Abstract
Huntington disease derives from a critically expanded polyglutamine tract in the huntingtin (Htt) protein; a similar polyglutamine expansion in the androgen receptor (AR) causes spinobulbar muscular atrophy. AR activity also plays an essential role in prostate cancer. Molecular mechanisms that regulate Htt and AR degradation are not well understood but could have important therapeutic implications. We find that a pentapeptide motif (FQKLL) within the Htt protein regulates its degradation and subcellular localization to cytoplasm puncta. Disruption of the motif by alanine substitution at the hydrophobic residues increases the steady state level of the protein. Pulsechase analyses indicate that the motif regulates degradation. A similar motif (FQNLF) has corresponding activities in the AR protein. Transfer of the Htt motif with five flanking amino acids on either side to YFP reduces the steady state YFP level by rendering it susceptible to proteasome degradation. This work defines a novel proteasome-targeting motif that is necessary and sufficient to regulate the degradation of two disease-associated proteins.
Many human diseases are linked to the activities of single proteins. Huntington disease is a devastating, progressive neurodegenerative disorder characterized by motor impairment, dementia, and psychiatric disturbances. It derives from a critically enlarged tract of glutamines, caused by CAG codon expansion in the huntingtin (Htt) gene (1). Spinobulbar muscular atrophy is a slowly progressive motor neuron disease and sensory neuronopathy caused by a similar CAG codon expansion in the androgen receptor (AR)3 gene (2-4). In each disease, the expanded polyglutamine form of the protein can form toxic aggregates (5), potentially as oligomeric species (6, 7). Full-length Htt and AR proteins are both subject to proteolysis that produces toxic polyglutamine-containing fragments (7-10), which are typically used to study protein biochemistry in vitro. These fragments reproduce essential aspects of neuronal toxicity in mouse models (11, 12) but do not replicate regional neuronal vulnerability, suggesting that their generation from full-length proteins is an important step in pathogenesis. In prostate cancer, the full-length, wild type AR is the principal therapeutic target (13). Metastatic prostate cancer is currently treated by androgen ablation therapy, and resistant tumors often show evidence of persistent AR signaling, such as mutations that increase AR responsiveness and up-regulation of AR expression levels (14). Selective reduction of target protein levels could lead to novel therapeutic approaches for Huntington disease, spinobulbar muscular atrophy, or prostate cancer, but the mechanisms that govern Htt and AR degradation are unclear.
Many studies have implicated proteasome inhibition in polyglutamine pathogenesis (15-19), and there is some evidence that Htt peptides can be a direct proteasome substrate (20-22). Other results point toward autophagy as an important mechanism for the clearance of aggregation-prone species (23). Induction of autophagy can reduce polyglutamine protein toxicity by improving protein clearance (23) and appears important in protecting against neurodegeneration (23-27). The specific molecular mechanisms that govern degradation of normal and expanded Htt and AR proteins thus have obvious therapeutic implications. Here we investigate the role of a motif common to Htt and AR that regulates their degradation.
EXPERIMENTAL PROCEDURES
Plasmids—HttN136-YFP was constructed by cloning PCR-amplified HttN136 into pEYFP-N1 (Clontech). cDNA fragments encoding ARN127 were subcloned from ps6R (31) into pEYFP-N1. The 3A mutation was introduced using QuikChange (Stratagene). YFP-15 and YFP-15 (3A) constructs were made by cloning annealed oligonucleotides into the EcoRI and BglII sites of pEYFP-C1. Full-length AR (3A) was described previously (30).
Knockdown of Rpt4—Rpt4 was knocked down using ON-TARGET plus SMART pool of small interfering RNA (catalog number L-009570-01; Dharmacon) using the manufacturer's protocol. ON-TARGET plus Nontargeting pool of small interfering RNA was used as a negative control. Rpt4 was detected using anti-subunit Rpt4 (S10b) monoclonal antibody (BioMol; catalog number PW 8830, 1:500 dilution).
Cell Culture and Protease Inhibition—HeLa cells were transfected using Lipofectamine 2000 (Invitrogen). 24 h after transfection cells were split as required. 24 h later the cells were treated with different proteasome inhibitors for a total of 6 h. The cells were washed, harvested, and lysed in SDS-PAGE buffer.
Protein Labeling and Immunoprecipitation—24 h posttransfection, the cells were washed twice in methionine-free Dulbecco's modified Eagle's medium and incubated in methionine-free Dulbecco's modified Eagle's medium for 20 min, followed by labeling of proteins in 150 mCi/ml [35S]methionine for 1 h. After labeling, the cells were washed once in methionine-free medium supplemented with 15 mg/l unlabeled methionine followed by incubation in Dulbecco's modified Eagle's medium for 0, 2, 4, and 16 h. At different time points, the cells were harvested and lysed in phosphate-buffered saline containing 0.1% Triton X-100 and Complete protease inhibitor mixture (Roche Applied Science). Approximately 1 mg of lysate was used for immunoprecipitation with 10 μl of agarose-conjugated mouse monoclonal antibody versus green fluorescent protein (Roche Applied Science). The precipitated products were resolved by SDS-PAGE, imaged by autoradiography, and quantified by densitometry using National Institutes of Health Image software.
Antibodies—EM48 monoclonal antibody (Chemicon) was used to detect Htt (1:2000). N-20 polyclonal (Santa Cruz) was used to detect ARN127 and AR (1:2000). Anti-green fluorescent protein polyclonal antibody (Santa Cruz) was used to detect YFP as well as YFP-conjugated proteins (1:2000). Anti-actin rabbit polyclonal antibody (Santa Cruz) was used to label actin. LAMP-1 and LAMP-2 antibodies were obtained from the Developmental Studies Hybridoma Bank.
RESULTS
FQKLL Motif Regulates Htt Turnover and Subcellular Localization—Studies have implicated a pentapeptide motif (23FQNLF27) in AR self-association (28, 29), and we have previously observed that this motif is required for proper intramolecular folding after hormone binding (30). Sequence evaluation of Htt revealed a similar motif in the N terminus (123FQKLL127) (Fig. 1). To test the role of this motif in Htt metabolism, we fused YFP to a 136-amino acid N-terminal fragment of Htt long enough to contain the FQKLL motif (HttN136YFP) (Fig. 1). Whole cell lysates from HeLa cells transiently expressing HttN136YFP with a normal length polyglutamine tract (Q25) were analyzed by EM48 antibody, which recognizes the Htt peptide, and confirmed expression of a peptide of appropriate size (Fig. 2A). We used site-directed mutagenesis to introduce sequential alanine substitutions into the motif (AQKLL (1A), AQKAL (2A), and AQKAA (3A)). Each successive substitution progressively increased the steady state levels of unexpanded HttN136YFP, with HttN136(3A)YFP being most abundant (Fig. 2A). Steady state levels of a HttN136YFP protein with a pathologically expanded polyglutamine tract (Q42) also increased when the three hydrophobic amino acids were substituted with alanine (Fig. 2A). The effect on protein steady state level was also evident when the peptides were expressed in HEK293 cells (data not shown).
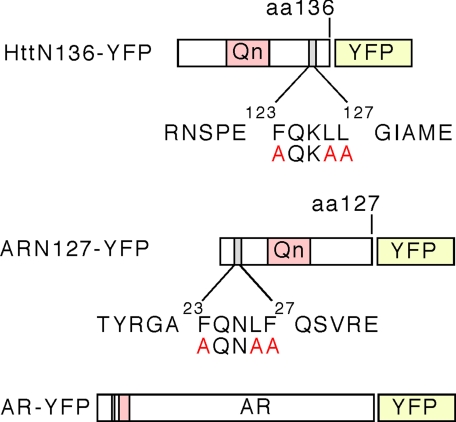
FIGURE 1.
Schematic map of Htt and AR constructs, with location of pentapeptide motifs. A construct containing the first 136 amino acids of the Htt protein was fused to YFP. A pentapeptide motif (123FQKLL127) is present C-terminal to the polyglutamine tract. A construct containing the first 127 amino acids of AR was fused to YFP. A pentapeptide motif (23FQNLF27) is present N-terminal to the polyglutamine tract. Amino acids immediately flanking the motifs are shown and have no homology. A full-length AR-YFP construct (not drawn to scale) is also pictured.
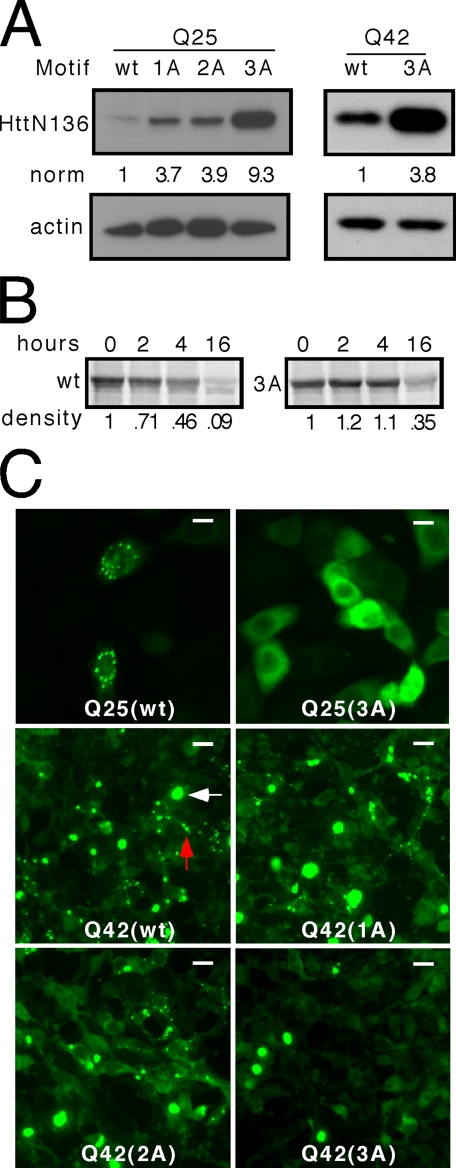
FIGURE 2.
The FQKLL motif in Htt regulates steady state, degradation rate, and subcellular localization. A, sequential mutation of hydrophobic amino acids to alanine (1A, AQKLL; 2A, AQKAL; 3A, AQKAA) produces a progressive increase in the steady state level of HttN136YFP peptide with unexpanded (Q25) or expanded (Q42) polyglutamine tracts. B, pulse-chase analysis with quantification using densitometry indicates that unexpanded (Q25) HttN136YFP has a shorter half-life (∼4 h) than HttN136(3A)-YFP (∼13 h), implicating the role of the motif in protein degradation. All of the gels are representative of at least three experiments. norm, normalized band intensity/actin. C, HeLa cells were transiently transfected with unexpanded (Q25) or expanded (Q42) HttN136YFP containing wt or mutant (1A, 2A, 3A) motifs. For unexpanded HttN136YFP protein (Q25), the motif causes subcellular localization in discrete puncta, and the 3A mutation leads to diffuse distribution. For the expanded protein (Q42), classical cytoplasm inclusions form (white arrow). However, the motif causes additional subcellular localization in cytoplasm puncta (red arrow). When the motif is mutated by single (1A), double (2A), or triple (3A) alanine substitutions, localization to these puncta is progressively disrupted, whereas the inclusions remain. Scale bars, 10 μminthe top row and 20 μminthe lower rows.
We next tested whether the difference in steady state levels of HttN136YFP and HttN136(3A)YFP was due to increased degradation. We exposed transfected cells for 1 h to [35S]methionine, followed by washout and harvesting at fixed time intervals for immunoprecipitation and resolution by SDS-PAGE and autoradiography. As determined by densitometry, HttN136YFP exhibited a t½ of ∼4 h, whereas that of HttN136(3A)YFP was ∼13 h (Fig. 2B). Thus the intact motif leads to a shorter protein half-life. We excluded the possibility that transcription efficiency accounted for the expression level differences by using quantitative reverse transcription-PCR to measure transcript levels, which were virtually identical (data not shown).
When studying both unexpanded and expanded forms of HttN136YFP, we noted that the protein exhibited unusual distribution within the cytoplasm and was localized in substantial numbers of cells to discrete cytoplasm puncta. These puncta were quite distinct from typical, large polyglutamine-dependent cytoplasm inclusions (Fig. 2C). We were unable to identify any organelles that colocalized with the puncta. In particular, we tested for colocalization with markers of the endosome/lysosome system (Lysotracker dye, Lamp2, and Lamp3), ubiquitin, the 20 S proteasome, and hsp70 (data not shown). To determine whether the FQKLL motif was required for this localization, we transiently transfected HeLa cells and evaluated the subcellular distribution of each construct (Fig. 2C). We observed a clear effect of successive mutations. Unexpanded (Q25) HttN136YFP had a diffuse distribution accompanied by marked cytoplasmic puncta. Upon mutation of the FQKLL motif (3A), these cytoplasmic puncta disappeared, leaving an exclusively diffuse distribution (Fig. 2C). Polyglutamine expanded (Q42) HttN136YFP exhibited both cytoplasm puncta, which disappeared with successive disruption of the FQKLL motif, and typical cytoplasm inclusions (Fig. 2C).
FQKLL Mediates Proteasome Degradation of HttN136—The relatively short half-life of HttN136YFP suggested that the proteasome might play an important role in its degradation. To investigate this possibility, we began by using RNA interference against a key proteasome component, Rpt4. HeLa cells were transfected for two rounds with a small interfering RNA construct against Rpt4, and HttN136YFP was cotransfected in the second round. Western blot using anti-Rpt4 antibody confirmed knockdown, and reprobing using ubiquitin likewise confirmed an increase in ubiquitinated protein levels, indicating that the construct was functional. RNA interference against Rpt4 increased the steady state level of HttN136YFP but not Htt N136(3A)YFP (Fig. 3A). We next tested the proteasome inhibitors MG132, ALLN, and lactacystin at 10 μm, along with a titration of lactacystin (Fig. 3, B and C), comparing their effects on wt and 3A mutants.6 h of treatment of cells transfected with HttN136YFP increased protein levels. In contrast, this had no effect on HttN136(3A)YFP (Fig. 3, B and C). Proteasome inhibition increased protein levels for HttN136YFP with both expanded (Q42) and unexpanded (Q25) polyglutamine sequences (Fig. 3B). Other protease inhibitors did not alter protein levels (data not shown).
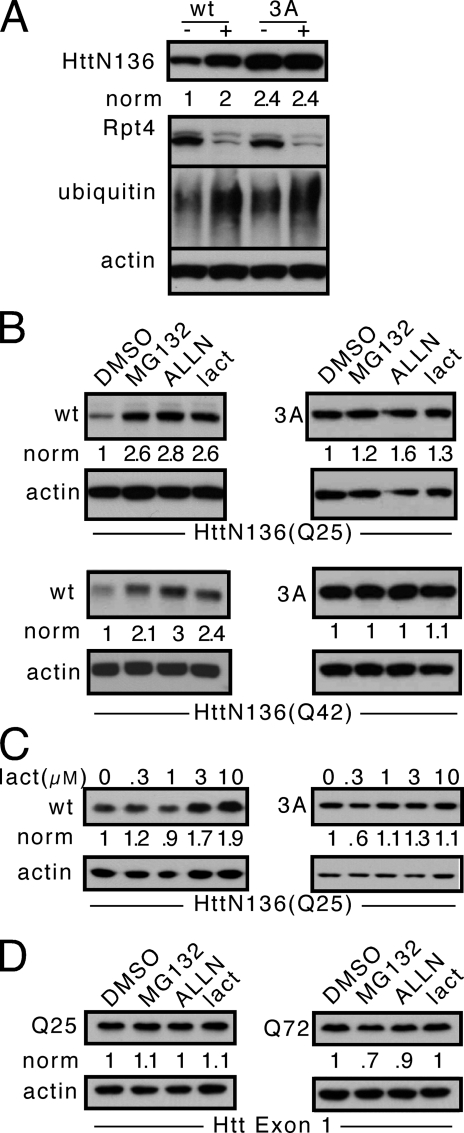
FIGURE 3.
FQKLF motif targets HttN136 to the proteasome. A, HeLa cells were transfected for two rounds with RNA interference constructs against Rpt4 (+), a proteasome component, or a negative control RNA interference pool (-) and were then transfected with HttN136YFP expression plasmid in the second round. Western blots were performed against HttN136YFP, Rpt4, ubiquitin, and actin. Knockdown of Rpt4 increases the steady state of wt protein but has no effect in the context of a 3A mutation. B, 6 h of treatment of cells expressing unexpanded (Q25) or expanded (Q42) HttN136YFP peptide with 10 μm proteasome inhibitors (MG132, ALLN, and lactacystin) versus vehicle control (dimethyl sulfoxide (DMSO)) increases the steady state of wt peptide but has no effect on the 3A mutant. C, lactacystin (lact) titration increases the steady state of HttN136YFP but has no effect on the 3A mutant. D, steady state levels of unexpanded (Q25) or expanded (Q72) Htt Exon 1-YFP, which lacks the FQNLF motif, are unaffected by 6 h of proteasome inhibition. All of the gels are representative of at least three experiments. norm, normalized band intensity/actin.
It has been unclear whether Htt exon 1, a truncated peptide used widely in vitro and in vivo, is degraded by the proteasome. Our experiments implicating the FQKLL motif in HttN136 degradation predicted that it would not be, because this motif is lacking in Htt exon 1. Thus we tested directly whether Htt exon 1 would be influenced by 6 h of treatment with proteasome inhibitors. We observed no effect of proteasome inhibition on Htt exon 1-YFP steady state (Fig. 3D). Taken together with our prior experiments, we concluded that the FQKLL motif in HttN136YFP is necessary for proteasome degradation.
A Similar Motif Functions in AR—AR carries a motif very similar to that found in Htt, 23FQNLF27, located N-terminal to the polyglutamine tract (Fig. 1). In full-length AR, the FQNLF motif has previously been linked both to intermolecular (29) and intramolecular conformational changes (30), but its role in regulating protein steady state or subcellular localization has not been reported. An N-terminal fragment consisting of the first 127 amino acids of the AR protein (ARN127) with an expanded polyglutamine tract is toxic when expressed in transgenic mice (12) and has been widely studied in models of spinobulbar muscular atrophy (7, 31, 32). In a manner similar to Htt, we disrupted the FQNLF motif both in full-length AR-YFP and ARN127YFP by replacing hydrophobic amino acids with alanine. Unexpanded ARN127(3A)YFP exhibited higher steady state levels compared with ARN127YFP (Fig. 4A). Similarly, full-length unexpanded AR(3A)YFP was detected at higher levels than full-length wt AR-YFP (Fig. 4A).
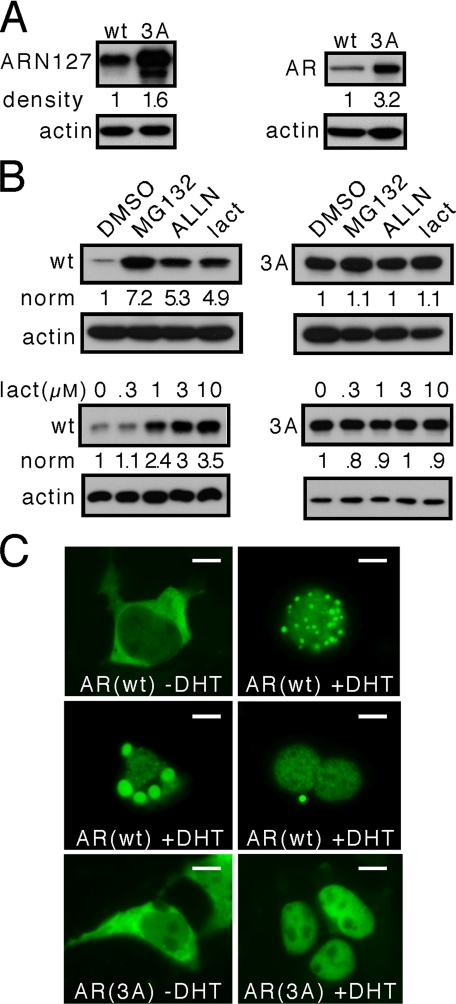
FIGURE 4.
A similar motif functions in AR. AR contains an FQNLF motif similar to that of Htt. This motif was mutated by alanine substitution to AQNAA (3A) in ARN127YFP and full-length AR-YFP proteins. These proteins were transiently transfected in HeLa cells, followed by Western blot or direct fluorescence imaging. A, the 3A mutation increases the steady state of both ARN127YFP and AR-YFP. B, 6 h of treatment of cells expressing ARN127YFP with 10 μm proteasome inhibitors (MG132, ALLN, and lactacystin) versus vehicle control (dimethyl sulfoxide (DMSO)) increases the steady state levels of wt protein but not the 3A mutant. Lactacystin (lact) titration also affects the steady state levels of wt ARN127YFP protein but not the 3A mutant. C, full-length AR-YFP localizes to the nucleus and to cytoplasm puncta in the presence of 10 nm DHT. The DHT-induced AR puncta have various appearances depicted in three panels: small versus large, single versus multiple. AR(3A)-YFP never localizes to these puncta and is localized exclusively in the cell nucleus after DHT treatment. All of the gels are representative of at least three experiments. norm, normalized band intensity/actin. Scale bars, 10 μm.
Given the effect of the motif on steady state levels of AR, we tested whether the motif rendered AR sensitive to proteasome inhibition. HeLa cells were transfected with ARN127YFP and treated with increasing amounts of 10 μm MG132, ALLN, or lactacystin for 6 h. This strongly increased ARN127YFP steady state but had no effect on ARN127(3A)-YFP, analogous to observations of the Htt peptide (Fig. 4B). These results suggest that the motif (FQXL(L/F)) regulates degradation of both AR and Htt by the proteasome. We did not observe localization of ARN127YFP to cytoplasm puncta as we did for HttN136YFP; however, full-length AR did exhibit such localization in ∼5% of cells after activation with 10 nm dihydrotestosterone (DHT) (Fig. 4C). The size and number of puncta varied among cells (Fig. 4C). The mutant full-length AR(3A) did not form these puncta and was restricted exclusively to the nucleus after activation by DHT (Fig. 4C).
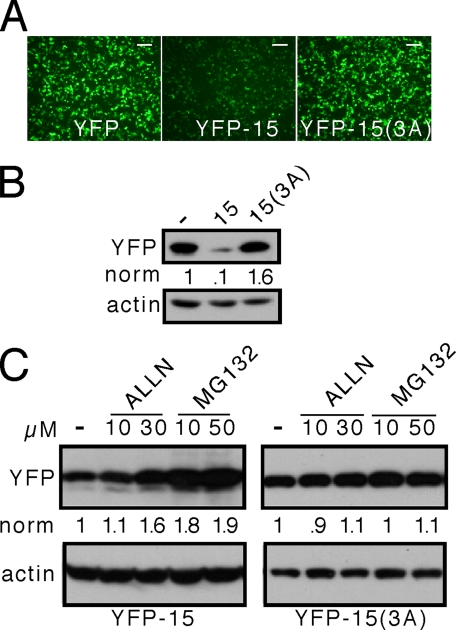
FQKLL Motif Is Sufficient to Direct Degradation of a Heterologous Protein—In full-length AR, the FQNLF motif has been implicated in normal ligand-induced intramolecular conformational change. We were thus uncertain whether the FQKLL motif is sufficient to direct a client protein to a degradation pathway or whether it is merely necessary for formation of a normal protein structure that is involved in proteasome targeting. YFP is not normally degraded by the proteasome (16), allowing us to test the sufficiency hypothesis. We cloned the FQKLL motif directly onto the C terminus of YFP but observed no discernable effect on protein steady state (data not shown). To test whether flanking amino acids might be required, we cloned a 15-amino acid-long fragment from Htt containing the motif in the center (RNSPEFQKLLGIAME) onto the C terminus of YFP (YFP-15). In this case, the steady state level of YFP-15 was much lower compared with that of YFP or YFP-15(3A) (Fig. 5, A and B). These results suggested that the motif functions in relative isolation, requiring only an appropriate local amino acid context.
FIGURE 5.
The FQKLL motif functions as a C-terminal fusion to YFP. A 15-amino acid sequence from Htt containing the FQKLL motif was fused in its wild type form (RNSPEFQKLLGIAME) or its 3A form (RNSPEAQKAAGIAME) to the C-terminal portion of YFP to produce YFP-15 or YFP-15(3A). These proteins were transiently expressed in HeLa cells and detected by fluorescence microscopy or Western blot. A, the 15-amino acid sequence causes a reduction in the apparent expression levels of YFP, but the 15(3A) sequence has no effect. B, Western blot demonstrates a reduction in YFP-15 expression versus YFP or YFP-15(3A). C, YFP-15 is sensitive to proteasome inhibition by ALLN or MG132, but YFP-15(3A) is not. All of the gels are representative of at least three experiments. norm, normalized band intensity/actin. Scale bars, 125 μm.
Next, we tested whether the motif would render YFP sensitive to the proteasome. We transfected HeLa cells with YFP-15 or YFP-15(3A) and exposed them for 6 h to proteasome inhibitors, followed by Western blot. ALLN and MG132 increased the steady state levels of YFP-15 but had no effect on YFP-15(3A) (Fig. 5C). These experiments indicated that within an appropriate local protein context, the pentapeptide motif is sufficient to target a heterologous protein for proteasome degradation.
DISCUSSION
We have identified a peptide motif with functional conservation between AR and Htt that targets each for proteasome-mediated degradation. We did not observe colocalization of AR and Htt peptides with the proteasome, which may be due to their diffuse distribution and rapid degradation once this occurs. The motif requires an appropriate amino acid context, because it is not functional when fused directly to YFP but is functional when flanking amino acids are preserved. Mutation of the motif by substituting the hydrophobic amino acids leucine or phenylalanine with alanine eliminates its activity. Steady state levels of proteins in which this motif is intact are increased by pharmacologic or genetic proteasome inhibition, whereas the steady state levels of peptides containing a mutated motif are insensitive to proteasome inhibition. We observed that the motif leads to localization of HttN136YFP and full-length AR (activated by DHT) to discrete cytoplasmic puncta whose identity remains unknown but is clearly tied to the motif. Interestingly, we have observed separate localization of HttN136YFP and CFP-AR in the cytoplasm puncta within a single cell (supplemental Fig. S1), suggesting that despite the common function of the motifs, protein-specific activities likely remain that lead to discrete subcellular targeting events.
Proteolytic Processing in the Polyglutamine Disease—It is now fairly well established that proteolytic cleavage of AR and Htt occurs in polyglutamine disease and likely generates toxic peptide fragments that are prone to misfolding, aggregation, and toxicity (7, 10, 33, 34). Our work suggests that peptide fragments of Htt that lack the proteasome targeting motif should be more prone to accumulation. The Htt exon 1 fragment, which is widely studied, is known to be particularly toxic in vitro and in vivo, and here we find that it is not degraded by the proteasome. Differential cleavage of Htt and AR proteins in distinct cell contexts could thus lead to proteolytic fragments of differential longevity and toxicity and consequently lead to specific cellular vulnerabilities.
A Novel Proteasome Degradation Signal—Hydrophobic amino acids that constitute signals for proteasome degradation have been reported previously. For example, a six-amino acid hydrophobic motif (19GMVAIL24) is required for proteasome-mediated degradation of SGK-1 (35), and the hydrophobic surface of an amphipathic helix at the N terminus of MATa-2 is an important determinant of its proteasomal degradation (36). We have identified key hydrophobic amino acids in both Htt and AR that are necessary for the degradation of each protein by the proteasome. These amino acids occur in a conserved pentapeptide pattern (FQXL(L/F)). Flanking amino acids are not conserved between these proteins yet were required for functional transfer of the minimal Htt FQKLL degradation signal onto YFP. In both AR and Htt the motifs are located near the N terminus, although a C-terminal fusion of the FQKLL motif to YFP was also functional.
The FQNLF motif in AR has been extensively studied but has not previously been linked to a degradation mechanism. It superficially resembles the LXXLL motif that mediates coactivator binding to nuclear receptors (37, 38). In AR, the FQNLF motif mediates interaction of the N terminus with a coactivator-binding pocket of the C-terminal ligand-binding domain (28, 30). The apparent dual role of this motif in AR may reflect its ability to mediate multiple protein-protein interactions within the cell, including those important for proteasome targeting. In Htt, there are multiple LXXLL motifs immediately C-terminal to the FQKLL motif we have studied here. Their roles in Htt function await further study. Ultimately, identification of factors that interact with this motif may increase our understanding of protein degradation specificity. This, in turn, may create novel therapeutic opportunities for diseases such as Huntington disease, spinobulbar muscular atrophy, and prostate cancer by increasing target protein degradation.
Supplementary Material
Acknowledgments
We thank Elizabeth Theusch for constructing some of the plasmids used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R01CA131226-01 and 1R01NS50284-01A1. This work was also supported by the Sandler Family Supporting Foundation, the Taube Family Foundation Program in Huntington's Disease Research, and funds from the Prostate Cancer Foundation (to M. I. D.), the National Science Foundation (to J. X. L.), and the Muscular Dystrophy Association (to J. S., M. L., and M. I. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: AR, androgen receptor; wt, wild type; YFP, yellow fluorescent protein; DHT, dihydrotestosterone.
References
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72 971-983 [DOI] [PubMed] [Google Scholar]
- 2.La Spada, A. R., Wilson, E. M., Lubahn, D. B., Harding, A. E., and Fischbeck, K. H. (1991) Nature 352 77-79 [DOI] [PubMed] [Google Scholar]
- 3.Kennedy, W. R., Alter, M., and Sung, J. H. (1968) Neurology 18671 -680 [DOI] [PubMed] [Google Scholar]
- 4.Harding, A. E., Thomas, P. K., Baraitser, M., Bradbury, P. G., Morgan-Hughes, J. A., and Ponsford, J. R. (1982) J. Neurol. Neurosurg. Psychiatry 451012 -1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross, C. A., and Poirier, M. A. (2004) Nat. Med. 10 (suppl.)S10 -S17 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi, T., Kikuchi, S., Katada, S., Nagai, Y., Nishizawa, M., and Onodera, O. (2007) Hum. Mol. Genet. 17 345-356 [DOI] [PubMed] [Google Scholar]
- 7.Li, M., Chevalier-Larsen, E. S., Merry, D. E., and Diamond, M. I. (2007) J. Biol. Chem. 2823157 -3164 [DOI] [PubMed] [Google Scholar]
- 8.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P., and Aronin, N. (1997) Sciences (N. Y.) 2771990 -1993 [DOI] [PubMed] [Google Scholar]
- 9.Hodgson, J. G., Agopyan, N., Gutekunst, C. A., Leavitt, B. R., LePiane, F., Singaraja, R., Smith, D. J., Bissada, N., McCutcheon, K., Nasir, J., Jamot, L., Li, X. J., Stevens, M. E., Rosemond, E., Roder, J. C., Phillips, A. G., Rubin, E. M., Hersch, S. M., and Hayden, M. R. (1999) Neuron 23 181-192 [DOI] [PubMed] [Google Scholar]
- 10.Li, M., Miwa, S., Kobayashi, Y., Merry, D. E., Yamamoto, M., Tanaka, F., Doyu, M., Hashizume, Y., Fischbeck, K. H., and Sobue, G. (1998) Ann. Neurol. 44 249-254 [DOI] [PubMed] [Google Scholar]
- 11.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L., and Bates, G. P. (1997) Cell 90 537-548 [DOI] [PubMed] [Google Scholar]
- 12.Abel, A., Walcott, J., Woods, J., Duda, J., and Merry, D. E. (2001) Hum. Mol. Genet. 10 107-116 [DOI] [PubMed] [Google Scholar]
- 13.Gelmann, E. P. (2002) J. Clin. Oncol. 203001 -3015 [DOI] [PubMed] [Google Scholar]
- 14.Culig, Z., Klocker, H., Bartsch, G., Steiner, H., and Hobisch, A. (2003) J. Urol. 1701363 -1369 [DOI] [PubMed] [Google Scholar]
- 15.Waelter, S., Boeddrich, A., Lurz, R., Scherzinger, E., Lueder, G., Lehrach, H., and Wanker, E. E. (2001) Mol. Biol. Cell 121393 -1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001) Sciences (N. Y.) 2921552 -1555 [DOI] [PubMed] [Google Scholar]
- 17.Chai, Y., Koppenhafer, S. L., Shoesmith, S. J., Perez, M. K., and Paulson, H. L. (1999) Hum. Mol. Genet. 8 673-682 [DOI] [PubMed] [Google Scholar]
- 18.Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T., and Zoghbi, H. Y. (1998) Nat. Genet. 19148 -154 [DOI] [PubMed] [Google Scholar]
- 19.Taylor, J. P., Tanaka, F., Robitschek, J., Sandoval, C. M., Taye, A., Markovic-Plese, S., and Fischbeck, K. H. (2003) Hum. Mol. Genet. 12749 -757 [DOI] [PubMed] [Google Scholar]
- 20.Venkatraman, P., Wetzel, R., Tanaka, M., Nukina, N., and Goldberg, A. L. (2004) Mol. Cell 14 95-104 [DOI] [PubMed] [Google Scholar]
- 21.Bhutani, N., Venkatraman, P., and Goldberg, A. L. (2007) EMBO J. 261385 -1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jana, N. R., Dikshit, P., Goswami, A., Kotliarova, S., Murata, S., Tanaka, K., and Nukina, N. (2005) J. Biol. Chem. 28011635 -11640 [DOI] [PubMed] [Google Scholar]
- 23.Ravikumar, B., Vacher, C., Berger, Z., Davies, J. E., Luo, S., Oroz, L. G., Scaravilli, F., Easton, D. F., Duden, R., O'Kane, C. J., and Rubinsztein, D. C. (2004) Nat. Genet. 36 585-595 [DOI] [PubMed] [Google Scholar]
- 24.Pandey, U. B., Batlevi, Y., Baehrecke, E. H., and Taylor, J. P. (2007) Autophagy 3 643-645 [DOI] [PubMed] [Google Scholar]
- 25.Pandey, U. B., Nie, Z., Batlevi, Y., McCray, B. A., Ritson, G. P., Nedelsky, N. B., Schwartz, S. L., DiProspero, N. A., Knight, M. A., Schuldiner, O., Padmanabhan, R., Hild, M., Berry, D. L., Garza, D., Hubbert, C. C., Yao, T. P., Baehrecke, E. H., and Taylor, J. P. (2007) Nature 447859 -863 [DOI] [PubMed] [Google Scholar]
- 26.Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E., and Tanaka, K. (2006) Nature 441880 -884 [DOI] [PubMed] [Google Scholar]
- 27.Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H., and Mizushima, N. (2006) Nature 441885 -889 [DOI] [PubMed] [Google Scholar]
- 28.He, B., Kemppainen, J. A., and Wilson, E. M. (2000) J. Biol. Chem. 27522986 -22994 [DOI] [PubMed] [Google Scholar]
- 29.He, B., and Wilson, E. M. (2002) Mol. Genet. Metab. 75293 -298 [DOI] [PubMed] [Google Scholar]
- 30.Schaufele, F., Carbonell, X., Guerbadot, M., Borngraeber, S., Chapman, M. S., Ma, A. A., Miner, J. N., and Diamond, M. I. (2005) Proc. Natl. Acad. Sci. U. S. A. 1029802 -9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond, M. I., Robinson, M. R., and Yamamoto, K. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 657-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merry, D. E., Kobayashi, Y., Bailey, C. K., Taye, A. A., and Fischbeck, K. H. (1998) Hum. Mol. Genet. 7 693-701 [DOI] [PubMed] [Google Scholar]
- 33.Kim, Y. J., Yi, Y., Sapp, E., Wang, Y., Cuiffo, B., Kegel, K. B., Qin, Z. H., Aronin, N., and DiFiglia, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 9812784 -12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leavitt, B. R., Wellington, C. L., and Hayden, M. R. (1999) Semin. Neurol. 19 385-395 [DOI] [PubMed] [Google Scholar]
- 35.Bogusz, A. M., Brickley, D. R., Pew, T., and Conzen, S. D. (2006) FEBS J. 2732913 -2928 [DOI] [PubMed] [Google Scholar]
- 36.Johnson, P. R., Swanson, R., Rakhilina, L., and Hochstrasser, M. (1998) Cell 94 217-227 [DOI] [PubMed] [Google Scholar]
- 37.Heery, D. M., Kalkhoven, E., Hoare, S., and Parker, M. G. (1997) Nature 387733 -736 [DOI] [PubMed] [Google Scholar]
- 38.Savkur, R. S., and Burris, T. P. (2004) J. Pept. Res. 63207 -212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.