Abstract
Carbohydrate response element-binding protein (ChREBP) is a glucose-responsive transcription factor that activates genes involved in de novo lipogenesis in mammals. The current model for glucose activation of ChREBP proposes that increased glucose metabolism triggers a cytoplasmic to nuclear translocation of ChREBP that is critical for activation. However, we find that ChREBP actively shuttles between the cytoplasm and nucleus in both low and high glucose in the glucose-sensitive β cell-derived line, 832/13. Glucose stimulates a 3-fold increase in the rate of ChREBP nuclear entry, but trapping ChREBP in the nucleus by mutagenesis or with a nuclear export inhibitor does not lead to constitutive activation. In fact, mutational studies targeting the nuclear export signal of ChREBP also identified a distinct function essential for glucose-dependent transcriptional activation. From this, we conclude that an additional event independent of nuclear translocation is required for activation. The N-terminal segment of ChREBP (amino acids 1-298) has previously been shown to repress activity under basal conditions. This segment has five highly conserved regions, Mondo conserved regions 1-5 (MCR1 to -5). Based on activating mutations in MCR2 and MCR5, we propose that these two regions act coordinately to repress ChREBP in low glucose. In addition, other mutations in MCR2 and mutations in MCR3 were found to prevent glucose activation. Hence, we conclude that both relief of repression and adoption of an activating form are required for ChREBP activation.
The mammalian liver plays a critical role in maintaining energy homeostasis of an organism in response to its dietary state. When food is abundant, excess dietary carbohydrates are converted to triglycerides in the liver through the pathway of de novo lipogenesis for long term energy storage. Lipogenic enzymes, such as L-type pyruvate kinase (1), acetyl-CoA carboxylase (2), fatty acid synthase (3), and stearoyl-CoA desaturase (4), involved in the conversion of glucose to triglycerides are induced upon feeding of a high carbohydrate diet. Transcriptional induction of these genes requires signals from insulin, acting through sterol response element-binding protein-1c (5-8), and a second signaling pathway initiated in response to increased metabolism of simple carbohydrates, such as glucose (9-12). Lipogenic genes responsive to glucose contain a DNA element called the carbohydrate response element (ChoRE)2 (13-17). The ChoRE consists of two E box sequences (CACGTG) separated by 5 base pairs and serves as the recognition site for two heterodimeric transcription factors: carbohydrate response element-binding protein (ChREBP) and Max-like protein X (Mlx) (18-22). Both ChREBP and Mlx are required for binding to the ChoRE, but recent evidence establishes ChREBP as the direct target of glucose signaling. ChREBP is highly expressed in glucose-responsive tissues, such as the liver, adipose, and pancreas, whereas Mlx expression is ubiquitous (23-25). High carbohydrate-fed ChREBP-/- mice do not induce de novo lipogenesis or lipogenic enzyme gene expression (25). Similarly, cultured mouse hepatocytes in which ChREBP expression has been inhibited by small interfering RNA do not induce lipogenic gene expression in response to glucose (26). Finally, a fusion protein of the Gal4 DNA binding domain and ChREBP can activate an appropriate reporter gene in response to glucose, even when the Mlx-interacting region of ChREBP is deleted (27-29). Hence, ChREBP is the major target of the glucose signaling pathway, whereas Mlx serves to help recruit ChREBP to appropriate target genes (30).
The originally proposed and widely accepted model for ChREBP activation suggests that its activity is regulated by reversible phosphorylation (20, 31). In this model, protein kinase A phosphorylates residues Ser-196, Ser-626, and Thr-666 of ChREBP in low glucose conditions, restricting it to the cytoplasm and inhibiting its DNA binding (19). An increase in glucose metabolism stimulates protein phosphatase 2A activity via direct binding to the glucose metabolite xyulose-5-phosphate (32). Dephosphorylation of ChREBP at these critical protein kinase A sites allows ChREBP to translocate to the nucleus and bind to the ChoRE, which is sufficient for activation of lipogenic gene expression.
Subsequent studies performed by our laboratory and others have questioned the current model for glucose regulation of ChREBP. The level of phosphorylation of ChREBP did not decrease in response to high glucose, as predicted by the current model (33). Mutating residues Ser-196, Ser-626, and Thr-666 to alanines did not lead to constitutive activation, suggesting that dephosphorylation of these residues is not sufficient for activation (27, 28, 33). Finally, inhibition of protein phosphatase 2A in 832/13 cells by the addition of cantharidic acid did not interfere with the activation of ChREBP by glucose (28). Thus, we conclude that although protein kinase A-mediated phosphorylation may repress ChREBP activity under fasting conditions, reversal of this phosphorylation is not sufficient for glucose activation.
To address the mechanism of ChREBP activation, we further evaluated the connection between ChREBP localization and glucose-stimulated activity. We present evidence that ChREBP shuttles between the nucleus and cytoplasm in both low and high glucose conditions, but accumulation in the nucleus occurs more rapidly in high glucose conditions than in low. However, nuclear localization is not sufficient for ChREBP activation. Thus, we suggest that there are additional events, independent of nuclear localization, required for glucose activation of ChREBP.
EXPERIMENTAL PROCEDURES
Construction of Mutant ChREBP Plasmids—Site-specific mutations of ChREBP were constructed with the QuikChange™ site-directed mutagenesis kit (Stratagene) using mouse FLAG-tagged ChREBP in the expression plasmid CMVS4 as a template (21). All mutations and coding sequences were confirmed by DNA sequencing. Immunoblotting of ChREBP from transfected HEK293 cells was performed using FLAG monoclonal antibody (Sigma) to ensure that each construct was expressed. Only mutants with comparable expression to wild-type (WT) ChREBP were used for subsequent analysis.
Transcriptional Reporter Gene Assays for Measuring ChREBP Activity—832/13 cells (a gift from Dr. C. Newgard, Duke University) were cultured in RPMI media containing 11 mm glucose in 24-well plates, as previously described (34). Cells were transduced with adenovirus expressing dominant negative ChREBP (29). The amount of virus used was determined empirically to give ∼90% inhibition of the glucose response. After 2 h of transduction, 832/13 cells were transfected using Lipofectamine 2000 reagent (Invitrogen) with a mixture of firefly luciferase reporter (700 ng) driven by the ACC ChoRE-containing promoter region (33) and a Renilla luciferase control plasmid (pRL-CMV; 15 ng; Promega). In addition, where indicated, cells were also co-transfected with 60 ng of each ChREBP expression plasmid and 30 ng of Mlx expression plasmid. After 18 h, cells were cultured in RPMI medium containing low (2.5 mm) or high (25 mm) glucose for 24 h and lysates were prepared in Passive Lysis Buffer (Promega). Dual luciferase assays were performed following the manufacturer's instructions. Values represent the ratio of firefly/Renilla luciferase from triplicate samples and are expressed as mean ± S.D.
Immunolocalization of ChREBP—832/13 cells were cultured in RPMI medium containing 11 mm glucose on glass slides and grown to 70% confluence (29). Cells were co-transfected with 0.15 μg each of FLAG-tagged WT or mutant ChREBP and hemagglutinin-tagged Mlx using Lipofectamine 2000 (Invitrogen) and 1.1 μl of Virofect (Targeting Systems, San Diego, CA). After overnight transfection, cells were refed RPMI medium containing 2.5 mm glucose for 4 h. Cells were then incubated in RPMI medium containing 2.5 or 25 mm glucose for 2 h. Where indicated, leptomycin B was added at 3.6 μm. After treatment, cells were fixed in a 1.6% formaldehyde solution containing phosphate-buffered saline and 0.2% Triton. Cells were subsequently washed in phosphate-buffered saline, 0.2% Triton and incubated overnight in a humidity chamber at 4 °C. The slides were washed two additional times and then blocked with 10 mg/ml bovine serum albumin in phosphate-buffered saline, 0.2% Triton, and 5 μl/ml donkey serum (Jackson ImmunoResearch Laboratories). Following the block, fixed cells were incubated with 0.25 μg of FLAG monoclonal antibody for 1 h at 37°C and washed five more times, followed by incubation with a secondary FITC-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories) for an additional 1 h. Nuclei were stained with TO-PRO3 (Molecular Probes) during the final wash steps. Cells were imaged using a multiphoton confocal microscope (Fluoview 1000; Olympus).
Co-immunoprecipitation—832/13 cells were co-transfected with FLAG-tagged ChREBP and Mlx in RMPI medium containing 11 mm glucose, as described above. After 36 h, cells were collected in 50 mm Tris-HCl, pH 8.0, 10% glycerol, 150 mm NaCl, 0.5% Triton X-100 with protease inhibitors (Roche Applied Science) and then lysed in a glass-Teflon homogenizer. Anti-FLAG-agarose beads (Sigma) were added, and samples were processed according to the manufacturer's instructions. Immunoadsorbed proteins were separated on a 10% polyacrylamide gel and immunoblotted with horseradish peroxidase-conjugated 14-3-3β antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Measurement of mRNA by Quantitative Reverse Transcription-PCR—Total cellular RNA was isolated from 832/13 cells using Trizol reagent (Invitrogen), and selected ChREBP target gene products were measured by quantitative reverse transcription-PCR performed by a two-step procedure described previously (35). Primers were designed using MacVector (Accelrys Software, Inc.). Reverse transcription-PCR results are expressed as -fold induction by normalizing the mean of the Ct values from the high glucose-treated cells relative to the mean of Ct values from low glucose cells. All samples were analyzed in triplicate and expressed as mean ± S.E.
RESULTS
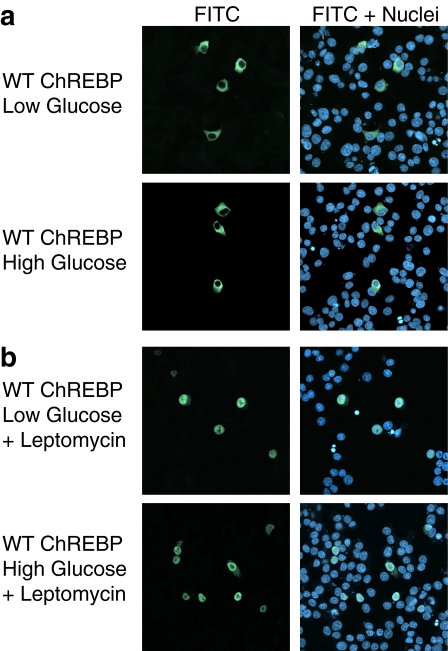
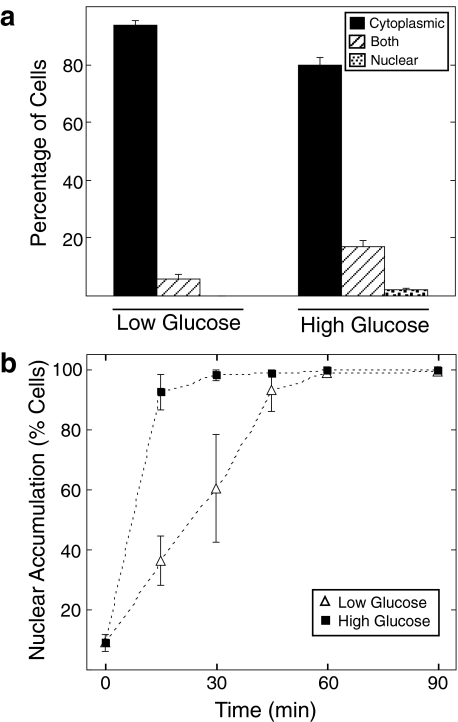
ChREBP Shuttles between the Nucleus and Cytoplasm under Low and High Glucose Conditions—The human homolog of ChREBP, Wbscr14 (36), and the ChREBP paralog, MondoA (37), have been shown to shuttle between the cytoplasm and nucleus in several cell lines, such as mouse 3T3, monkey COS7, and human 293 cells (38, 39). However, these cell lines are not known to respond transcriptionally to changes in glucose metabolism. Thus, we decided to examine ChREBP nucleocytoplasmic shuttling and the effect of glucose on this process in a glucose-sensitive cell line. Immunofluorescence was performed in 832/13 cells, an INS-1-derived cell line with robust glucose-stimulated insulin secretion (34). These cells express ChREBP and support a transcriptional activation of ChREBP in response to elevated glucose (24, 27, 28). 832/13 cells were co-transfected with plasmids expressing FLAG-tagged ChREBP and Mlx. Mlx was incorporated in these studies because of its known interaction with ChREBP (21, 30), its requirement for ChREBP-dependent gene transcription (22, 35), and its requirement for ChREBP nuclear entry (see Fig. S1). Transfected cells were incubated in low glucose medium for 4 h and then treated with either low or high glucose medium for 2 h. At this time, optimal induction of several ChREBP target genes is observed (28, 35). Immunofluorescence was performed with anti-FLAG primary and FITC-conjugated secondary antibodies and visualized by confocal microscopy. As expected, ChREBP was predominantly localized to the cytoplasm under low glucose conditions (Fig. 1a). However, as previously reported (29), treatment of cells with high glucose to induce ChREBP activity did not result in a major accumulation of ChREBP in the nucleus. The majority of fluorescent signal in these cells was still found within the cytoplasm. Quantification of signals from a large number of cells revealed that 94% displayed cytoplasmic localization and 6% had signals from both cytoplasm and nucleus in low glucose (Fig. 2a). In high glucose, ChREBP was cytoplasmic in 78%, both cytoplasmic and nuclear in 18%, and nuclear in 2% of cells. Hence, although there was a shift to a more nuclear pattern in high glucose, this trend was only observed in a small fraction of the total cells. Similar results were reported for a green fluorescent protein-tagged fusion of ChREBP in 832/13 cells at 6 h of glucose treatment (28).
FIGURE 1.
ChREBP shuttles between the cytoplasm and nucleus under both low and high glucose conditions. a, 832/13 cells were co-transfected with expression plasmids for FLAG-tagged ChREBP and Mlx overnight in 11 mm glucose and subsequently incubated in RPMI medium containing 2.5 mm (low) glucose for 4 h. Cells were then continued in the same medium or switched to RPMI medium containing 25 mm (high) glucose for 2 h. Immunofluorescence was performed using an anti-FLAG primary antibody and a FITC-conjugated secondary antibody, and images were obtained by confocal microscopy. FITC immunofluorescence represents ChREBP localization and is shown in green. Nuclei were stained with TO-PRO3 and are shown in blue. The panels labeled FITC represent ChREBP localization, whereas the panels labeled FITC + Nuclei show both ChREBP localization and nuclear staining. b, cells were treated as described above, except that leptomycin B was added simultaneously with the glucose treatment for 2 h.
FIGURE 2.
The rate of ChREBP nuclear entry is increased in high glucose conditions. a, 832/13 cells were co-transfected with FLAG-tagged ChREBP and Mlx overnight in RPMI medium containing 11 mm glucose and were then incubated in low glucose medium for 4 h. Cells were then treated with either low or high glucose for 2 h, and immunofluorescence was performed as described under “Experimental Procedures.” Over 100 cells in each treatment were scored as predominantly cytoplasmic, both cytoplasmic and nuclear, or predominantly nuclear by two observers. Results represent the means ± S.E. for three experiments. b, cells were treated with low (open triangles) or high glucose (closed squares) medium with the addition of leptomycin B at various time points as indicated. Immunofluorescence and quantification was performed as described above. Cells displaying either predominantly nuclear or both nuclear and cytoplasmic localization were combined and expressed as a percentage of the total cells.
Since ChREBP was located in the cytoplasm under high glucose conditions in the majority of cells, shuttling between the cytoplasm and the nucleus must occur to support its transcriptional function. The domains of Wbscr14 and MondoA found to contain the nuclear export signal (NES) are highly conserved in ChREBP, suggesting that it may be subject to Crm-1-dependent export. To confirm this, localization of ChREBP was observed in 832/13 cells treated with low or high glucose conditions for 2 h with the addition of leptomycin B, a Crm-1 nuclear export inhibitor (Fig. 1b). ChREBP accumulated in the nucleus in high glucose conditions with leptomycin B treatment, as expected. However, ChREBP was found predominantly in the nucleus in low glucose as well. The observation that ChREBP actively shuttles under low glucose conditions in which it is not transcriptionally active suggests that an additional event is required for its transcriptional activation.
The Rate of ChREBP Nuclear Entry Is Increased under High Glucose Conditions—The fact that ChREBP is predominantly cytoplasmic under stimulating conditions indicates that only a fraction of total cellular ChREBP needs to be in the nucleus for gene activation. Therefore, it seemed plausible that glucose might act by transiently increasing the rate of ChREBP entry to the nucleus without a dramatic shift in the cellular pool of ChREBP to the nuclear compartment. To measure its rate of nuclear accumulation, 832/13 cells were co-transfected with ChREBP and Mlx. Subsequently, cells were treated with leptomycin B, and the time course of ChREBP nuclear accumulation in low or high glucose conditions was compared. ChREBP localization was classified as cytoplasmic, both cytoplasmic and nuclear, or nuclear. Cells that displayed either nuclear or both nuclear and cytoplasmic localization were combined to represent cells that had initiated nuclear accumulation. An increased nuclear accumulation was observed in high glucose conditions at the 15 and 30 min time points relative to cells maintained in low glucose (Fig. 2). At 45 min, the difference in ChREBP accumulation between treatments was less evident, and at 1 h, ChREBP was predominantly nuclear in both low and high glucose conditions. From the time required to achieve 50% nuclear accumulation, we estimate that the rate of nuclear entry was 3-fold greater in high glucose compared with low glucose. Thus, we conclude that glucose regulates ChREBP at the level of nuclear entry.
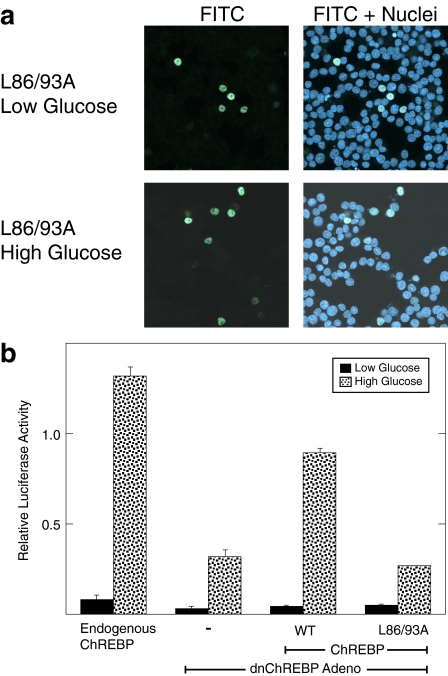
The L86A/L93A ChREBP Mutant Is Deficient in Nuclear Export and Is Not Transcriptionally Active—The observation that ChREBP enters the nucleus more rapidly in high glucose raises the question of whether this event is sufficient to account for the elevated ChREBP activity in these conditions. To further examine this question, a mutant ChREBP deficient in nuclear export was constructed and tested for functional activity. This ChREBP mutant was prepared by mutating two conserved leucine residues (Leu-86 and Leu-93) in the NES to alanines. The comparable mutations in Wbscr14 and MondoA have been shown to accumulate in the nucleus (38, 39). To test whether the L86A/L93A ChREBP mutant is deficient in nuclear export, we performed immunofluorescence in 832/13 cells (Fig. 3a). The L86A/L93A ChREBP mutant localizes to the nucleus under both low and high glucose conditions without the addition of leptomycin B, indicating that this mutant was indeed deficient in nuclear export. We then tested the activity of the L86A/L93A mutant using a functional rescue assay that our laboratory has previously established (33). To accomplish this, 832/13 cells were transduced with an adenoviral construct expressing a dominant negative form of ChREBP. This form of ChREBP is able to dimerize with Mlx, but the resultant dimer cannot bind to DNA. Hence, this mutant ChREBP competes with endogenous ChREBP for Mlx heterodimerization. Subsequently, 832/13 cells were transfected with a plasmid expressing WT or mutant ChREBP, together with a ChoRE-containing reporter construct. The activity of the reporter gene reflects the ability of the overexpressed ChREBP to support a glucose response.
FIGURE 3.
The L86A/L93A mutant of ChREBP is trapped in the nucleus but is transcriptionally inactive. a, immunofluorescence images of 832/13 cells co-transfected with FLAG-tagged L86A/L93A ChREBP mutant and Mlx in low and high glucose conditions are shown. See the legend to Fig. 1 for details. b, a functional assay for ChREBP activity was performed in 832/13 cells. 832/13 cells were transduced with an adenoviral vector expressing dominant negative ChREBP. Cells were then co-transfected with a luciferase reporter plasmid containing two copies of the ACC ChoRE, a Renilla luciferase reporter, and expression plasmids for either WT ChREBP or the L86A/L93A ChREBP mutant and WT Mlx. After 18 h, cells were treated with low or high glucose for 24 h, and extracts were prepared. Values are in relative light units (firefly/Renilla) and represent the means ± S.D. for triplicate samples.
As shown in Fig. 3b, overexpressing WT ChREBP rescues the glucose response, although not to the level of endogenous ChREBP. The extent of rescue in different experiments varied from 30 to 80%, depending on the effectiveness of the viral transduction of dominant negative ChREBP. However, the L86A/L93A ChREBP mutant did not rescue the glucose response, indicating that it is not functional for transcriptional activation. Western blotting showed that L86A/L93A ChREBP is expressed equivalently to WT ChREBP (see Fig. S2) and bound to a ChoRE-containing oligonucleotide in an electro-phoretic mobility shift assay.3 Thus, trapping ChREBP within the nucleus did not result in constitutive transcriptional activation, as might have been expected, but rather a loss of function.
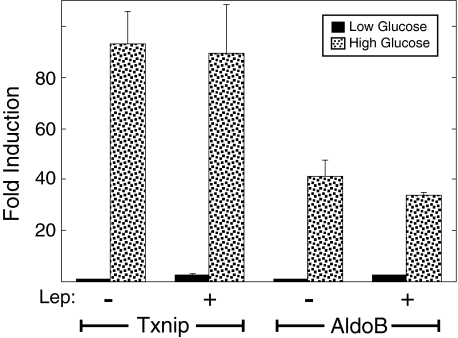
Preventing Nucleocytoplasmic Shuttling of ChREBP Does Not Inhibit Glucose Activation—We considered two possible explanations for why the L86A/L93A mutant is inactive despite its nuclear localization: (a) ChREBP may need to be activated by an event in the cytoplasm and nucleocytoplasmic shuttling may be required to maintain this active state, or (b) a separate event, independent of nuclear localization and shuttling, may be required to activate ChREBP and stimulate gene transcription. In the latter circumstance, the L86A/L93A mutant would have altered both NES function and the distinct event required for activation. To test whether nucleocytoplasmic shuttling is required to maintain activity, mRNA levels of ChREBP target genes were measured in 832/13 cells pretreated with leptomycin B for 90 min in low glucose to trap ChREBP in the nucleus. Cells were subsequently maintained in low glucose or shifted to high glucose for 4 h. A short time point was selected to avoid potential secondary effects of inhibiting Crm-1-dependent export. If nucleocytoplasmic shuttling were essential, then leptomycin B treatment should inhibit ChREBP activation in high glucose conditions. mRNA from two ChREBP-responsive genes, thioredoxin-interacting protein and aldolase B, were measured. Expression of both gene products was higher in control cells treated with high glucose compared with low glucose at 4 h (Fig. 4). In cells treated with leptomycin B, the ability of high glucose to induce the ChREBP target mRNAs was not diminished. Therefore, preventing cytoplasmic shuttling did not inhibit ChREBP activity. It is also note-worthy that mRNA expression levels in low glucose plus leptomycin B were not different from those observed in the low glucose alone. These observations are consistent with the earlier conclusion that localizing ChREBP to the nucleus is not sufficient for activation.
FIGURE 4.
Leptomycin B does not inhibit the induction of ChREBP target genes in 832/13 cells. 832/13 cells were pretreated in low glucose RPMI medium for 4 h with or without leptomycin B (Lep) for the last 90 min. Cells from both experimental groups were then incubated with low or high glucose for 4 h. RNA was isolated and converted into cDNA using reverse transcriptase. mRNA levels of ChREBP target genes, thioredoxin-binding protein (Txnip) and aldolase B (AldoB), were measured by quantitative reverse transcription-PCR. mRNA levels in low glucose without the addition of leptomycin B were set to 1, and all values are normalized to this group. Values represent the mean ± S.E. of triplicate samples.
Glucose-stimulated ChREBP Activation Requires an Event Independent of Nuclear Localization—The inability of the L86A/L93A mutant of ChREBP to support a glucose response suggested that this region is involved in two separate functions, nuclear export and glucose-activation, both of which were inactivated in the mutant. If this is the case, then it should be possible to find other mutations in this region that effect only one or the other of these two functions. Consequently, conserved residues within the NES region were individually mutated, and these mutants were tested for nuclear export and transcriptional activation.
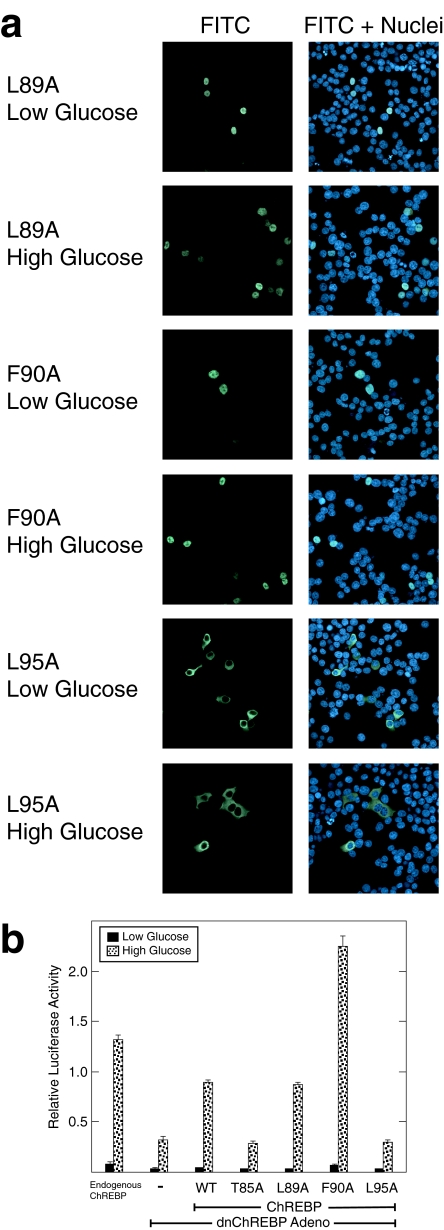
Of the ChREBP mutants in this region, several yielded informative results. For example, ChREBP mutants at Thr-85 and Leu-95 localized predominantly to the cytoplasm in both low and high glucose-treated 832/13 cells, similar to wild-type ChREBP (Fig. 5a).3 To ensure that these mutations did not interfere with normal nuclear accumulation, their localization in the presence of leptomycin B was analyzed. Both T85A and L95A ChREBP mutants accumulated in the nucleus by 60 min in low and high glucose conditions (see Fig. S3). When the function of T85A and L95A ChREBP mutants was tested, however, they did not successfully rescue ChREBP activity (Fig. 5b). Thus, the T85A and L95A ChREBP mutants shuttle between the nucleus and cytoplasm but are transcriptionally inactive, separating nuclear export function from activation. In contrast, mutations at residues Leu-89 and Phe-90 resulted in nuclear accumulation of ChREBP regardless of glucose treatment (Fig. 5a). Both L89A and F90A ChREBP mutants were able to rescue ChREBP activity in high glucose conditions (Fig. 5b). It is note-worthy that the F90A ChREBP mutant consistently displayed a 2-3-fold higher increase in high glucose conditions than WT ChREBP. This increased activity of the F90A mutant was also observed when tested in the context of rat primary hepatocytes (see Fig. S4). Thus, L89A and F90A ChREBP mutants are nuclear export-deficient but maintain transcriptional activity, again separating the two functions. Together, these data support the conclusion that the NES region of ChREBP plays distinct roles in both nuclear export and activation.
FIGURE 5.
NES region mutants of ChREBP separate NES function from transcriptional activation. a, immunofluorescence images of 832/13 cells transfected with FLAG-tagged L89A, F90A, and L95A ChREBP mutants and Mlx in low and high glucose conditions are shown. See the legend to Fig. 1 for details. b, functional activities of ChREBP mutants were tested using the reporter assay as described in the legend to Fig. 3b. Values are shown as relative light units (firefly/Renilla) and represent the means ± S.D. for triplicate samples.
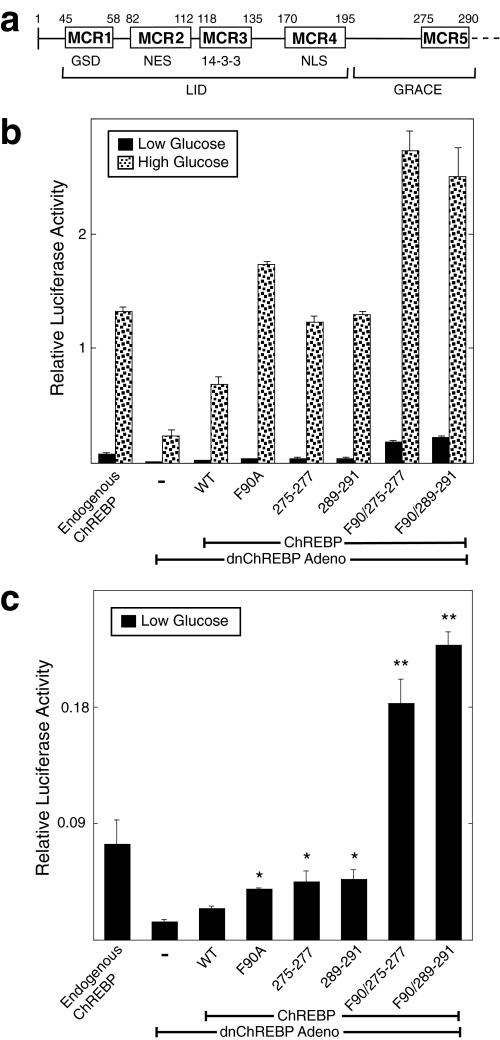
Roles of Conserved MCR Domains in Repression and Activation of ChREBP—Previous work from Li et al. (28) used deletion mutagenesis to distinguish two separable functions in the N-terminal segment (amino acids 1-298) of ChREBP. Amino acids 37-196 were found to be responsible for repression of ChREBP activity in low glucose, since deletion of these residues resulted in constitutive activation of ChREBP. This region was termed the LID (for low glucose inhibitory domain). Amino acids 197-298 were found to contain transcriptional activation function that is repressed by the LID in low glucose and was designated GRACE (for glucose response activation conserved element). The N-terminal segment of ChREBP contains five highly conserved domains designated MondoA conserved regions 1-5 (MCR1 to -5) based on their striking homology (>90% identity) with the ChREBP paralog, MondoA (Fig. 6a). These conserved domains range in size from 14 to 31 amino acids. MCR1 to -4 are located in the LID of ChREBP, whereas MCR5 is found in the GRACE. We were interested in further exploring the role of each of these highly conserved domains.
FIGURE 6.
MCR5 and MCR2/MCR5 mutants of ChREBP display increased transcriptional activity. a, conserved regions in the amino-terminal segment of ChREBP. The locations of five highly conserved regions in the amino-terminal segment of ChREBP are indicated. Numbering is based on mouse ChREBP. These regions are highly conserved (>90% identity) with the ChREBP paralog, MondoA, as well as with ChREBP orthologs from pufferfish to human. Previously proposed functions for these conserved domains are indicated below. GSD, glucose-sensing domain; NLS, nuclear localization signal; LID, low glucose inhibitory domain; GRACE, glucose response activation conserved element. b, functional activity of ChREBP was tested using the rescue assay described in the legend to Fig. 3b. WT, F90A, Y275A/V276A/G277A (275-277), L289A/Q290A/P291A (289-291), F90A/275-277, and F90A/289-291 were introduced and tested for activity. Values are shown as relative light units (firefly/Renilla) and represent means ± S.D. for triplicate samples. c, luciferase values measured in low glucose conditions of the mutants above are compared. The data presented in this panel is from the experiment shown in b.*, p < 0.05 compared with WT ChREBP; **, p < 0.01 compared with WT ChREBP.
We have previously shown that deletion of MCR1 alone results in a form of ChREBP that cannot be activated, suggesting that this domain is critical for receiving the glucose signaling event (29). The MCR4 domain contains the nuclear localization signal of ChREBP, and a mutation introduced into this region blocked nuclear import and correspondingly gave an inactive form of ChREBP.3 The NES function of ChREBP is found in MCR2. However, the mutations in MCR2 described above indicated that this domain is also involved in glucose-dependent transcriptional activation. To evaluate the roles of MCR3 and MCR5 domains, several additional mutants of ChREBP were constructed.
Two mutants in the MCR5 domain gave a particularly interesting phenotype. Residues Tyr-275/Val-276/Gly-277 (275-277) or Leu-289/Gln-290/Pro-291 (289-291) were mutated to alanines, resulting in two separate ChREBP triple mutants. When functionally tested in 832/13 cells, these mutants displayed increased activity in both low and high glucose (Fig. 6b). These mutants are found in the nucleus in a greater percentage of cells than wild type ChREBP (Fig. S5). This result indicates that the MCR5 domain is not an essential part of the trans-activation function of the GRACE region but instead may be involved in repression of transcriptional activity. The increased activity of the two MCR5 mutants was, in fact, similar to that observed with the F90A mutation of the MCR2 domain. These results suggested that these two domains may be involved in the same event to repress ChREBP activity. If these two domains act together in a repressive manner, then mutating both regions simultaneously should result in a further increase in activity. To address this possibility, the F90A mutation was combined with either the ChREBP mutations at 275-277 or 289-291 and tested functionally. Both combined mutant forms showed a further increase in activity in low and high glucose conditions when compared with the F90A, 275-277, and 289-291 ChREBP mutant counterparts (Fig. 6b). The increased activity observed with both combined mutants in the low glucose treatment was particularly striking and was synergistic compared with the effects of the individual mutants (Fig. 6c). These data suggest that MCR2 and MCR5 act in a coordinate manner to repress ChREBP activity.
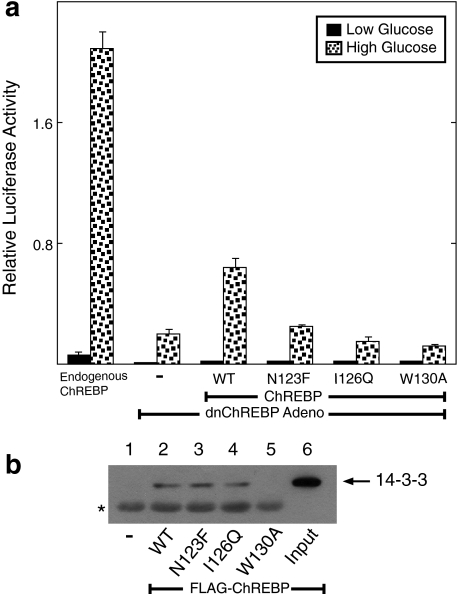
In addition to the mutations made in MCR5, we also targeted the MCR3 domain using site-directed mutagenesis. Mutations were made at residues Asn-123, Ile-126, and Trp-130. When these mutants were tested using the rescue assay, all three failed to support a glucose response (Fig. 7a). Hence, this domain appears to play an important role in activation of ChREBP. However, this domain has previously been shown to interact with 14-3-3 proteins in Wbscr14 and MondoA (38, 39). It was suggested that the interaction with 14-3-3 might be responsible for the predominantly cytoplasmic localization of ChREBP. We therefore asked whether the MCR3 mutations disrupted the ability of ChREBP to interact with 14-3-3. In co-immunoprecipitation experiments, WT ChREBP did interact with 14-3-3β (Fig. 7b). The W130A mutation disrupted the interaction between ChREBP and 14-3-3β, confirming the importance of this region for interaction. This disruption corresponded to a greater localization of N130A to the nucleus in both low and high glucose, suggesting that the 14-3-3 interaction may be important for cytoplasmic retention of WT ChREBP (Fig. S5). However, neither of the other two ChREBP mutants (N123F and I126Q) altered the binding of ChREBP to 14-3-3. Hence, the inactivity of these two MCR3 mutants cannot be attributed to their inability to bind to 14-3-3. These data suggest that the MCR3 domain plays an essential role in supporting glucose activation that is independent of its interaction with 14-3-3 proteins.
FIGURE 7.
MCR3 mutants of ChREBP inhibit glucose activation. a, functional activity of ChREBP was tested using the reporter assay described in the legend to Fig. 3b. WT ChREBP or ChREBP mutants N123F, I126Q, and W130A were introduced and tested for activity. Values are shown as relative light units (firefly/Renilla) and represent the means ± S.D. for triplicate samples. b, 832/13 cells were co-transfected with FLAG-tagged WT or mutant ChREBP and Mlx and incubated overnight in RPMI medium with 11 mm glucose. Cell extracts were prepared, and co-immunoprecipitations were performed with anti-FLAG immunobeads as described under “Experimental Procedures.” Immunoblotting was carried out with a 14-3-3β antibody. Lane 1, extracts from untransfected 832/13 cells. Lanes 2-5, cells transfected with WT ChREBP and MCR3 mutations N123F, I126Q, and W130A, respectively. Lane 6, an aliquot of the cell extract without co-immunoprecipitation. The arrow indicates the position of 14-3-3β protein, and the asterisk indicates a background band that cross-reacts with the 14-3-3β antibody.
DISCUSSION
ChREBP, a glucose-responsive transcription factor, plays a critical role in converting excess carbohydrates to triglycerides through de novo lipogenesis. Although the importance of ChREBP in de novo lipogenesis and hepatic energy utilization is strongly supported (25, 40-43), the mechanism driving its activation remains controversial and not fully understood. To address the mechanism of ChREBP activation, we focused on the importance of cellular localization. Under both low and high glucose conditions, ChREBP localized to the cytoplasm in the majority of 832/13 cells. Li et al. (28) also found that a green fluorescent protein-fused form of ChREBP was primarily cytoplasmic in these cells. However, leptomycin B treatment trapped ChREBP in the nucleus under either basal or stimulating conditions. These results demonstrate that ChREBP continuously shuttles between the cytoplasmic and nuclear compartments. The ability of ChREBP to transit through the nucleus in low glucose conditions, in which lipogenic gene expression is minimal, suggests that nuclear localization is not solely responsible for its activation. However, one potential explanation for activation of ChREBP could be that glucose stimulates a transient increase in the rate of nuclear entry. Indeed, ChREBP nuclear entry was 3-fold greater in high glucose conditions than in low glucose, consistent with a modest increase in nuclear localization observed at 2 h (Fig. 2a). Previous studies using cell fractionation also reported increased levels of ChREBP in the nucleus acutely after glucose stimulation (27, 40, 44). Thus, we conclude that glucose regulates the rate of nuclear entry.
Glucose control of ChREBP trafficking could be due to an altered association with a cytoplasmic anchor protein, such as 14-3-3. Increased glucose metabolism could reduce the strength of this interaction, perhaps in response to post-translational modifications of ChREBP (29). Both the human homolog of ChREBP, Wbscr14, and the paralog, MondoA, have been shown to interact with multiple 14-3-3 isoforms (36, 38). Disruption of this interaction by mutating the 14-3-3 binding site of these proteins led to increased nuclear localization, as we have also observed for the W130A ChREBP mutant. Alternatively, glucose could regulate nuclear entry by promoting the interaction of ChREBP with its nuclear import receptor. Characterization of the pathway of nuclear translocation will clearly be required to further elucidate this mechanism and its importance to the overall regulation of ChREBP.
Although glucose regulates the nuclear entry of ChREBP, we cannot conclude that this step is sufficient for its activation. In fact, mutation of two residues within the NES not only trapped ChREBP in the nucleus; it also prevented glucose activation. This result indicates that nuclear localization is not sufficient for activation and that the NES region plays a critical role distinct from its nuclear export function. In addition, trapping ChREBP in the nucleus with leptomycin B did not interfere with the induction of target genes. Hence, active shuttling is not required for glucose stimulation. Together, these data indicate that an additional event independent of ChREBP trafficking is required for activation.
If subcellular localization is not the key event in regulating ChREBP activity, then what is the purpose of nucleocytoplasmic shuttling? In fact, many transcription factors actively shuttle under both basal and stimulating conditions. For example, despite nuclear localization being the primary means of controlling their activity, STAT and SMAD transcription factors shuttle in the absence of ligand binding to their activating receptors (45, 46). In the case of SMADs, this shuttling has been suggested to increase the dynamic regulation affecting the duration and magnitude of signaling (47). For STATs, shuttling has been suggested to be important for cytokine sensitivity (48). In the case of ChREBP, it is yet unknown whether the critical cell signal that triggers activation in response to glucose metabolism occurs in the cytoplasm, nucleus, or both. However, all of our studies have been carried out under conditions where glucose levels were saturating with respect to activation. Under conditions of intermediate and changing glucose levels that normally occur, nuclear-cytoplasmic shuttling may be important for sensitization of ChREBP to the metabolic status of the cell.
The N-terminal segment of ChREBP (amino acids 1-298) contains five highly conserved domains (MCR1 to -5) and is critical for its ability to respond to glucose (28, 29). Deletion of MCR1 to -3 or MCR1 to -4 yields constitutively active forms of ChREBP, indicating that regulation involves repression in basal conditions mediated through this region. In the present study, the behavior of certain mutations in MCR2 and MCR5 suggested a possible mechanism for the repressive event. Mutation of residue Phe-90 in the NES region (MCR2) increased ChREBP activity while also blocking nuclear export. A similar mutation of residue Leu-89 blocked nuclear export but had normal activity, suggesting that the behavior of the F90A mutant was not simply due to its nuclear accumulation. Interestingly, two independent mutations made in MCR5 (residues 275-277 and 289-291) gave a similar phenotype to the F90A mutation. Thus, we speculate that MCR2 and MCR5 function coordinately to repress ChREBP activation. Repression could result from an intramolecular interaction of MCR2 and MCR5 or from a simultaneous interaction of these domains with an independent repressor. In either case, we would predict that a combined mutation would further disrupt the repressive interactions of MCR2 and MCR5 and amplify activity. In fact, both combined mutations yielded superactive forms of ChREBP. This result was particularly dramatic in low glucose conditions in which a synergistic effect of combining mutations was observed. Thus, MCR2 and MCR5 play a role in the repressive mechanism of ChREBP under basal conditions. It is worth noting that deletion of MCR1 to -5 results in an inactive form of ChREBP, whereas deletion of MCR1 to -4 yields a constitutively active protein, as previously mentioned. Hence, we also conclude that MCR5 must have a dual role in promoting both inactivating and activating conformations.
Although several MCR mutations resulted in increased ChREBP activity, others yielded forms that could not be activated by glucose. These mutations were found in both MCR2 and MCR3. Since deletion of the N-terminal segment that includes MCR2 and MCR3 yielded a constitutively active protein, these observations seem paradoxical. If the only function of the N-terminal region were in repression, then one would expect most point mutations would yield active forms of ChREBP. These data suggest that the relief of repression is not sufficient for activation, and a second step involving recruitment of co-activating factors is required. Inactivating mutations in MCR2 and MCR3 could directly or indirectly prevent recruitment of these co-activating factors.
These studies were carried out in 832/13 cells, a β cell-derived line. An important question is whether the mechanism of regulation in 832/13 cells is the same as that in hepatocytes or adipocytes, two major target organs for glucose control. Previous studies have reported increased accumulation of green fluorescent protein-tagged ChREBP in the nucleus of cultured hepatocytes treated with high glucose (19, 32). This accumulation, however, occurs with relatively slow kinetics (lag of 3 h, half-maximal at 5 h) compared with the rapid activation of ChREBP-targeted genes following the addition of high glucose. Thus, the importance of increased nuclear accumulation of ChREBP in hepatocytes to activation is uncertain. Because of the high autofluorescence of cultured hepatocytes, we have been unable to examine the effects of glucose on nuclear entry and shuttling in these cells. However, mutating Ser-196 of ChREBP does not result in increased activity in low glucose conditions despite its proposed role in controlling localization (28, 33, 49). We have also tested the functional activity of each of the ChREBP mutants used in the present study in primary hepatocytes and found that all behave identically in the two cell types. Based on these observations, we surmise that control of ChREBP activity in response to glucose in the two cells is likely to be similar.
While this work was in revision, a paper from the laboratory of L. Chan (50) appeared that reached several similar conclusions regarding the regulation of ChREBP. This work, which was also performed in 832/13 cells, showed that MCR2 and -3 were involved in critical functions for activation independent of their role in controlling subcellular localization.
In conclusion, glucose regulation of ChREBP involves at least two distinct processes. One event is to accelerate the rate of nuclear entry. This may contribute to rapid effects of ChREBP in stimulating transcription of many metabolic enzyme genes but is not sufficient for ChREBP activation. In addition, high glucose triggers a second process involving both relief of repression and adoption of an activating form. The MCR2 domain plays a particularly important role in both supporting NES function and in the transition from the repressive to the active state.
Supplementary Material
Acknowledgments
The assistance of Eric Refsland in preparing adenoviruses and the University of Minnesota Biomedical Image and Processing Laboratory in confocal microscopy are greatly appreciated. We thank Drs. Nikolas Tsatsos, Do-Hyung Kim, and Douglas Mashek for helpful comments in the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK26919 and P30 DK50456 (to the Minnesota Obesity Center). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains Figs. S1-S5.
Footnotes
The abbreviations used are: ChoRE, carbohydrate response element; ChREBP, carbohydrate response element-binding protein; Mlx, Max-like factor X; NES, nuclear export signal; WT, wild type; FITC, fluorescein isothiocyanate; STAT, signal transducers and activators of transcription.
M. N., Davies, and H. C., Towle, unpublished results.
References
- 1.Vaulont, S., Munich, A., Decaux, J. F., and Kahn, A. (1986) J. Biol. Chem. 261 7621-7625 [PubMed] [Google Scholar]
- 2.Katsurada, A., Iritani, N., Fukuda, H., Matsumara, Y., Nishimoto, N., Nogushi, T., and Tanaka, T. (1990) Eur. J. Biochem. 190 427-433 [DOI] [PubMed] [Google Scholar]
- 3.Katsurada, A., Iritani, N., Fukuda, H., Matsumura, Y., Noguchi, T., and Tanaka, T. (1989) Biochim. Biophys. Acta 1004 103-107 [DOI] [PubMed] [Google Scholar]
- 4.Ntambi, J. (1992) J. Biol. Chem. 267 10925-10930 [PubMed] [Google Scholar]
- 5.Horton, J. D., Bashmakov, Y., Shimomura, I., and Shimano, H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5987-5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foretz, M., Guichard, C., Ferre, P., and Foufelle, F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12737-12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimano, H., Yahagi, N., Amemiya-Kudo, M., Hasty, A. H., Osuga, J.-I., Tamura, Y., Shionoiri, F., Iizuka, Y., Ohashi, K., Harada, K., Gotoda, T., Ishihashi, S., and Yamada, N. (1999) J. Biol. Chem. 274 35832-35839 [DOI] [PubMed] [Google Scholar]
- 8.Osborne, T. F. (2001) J. Biol. Chem. 275 32379-32382 [DOI] [PubMed] [Google Scholar]
- 9.Vaulont, S., Vasseur-Cognet, M., and Kahn, A. (2000) J. Biol. Chem. 275 31555-31558 [DOI] [PubMed] [Google Scholar]
- 10.Koo, S.-H., Dutcher, A. K., and Towle, H. C. (2001) J. Biol. Chem. 276 9437-9445 [DOI] [PubMed] [Google Scholar]
- 11.Towle, H. C. (2005) Trends Endocrinol. Metab. 16 489-494 [DOI] [PubMed] [Google Scholar]
- 12.Dentin, R., Girard, J., and Postic, C. (2005) Biochimie (Paris) 87 81-86 [DOI] [PubMed] [Google Scholar]
- 13.Bergot, M.-O., Diaz-Guerra, M.-J. M., Puzenat, N., Raymondjean, M., and Kahn, A. (1992) Nucleic Acids Res. 20 1871-1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, Z., Thompson, K. S., and Towle, H. C. (1993) J. Biol. Chem. 268 12787-12795 [PubMed] [Google Scholar]
- 15.Shih, H.-M., and Towle, H. C. (1994) J. Biol. Chem. 269 9380-9387 [PubMed] [Google Scholar]
- 16.O'Callaghan, B. L., Koo, S.-H., Wu, Y., Freake, H. C., and Towle, H. C. (2001) J. Biol. Chem. 276 16033-16039 [DOI] [PubMed] [Google Scholar]
- 17.Rufo, C., Teran-Garcia, M., Makamura, M., Koo, S.-H., Towle, H. C., and Clarke, S. D. (2001) J. Biol. Chem. 276 21969-21975 [DOI] [PubMed] [Google Scholar]
- 18.Yamashita, H., Takenoshita, M., Sakurai, M., Bruick, R. K., Henzel, W. J., Shillinglaw, W., Arnot, D., and Uyeda, K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9116-9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi, T., Takenoshita, M., Li, Y., Kabashima, T., and Uyeda, K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13710-13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii, S., Iizuka, K., Miller, B. C., and Uyeda, K. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15597-15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeckman, A. K., Ma, L., and Towle, H. C. (2004) J. Biol. Chem. 279 15662-15669 [DOI] [PubMed] [Google Scholar]
- 22.Ma, L., Tsatsos, N. G., and Towle, H. C. (2005) J. Biol. Chem. 280 12019-12027 [DOI] [PubMed] [Google Scholar]
- 23.Billin, A. N., Eilers, A. L., Queva, C., and Ayer, D. E. (1999) J. Biol. Chem. 274 36344-36350 [DOI] [PubMed] [Google Scholar]
- 24.Wang, H., and Wollheim, C. B. (2002) J. Biol. Chem. 277 32746-32752 [DOI] [PubMed] [Google Scholar]
- 25.Iizuka, K., Bruick, R. K., Liang, G., Horton, J. D., and Uyeda, K. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7281-7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dentin, R., Pegorier, J. P., Benhamed, F., Foufelle, F., Ferre, P., Fauveau, V., Magnuson, M. A., Girard, J., and Postic, C. (2004) J. Biol. Chem. 279 20314-20326 [DOI] [PubMed] [Google Scholar]
- 27.Collier, J. J., Zhang, P., Pedersen, K. B., Burke, S. J., Haycock, J. W., and Scott, D. K. (2007) Am. J. Physiol. 293 E48-E56 [DOI] [PubMed] [Google Scholar]
- 28.Li, M. V., Chang, B., Imamura, M., Poungvarin, N., and Chan, L. (2006) Diabetes 55 1179-1189 [DOI] [PubMed] [Google Scholar]
- 29.Tsatsos, N. G., Davies, M. N., O'Callaghan, B. L., and Towle, H. C. (2008) Biochem. J. 411 261-270 [DOI] [PubMed] [Google Scholar]
- 30.Ma, L., Sham, Y. Y., Walters, K. J., and Towle, H. C. (2007) Nucleic Acids Res. 35 35-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uyeda, K., and Repa, J. J. (2006) Cell Metab. 4 107-110 [DOI] [PubMed] [Google Scholar]
- 32.Kabashima, T., Kawaguchi, T., Wadzinski, B. E., and Uyeda, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5107-5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsatsos, N. G., and Towle, H. C. (2006) Biochem. Biophys. Res. Commun. 340 449-456 [DOI] [PubMed] [Google Scholar]
- 34.Hohmeier, H. E., Mulder, H., Chen, G., Henkel-Rieger, R., Prentki, M., and Newgard, C. B. (2000) Diabetes 49 424-430 [DOI] [PubMed] [Google Scholar]
- 35.Ma, L., Robinson, L. N., and Towle, H. C. (2006) J. Biol. Chem. 281 28721-28730 [DOI] [PubMed] [Google Scholar]
- 36.de Luis, O., Valero, M. C., and Jurado, L. A. (2000) Eur. J. Hum. Genet. 8 215-222 [DOI] [PubMed] [Google Scholar]
- 37.Billin, A. N., Eilers, A. L., Coulter, K. L., Logan, J. S., and Ayer, D. E. (2000) Mol. Cell Biol. 20 8845-8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eilers, A. L., Sundwell, E., Lin, M., Sullivan, A. A., and Ayer, D. E. (2002) Mol. Cell Biol. 22 8514-8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merla, G., Ucla, C., Guipponi, M., and Reymond, A. (2002) Hum. Genet. 110 429-438 [DOI] [PubMed] [Google Scholar]
- 40.Dentin, R., Benhamed, F., Hainault, I., Fauveau, V., Foufelle, F., Dyck, J. R., Girard, J., and Postic, C. (2006) Diabetes 55 2159-2170 [DOI] [PubMed] [Google Scholar]
- 41.He, Z., Jiang, T., Wang, Z., Levi, M., and Li, J. (2004) Am. J. Physiol. 287 E424-E430 [DOI] [PubMed] [Google Scholar]
- 42.Iizuka, K., Miller, B., and Uyeda, K. (2006) Am. J. Physiol. 291 E358-E364 [DOI] [PubMed] [Google Scholar]
- 43.Burgess, S. C., Iizuka, K., Jeoung, N. H., Harris, R. A., Kashiwaya, Y., Veech, R. L., Kitazume, T., and Uyeda, K. (2008) J. Biol. Chem. 283 1670-1678 [DOI] [PubMed] [Google Scholar]
- 44.da Silva Xavier, G., Rutter, G. A., Diraison, F., Andreolas, C., and Leclerc, I. (2006) J. Lipid Res. 47 2482-2491 [DOI] [PubMed] [Google Scholar]
- 45.Meyer, T., and Vinkemeier, U. (2004) Eur. J. Biochem. 271 4606-4612 [DOI] [PubMed] [Google Scholar]
- 46.Pierreux, C. E., Nicolas, F. J., and Hill, C. S. (2000) Mol. Cell Biol. 20 9041-9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmierer, B., Tournier, A. L., Bates, P. A., and Hill, C. S. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6608-6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lodige, I., Marg, A., Wiesner, B., Malecova, B., Oelgeschlager, T., and Vinkemeier, U. (2005) J. Biol. Chem. 280 43087-43099 [DOI] [PubMed] [Google Scholar]
- 49.Collier, J. J., Doan, T.-T. T., Daniels, M. C., Schurr, J. R., Kolls, J. K., and Scott, D. K. (2003) J. Biol. Chem. 278 6588-6595 [DOI] [PubMed] [Google Scholar]
- 50.Li, M. V., Chen, W., Poungvarin, N., Imamura, M., and Chan, L. (2008) Mol. Endocrinol. 22 1658-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.