hofman, P. (2003) Infect. Immun. 71 1068-1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi, M., Lee, H., Schaffer, L., Gilmartin, T. J., Head, S. R., Takaishi, S., Wang, T. C., Nakayama, J., and Fukuda, M. (2007) J. Histochem. Cytochem. 55 263-274 [DOI] [PubMed] [Google Scholar]
- 60.Steele, I. A., Dimaline, R., Pritchard, D. M., Peek, R. M., Jr., Wang, T. C., Dockray, G. J., and Varro, A. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 293 347-354 [DOI] [PubMed] [Google Scholar]
- 61.Ohtani, M., Garcia, A., Rogers, A. B., Ge, Z., Taylor, N. S., Xu, S., Watanabe, K., Marini, R. P., Whary, M. T., Wang, T. C., and Fox, J. G. (2007) Carcinogenesis 28 2597-2604 [DOI] [PubMed] [Google Scholar]
- 62.Gelderman, K. A., Tomlinson, S., Ross, G. D., and Gorter, A. (2004) Trends Immunol. 25 158-164 [DOI] [PubMed] [Google Scholar]
- 63.Fishelson, Z., Donin, N., Zell, S., Schultz, S., and Kirschfink, M. (2003) Mol. Immunol. 40 109-123 [DOI] [PubMed] [Google Scholar]
- 64.Jurianz, K., Ziegler, S., Garcia-Schuler, H., Kraus, S., Bohana-Kashtan, O., Fishelson, Z., and Kirschfink, M. (1999) Mol. Immunol. 36 929-939 [DOI] [PubMed] [Google Scholar]
- 65.Kiso, T., Mizuno, M., Nasu, J., Shimo, K., Uesu, T., Yamamoto, K., Okada, H., Fujita, T., and Tsuji, T. (2002) Histopathology 40 339-347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.