Abstract
Dysregulation of protein synthesis has been implicated in oncogenesis through a mechanism whereby “weak” mRNAs encoding proteins involved in cell proliferation are strongly translated when the protein synthesis apparatus is activated. Previous work has determined that many cancer cells contain high levels of eIF3h, a protein subunit of translation initiation factor eIF3, and overexpression of eIF3h malignantly transforms immortal NIH-3T3 cells. This is a general feature of eIF3h, as high levels also affect translation, proliferation, and a number of malignant phenotypes of CHO-K1 and HeLa cells and, most significantly, of a primary prostate cell line. Furthermore, overexpressed eIF3h inhibits Myc-dependent induction of apoptosis of primary prostate cells. eIF3h appears to function through translation, as the initial appearance of overexpressed eIF3h in rapidly induced NIH-3T3 cells correlates tightly with the stimulation of protein synthesis and the generation of malignant phenotypes. This oncogenic potential of eIF3h is enhanced by phosphorylation at Ser183. Finally, reduction of eIF3h levels in breast and prostate cancer cell lines by short interfering RNA methods reduces their rates of proliferation and anchorage-independent growth in soft agar. The results provide compelling evidence that high eIF3h levels directly stimulate protein synthesis, resulting in the establishment and maintenance of the malignant state in cells.
The failure to down-regulate protein synthesis leads to an overproduction of oncogenic proteins, resulting in malignant transformation of cells (reviewed in Ref. 1). This view is based on the fact that mRNAs encoding oncogenic proteins are weak competitors for the translational apparatus relative to other mRNAs. Their translation is inefficient when overall protein synthesis is mildly inhibited, although it is greatly stimulated when overall protein synthesis is activated. Because many of the weak oncogenic proteins, such as growth and transcription factors, are involved in promoting cell proliferation, activation of protein synthesis leads to their overproduction and to rapid cell growth, whereas down-regulation impairs their synthesis and enables cells to maintain tight control on the rate of proliferation. Changing the level or specific activity of elements of the translational apparatus, in particular the initiation factors, can alter the overall activity of protein synthesis. For example, overexpression of eIF4E (2, 3) or eIF4G (4) results in a mild stimulation of global protein synthesis and the malignant transformation of immortal cells. Reducing or preventing eIF2 phosphorylation, which normally leads to translational inhibition, interferes with the down-regulation of protein synthesis and also causes malignancy (5, 6). These findings point to the importance of regulating the rate of protein synthesis in maintaining cell growth control.
Recently we reported that the individual overexpression of 5 of the 13 subunits of eIF3 causes the malignant transformation of immortal NIH-3T3 cells (7). eIF3 is the largest and most complex of the mammalian initiation factors and plays an important role in the initiation pathway. The eIF3 complex binds to the solvent side of the 40 S ribosomal subunit (8), yet promotes the binding of methionyl-tRNAi and mRNA (reviewed in Ref. 9), by reaching around the ribosome with its long appendages (10). How some of the eIF3 subunits function as apparent oncogenic proteins is not well established, however. Three of the five mutagenic proteins, eIF3a, eIF3b, and eIF3c, when individually overexpressed, cause a substantial elevation of the whole eIF3 complex, whereas other initiation factor levels are not changed (7). The increase in eIF3 level may therefore be the cause of the stimulation of protein synthesis and the preferential activation of oncogenic mRNAs. In contrast, overexpression of eIF3h or eIF3i does not lead to elevated eIF3, yet the rate of protein synthesis is enhanced, as is the translation of oncogenic mRNAs (7). It remains unclear how these two eIF3 subunits stimulate protein synthesis and promote malignant transformation of immortal cells. One possibility is that eIF3h and eIF3i act indirectly on protein synthesis by stimulating cell proliferation through some other pathway, with the increase in protein synthesis being a consequence of more rapid growth. Our study addresses this issue.
eIF3h has been implicated in a large number of cancerous states. A common chromosomal aberration found in many solid tumors results in an elevated level of the EIF3h gene (11-14), and fluorescence in situ hybridization analysis shows high eIF3h mRNA levels in primary breast cancers, hormone-refractory prostate tumors, and hepatocellular carcinomas (14, 15). In a recent genome-wide association study, Tomlinson et al. (16) identified eIF3h as a causative factor in colorectal cancer. These observations, together with our finding that eIF3h overexpression is oncogenic, indicate that eIF3h may indeed play a crucial role in establishing and maintaining the cancerous state. Here we have quantitated the levels of eIF3h mRNA and protein in a number of cancer cell lines and tissues. We also have created a cell line where eIF3h expression can be rapidly induced, enabling determination of early consequences of the increase in cell level. Finally, we demonstrate using siRNA methods that depletion of eIF3h in cancer cell lines that overexpress eIF3h results in a reduction in their growth rates and in altered malignant phenotypes. The results better establish the view that eIF3h plays a direct role in the etiology and maintenance of cancer through its effect on protein synthesis.
EXPERIMENTAL PROCEDURES
Cell Lines and Tumor Samples—All of the following cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA), including two immortal cell lines (NIH-3T3, CHO-K1), HeLa cells, three prostate cancer cell lines (PC-3, LNCaP, and 22Rv1), and three breast cancer cell lines (MCF-7, MDA436, and SK-Br-3). Cells were cultured under the recommended conditions; all cell culture materials were obtained from Mediatech Inc. (Herndon, VA). The 22Rv1 cell line was derived from an androgen-independent outgrowth of the CWR22 transplantable tumor, which was in turn established from a primary prostate adenocarcinoma (17). Human prostate epithelial cells (PrEC)4 were purchased from Cambrex/Lonza (Walkersville, MD), maintained with the PrEGM BulletKit (Cambrex/Lonza), and passaged with ReagentPack subculture reagents (Cambrex/Lonza), following the manufacturer's instructions. Benign prostate hyperplasia (from transurethral resection), untreated primary prostate carcinomas (from prostatectomies), and locally recurrent hormone-refractory prostate carcinomas (from transurethral resection) were obtained from the University of California, Davis Cancer Center. All samples have been histologically confirmed to contain more than 60% of hyperplastic or cancerous tissue using hematoxylin-eosin staining. 22Rv1 cells were injected subcutaneously into athymic nude mice to initiate tumor growth. Both original tumors at the injection sites and autonomic metastatic tumors were excised and fixed for further study.
Plasmids and Cloning Strategies—pcDNA5/eIF3h, described previously (7), contains an N terminal hemagglutinin (HA) tag upstream of the eIF3h coding region (GenBank™ accession number U54559). Full-length HA-eIF3h was released from pcDNA5/eIF3h by HindIII/XhoI digestion and ligated into the same sites of pcDNA3 to produce pcDNA3/HA-eIF3h. Untagged eIF3h was modified from pAMV/eIF3h by PCR to incorporate HindIII/XhoI sites using the following primers: 5′-AAGCTTATGGCGTCCCGCAAGGAAGGTAC-3′ (sense) and 5′-CTCGAGTTAGTTGTTGTATTCTTGAA-3′ (antisense), and ligated into the same sites of pcDNA3 to produce pcDNA3/eIF3h. The tet-responsive promoter PhCMV-1 of pTRE2 (Clontech) was modified by PCR so that its sequence is released by BglII/HindIII digestion for insertion at the BglII/HindIII sites of pcDNA5/eIF3h. Primers used were 5′-AGATCTCGTATCACGAGGCCCTTTCG-3′ (sense) and 5′-AAGCTTCCGGGTACCGAGCTCGAATT-3′ (antisense). Thus, the constitutive promoter PCMV of pcDNA5/eIF3h is replaced by the inducible promoter PhCMV-1. pMIG/Myc expressing human c-Myc was obtained from JM Bishop (University of California, San Francisco). Site-directed mutagenesis was performed in pcDNA5/eIF3h to insert mutations at Ser183 of eIF3h to encode alanine, glutamate, or aspartate substitutions by using the following primers: S183A sense, 5′-GTAAAGAAAAGGATTTTGCCCCTGAAGCATTGAAA-3′, and antisense, 5′-TTTCAATGCTTCAGGGGCAAAATCCTTTTCTTTAC-3′; S183D sense, 5′-GTAAAGAAAAGGATTTTGACCCTGAAGCATTGAAA-3′, and antisense, 5′-TTTCAATGCTTCAGGGTCAAAATCCTTTTCTTTAC-3′; S183E sense, and 5′-GTAAAGAAAAGGATTTTGAGCCTGAAGCATTGAAA-3′, and antisense, 5′-TTTCAATGCTTCAGGCTCAAAATCCTTTTCTTTAC-3′. All of the above PCR constructs were confirmed by double strand DNA sequencing.
Construction of Constitutive CHO-K1-eIF3h and HeLa-eIF3h Expression Systems—CHO-K1 and HeLa cells were transfected with pcDNA3/HA-eIF3h, pcDNA3/eIF3h, and pcDNA3 by using the calcium phosphate transfection kit (Invitrogen). G418-resistant clones were pooled to make CHO-K1/HA-eIF3h, CHO-K1/eIF3h, CHO-K1/vector, HeLa/HA-eIF3h, HeLa/eIF3h, and HeLa/vector cell lines.
Construction of PrEC Cells Overproducing eIF3h—PrEC cells were transfected with pcDNA3/HA-eIF3h and pcDNA3 by electroporation with the human PrEC nucleofector kit (Amaxa Biosystems, Gaithersburg, MD), following the manufacturer's instructions. G418-resistant clones were pooled to make PrEC/eIF3h and PrEC/vector cells. For expression of c-Myc, PrEC cells were transfected with pMIG/Myc alone or with pcDNA3/eIF3h by electroporation as above, and used for further experiments at 48 h post-transfection.
Construction of an Inducible FlpIn-Tet-On eIF3h Expression System—A pTet-On plasmid (Clontech) expressing a reversible tet-responsive transcriptional activator (rtTA) was inserted into the genome of Flp-In-3T3 host cells (7) to construct an inducible FlpIn-Teton-3T3 host cell line by following the manufacturer's instructions. Stable clones were selected and tested for doxycycline (Dox) inducibility by transient transfection with the luciferase expression plasmid and assay system (Promega, Madison, WI). Subsequently, pcDNA5/PhCMV-1/HA-eIF3h expressing HA-eIF3h and pOG44 (Invitrogen) expressing a Flp recombinase were cotransfected into the FlpIn-Teton-3T3 host cells. A site-specific recombination event integrates the PhCMV-1/eIF3h expression cassette into the genomic FRT site. Similarly, the empty vector pcDNA5/PhCMV-1 was transfected into the host cells to construct a FlpIn-Tet-On vector cell line. From the resulting FlpIn-Teton-eIF3h cell lines, 30 were selected and tested for induction of HA-eIF3h by Dox. Six clones representing mild, moderate, and robust expression were studied.
Construction of FlpIn-3T3 Cells Expressing eIF3h Mutants—FlpIn-eIF3h(S183A), FlpIn-eIF3h(S183D), and FlpIn-eIF3h(S183E) cell lines were constructed by cotransfecting the FlpIn-3T3 host cells (7) with pOG44 and pcDNA5/eIF3h(S183A), pcDNA5/eIF3h(S183D), pcDNA5/eIF3h(S183E), and screening for hygromycin resistance, according to the manufacturer's instructions (FlpIn system, Invitrogen). For coexpression of eIF3h mutants and c-Myc, pMIG/Myc was transfected with the FuGENE 6 transfection reagent (Roche Applied Science) into the above cells and used for further experiments at 48 h post-transfection.
Immunoprecipitation and Immunoblotting—For immunoprecipitation, cells were washed with phosphate-buffered saline and lysed in RIPA buffer. Immunoprecipitations were prepared by incubating the lysates with anti-HA-agarose (Sigma) and analyzed by SDS-PAGE. For immunoblotting, proteins in SDS gels were transferred to polyvinylidene difluoride membranes and subjected to immunoblotting with various antibodies as indicated in the figures, followed by incubation with the corresponding secondary antibodies (Sigma). Signals were detected by incubating the blots with a 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (Sigma) solution. The blots were scanned with a photoscanner and quantified with Scion Image software. Antibodies used were as follows: monoclonal anti-eIF3a (J. T. Parsons, University of Virginia); anti-eIF3b, -eIF3f, and -eIF3h (Santa Cruz Biotechnology); anti-eIF3c (gift of D. R. Scoles, UCLA); anti-eIF3j (gift of C. S. Fraser, University of California, Berkeley); anti-actin and anti-HA (Sigma); and anti-poly(ADP-ribose) polymerase (anti-PARP) and anti-Myc (Cell Signaling Technology).
Replicative Life Span and Senescence-associated β-Galactosidase Staining—Population doublings (PDs) per passage were determined by log2 (number of cells at time of subculture divided by number of cells plated). Cumulative PDs were calculated and plotted against days after transfection. When the cells stopped dividing and were not passaged for about 2 weeks, they were regarded as growth-arrested. Senescence-associated β-galactosidase activity was detected using the β-galactosidase staining kit (Invitrogen).
eIF3h Knockdown by siRNA—Pre-designed eIF3h siRNA (sieIF3h) and negative control siRNA (siNC) were purchased from Ambion (Austin, TX). Transient transfections were performed with siPORT NeoFX (Ambion) following the manufacturer's instructions. For growth curves and immunoblot analysis, 2 μl of siPORT NeoFX was used for 2 × 104 cells/well in 24-well plates. For soft agar assays, 5 μl of siPORT NeoFX was used for 2500 cells/well in 6-well plates. The siRNAs were added at a final concentration of 100 nm. The media were changed every 24 h with repeated additions of the siRNAs.
General Protein Synthesis Rate—1 × 105 cells/well were plated in 12-well plates in triplicate and incubated for 24 h. The cells were pulse-labeled for 30 min with 20 mCi/ml [35S]methionine (>1000 Ci/mmol), washed, and trypsinized. The cells were counted and precipitated with 10% trichloroacetic acid. The acid-insoluble material was collected on a filter by rapid filtration, and radioactivity was determined by scintillation counting.
Polysome Profile Analysis—The cells freshly fed with serum were lysed with buffer containing 20 mm HEPES-KOH (pH 7.5), 100 mm KCl, 10 mm MgCl2, 1 mm dithiothreitol, 0.25% Nonidet P-40, 1× protease inhibitor mixture solution, and 100 μg/ml cycloheximide. The lysates were clarified by centrifugation, and an aliquot (10 A260 units) was layered on a 15-45% sucrose gradient. After centrifugation at 4 °C in a Beckman SW60 rotor for 2 h at 38,000 rpm, gradient fractions were scanned for absorbance at 254 nm with a UV monitor and collected into 10 fractions with an ISCO gradient fractionator.
Quantitative Real Time RT-PCR—Total RNA from the cell lines was extracted with the RNeasy mini kit (Qiagen, Valencia, CA). Total RNA from the gradient fractions was extracted with the TRIzol reagent (Invitrogen) and adjusted to the same volume. cDNA synthesis was performed with the SuperScript III cDNA synthesis kit (Invitrogen). Specific primers for mouse GAPDH, cyclin D1, FGF2, and ornithine decarboxylase (Applied Biosystems, Foster City, CA) and specific primers for human GAPDH, cyclin D1, FGF2, ornithine decarboxylase, and eIF3h (Primer Bank, Xiaowei Wang, Massachusetts General Hospital, Boston) were used. Real time PCR analysis was performed with an automated sequence detection system (MyiQ, Bio-Rad). eIF3h expression in the cell lines was normalized against the expression of the housekeeping gene for GAPDH. The results of the gradient fractions are shown as the percentage of total signal for each fraction.
Statistical Analysis—The statistical significance of the data is presented as means ± S.E. as analyzed by Student's t test. A p value below 0.05 is considered to be significant.
RESULTS
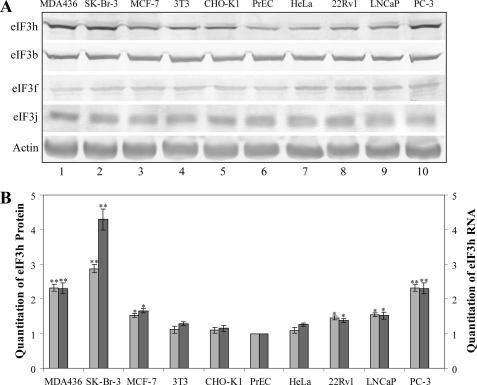
eIF3h Is Up-regulated in Breast and Prostate Cancers—Although enhanced levels of eIF3h transcripts have been reported for a number of cancer cell lines, there has been no measure of the protein itself. We therefore investigated eIF3h protein abundance by Western immunoblotting in three breast cancer (MDA436, SK-Br-3, and MCF-7), three prostate cancer (PC-3, LNCap, and 22Rv1), one transformed (HeLa), and two immortal (NIH-3T3 and CHO-K1) cell lines, as well as in primary prostate cells (PrEC). The immunoblots indicate 1.4-3-fold higher eIF3h levels in the prostate and breast cancer cell lines compared with primary PrEC cells, whereas little or no increase is seen with the other cell lines (Fig. 1A). The levels of three other eIF3 subunits do not vary in any of these cell lines (Fig. 1A), indicating that the ratio of eIF3h to the eIF3 complex is enhanced in the overproducing cell lines. The eIF3h immunoblot bands were quantitated and plotted as a bar graph, together with eIF3h mRNA levels determined by real time RT-PCR (Fig. 1B). A close correspondence of mRNA and protein levels is seen, suggesting that the overexpression of eIF3h does not involve a change in the translational efficiency of its mRNA.
FIGURE 1.
eIF3h overexpression in human breast and prostate cancer cell lines. A, Western blot analysis of lysates from breast cancer cell lines (MDA436, SK-Br-3, and MCF-7), immortal cell lines (3T3 and CHO-K1), primary culture (PrEC), HeLa cells, and prostate cancer cell lines (PC-3, LNCaP, and 22Rv1). The blots were probed with the antisera identified on the left; the anti-actin blot is a loading control. B, immunoblots obtained from A were quantitated and graphed as light gray bars. The mRNAs from the above cells were subjected to real time RT-PCR with eIF3h-specific primers, and quantitation of eIF3h RNA is graphed as dark bars. The results are expressed relative to that determined in PrEC cells. The values are the average of three independent experiments; **, p < 0.01; *, p < 0.05 by Student's t test.
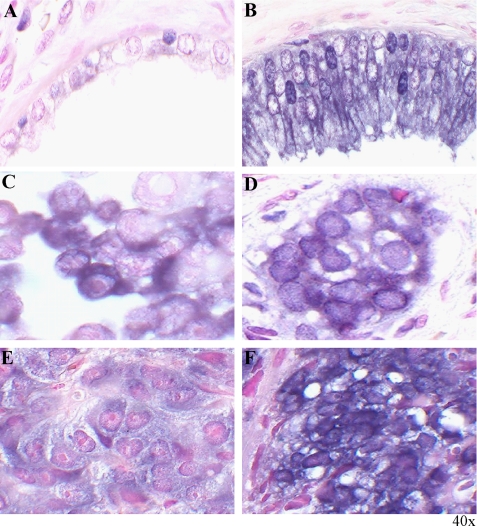
To identify the types of cells overexpressing eIF3h in prostate cancer, we performed RNA in situ hybridization with an eIF3h cRNA probe on normal prostate gland and prostate carcinoma (CaP) tissues. The epithelial cells from prostate carcinomas display strong cytoplasmic staining of eIF3h mRNA (Fig. 2B), whereas the eIF3h signal is below the detection limit in normal prostate controls (Fig. 2A). Cytoplasmic eIF3h mRNA signals are also observed in PC-3 cells (Fig. 2C) and in the invasive prostate cancer cell lesions called “cancer nests” (Fig. 2D). The strong signals generated from this small sample of human CaP tissue specimens also indicate that high eIF3h mRNA levels correlate with higher Gleason scores of prostate cancer (results not shown). Overexpression of eIF3h mRNA was observed in CaP xenografts by in situ hybridization analysis, and even higher levels were seen in androgen-refractory CWR22R compared with androgen-sensitive CWR22 (Fig. 2, E and F). These analyses suggest that overexpression of eIF3h is associated with the more aggressive malignant potential of human prostate cancers. They also suggest that eIF3h may be involved in the regulation of prostate cell growth.
FIGURE 2.
In situ hybridization analysis of eIF3h mRNAs in human prostate carcinoma specimens and CWR22 xenografts. A digoxigenin-labeled antisense eIF3h RNA (cRNA) probe was made from human eIF3h cDNA with the digoxigenin RNA labeling kit (Roche Applied Science). Formalin-fixed, paraffin-embedded specimens (4 μm thick) were deparaffinized, rehydrated, and hybridized to 200 ng/ml of eIF3h probe at 55 °C overnight. The slides were then incubated with alkaline phosphatase-conjugated anti-digoxigenin, stained with 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium solution, and counterstained with Nuclear Fast Red as described (14, 38, 39). A, normal prostate gland tissue. B, malignantly transformed prostate epithelial tissue. C, PC-3 cells. D, cancer cell nest formed by invasive prostate carcinoma. E, human prostate cancer xenograft CWR22 tumors grown in mice androgen-sensitive, nonmalignant. F, human prostate cancer xenograft CWR22R androgen-resistant, malignant.
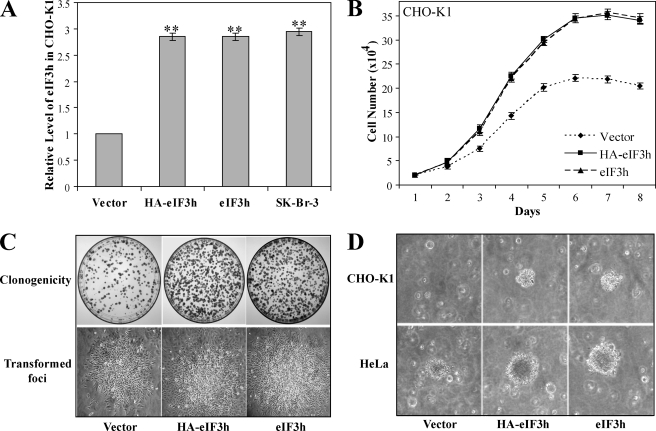
Overexpression of eIF3h Affects the Growth Characteristics of a Variety of Cell Types—We have shown recently that overexpression of eIF3h in a stable transfectant of immortal NIH-3T3 cells results in their malignant transformation (7). However, the oncogenic effects of eIF3 subunit overexpression may require the particular genetic background provided by the NIH-3T3 cell line. To strengthen the view that eIF3h is oncogenic, we wished to determine whether or not overexpression causes malignancy in another immortal cell line, Chinese hamster ovary cells (CHO-K1). CHO-K1 cells were stably transfected with pcDNA3 plasmids expressing HA-tagged eIF3h (HA-eIF3h), untagged eIF3h (eIF3h), or nothing (empty vector control), and neomycin-resistant clones were pooled as described under “Experimental Procedures.” Compared with vector control cells, the CHO-K1/HA-eIF3h and CHO-K1/eIF3h cell lines overexpress eIF3h about 3-fold (Fig. 3A and supplemental Fig. S1A), comparable with the overexpression seen in the breast cancer cell line, SK-Br-3. They grow faster (Fig. 3B), show about 2.2-fold greater clonogenicity (Fig. 3C and supplemental Fig. S1B), and exhibit growth in soft agar (Fig. 3D). The 13-fold higher efficiency of colony formation in soft agar compared with vector control cells (supplemental Fig. S1C), together with the other characteristics, indicate that the CHO-K1 cells have been malignantly transformed. These results are strikingly similar to those obtained by overexpression of eIF3h in NIH-3T3 cells (7), suggesting that malignant transformation by high levels of eIF3h is a general response of immortal cells. Furthermore, the near-identical phenotype of CHO-K1 cells that overexpress either the HA-tagged or untagged eIF3h means that the HA tag does not contribute to the oncogenesis.
FIGURE 3.
Overexpression of eIF3h in CHO-K1 and HeLa cells. A, cellular levels of eIF3h in eIF3h-transfected CHO-K1 cells. Band intensities from the immunoblots (shown in supplemental Fig. S1A) of transfected (HA-eIF3h and eIF3h), control (empty vector), and SK-Br-3 cell extracts were measured and normalized to that of the empty vector control cells. The values shown are means ± S.D. of three independent experiments. **, p < 0.01. B, growth curves. eIF3h-transfected CHO-K1 (HA-eIF3h, squares; eIF3h, triangles) and vector control cells (diamonds) were seeded into 24-well plates at a density of 2 × 104 cells/well, fed, and counted every 24 h with a hemocytometer. C, clonogenicity. Transfected CHO-K1 cells (1000) were seeded in 10-cm plates, incubated for 2-3 weeks to form colonies, and photographed (upper row). The number of colonies was counted and reported in supplemental Fig. S1B. One of the transformed foci for each cell type is shown at ×20 magnification (lower row). D, colonies growing in soft agar. Anchorage-independent growth was shown by suspending 2500 cells in 0.5 ml of 0.4% top agar and overlaying onto 0.8% basal agar in 6-well plates. Cells were fed twice weekly for 3 weeks, and a typical colony was photographed. Transfected CHO-K1 cells (upper row); transfected HeLa cells (lower row). The number of transformed cells for each cell type was counted with a cell transformation detection assay kit (Chemicon), and is reported in supplemental Fig. S1C.
Similar experiments were performed with HeLa cells, which normally exhibit low eIF3h levels (Fig. 1A). In this case, 3-fold enhanced eIF3h levels (supplemental Fig. S1D) cause similar, although somewhat less robust, effects compared with those in CHO-K1 and NIH-3T3 cells as follows: ∼20% reduced doubling times and 1.3-fold enhanced saturation density (supplemental Fig. S1E); 1.9-fold higher clonogenicity (supplemental Fig. S1, B and F); and a more refractile and rounded-up cell morphology (supplemental Fig. S1F). These phenotypes, together with the fact that the efficiency of anchorage-independent growth in soft agar is 1.9-fold greater (Fig. 3D and supplemental Fig. S1C), indicate that stable overexpression of eIF3h in HeLa cells also stimulates cell proliferation.
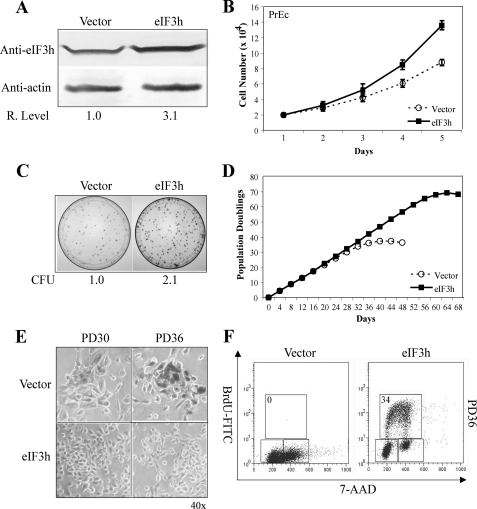
To study the effects of eIF3h overexpression on normal cells, primary prostate cells (PrEC) stably overexpressing eIF3h (PrEC/eIF3h) or vector (PrEC/vector) were generated as described under “Experimental Procedures.” We observed a 3.1-fold increase in total eIF3h protein, compared with endogenous eIF3h levels (Fig. 4A). PrEC/eIF3h cells grow faster than PrEC/vector cells during exponential growth (Fig. 4B) and exhibit about 2.1-fold higher clonogenic ability (Fig. 4C). Parental PrEC and empty vector cells have a finite life span in culture of ∼35 PDs, whereas overexpression of eIF3h results in an extension of proliferative activity up to ∼70 PDs (Fig. 4D). The control cells adopt a flat enlarged morphology and about 90% stain positive at ∼36 PD for senescence-associated β-galactosidase. In contrast, PrEC/eIF3h cells exhibit a normal epithelial phenotype and only 2-5% senescence (Fig. 4E). BrdUrd incorporation and cell cycle analysis confirm that the control cells cease proliferation at subconfluent densities at ∼ 36 PD, whereas PrEC/eIF3h cells reveal a large population of BrdUrd-positive cells (Fig. 4F) at the same PD. The experiments above show that high levels of eIF3h affect the growth properties of all three cell types: transformed, immortal, and primary cells.
FIGURE 4.
Overexpression of eIF3h increases the proliferation potential of PrEC cells. A, cell extracts of PrEC/eIF3h and PrEC/vector cells analyzed by immunoblots probed with anti-eIF3h and anti-actin as described under “Experimental Procedures.” Quantitations of eIF3h levels in PrEC/eIF3h relative to vector cells (assigned the value of 1) are shown below the blots. B, growth curves of PrEC/eIF3h (squares) and PrEC/vector (circles). C, clonogenicity of eIF3h- and vector-expressing PrEC cells. Colony-forming units of PrEC/eIF3h relative to control cells (normalized to 1) are shown at the bottom of the panel. D, PD of PrEC/eIF3h (squares) and PrEC/vector (circles) are plotted against time (days). E, senescence-associated β-galactosidase (SA-β-gal) staining of PrEC/eIF3h and vector cells at PD ∼ 30 and PD ∼ 36. Photographs are at the same ×40 magnification. F, cell cycle distribution of PrEC/eIF3h and control cells was measured by BrdUrd incorporation and DNA-content (7-aminoactinomycin D (7-AAD)) flow cytometry analysis at PD ∼ 36 as described previously (7). The upper box identifies cells incorporating BrdUrd (∼S phase), the lower left box identifies G0/G1 cells, and the lower right box displays G2/M cells. The percentage of S phase cells is indicated in the upper left corner of the upper box. The results in all panels are representative of three independent experiments.
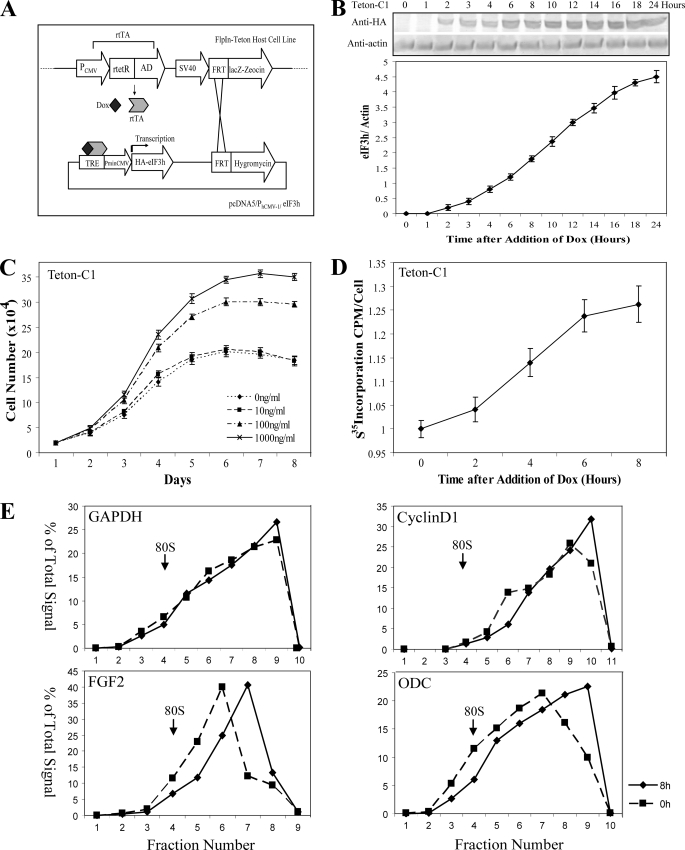
Enhanced Protein Synthesis Rates Occur Soon after Rapid Induction of eIF3h Expression—The correlation of high eIF3h levels in cancer cells and the effects of overexpression on cell growth suggest, but do not prove, that eIF3h is oncogenic. Instead, high eIF3h levels may be a consequence rather than a cause of the malignant state. To address this issue, we constructed and characterized a number of stable NIH-3T3 cell lines where eIF3h overexpression can be rapidly induced. If the effects on protein synthesis and growth properties occur at the same time that higher levels of eIF3h are first produced, this would lend support for the view that eIF3h plays a direct role in oncogenesis. The inducible cell lines (called Teton-C1, Teton-C2, etc.) were generated as described in Fig. 5A and “Experimental Procedures.” Six clones were studied that show mild, moderate, and robust overexpression of eIF3h from the PhCMV-1 promoter in the presence of the inducer Dox (supplemental Fig. S2A). Half-maximal levels of eIF3h are seen with Teton-C1 cells after 10 h of induction (Fig. 5B). A Dox dose-response curve involving induction for 24 h was determined (supplemental Fig. S2B), with nearly full induction achieved at 100 ng/ml. Following long term Dox treatment, the cells expressing higher levels of eIF3h grow faster (Fig. 5C and supplemental Fig. S2C) and exhibit a malignant phenotype (supplemental Fig. S2, D and E), and therefore are suitable for the examination of early events in their transformation.
FIGURE 5.
Effects of rapid induction of eIF3h expression in NIH-3T3 cells. A, schematic for the construction of an inducible FlpIn-Teton-eIF3h expression system. The FlpIn-Teton-3T3 host cells contain a Flp recombination target (FRT) site and express the reversible tet-responsive transcriptional activator (rtTA). The inducible eIF3h expression cassette (or the empty vector) is integrated into the FRT site through a site-specific recombination event. After addition of doxycycline (Dox), rtTA binds to the upstream tet-responsive element (40) and enables the minimal cytomegalovirus promoter (PminCMV) to express HA-tagged eIF3h. B, time course of expression of HA-eIF3h in Teton-C1 cells induced with 1 μg/ml Dox. Cell lysates were prepared at the indicated times and analyzed by immunoblotting with anti-HA and anti-actin antibodies (upper panel). The HA-eIF3h/actin ratio at each time point was calculated and graphed below. C, growth curves of Teton-C1 cells maintained in 0, 10, 100, and 1000 ng/ml of Dox were determined as described in the legend to Fig. 3B. D, rate of [35S]methionine incorporation into protein in Teton-C1 cells measured at the indicated times after treatment with 1 μg/ml Dox. Incorporation at 0 Dox is 1.22 cpm per cell, normalized to 1. The data in B-D are means ± S.D. of three independent experiments, each done in triplicate. E, Teton-C1 cells treated0h(squares) and 8 h(diamonds) with 1 μg/ml Dox were lysed and subjected to sucrose gradient centrifugation as described previously (7). Ten gradient fractions were collected, and total RNA was isolated from each fraction and analyzed by Taqman real time RT-PCR using specific primer and probe sets as described previously (7). Quantitation of mRNA in each gradient fraction is reported as the percentage of the total amount present in all fractions. Sedimentation is from left to right; the sedimentation position of 80 S ribosomes is indicated by an arrow.
Because high levels of eIF3h stimulate protein synthesis (7), we measured [35S]methionine incorporation into protein at different times following the addition of Dox to Teton-C1 cells. The translation rate begins to increase after only2hofDox treatment, when eIF3h overproduction first begins to appear, and is stimulated by ∼25% by 6 h (Fig. 5D), approaching the stimulation (35%) seen previously with long term overexpression of eIF3h (7). An apparent plateau in the translation rate is reached in about 8 h, whereas eIF3h levels continuously increase up to about 24 h. This suggests that the amount of eIF3h required to stimulate protein synthesis may be only about a third of that generated at full overexpression. A Dox dose-response curve for protein synthesis following8hof induction was determined (supplemental Fig. S2F) and found to resemble that for eIF3h induction (supplemental Fig. S2B), with ∼20% stimulation of protein synthesis at 100 ng/ml. The increase in protein synthesis rate also is seen in polysome profiles (supplemental Fig. S3A), where the polysome to monosome ratio (P/M) increases from 0.87 with no Dox treatment to 0.97 after 4 h and 1.12 after 8 h of Dox treatment. The shift of ribosomes from monosomes into polysomes is consistent with the [35S]methionine incorporation data. The immediate response of protein synthesis to the rise in eIF3h levels implies a direct effect of the protein on the process of translation. This effect is amplified in the translational efficiencies of weak oncogenic mRNAs such as cyclin D1, FGF2, and ornithine decarboxylase mRNAs determined by polysome profile analysis after 8 h of Dox induction (Fig. 5E). A distinct shift to heavier polysomes is seen for the FGF2 and ornithine decarboxylase, whereas the cyclin D1 mRNA shifts more modestly and the control GAPDH mRNA does not shift at all. Thus, eIF3h stimulates the translation of oncogenic mRNAs at an early time following its induction. We propose that the translational stimulation, in turn, results in the overproduction of proteins that support cell proliferation, which may be the actual cause of the malignancy.
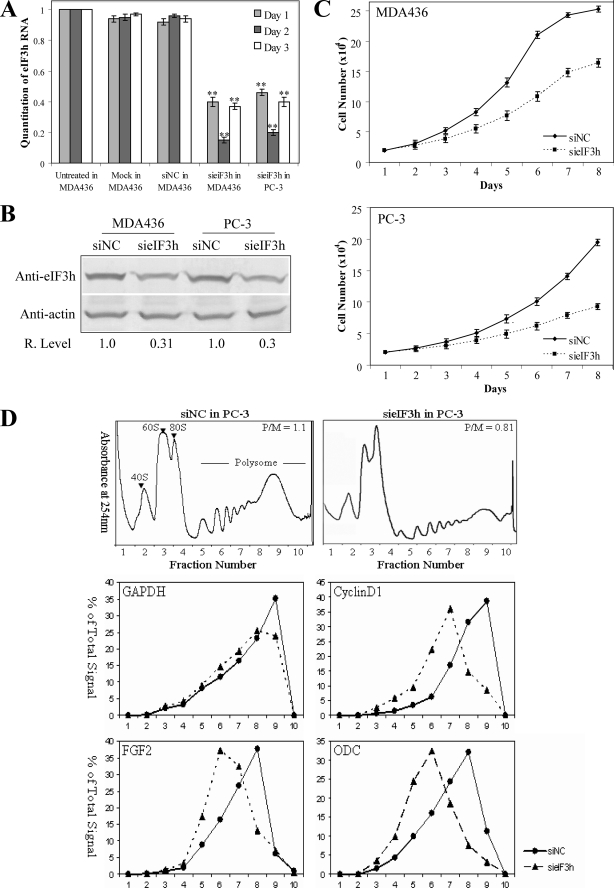
Down-regulation of eIF3h with siRNA Reduces the Malignant Phenotype of MDA436 and PC-3 Cells—If the high level of eIF3h in MDA436 and PC-3 cells (Fig. 1A) contributes importantly to the malignant phenotype of these cells, lowering its level might reduce or reverse the malignant phenotype. The cells were transfected with a duplex synthetic siRNA directed against eIF3h (sieIF3h) as described under “Experimental Procedures.” Two controls were run as follows: transfection of a negative control duplex RNA (siNC) with a scrambled sequence that has no known homology to human, mouse, or rat sequences; and a mock transfection to determine the possible cytotoxic effects of the reagents. eIF3h mRNA levels were measured by real time RT-PCR over 3 consecutive days following transfection. A substantial reduction of mRNA level is seen after 1 day for both cell lines, with maximal silencing to levels under 15% seen after 2 days (Fig. 6A). Because mRNA levels begin to increase at 3 days post-transfection, cells were routinely fed siRNA daily. As expected, at 48 h post-transfection the reduced mRNA levels result in lower eIF3h protein levels, to ∼30% of the siNC control (Fig. 6B). The growth rate of MDA436 and PC-3 cells fed (daily) with sieIF3h is reduced compared with those with siNC (Fig. 6C), with doubling times increased to 34.2 h from 30.1 h for the control. Reduced eIF3h levels also affect anchorage-independent growth, as MDA436 or PC-3 cells plated at 48 h post-transfection show an ∼2.7-fold decrease in colony formation after 10 days (supplemental Fig. S3C). The effects of eIF3h silencing support the view that eIF3h plays an important role in maintaining the malignant phenotype of the MDA436 and PC-3 cells, although we cannot rule out that reduced eIF3h simply makes cells grow less efficiently.
FIGURE 6.
Induced depletion of eIF3h in MDA436 and PC-3 cells. A, siRNA-directed reduction of eIF3h mRNA levels in MDA436 and PC-3 cells. Cells were transected with a duplex RNA designed to deplete eIF3h mRNA (sieIF3h) or with a negative siRNA control (siNC) or were mock-transfected, and eIF3h mRNA levels in cell lysates were analyzed by real time RT-PCR as described under “Experimental Procedures.” The panel shows relative eIF3h mRNA levels determined on 3 consecutive days after transfection. **, p < 0.01. B, eIF3h levels relative to actin were determined by immunoblotting of the lysates of the cells described in A on day 2. C, growth curves for MDA436 and PC-3 cells transfected with sieIF3h (squares) or siNC control (diamonds) were determined as described in the legend to Fig. 3B. The data reported in A and C are from three independent experiments, each done in triplicate. D, representative polysome profiles of PC-3 cells treated with siNC and sieIF3h RNA (upper panels). Ten fractions (numbered below the figure) were collected for analysis, with fraction 1 representing the top of the gradient. The sedimentation positions of 40 S, 60 S, and 80 Sribosomes and polysomes are labeled. RNA was extracted from individual fractions and analyzed by Taqman real time RT-PCR using specific primers and probes for GAPDH, cyclin D1, FGF2, and ornithine decarboxylase (middle and lower panels). mRNA quantitations and reporting are described under “Experimental Procedures.”
Do the reduced eIF3h levels affect protein synthesis? The rate of protein synthesis was determined at 48 h post-transfection, when eIF3h protein levels are reduced to ∼30% of the siNC control. Such reduction results in a somewhat modest inhibition of protein synthesis, with only 23 and 32% inhibition seen with the transfected MDA436 and PC-3 cells, respectively (supplemental Fig. S3D). This result suggests that the level of eIF3h is not strictly limiting for protein synthesis in normal cells. The inhibition of protein synthesis by sieIF3h in PC-3 cells also is seen in their polysome profiles (Fig. 6D, upper panels), where the amount of polysomes clearly is reduced. We also measured the effect of eIF3h depletion on the translational efficiencies of cyclin D1, FGF2, and ornithine decarboxylase mRNAs. Transfection with sieIF3h RNA clearly reduces the polysome size of all three mRNAs, as evidence by a shift of the polysome peak to smaller polysomes, although it has little or no effect on the nononcogenic mRNA encoding GAPDH (Fig. 6D, lower panels). The specific sensitivity of the oncogenic mRNAs to eIF3h levels mirrors that observed when eIF3h is overproduced.
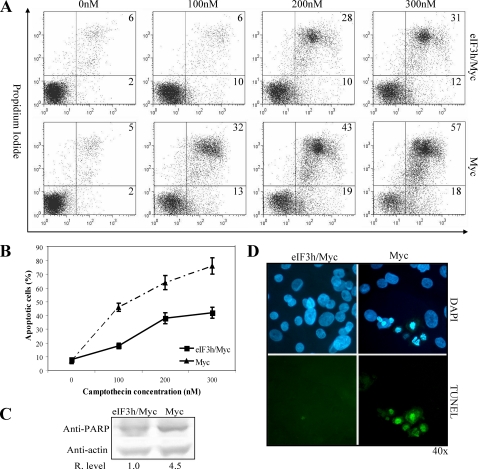
eIF3h Rescues PrEC Cells from Myc-dependent Apoptosis—eIF3h and c-Myc are coamplified in about one-third of locally recurrent hormone-refractory prostate carcinomas (15). Primary PrEC cells were transiently transfected by electroporation to express Myc alone (PrEC/Myc) or Myc and eIF3h (PrEC/eIF3h/Myc) simultaneously. Western blots probed with anti-Myc antibodies indicate a 3-4-fold increase in Myc in PrEC/Myc and PrEC/Myc/eIF3h cells (supplemental Fig. S4A). The above cells were treated with different concentrations of camptothecin for 24 h to induce apoptosis, which was analyzed by flow cytometry with annexin-V-fluorescein isothiocyanate and propidium iodide. PrEC/eIF3h/Myc cells are about 2-fold more resistant to induction of apoptosis at the various concentrations of camptothecin (Fig. 7, A and B). Similar results are seen when apoptosis is measured by PARP cleavage and TUNEL staining of apoptotic bodies containing condensed chromatin and nuclear fragments (Fig. 7, C and D, and supplemental Fig. S4B). These data demonstrate that eIF3h substantially inhibits Myc-dependent apoptosis in response to camptothecin treatment. eIF3h rescue of these cells from Myc-dependent apoptosis appears to be similar to that by eIF4E, where eIF4E overexpression inhibits cytochrome c release from mitochondria through its translational activation of bcl-x (18). It will be interesting to determine whether eIF3h functions similarly in regulating apoptosis.
FIGURE 7.
Ectopic expression of eIF3h rescues PrEC/Myc cells from apoptosis. A, PrEC/Myc and PrEC/eIF3h/Myc cells were incubated with the indicated concentrations of camptothecin for 24 h. Flow cytometric analyses with annexin-V-fluorescein isothiocyanate and propidium iodide detect early apoptotic cells (lower right panel) and late apoptotic cells (upper right panel). The percent of cells in each panel is indicated. B, quantitation of apoptotic cells (early + late) in PrEC/Myc (triangles) and PrEC/eIF3h/Myc (squares) cells obtained from A. C, immunoblot analysis of PARP cleavage after treatment of PrEC/Myc and PrEC/eIF3h/Myc cells with 300 nm camptothecin for 24 h. The relative PARP band intensities are shown below the blots. D, TUNEL analysis of PrEC/Myc and PrEC/eIF3h/Myc cells after treatment with 300 nm camptothecin for 24 h. 4′,6-Diamidino-2-phenylindole (DAPI) staining cells (blue) are shown in the upper panel, and TUNEL-positive staining cells (green) in the same visual field are shown in the lower panel. The results in the entire figure are representative of three independent experiments.
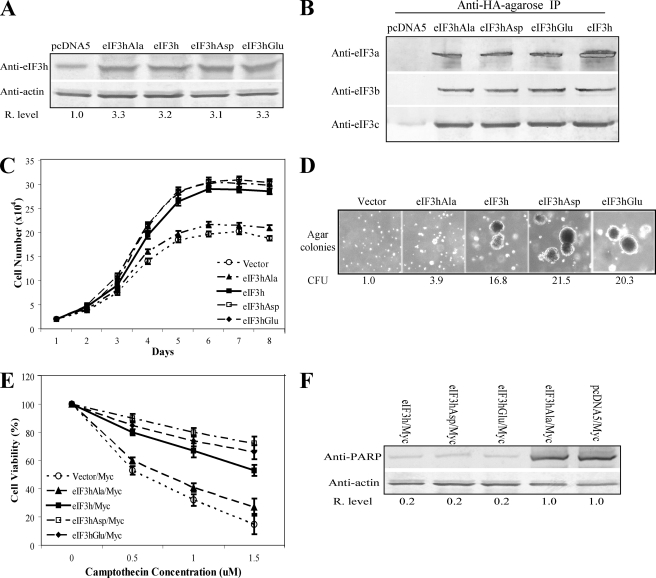
Phosphorylated eIF3h Is the Activated Isoform of eIF3h—eIF3h is phosphorylated in vivo at residue Ser183 in HeLa cells (19). Many initiation factors are phosphoproteins, and phosphorylation has been shown to function as a general mechanism for the regulation of translation initiation (20, 21). To study the role of eIF3h phosphorylation at Ser183 in cell growth control, three mutant forms of eIF3h were made by Ala, Asp, or Glu substitutions at Ser183 as follows: eIF3h(S183A), which cannot be phosphorylated; and eIF3h(S183D) and eIF3h(S183E), which mimic phosphorylation. The FlpIn-3T3 system described previously (7) was used to express vector control, wild type eIF3h, and the three eIF3h mutant forms in NIH-3T3 cells. Because all expression cassettes are regulated by the same promoter and are inserted into the same recombination site in the genome, equal amounts of eIF3h and eIF3h mutants are produced (Fig. 8A). All forms of eIF3h are incorporated into the eIF3 complex, as shown in Fig. 8B by co-immunoprecipitation of three other eIF3 subunits (a, b, and c). The stably transfected cells expressing eIF3h(S183D), eIF3h(S183E), and wild type eIF3h grow faster during exponential growth phase and show greater survival potential after reaching confluence in comparison with those expressing the vector control and eIF3h(S183A) (Fig. 8C). The former group of cells produce 16.8-21.5-fold more colonies in soft agar than the vector control cells, whereas eIF3h(S183A)-expressing cells display only a 3.9-fold increase (Fig. 8D and supplemental Fig. S4C). The stronger effects on cell growth caused by overexpression of eIF3h(S183D) and eIF3h(S183E) relative to eIF3h(S183A) strongly suggests that phosphorylation of eIF3h at Ser183 enhances the activity of the protein. This difference in activity is not because of different levels of expression or incorporation into the eIF3 complex but rather reflects an altered activity of the subunit, presumably by phosphorylation at Ser183.
FIGURE 8.
Phosphorylated eIF3h is likely the activated isoform of eIF3h. A, cell extracts of 3T3/pcDNA5, 3T3/eIF3h(S183A), 3T3/eIF3h, 3T3/eIF3h(S183D), and 3T3/eIF3h(S183E) cells were analyzed by immunoblots probed with anti-eIF3h and anti-actin antibodies. Relative eIF3h band intensities are shown below the blots. B, 3T3 cells as in A were lysed and subjected to immunoprecipitation (IP) with anti-HA resin as described under “Experimental Procedures.” The immunoprecipitations were analyzed by SDS-PAGE and immunoblotting with antiserum specific for eIF3a, eIF3b, and eIF3c. C, growth curves of the indicated cell lines, determined as described in the legend to Fig. 3B. D, colonies formed in soft agar with the same cell lines. Colony forming units (CFU) measured as described in the legend to Fig. 3D are shown below. E, indicated cell lines were transfected with a vector expressing Myc; 48 h post-infection the cells were treated with the indicated concentrations of camptothecin for 24 h. Cell viability was measured as described in Ref. 7. F, immunoblot analysis with anti-PARP after treatment of the Myc-transfected cells with 1.5 μm camptothecin for 24 h. The relative PARP levels are shown below the blots. The results shown above are a representative of three independent experiments.
The vector expressing Myc was used to transfect the NIH-3T3 cell lines overexpressing the various mutant forms of eIF3h. At ∼48 h post-transfection, different concentrations of camptothecin were used to induce apoptosis. As expected, apoptosis is reduced in the cell lines expressing wild type eIF3h, eIF3h(S183D), and eIF3h(S183E), although it is not in cells expressing eIF3h(S183A) and the vector control (Fig. 8E). Consistently, more 85-kDa PARP fragments are produced in the eIF3h(S183A) and vector control cells after camptothecin treatment (Fig. 8F). Taken together, the results indicate that phosphorylation of eIF3h at Ser183 inhibits camptothecin-induced apoptosis and enhances the oncogenic potential of this eIF3 subunit.
DISCUSSION
A broad array of data and analyses support the view that eIF3h is involved in cancer. The gene encoding eIF3h is located in the long arm of chromosome 8 (specifically, 8q23-24), a region that is frequently amplified in many tumor types (22-24). For example, gain at 8q has been detected in bladder (25), endometrial (26), ovarian (27), and hepatocellular tumors (28). Although other genes, such as MYC and TRPSI, are also associated with the 8q23-24 region, only the EIF3h gene is consistently overexpressed in breast, prostate, and hepatocellular cancers (11-15). These correlations of high eIF3h mRNA levels with malignancy clearly implicate eIF3h in cancer.
A role for this initiation factor in growth control was demonstrated more convincingly by our finding that eIF3h overexpression leads to the malignant transformation of immortal NIH-3T3 cells. Here we have strengthened this finding by showing that the oncogenic consequences of eIF3h overexpression are more general, applying not only to NIH-3T3 cells but also to CHO-K1 and HeLa cells. Most compelling is the observation that when eIF3h is overexpressed in normal primary prostate (PrEC) cells, the cells exhibit more rapid proliferation and increased clonogenicity. Furthermore, these cells continue to proliferate at subconfluent densities, well beyond the 36 doublings associated with the senescence of nonoverexpressing PrEC cells. In addition, eIF3h overexpression rescues PrEC cells from Myc-dependent apoptosis. Overexpression of eIF4E also rescues cells from Myc-dependent apoptosis (18) that is dependent on cyclin D1 (29). Because both eIF3h and eIF4E overexpression stimulate protein synthesis and the translation of cyclin D1 mRNA, it is tempting to speculate that these two initiation factor proteins function similarly in reducing apoptosis.
Because an important role in oncogenesis has been established for eIF3h, we asked if its overexpression is required to maintain the malignant phenotype. Using siRNA knockdown of eIF3h expression in a breast cancer (MDA436) and a prostate (PC3) cell line, we show that lowering the level of eIF3h affects their malignant phenotypes. Both their rates of proliferation and their anchorage-independent growth in soft agar are reduced. The results support the conclusion that high eIF3h levels help to establish and maintain the malignant state, although other interpretations are possible.
How does a high level of eIF3h affect cell growth? Our earlier studies indicated that elevated levels of eIF3h result in stimulation of the rate of protein synthesis (7). However, the correlation of high eIF3h and protein synthesis rate does not demonstrate convincingly that eIF3h plays a causal role, as the higher protein synthesis rate may have been a consequence of the higher rate of cell proliferation. To address this issue, we investigated the effect of induced overexpression of eIF3h on protein synthesis at early times. The tight correlation of rising eIF3h levels and stimulation of protein synthesis indicates that eIF3h plays a direct role in regulating translation. Therefore, eIF3h is similar to eIF4E and eIF4G, initiation factor proteins whose levels also affect protein synthesis and the malignant state of cells (2-4).
How eIF3h affects the rate of protein synthesis is not clear, however. eIF3h is absent in Saccharomyces cerevisiae and indeed is dispensable in Arabidopsis thaliana, although its level in plant cells affects the translation of specific mRNAs (30). In contrast, eIF3h was shown to be present in a minimal eIF3 complex capable of functioning in the in vitro binding of mRNA to ribosomes (31). Furthermore, siRNA screening identified the human EIF3h gene (among 37 others) as essential for cell division in HeLa cells (32). In our knockdown experiments in PC-3 cells, lowering eIF3h levels to about 30% of normal has a rather mild effect on protein synthesis, suggesting either that the protein is in significant excess in cells or that it is not absolutely essential for global protein synthesis. One possible mechanism for eIF3h stimulation of translation is that the subunit is essential for optimal protein synthesis, but its normal cellular level is deficient. However, the level of eIF3h relative to other eIF3 subunits in human cells is not known, nor is it certain that all eIF3 complexes contain the h-subunit. A second mechanism postulates that eIF3h interacts with a negative regulator of protein synthesis, affecting eIF3 activity. High levels of eIF3h outside the eIF3 complex would bind the regulator, preventing its interaction with eIF3h in eIF3. For example, rabies virus M protein has been shown to inhibit protein synthesis and to interact specifically with eIF3h (33). Such a mechanism is reminiscent of that for interferon-induced p56 which binds to eIF3 and inhibits the initiation phase of protein synthesis (34, 35). Clearly additional work is needed to elucidate the role of eIF3h in protein synthesis and how its levels affect this process.
eIF3h-promoted oncogenesis is stimulated by phosphorylation of eIF3h at Ser183. This conclusion is based on the observation that phosphomimetic substitution forms (Asp and Glu) at Ser183 are as active as wild type eIF3h, whereas the Ala substitution form exhibits little transforming activity. This is the first demonstration that phosphorylation of a specific mammalian eIF3 subunit alters the activity of the protein. It is tempting to speculate that mammalian target of rapamycin may be involved, because its activation correlates with stimulation of protein synthesis. Furthermore, the eIF3h phosphorylation site possesses the mammalian target of rapamycin consensus sequence, (S/T)P (36), and the kinase associates with eIF3-40S initiation complexes (37). Further work is required to identify the actual protein kinase involved and to elucidate how temporal regulation of eIF3h phosphorylation occurs. As an aside, the differences in oncogenic activity for the various eIF3h substitutions at Ser183 make unlikely the possibility that the oncogenesis is because of titration of a small noncoding RNA (miRNA) by the overexpressed mRNA, rather than to overexpression of the eIF3h protein.
It is now clear that high eIF3h levels affect protein synthesis and thereby cause malignant transformation of immortal cells, although the precise mechanism is not yet known. Thus eIF3h, along with four other eIF3 subunits as well as a number of other initiation factors, is implicated in growth control through protein synthesis. The findings provide further support for the hypothesis that regulation of the rate of translation is essential for proper growth control. Here, as well as in other studies (reviewed in Ref. 1), activation of protein synthesis results in a large increase in the translational efficiencies of oncogenic mRNAs involved in growth control. The increased levels of the resulting proteins then lead to the malignant state. It is reasonable to anticipate that initiation factors will be useful therapeutic targets for the treatment of a wide variety of cancer cells.
Supplementary Material
Acknowledgments
We thank Drs. R. Gandour-Edwards, P. Gumerlock, and R. deVere White for generously making available experimental materials for in situ analysis, and Eugen Damoc and Julie Leary for sharing knowledge of the eIF3h phosphorylation site prior to publication.
This work was supported, in whole or in part, by National Institutes of Health Grants GM22135 and GM07373 (to J. W. B. H.). This work was also supported by American Cancer Society Grant IRG 95-125-04 from the Institutional Research Grant Program (to Z. S. M.) and by the Dean, University of California Davis School of Medicine. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4 and additional references.
Footnotes
The abbreviations used are: PrEC, prostate epithelial cells; HA, hemagglutinin; Dox, doxycycline; PD, population doubling; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, short interfering RNA; RT, reverse transcription; PARP, poly(ADP-ribose) polymerase; TUNEL, terminal dUTP nick-end labeling; BrdUrd, bromodeoxyuridine; siNC, negative control siRNA; tet, tetracycline.
References
- 1.Schneider, R. J., and Sonenberg, N. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J. W. B., eds), pp.401 -431, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 2.Lazaris-Karatzas, A., Montine, K. S., and Sonenberg, N. (1990) Nature 345544 -547 [DOI] [PubMed] [Google Scholar]
- 3.De Benedetti, A., and Graff, J. R. (2004) Oncogene 233189 -3199 [DOI] [PubMed] [Google Scholar]
- 4.Fukuchi-Shimogori, T., Ishii, I., Kashiwagi, K., Mashiba, H., Ekimoto, H., and Igarashi, K. (1997) Cancer Res. 575041 -5044 [PubMed] [Google Scholar]
- 5.Koromilas, A. E., Roy, S., Barber, G. N., Katze, M. G., and Sonenberg, N. (1992) Science 2571685 -1689 [DOI] [PubMed] [Google Scholar]
- 6.Donze, O., Jagus, R., Koromilas, A. E., Hershey, J. W. B., and Sonenberg, N. (1995) EMBO J. 143828 -3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, L., Pan, X., and Hershey, J. W. B. (2007) J. Biol. Chem. 2825790 -5800 [DOI] [PubMed] [Google Scholar]
- 8.Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A., and Nogales, E. (2005) Science 3101513 -1515 [DOI] [PubMed] [Google Scholar]
- 9.Pestova, T. V., Lorsch, J. R., and Hellen, C. U. T. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J. W. B., eds) pp. 87-128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 10.Fraser, C. S., Berry, K. E., Hershey, J. W. B., and Doudna, J. A. (2007) Mol. Cell 26 811-819 [DOI] [PubMed] [Google Scholar]
- 11.Nupponen, N. N., Porkka, K., Kakkola, L., Tanner, M., Persson, K., Borg, A., Isola, J., and Visakorpi, T. (1999) Am. J. Pathol. 1541777 -1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savinainen, K. J., Linja, M. J., Saramaki, O. R., Tammela, T. L., Chang, G. T., Brinkmann, A. O., and Visakorpi, T. (2004) Br. J. Cancer 901041 -1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto, H., Yasui, K., Zhao, C., Arii, S., and Inazawa, J. (2003) Hepatology 381242 -1249 [DOI] [PubMed] [Google Scholar]
- 14.Nupponen, N. N., Isola, J., and Visakorpi, T. (2000) Genes Chromosomes Cancer 2 203-210 [PubMed] [Google Scholar]
- 15.Saramaki, O., Willi, N., Bratt, O., Gasser, T. C., Koivisto, P., Nupponen, N. N., Bubendorf, L., and Visakorpi, T. (2001) Am. J. Pathol. 1592089 -2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlinson, I. P., Webb, E., Carvajal-Carmona, L., Broderick, P., Howarth, K., et al. (2008) Nat. Genet. 40623 -630 [DOI] [PubMed] [Google Scholar]
- 17.Sramkoski, R. M. (1999) In Vitro Cell Dev. Biol. Anim. 35403 -409 [DOI] [PubMed] [Google Scholar]
- 18.Li, S., Takasu, T., Perlman, D. M., Peterson, M. S., Burrichter, K., Avdolulov, S., Bitterman, P. B., and Polunovsky, V. A. (2003) J. Biol. Chem. 2783015 -3022 [DOI] [PubMed] [Google Scholar]
- 19.Damoc, E., Fraser, C. S., Zhou, M., Videler, H., Mayeur, G. L., Hershey, J. W. B., Doudna, J. A., Robinson, C. V., and Leary, J. A. (2007) Mol. Cell. Proteomics 61135 -1146 [DOI] [PubMed] [Google Scholar]
- 20.Hershey, J. W. B. (1989) J. Biol. Chem. 26420823 -20826 [PubMed] [Google Scholar]
- 21.Dever, T. E. (2002) Cell 108545 -556 [DOI] [PubMed] [Google Scholar]
- 22.Forozan, F., Karhu, R., Kononen, J., Kallioniemi, A., and Allioniemi, O. P. (1997) Trends Genet. 13 405-409 [DOI] [PubMed] [Google Scholar]
- 23.Rooney, P. H., Murray, G. I., Stevenson, D. A., Haites, N. E., Cassidy, J., and McLeod, H. L. (1999) Br. J. Cancer 80862 -873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nupponen, N. N., Kakkola, L., Koivisto, P., and Visakorpi, T. (1998) Am. J. Pathol. 153141 -148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo, S. H., Kwon, K. C., Ihm, C. H., Jeon, Y. M., Park, J. W., and Sul, C. K. (1999) Cancer Genet. Cytogenet. 11087 -93 [DOI] [PubMed] [Google Scholar]
- 26.Sonoda, G., du Manoir, S., Godwin, A. K., Bell, D. W., Liu, Z., Hogan, M., Yakushiji, M., and Testa, J. R. (1997) Genes Chromosomes Cancer 18 115-125 [DOI] [PubMed] [Google Scholar]
- 27.Arnold, N., Hagele, L., Walz, L., Schempp, W., Pfisterer, J., Bauknecht, T., and Kiechle, M. (1996) Genes Chromosomes Cancer 1646 -54 [DOI] [PubMed] [Google Scholar]
- 28.Wong, N., Lai, P., Lee, S. W., Fan, S., Pang, E., Liew, C. T., Sheng, Z., Lau, J. W., and Johnson, P. L. (1999) Am. J. Pathol. 15437 -43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan, A., Bitterman, P. B., Sonenberg, N., Peterson, M., and Polunovsky, V. A. (2000) Oncogene 191437 -1447 [DOI] [PubMed] [Google Scholar]
- 30.Kim, T. H., Kim, B. H., Yahalom, A., Chamovitz, D. A., and von Arnim, A. G. (2004) Plant Cell 163341 -3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mamiko, M., Sonenberg, N., Yokoyama, S., and Imataka, H. (2007) EMBO J. 263373 -3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kittler, R., Putz, G., Pelletier, L., Poser, I., Heninger, A. K., Drechsel, D., Fischer, S., Konstantinova, I., Habermann, B., Grabner, H., Yaspo, M. L., Himmelbauer, H., Korn, B., Neugebauer, K., Pisabarro, M. T., and Buchholz, F. (2004) Nature 4321036 -1040 [DOI] [PubMed] [Google Scholar]
- 33.Komarova, A. V., Real, E., Borman, A. M., Brocard, M., England, P., Tordo, N., Hershey, J. W. B., Kean, K. M., and Jacob, Y. (2007) Nucleic Acids Res. 351522 -1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hui, D. J., Bhasker, C. R., Merrick, W. C., and Sen, G. C. (2003) J. Biol. Chem. 27839477 -39482 [DOI] [PubMed] [Google Scholar]
- 35.Hui, D. J., Terenzi, F., Merrick, W. C., and Sen, G. C. (2005) J. Biol. Chem. 2803433 -3440 [DOI] [PubMed] [Google Scholar]
- 36.Raught, B., and Gingras, A.-C. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J. W. B., eds) pp.369 -400, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 37.Holz, M. K., Ballif, B. A., Gygi, S. P., and Blenis, J. (2005) Cell 123569 -580 [DOI] [PubMed] [Google Scholar]
- 38.Zhou, C., Arslan, F., Wee, S., Krishnan, S., Ivanov, A. R., Oliva, A., Leatherwood, J., and Wolf, D. A. (2005) BMC Biol. 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit-McBride, Z., Mattapallil, J. J., McChesney, M., Ferrick, D., and Dandekar, S. (1998) J. Virol. 726646 -6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anzick, S. L., Kononen, J., Walker, R. L., Azorsa, D. O., Tanner, M. M., Guan, X. Y., Sauter, G., Kallioniemi, P. P., Trent, J. M., and Meltzer, P. S. (1997) Science 277965 -968 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.