Abstract
β-Catenin functions both as an adherens junction adhesion protein and as an essential mediator of the canonical Wnt signaling pathway. Wnts stabilize β-catenin and promote its accumulation in the nucleus, where it regulates transcription of the target genes. Here we show that Smad7 promotes cell-cell adhesion by stabilizing β-catenin and consequently increases the β-catenin-E-cadherin complex level at the plasma membrane. A Smad7-Axin interaction disassociates GSK-3β and β-catenin from Axin, as well as inhibits the recruitment of Smurf2, an E3 ligase, to β-catenin, thus protecting β-catenin from phosphorylation and degradation. Smad7 increases the stabilized β-catenin to form a complex with E-cadherin and stabilizes the E-cadherin-β-catenin complex. Thereby, rather than being translocated to the nucleus for regulating the target gene transcription, Smad7-stabilized-β-catenin is shunted to the E-cadherin complex to modulate cell-cell adhesion.
Cell-cell communication is critical for the regulation of cell proliferation, differentiation, and embryonic patterning. Loss of cell-cell adhesion is a hallmark of carcinogenesis and tumor progression (1, 2). In particular, the epithelial-mesenchymal transition that occurs during the development of carcinomas is characterized by a loss of cell-cell adhesion and an increase in cell motility (3). The precise mechanisms responsible for the loss of adhesion have not yet been identified, but the loss of E-cadherin-β-catenin-mediated cell-cell adhesion appears to be an important component (1, 2).
β-Catenin has been shown to have two distinct functions. It can act either as a cytoplasmic partner of cadherins junction proteins, in which capacity it is essential for cell-cell adhesion, or as a nuclear co-factor that regulates gene expression (2, 4). For β-catenin to accumulate, it requires protection from phosphorylation by glycogen synthase kinase 3β (GSK-3β),3 which normally targets the protein for ubiquitination and degradation by the 26 S proteasome. It has been shown that Axin and adenomatous polyposis coli (APC) function as scaffolds that facilitate the phosphorylation of β-catenin by GSK-3β (1, 5). In the presence of Wnt, Wnt-activated Dishevelled causes dissociation of GSK-3β from Axin, which prevents the phosphorylation of β-catenin by GSK-3β. The stabilized β-catenin is trafficked to the nucleus where it associates with leukocyte enhancer factor/T-cell factor to regulate transcription of target genes such as cyclin D1 and c-myc (6, 7). β-Catenin also can bind with cadherins to promote cell-cell adhesion. The association of β-catenin with type I cadherins links cadherins through α-catenin to actin in a process that is required for the role of β-catenin in the structural organization and signaling of cadherins (8, 9).
Smad7, which is the inhibitory Smad in the transforming growth factor β (TGFβ) signaling pathway, is not only induced by TGFβ and bone morphogenetic protein (BMP) but also can be regulated by other stimuli including shear stress, as well as epidermal growth factor, interferon-γ, and tumor necrosis factor α, independently of TGFβ/BMP (10-14). Recent work has identified functional interactions between Smad7 and β-catenin. In human prostate cancer cells, Smad7 can interact with β-catenin to modulate TGFβ-induced apoptosis (15, 16). In this case, the association of Smad7 with β-catenin to form a complex with T-cell factor regulates the transcription of c-myc. During the development of skin, Smad7 has been shown to recruit Smurf2, an E3 ligase, to β-catenin, resulting in the degradation of β-catenin (17). Overexpression of Smad7 also has been found to block the metastasis of mouse mammary carcinomas (18). Collectively, these studies indicate that Smad7 could act as an alterative regulator of β-catenin. However, the mechanism by which Smad7 regulates β-catenin and affects its role at the plasma membrane is still not clear.
In these studies, we show that Smad7 stabilizes β-catenin, resulting in the increase of an E-cadherin-β-catenin complex, thereby promoting cell-cell adhesion. We find that Smad7 increases E-cadherin by up-regulation of β-catenin. Smad7 inhibits the phosphorylation of β-catenin and, thus, proteasomal degradation. We further show that Smad7 inhibits the phosphorylation of β-catenin by interacting with Axin, which results in the dissociation of GSK-3β and β-catenin from the Axin scaffold. The Smad7-Axin interaction also inhibits the recruitment of Smuf2 to β-catenin. In breast cancer cells, the Smad7-stabilized β-catenin is not translocated to the nucleus but rather is delivered to E-cadherin to form the adherens junction complex. These results suggest a novel mechanism for Smad7 in modulating cell-cell adhesion and provide an explanation for the in vivo observations of Smad7 inhibition of tumor metastasis.
EXPERIMENTAL PROCEDURES
Primary Antibodies and Reagents—Mouse monoclonal antibodies against β-catenin, E-cadherin, APC, Lamin A/C, and rabbit anti-ubiquitin antibody, as well as goat polyclonal antibody against Smad7, were obtained from Santa Cruz Biotechnology, Inc. Rabbit anti-Axin antibody was purchased from Zymed Laboratories, Inc. Rabbit antibodies against GSK-3β, β-catenin, phospho-β-catenin (Thr41/Ser45), phospho-β-catenin (Ser33/37/Thr41), and rabbit monoclonal antibody against E-cadherin were obtained from Cell Signaling Technology. Mouse anti-α-tubulin, FLAG, and HA antibodies as well as rabbit anti-HA antibody were purchased from Sigma-Aldrich. TGFβ1 from human platelets, MG132, and mouse recombinant E-cadherin were obtained from Sigma-Aldrich.
Cell Culture, Retroviral Constructs, and Infection—T47D, MDA-MB-468, MDA-MB-231, and MDA-MB-435 cells were cultured as described previously (19). NMuMG (ATCC) (mouse nontransformed breast epithelial cells) cells were maintained in DMEM with 10% fetal calf serum and 10 μg/ml insulin. HEK-293 cells were cultured in DMEM with 10% fetal calf serum. L-cells and Wnt-3a producing cells (ATCC) were maintained in DMEM with 10% fetal calf serum, and the conditioned medium was harvested as described by the provider.
Recombinant pMSCVneo retroviral vectors obtained from BD Biosciences carrying cDNA encoding FLAG-Smad7, Axin-HA, and GFP genes were generated and packed in 293-GPG cells. After harvesting the retrovirus, the cells were infected with the virus and selected with G418 to ensure infection of all cells in culture.
Aggregation Assay—The cells were detached with 2 mm EDTA/PBS, washed twice in PBS, and resuspended at 5 × 105 cells/ml in serum-free medium. 0.5 ml/well of cells were seeded in 24-well culture plates previously coated with 2% BSA in PBS and allowed to aggregate for 180 min at 37 °C on a rotating shaker (80 rpm). For quantification, the number of cells in aggregates of more than three cells, as well as the total number of cells, was counted in four 1-mm squares of the hematocytometer grid. At least 500 cells were counted from each sample. Quantification of aggregation was estimated by the following formula: % aggregation = (N0 - Nt)/N0 ×100, where Nt is the total number of aggregates at the incubation time t, and N0 is the total number of cells.
Adhesion Assays—The cell adhesion assay was performed as described previously (20). Briefly, the cells were suspended at 2 × 106/ml in PBS, 0.1% BSA and incubated with 2.5 μm fluorescent dye (calcein) at 37 °C for 30 min. After washing, the cells were preincubated with E-cadherin antibody (50 μg/ml) or IgG control (50 μg/ml) for 20 min with EDTA (2 mm) for 10 min or left untreated. The labeled T47D cells were added to a monolayer of unstained cells in 24-well plates at 5 × 104 cells/well. The fluorescence signal from the adherent cells was measured after 30 min of incubation before and after PBS washing, using a fluorescence plate reader.
For analysis of cell substrate adhesion, 96-well plates were coated with 5 μg/ml mouse recombinant E-cadherin or 5 μg/ml BSA in 100 μl of PBS at 4 °C overnight. Calcein-labeled cells were seeded at a density of 5 × 104 cells/well, incubated for 30 min, and analyzed as described above.
Immunoprecipitation and Immunoblotting—The cells were lysed in buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet-P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitors. The lysates were immunoprecipitated by incubating with the appropriate antibodies, followed by absorption to protein G-Sepharose. The immunoprecipitates were separated by SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and visualized using enhanced chemiluminescence.
Reverse Transcription-Polymerase Chain Reaction Analysis—Total RNAs isolated from cells were used for synthesis of cDNA using an iScript cDNA Synthesis kit (Bio-Rad). 1 μl of each of the cDNAs was used as a template for quantitative real time PCR using iQ SYBR Green Supermix (Bio-Rad). The primers for human β-catenin were: forward, GTTCGTGCACATCAGGATAC, and reverse, CGATAGCTAGGATCATCCTG; for human E-cadherin were: forward, CCACCAAAGTCACGCTGAATACAGT, and reverse, GTTGTCATTCTGATCGGTTACCGTG; and for human β-actin were: forward, TGACTACCTCATGAAGATCCTCACCG, and reverse, TTGCTGATCCACATCTGCTGGAAGGT.
siRNA—The siRNAs of GFP, human or mouse Smad7, human β-catenin, and E-cadherin were purchased from Santa Cruz Biotechnology, and the knockdown assays were performed according to the manufacturer's directions.
Luciferase Assay—The cells were transfected with TOPFLASH reporter and Prl plasmids as a co-transfection control. Twenty-four hours after transfection, the cells were starved overnight and then treated with Wnt-3a conditioned medium for 5 h. Luciferase activities were assayed with a dual luciferase assay kit (Promega) according to the manufacturer's directions.
Nuclear and Cytoplasmic Fraction—Nuclear and cytoplasmic proteins of cells were extracted as described previously (21).
Immunofluorescence Assay—The cells were fixed with methanol at -20 °C for 10 min, blocked with 3% BSA in PBS for 30 min, incubated with primary antibody for 1 h at room temperature, washed three times with PBS, and incubated with second antibodies. Fluorescence localization was analyzed by confocal microscopy (Leica TCS SP2) with LCS software.
Pulse-Chase Analysis—The cells were pulse-labeled with [35S]methionine and chased as described previously (22).
Triton X-100 Extraction Assay—Triton X-100-soluble and -insoluble proteins of cells were extracted as described previously (23, 24).
RESULTS
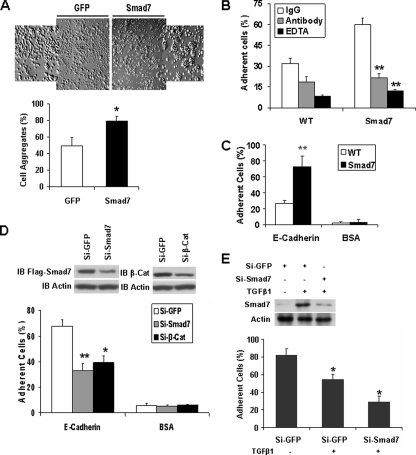
Smad7 Increases E-cadherin-β-Catenin-mediated Cell-Cell Adhesion—To determine whether Smad7 regulates cell-cell adhesion, we established a cell line that expresses Smad7 in TGFβ signaling-deficient T47D cells (19, 25). In a cell aggregation assay, Smad7 enhanced cell aggregation by ∼30% in comparison with cells expressing GFP (Fig. 1A). Cell-cell adhesion also was examined by seeding calcein-stained wild-type and Smad7-expressing T47D cells onto unstained monolayers of the corresponding cells in the presence or absence of EDTA, an anti-E-cadherin antibody, or the corresponding rabbit IgG. The Smad7-induced cell-cell adhesion was inhibited by both treatment with EDTA and addition of the antibody against the extracellular region of E-cadherin (Fig. 1B). Thus, Smad7 appears to enhance cell-cell adhesion through E-cadherin. This was further confirmed in a cell-substrate adhesion assay. E-cadherin-mediated adhesion of wild-type T47D cells or Smad7-expressing cells was analyzed in 96-well plates coated with recombinant E-cadherin or BSA. In this assay, Smad7 enhanced E-cadherin-mediated cell adhesion (Fig. 1C). In a reverse approach, reduction of Smad7 or β-catenin by siRNAs inhibited cell adhesion to E-cadherin (Fig. 1D). Furthermore, TGFβ1 reduced cell adhesion to E-cadherin in TGFβ-responsive NMuMG cells, and reduction of endogenous Smad7 by siRNA further decreased the cell adhesion (Fig. 1E). Taken together, these data indicate that Smad7 promotes E-cadherin-β-catenin-mediated cell-cell adhesion.
FIGURE 1.
Smad7 promotes E-cadherin-β-catenin-mediated cell-cell adhesion. A, single cell suspensions of T47D cells expressing GFP or FLAG-Smad7 were added to 24-well plates coated with 2% BSA; cell aggregates were photographed; and the percentage of aggregated cells compared with the total number of cells is shown. *, p < 0.05. B, calcein-labeled T47D cells with expression of FLAG-Smad7 or their parental cells were seeded on top of unlabeled monolayer of the corresponding cells in the presence or absence of EDTA (2 mm), the inhibitory anti-E-cadherin antibody, or the corresponding IgG. The cells were washed after a 30-min incubation, and the remaining fluorescence was calculated as the percentage of total fluorescence without washing. **, p < 0.01. C, calcein-stained wild-type (WT) or FLAG-Smad7 expressing T47D cells were seeded into E-cadherin- or BSA-coated wells of 96-well plate in triplicates. After a 30-min incubation, nonadherent cells were washed away with PBS, and the fluorescence of the remaining cells was quantified as a percentage of total fluorescence. D, FLAG-Smad7 expressing T47D cells were transfected with GFP siRNA, human Smad7 siRNA, or human β-catenin (β-Cat) siRNA, and 48 h later, Smad7 and β-catenin were detected by immunoblotting (IB). Analysis of cell adhesion to recombinant E-cadherin was performed as described in C. Knockdown of Smad7 or β-catenin reduced cell adhesion. E, NMuMG cells were transfected with GFP siRNA or mouse Smad7 siRNA, and 24 h later, the cells were treated with or without 10 ng/ml TGFβ1 overnight. Smad7 and Actin were then detected by Western blotting. Cell adhesion to recombinant E-cadherin was performed as described in C.
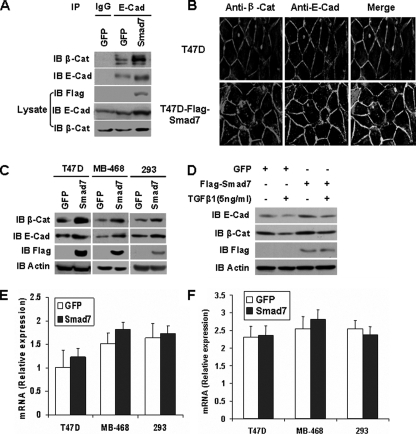
Smad7 Increases the Abundance of β-Catenin-E-cadherin Complexes—We next examined the potential role of Smad7 in regulating β-catenin-E-cadherin adherens junction complex formation. Cell lysates from T47D cells expressing either GFP or Smad7 were subjected to immunoprecipitation with an antibody against E-cadherin, and the resulting complexes were immunoblotted with anti-β-catenin antibody. The amount of β-catenin that co-immunoprecipitated with E-cadherin was increased significantly in cells expressing Smad7 in comparison with those expressing GFP (Fig. 2A). Indeed, Smad7 enhanced the co-localization of β-catenin-E-cadherin at the plasma membrane (Fig. 2B). Smad7 also up-regulated E-cadherin and β-catenin protein levels in both TGFβ signaling-deficient T47D and MB-468 breast cancer cells and the TGFβ-responsive human embryonic kidney 293 cells (19, 25) (Fig. 2C). Moreover, overexpression of Smad7 in NMuMG cells inhibited the loss of E-cadherin and β-catenin induced by TGFβ1 (Fig. 2D). However, mRNA levels of β-catenin and E-cadherin remained unchanged in Smad7-expressing cells (Fig. 2, E and F). These results suggest that Smad7 promotes cell-cell adhesion by increasing the abundance of E-cadherin-β-catenin complexes.
FIGURE 2.
Smad7 increases the E-cadherin-β-catenin complexes. A, the interaction between β-catenin (β-Cat) and E-cadherin (E-cad) was examined by immunoprecipitation (IP) of GFP or FLAG-Smad7 expressing T47D cells. B, T47D cells expressing FLAG-Smad7 or their parental cells were immunostained with anti-E-cadherin (green) and anti-β-catenin (red) antibodies. The merged images are on the right. C, Western blotting analysis of E-cadherin and β-catenin protein levels in TGFβ signaling-deficient T47D or MDA-MB-468 (MB-468) cells, and TGFβ-responsive HEK-293 (293) cells infected with pMSCV-GFP or pMSCV-FLAG-Smad7. Positive infected cells were selected with G418. Cell extracts were resolved by SDS-PAGE and subjected to immunoblotting (IB) analysis. D, Western blotting analysis of E-cadherin and β-catenin protein levels in NMuMG cells expressing GFP or Smad7 treated with or without 5 ng/ml TGFβ1 for 24 h. E and F, β-catenin (E) and E-cadherin (F) mRNA levels were measured by quantitative real time reverse transcription-PCR in three independent experiments. Total RNA was extracted from cells treated as described for C.
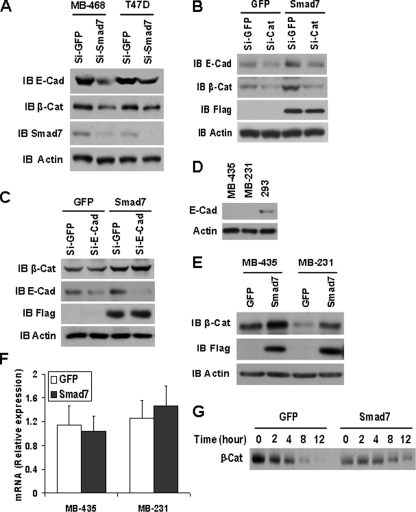
Smad7 Increases E-cadherin-β-Catenin Complexes through Stabilizing β-Catenin—To determine how Smad7 increased the abundance of E-cadherin-β-catenin complexes, MDA-MB-468 and T47D cells were treated with siRNAs to knock down endogenous Smad7 expression. Reduction of Smad7 expression significantly decreased both β-catenin and E-cadherin levels (Fig. 3A). Also, siRNA induced reduction of β-catenin expression in T47D cells resulted in a decrease in E-cadherin levels, whereas overexpression of Smad7 was not sufficient to restore E-cadherin levels in the cells treated with β-catenin siRNA (Fig. 3B). However, the reduction of E-cadherin in T47D cells had no effect on the β-catenin levels, and overexpression of Smad7 still up-regulated β-catenin in cells knocked down by E-cadherin siRNA (Fig. 3C). In E-cadherin-deficient MDA-MB-435 and MDA-MB-231 cells (Fig. 3D), overexpression of Smad7 also increased the β-catenin protein levels (Fig. 3E), whereas the mRNA levels of β-catenin remained unchanged (Fig. 3F). As additional confirmation for stabilization of β-catenin by Smad7, we performed a pulse-chase analysis in T47D cells. As shown in Fig. 3G, overexpression of Smad7 increased the stabilization of β-catenin. These data further suggest that Smad7 up-regulates β-catenin independent of E-cadherin, and the stabilization of β-catenin by Smad7 results in the increase of E-cadherin-β-catenin complexes.
FIGURE 3.
Smad7 increases E-cadherin-β-catenin complexes by up-regulation of β-catenin. A, MDA-MB-468 and T47D cells were transfected with GFP siRNA or human Smad7 siRNA, and 48 h after transfection, E-cadherin (E-cad) and β-catenin (β-Cat) were detected by Western blotting. B, T47D cells expressing GFP or Smad7 were transfected with GFP siRNA or human β-catenin siRNA; then E-cadherin and β-catenin were analyzed the same as A. C, T47D cells expressing GFP or Smad7 were transfected with GFP siRNA or human E-cadherin siRNA, and then E-cadherin and β-catenin were analyzed the same as A. D, Western blotting of E-cadherin in MDA-MB-435 (MB-435), MDA-MB-231 (MB-231), and 293 cells. E, immunoblotting (IB) of β-catenin in MDA-MB-435 and MDA-MB-231 with Smad7 or GFP expression. F, β-catenin mRNA levels were measured by quantitative real time reverse transcription-PCR in three independent experiments. Total RNA was extracted from MDA-MB-435 and MDA-MB-231 with Smad7 or GFP expression. G, pulse-chase analysis of β-catenin. T47D cells expressing GFP or Smad7 were pulse-labeled with 150 μCi/ml [35S]methionine for 30 min and chased in the absence of label for the indicated time period. The cell extracts were prepared at each time point, and protein-equivalent aliquots were immunoprecipitated with an anti-β-catenin antibody. Si, small interfering RNA.
Smad7 Stabilizes β-Catenin by Inhibiting Its Phosphorylation and Ubiquitination—Under unstimulated conditions, β-catenin resides in a complex consisting of APC, GSK-3β, CKIα, and Axin. In this “destruction” complex, β-catenin is constitutively phosphorylated by GSK-3β and CKIα on serine and threonine residues at its N terminus (1, 5). The phosphorylated β-catenin is then removed by the ubiquitination-proteasome system. Mutations of β-catenin at these N-terminal phosphorylation sites, such as serine/threonine residues 33, 37, 41, and 45, block the ubiquitination of β-catenin and cause its accumulation in cells (26-28). Therefore, we examined the effect of Smad7 on the phosphorylation of β-catenin in TGFβ signaling-deficient T47D or MDA-MB-468 cells, and TGFβ-responsive NMuMG cells. In each of these cell types, Smad7 inhibited the phosphorylation of β-catenin, whereas Axin enhanced the phosphorylation of β-catenin, as expected (Fig. 4A). Furthermore, reduction of endogenous Smad7 by siRNAs in NMuMG and T47D cells resulted in enhanced phosphorylation of β-catenin (Fig. 4B).
FIGURE 4.
Smad7 inhibits phosphorylation and ubiquitination of β-catenin. A, Western blotting analysis of phosphorylated β-catenin in T47D, MDA-MB-468, and NMuMG cells with expression of GFP, FLAG-Smad7, or Axin-HA, treated with 25 μm MG132 for 3 h. The cell extracts were immunoblotted (IB) for phosphorylated β-catenin with antibodies recognizing Thr41/Ser45 (2P-β-Cat) or Ser33/37/Thr41 (3P-β-Cat). B, Western blotting analysis of the effect of Smad7 siRNA on phosphorylated β-catenin in NMuMG and T47D cells. Immunoblotting was done as described for A. C, immunoprecipitation analysis of β-catenin ubiquitination. T47D cells with expression of GFP, FLAG-Smad7, or Axin-HA were incubated with 25 μm MG132 for 3 h, and the cell extracts were immunoprecipitated with anti-β-catenin antibody and immunoblotted with anti-ubiquitin (Ubi) antibody. D, FLAG-Smad7, Axin-HA alone, or both were co-transfected with β-catenin in 293 cells. Ubiquitination of β-catenin was analyzed 36 h after transfection as described for C.
Because inhibition of phosphorylation of β-catenin by Smad7 likely affects the ubiquitination of β-catenin, T47D cells expressing Smad7, Axin, or GFP were treated with MG132 for the ubiquitination analysis. Smad7 expression reduced the ubiquitination of β-catenin in T47D cells, whereas Axin enhanced the ubiquitination of β-catenin in comparison with cells expressing GFP (Fig. 4C). Smad7 also inhibited Axin-induced ubiquitination of β-catenin in 293 cells (Fig. 4D). Collectively, these results indicate that Smad7 stabilizes β-catenin through inhibition of its phosphorylation, which in turn reduces its ubiquitination.
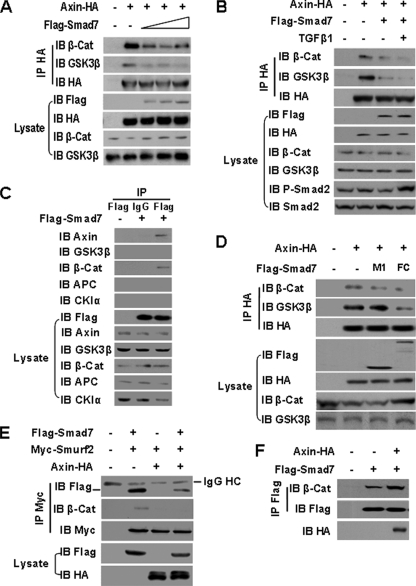
Interaction of Smad7-Axin Disassociates the β-Catenin Destruction Complex and Inhibits Recruitment of Smurf2 to β-Catenin—The ability of Smad7 to interfere with the phosphorylation of β-catenin suggested a specific effect of Smad7 on the β-catenin destruction complex. We therefore examined whether Smad7 regulates the association of Axin with β-catenin and GSK-3β. This was accomplished by co-transfection of HA-tagged Axin with increasing amounts of Smad7 encoding plasmid DNA and a subsequent immunoprecipitation assay. Increased Smad7 expression was associated with a decrease in the association of Axin with both GSK-3β and β-catenin (Fig. 5A). However, TGFβ1 had no significant effect on the disruption of the destruction complex by Smad7 (Fig. 5B).
FIGURE 5.
Interaction of Smad7-Axin disassociates theβ-catenin destruction complex and inhibits the recruitment of Smurf2 to β-catenin. A, immunoprecipitation (IP) analysis of the effects of Smad7 on dissociation of the β-catenin (β-Cat) destruction complex. Different amounts of FLAG-Smad7 plasmids were co-transfected with Axin-HA into 293 cells. Cell lysates were subjected to immunoprecipitation with anti-HA antibody. The endogenous β-catenin or GSK3β in the precipitated complex was blotted as indicated. B, the cells were treated with or without 5 ng/ml TGFβ1 for 1 h before harvest. Immunoprecipitation and immunoblotting (IB) were performed as described in A. C, immunoprecipitation analysis of the interaction of FLAG-Smad7 with endogenous components of the destruction complex. 293 cells were transfected with FLAG-Smad7 or empty vector, and the cell extracts were immunoprecipitated with anti-FLAG antibody or mouse IgG and then immunoblotted with anti-β-catenin, GSK3β, APC, CKIα, or Axin antibodies. D, the effects of the Smad7 interaction domain M1 and FC on formation of the β-catenin destruction complex. M1 and FC plasmids were co-transfected with Axin-HA. The cell lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-β-catenin or anti-GSK3β antibodies. E, immunoprecipitation assay of the interaction of Smad7-Smurf2 and Smurf2-β-catenin under the influence of Axin. FLAG-Smad7 and Myc-Smurf2 were co-transfected with or without Axin-HA into 293 cells, and the cell extracts were immunoprecipitated with anti-Myc antibody and then immunoblotted with anti-FLAG and anti-β-catenin antibodies. F, immunoprecipitation analysis of the interaction of Smad7-β-catenin under the influence of Axin. 293 cells were co-transfected with Smad7 and Axin or empty vector, and the cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with anti-β-catenin antibody.
To examine the mechanism by which Smad7 disrupts the β-catenin destruction complex, we assessed the interactions between Smad7 and the components of the destruction complex. Cell extracts of 293 cells transfected with FLAG-Smad7 were immunoprecipitated with anti-FLAG antibody followed by immunoblotting with the antibodies against Axin, β-catenin, CKIα, GSK-3β, and APC. Both Axin and β-catenin were immunoprecipitated with FLAG-Smad7 (Fig. 5C). Because Smad7 interacts with β-catenin at its MH1 domain (M1, amino acids 1-159) and with Axin in its MH2 and Linker domain (FC, amino acids 204-426) (16, 29), the interaction domains were tested for their potential involvement in the disruption of β-catenin destruction complex. As shown in Fig. 5D, the FC domain, but not the M1 domain, of Smad7 was required for the disassociation of the β-catenin destruction complex. Recruitment of Smurf2, an E3 ligase, to β-catenin by Smad7 has been reported to be an alterative pathway for β-catenin degradation (17). We determined whether the interaction of Smad7 and Axin interferes with the Smad7-Smurf2-β-catenin complex in an immunoprecipitation assay. Axin expression inhibited the association of both Smad7-Smurf2 and Smurf2-β-catenin (Fig. 5E), but it had no inhibitory effect on the interaction of Smad7-β-catenin (Fig. 5F). Thus, the Smad7-Axin interaction also inhibits the recruitment of Smurf2 to β-catenin by Smad7.
Smad7 Co-localizes with β-Catenin and E-cadherin at the Plasma Membrane—To investigate whether Smad7 regulates the subcellular localization of β-catenin, cytosolic and nuclear fractions were prepared from T47D cells expressing Smad7 or GFP, and the levels of β-catenin in both fractions were assessed by Western blotting. Smad7 expression was associated with an increase in the levels of β-catenin in the cytosolic fractions but not in the nuclear fractions (Fig. 6A). Consistent with this observation, Smad7 failed to alter the transcriptional activity induced by Wnt-3a in a TOPFLASH luciferase assay (Fig. 6B) or enhance the protein expression of cyclin D1, a β-catenin target gene (Fig. 6C). The subcellular localization of β-catenin regulated by Smad7 was also examined by immunostaining of 293 cells transfected with Smad7. In these cells, β-catenin was also found to co-localize with Smad7 mainly at the plasma membrane (Fig. 6D). In a similar immunostaining experiment in T47D cells, Smad7 was found to co-localize with E-cadherin (Fig. 6E). Together, these results suggest that Smad7 directs β-catenin to E-cadherin adherens junction complexes and away from the nucleus.
FIGURE 6.
Smad7 co-localizes with β-catenin at the plasma membrane. A, Western blotting analysis of the effect of Smad7 on translocation of β-catenin (β-Cat). Cytosolic and nuclear protein fractions were isolated in T47D cells expressing GFP or FLAG-Smad7. B, the effect of Smad7 on Wnt canonical signaling was examined by using a TOPFLASH luciferase reporter assay. 293 cells expressing FLAG-Smad7 or GFP were transfected with TOPFLASH reporter and treated with Wnt-3a- or L-cell-conditioned medium. The mean of the relative luciferase activity unit ± S.D. represents triplicates for each treatment with repeat of three times. C, Western blotting analysis of the effect of Smad7 on cyclin D1. The cell lysates were prepared from T47D and MDA-MB-468 cells expressing GFP or FLAG-Smad7. D, Immuno-co-localization images of FLAG-Smad7 (green) and β-catenin (red) in 293 cells transfected with FLAG-Smad7. E, immuno-co-localization images of FLAG-Smad7 (red) and E-cadherin (green) in T47D cells. IB, immunoblot.
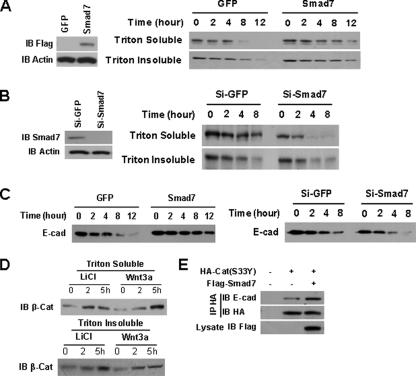
Smad7 Stabilizes E-cadherin-β-Catenin Complex and Increases β-Catenin Binding with E-cadherin—Given that Smad7 stabilized β-catenin, resulting in the increase of the E-cadherin-β-catenin complexes, we wonder whether Smad7 regulates the stability of E-cadherin-bound β-catenin. NMuMG cells expressing either GFP or Smad7 were treated with a protein synthesis inhibitor cycloheximide (30, 31) and then subjected to Triton X-100 extraction, because the E-cadherin-β-catenin complex is resistant to Triton X-100 extraction (23, 24). Smad7 expression increased the stability of cadherin-bound β-catenin in the Triton X-100-insoluble fraction and that of free cytoplasmic β-catenin in the Triton X-100-soluble fraction (Fig. 7A). Conversely, knockdown of Smad7 expression using siRNA decreased the stability of β-catenin in both Triton X-100-soluble and -insoluble fractions (Fig. 7B). Smad7 also increased the stability of E-cadherin in NMuMG cells, whereas knockdown of Smad7 expression in these cells increased the turnover of E-cadherin (Fig. 7C).
FIGURE 7.
Smad7 stabilizes E-cadherin-β-catenin complex and increases β-catenin binding to E-cadherin. A, Smad7 increases the stability of β-catenin (β-Cat). NMuMG cells expressing GFP or Smad7 were treated with 50 μg/ml cycloheximide for the indicated lengths of time, and the cells were extracted with Triton X-100. Then β-catenin in Triton X-100-soluble or -insoluble fractions was detected by Western blotting. Equal amounts of protein from each group were loaded for SDS-PAGE. B, knockdown of Smad7 in NMuMG cells decreases the stability of β-catenin. NMuMG cells were transfected with Si-GFP or Si-Smad7 siRNA, and the stability of β-catenin was analyzed as described in A. C, the stability of E-cadherin (E-cad) is regulated by Smad7. NMuMG cells were infected with GFP or Smad7, as well as were transfected with siRNAs the same as B, and then treated with 50 μg/ml cycloheximide for the indicated lengths of time. Whole cell lysates were detected by Western blotting. Equal amounts of protein from each group were loaded for SDS-PAGE. D, Western blotting analysis of β-catenin in Triton X-100-soluble and -insoluble extraction fractions prepared from T47D cells treated with LiCl (30 mm/liter) or Wnt3a for 2 or 5 h. Equal amounts of protein from each group were loaded for SDS-PAGE. E, immunoprecipitation analysis of the effects of Smad7 on the association of β-catenin (S33Y) with E-cadherin. FLAG-Smad7 plasmids were co-transfected with HA-β-catenin (S33Y) into 293 cells. The cell lysates were subjected to immunoprecipitation with anti-HA antibody. The endogenous E-cadherin in the precipitated complexes was blotted as indicated. IB, immunoblot. Si, small interfering RNA.
To determine whether β-catenin stabilized by disruption of the β-catenin destruction complex can be delivered to E-cadherin complex, T47D cells were treated with LiCl, a GSK3β inhibitor, or Wnt-3a, and then E-cadherin-bound β-catenin levels were detected by Western blotting. The abundance of β-catenin in Triton X-100-soluble fraction was increased by the treatment with either LiCl, or Wnt-3a. Importantly, the increase of cadherin-bound β-catenin in Triton X-100-insoluble fraction also was observed (Fig. 7D). To determine the potential role of Smad7 in recruiting the cytoplasmic β-catenin to E-cadherin, a mutant HA-tagged β-catenin (S33Y), which is resistant to phosphorylation by β-catenin destruction complex (28), was co-transfected with or without a Smad7 plasmid DNA into 293 cells, and cellular extracts were analyzed by an immunoprecipitation assay. As shown in Fig. 7E, E-cadherin-β-catenin (S33Y) complexes were enhanced by Smad7 expression. Likely, these data suggest that Smad7 facilitates the binding of β-catenin to E-cadherin, resulting in stabilization of the E-cadherin-β-catenin complex.
DISCUSSION
Previous studies support a role for Smad7 in the progression and metastatic potential of a number of malignancies. For example, in a genome-wide survey, Broderick et al. (32) identified three single nucleotide polymorphisms of Smad7 to be associated with a greater risk for colorectal cancer. In addition, enforced expression of Smad7 inhibits the metastasis of melanoma and breast cancer (18, 33, 34). Furthermore, increased β-catenin and E-cadherin expression by Smad7 in breast cancer and hepatocellular carcinoma also has been noted (18, 35). In this study, we present evidence for a novel mechanism through which Smad7 stabilizes β-catenin and promotes complex formation with E-cadherin, thereby promoting cell-cell adhesion. Such a mechanism could provide a link between Smad expression and the metastatic behavior of these cancer cell types.
β-Catenin is well recognized for its role as a transcriptional co-factor in the Wnt signaling, but its precise functions depend on where the protein is trafficked within the cell. In this study, we showed that Smad7-stabilized-β-catenin is shunted to E-cadherin rather than being transported into the nucleus to activate Wnt signaling. Thus, neither TOPFLASH-luciferase reporter nor the target gene of β-catenin is activated by Smad7. Rather, Smad7 is found to interact with β-catenin and co-localize at the plasma membrane. Moreover, Smad7 increases cytosolic β-catenin binding with E-cadherin and facilitates the formation of E-cadherin-β-catenin complexes. It has been reported that an association of β-catenin with E-cadherin occurs in endoplasmic reticulum prior to being transported to plasma membrane (36). The cytosolic β-catenin pool also has been shown to exchange with the E-cadherin-bound β-catenin pool (37). Our findings suggest that Smad7 might regulate the assembly of cytoplasmic β-catenin with the adherens junction complex.
E-cadherin has a high affinity for β-catenin (38). Expression of E-cadherin in L-cells (E-cadherin-deficient cells) increases β-catenin levels and increases the formation of E-cadherin-β-catenin complexes (39). Our data show that Smad7 stabilizes both E-cadherin and β-catenin, but Smad7 increases the E-cadherin-β-catenin complex level depending on the stabilization of β-catenin. This suggests the possibility of a multi-staged mechanism through which Smad7 alters β-catenin stability and increases E-cadherin-β-catenin complex formation. In support of this idea, Smad7 fails to increase E-cadherin expression in cells knocked down by β-catenin siRNA. In addition, when E-cadherin expression is inhibited by siRNA in T47D cells or in E-cadherin-deficient cells, Smad7 is still able to up-regulate β-catenin levels. Finally, and importantly, Wnt1-stabilized-β-catenin is trafficked to cadherins to form the cadherin-β-catenin complex (22).
In prostate cancer cells, Smad7 has been shown to interact with β-catenin and direct it to the nucleus where it acts as a transcriptional co-factor by associating with T-cell factor (16). Alternatively, during skin development, Smad7 interacts with β-catenin and recruits Smurf2, a ubiquitin E3 ligase, that degrades β-catenin in a process that most likely represents a mechanism specific to hair follicle morphogenesis (17). In the current study, we find that the interaction of Axin and Smad7 disassociates the destruction complex of β-catenin and inhibits the recruitment of Smurf2 to β-catenin, thereby stabilizing β-catenin. The stabilized β-catenin is delivered to E-cadherin, thus strengthening cell-cell adhesion. Therefore, our findings provide additional support for the concept that Smad7 regulates the fate and function of β-catenin differently in different cell contexts.
Smad7 is an inhibitory Smad and antagonizes TGFβ signaling in a negative feedback loop (40, 41). However, Smad7 can also be induced independently of TGFβ by factors such as shear stress, epidermal growth factor, interferon-γ, and tumor necrosis factor α (10-14). Consistent with this concept, our studies show that Smad7 can stabilize β-catenin in both TGFβ signaling-deficient and -responsive epithelial cells, as well as promote cell-cell adhesion in TGFβ signaling-deficient T47D cells. In addition, EGF and TGFβ are important factors in the induction of the epithelial-mesenchymal transition, which is associated with the loss of E-cadherin and a gain in invasive properties (42, 43). Consequently, the induction of Smad7 by these factors may constitute a feedback response that maintains cell-cell adhesion. Consistent with this conception, we found that an siRNA-mediated reduction in Smad7 expression further decreases the inhibition of E-cadherin-mediated cell-cell adhesion induced by TGFβ1. Overall, our studies provide additional insights into the mechanism through which Smad7 operates as an integrator of multiple signal pathways.
Acknowledgments
We thank Dr. Bert Vogelstein for TOPFLASH reporter construct and Dr. Jeff Wrana for Smurf2 constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant DK57501 (to X. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GSK-3β, glycogen synthase kinase 3β; APC, adenomatous polyposis coli; TGFβ, transforming growth factor β; GFP, green fluorescent protein; BSA, bovine serum albumin; CKI, casein kinase I; E3, ubiquitin-protein isopeptide ligase; HA, hemagglutinin; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; BSA, bovine serum albumin; siRNA, small interfering RNA.
References
- 1.Nelson, W. J., and Nusse, R. (2004) Science 3031483 -1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brembeck, F. H., Rosario, M., and Birchmeier, W. (2006) Curr. Opin. Genet. Dev. 16 51-59 [DOI] [PubMed] [Google Scholar]
- 3.Thiery, J. P. (2003) Curr. Opin. Cell Biol. 15740 -746 [DOI] [PubMed] [Google Scholar]
- 4.Bienz, M. (2005) Curr. Biol. 15 R64-R67 [DOI] [PubMed] [Google Scholar]
- 5.Peifer, M., and Polakis, P. (2000) Science 2871606 -1609 [DOI] [PubMed] [Google Scholar]
- 6.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B., and Kinzler, K. W. (1998) Science 2811509 -1512 [DOI] [PubMed] [Google Scholar]
- 7.Tetsu, O., and McCormick, F. (1999) Nature 398422 -426 [DOI] [PubMed] [Google Scholar]
- 8.Jamora, C., and Fuchs, E. (2002) Nat. Cell Biol. 4E101 -E108 [DOI] [PubMed] [Google Scholar]
- 9.Gumbiner, B. M. (2000) J. Cell Biol. 148399 -403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derynck, R., and Zhang, Y. E. (2003) Nature 425577 -584 [DOI] [PubMed] [Google Scholar]
- 11.Topper, J. N., Cai, J. X., Qiu, Y. B., Anderson, K. R., Xu, Y. Y., Deeds, J. D., Feeley, R., Gimeno, C. J., Woolf, E. A., Tayber, O., Mays, G. G., Sampson, B. A., Schoen, F. J., Gimbrone, M. A., and Falb, D. (1997) Proc. Natl. Acad. Sci. U. S. A. 949314 -9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsunobuchi, H., Ishisaki, A., and Imamura, T. (2004) Biochem. Biophys. Res. Commun. 316712 -719 [DOI] [PubMed] [Google Scholar]
- 13.Bitzer, M., Von Gersdorff, G., Liang, D., Dominguez-Rosales, A., Beg, A. A., Rojkind, M., and Bottinger, E. P. (2000) Genes Dev. 14187 -197 [PMC free article] [PubMed] [Google Scholar]
- 14.Ulloa, L., Doody, J., and Massague, J. (1999) Nature 397710 -713 [DOI] [PubMed] [Google Scholar]
- 15.Landstrom, M., Heldin, N. E., Bu, S. Z., Hermansson, A., Itoh, S., ten Dijke, P., and Heldin, C. H. (2000) Curr. Biol. 10535 -538 [DOI] [PubMed] [Google Scholar]
- 16.Edlund, S., Lee, S. Y., Grimsby, S., Zhang, S. H., Aspenstrom, P., Heldin, C. H., and Landstrom, M. (2005) Mol. Cell Biol. 251475 -1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, G. W., Li, A. G., Liang, Y. Y., Owens, P., He, W., Lu, S. L., Yoshimatsu, Y., Wang, D., ten Dijke, P., Lin, X., and Wang, X. J. (2006) Dev. Cell 11 301-312 [DOI] [PubMed] [Google Scholar]
- 18.Azuma, H., Ehata, S., Miyazaki, H., Watabe, T., Maruyama, O., Imamura, T., Sakamoto, T., Kiyama, S., Kiyama, Y., Ubai, T., Inamoto, T., Takahara, S., Itoh, Y., Otsuki, Y., Katsuoka, Y., Miyazono, K., and Horie, S. (2005) J. Natl. Cancer Inst. 971734 -1746 [DOI] [PubMed] [Google Scholar]
- 19.Kalkhoven, E., Roelen, B. A. J., Dewinter, J. P., Mummery, C. L., Vandeneijndenvanraaij, A. J. M., Vandersaag, P. T., and Vanderburg, B. (1995) Cell Growth Differ. 61151 -1161 [PubMed] [Google Scholar]
- 20.Reiss, K., Maretzky, T., Ludwig, A., Tousseyn, T., De Strooper, B., Hartmann, D., and Saftig, P. (2005) EMBO J. 24742 -752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jian, H. Y., Shen, X., Liu, I., Sernenov, M., He, X., and Wang, X. F. (2006) Genes Dev. 20 666-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinck, L., Nelson, W. J., and Papkoff, J. (1994) J. Cell Biol. 124729 -741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barth, A. I. M., Pollack, A. L., Altschuler, Y., Mostov, K. E., and Nelson, W. J. (1997) J. Cell Biol. 136693 -706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perego, C., Vanoni, C., Massari, S., Longhi, R., and Pietrini, G. (2000) EMBO J. 193978 -3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fink, S. P., Mikkola, D., Willson, J. K. V., and Markowitz, S. (2003) Oncogene 221317 -1323 [DOI] [PubMed] [Google Scholar]
- 26.Yost, C., Torres, M., Miller, R. R., Huang, E., Kimelman, D., and Moon, R. T. (1996) Genes Dev. 101443 -1454 [DOI] [PubMed] [Google Scholar]
- 27.Munemitsu, S., Albert, I., Rubinfeld, B., and Polakis, P. (1996) Mol. Cell Biol. 164088 -4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morin, P. J. (1999) Bioessays 211021 -1030 [DOI] [PubMed] [Google Scholar]
- 29.Liu, W., Rui, H. L., Wang, J. F., Lin, S. Y., He, Y., Chen, M. L., Li, Q. X., Ye, Z. Y., Zhang, S. P., Chan, S. C., Chen, Y. G., Han, J. H., and Lin, S. C. (2006) EMBO J. 251646 -1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, X., Ramirez, A., Waddell, D. S., Li, Z. Z., Liu, X. D., and Wang, X. F. (2008) Genes Dev. 22 106-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujimuro, M., Wu, F. Y., ApRhys, C., Kajumbula, H., Young, D. B., Hayward, G. S., and Hayward, S. D. (2003) Nat. Med. 9300 -306 [DOI] [PubMed] [Google Scholar]
- 32.Broderick, P., Carvajal-Carmona, L., Pittman, A. M., Webb, E., Howarth, K., Rowan, A., Lubbe, S., Spain, S., Sullivan, K., Fielding, S., Jaeger, E., Vijayakrishnan, J., Kemp, Z., Gorman, M., Chandler, I., Papaemmanuil, E., Penegar, S., Wood, W., Sellick, G., Qureshi, M., Teixeira, A., Domingo, E., Barclay, E., Martin, L., Sieber, O., Kerr, D., Gray, R., Peto, J., Cazier, J. B., Tomlinson, I., and Houlston, R. S. (2007) Nat. Genet. 391315 -1317 [DOI] [PubMed] [Google Scholar]
- 33.Javelaud, D., Mohammad, K. S., McKenna, C. R., Fournier, P., Luciani, F., Niewolna, M., Andre, J., Delmas, V., Larue, L., Guise, T. A., and Mauviel, A. (2007) Cancer Res. 672317 -2324 [DOI] [PubMed] [Google Scholar]
- 34.Javelaud, D., Delmas, V., Moller, M., Sextius, P., Andre, J., Menashi, S., Larue, L., and Mauviel, A. (2005) Oncogene 247624 -7629 [DOI] [PubMed] [Google Scholar]
- 35.Mikula, M., Proell, V., Fischer, A. N. M., and Mikulits, W. (2006) J. Cell Physiol. 209560 -567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, Y. T., Stewart, D. B., and Nelson, W. J. (1999) J. Cell Biol. 144687 -699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klingelhofer, J., Troyanovsky, R. B., Laur, O. Y., and Troyanovsky, S. (2003) Oncogene 221181 -1188 [DOI] [PubMed] [Google Scholar]
- 38.Huber, A. H., and Weis, W. I. (2001) Cell 105391 -402 [DOI] [PubMed] [Google Scholar]
- 39.Orsulic, S., Huber, O., Aberle, H., Arnold, S., and Kemler, R. (1999) J. Cell Sci. 1121237 -1245 [DOI] [PubMed] [Google Scholar]
- 40.Hayashi, H., Abdollah, S., Qiu, Y. B., Cai, J. X., Xu, Y. Y., Grinnell, B. W., Richardson, M. A., Topper, J. N., Gimbrone, M. A., Wrana, J. L., and Falb, D. (1997) Cell 891165 -1173 [DOI] [PubMed] [Google Scholar]
- 41.Nakao, A., Afrakhte, M., Moren, A., Nakayama, T., Christian, J. L., Heuchel, R., Itoh, S., Kawabata, N., Heldin, N. E., Heldin, C. H., and ten-Dijke, P. (1997) Nature 389631 -635 [DOI] [PubMed] [Google Scholar]
- 42.Lu, Z. M., Ghosh, S., Wang, Z. Y., and Hunter, T. (2003) Cancer Cell 4 499-515 [DOI] [PubMed] [Google Scholar]
- 43.Oft, M., Heider, K. H., and Beug, H. (1998) Curr. Biol. 81243 -1252 [DOI] [PubMed] [Google Scholar]