Abstract
Telomerase adds telomeric repeat sequences to chromosome ends using a short region of its RNA subunit as a template. Telomerase RNA subunits are phylogenetically highly divergent, and different strategies have evolved to demarcate the boundary of the template region. The recent identification of the gene encoding telomerase RNA in the fission yeast Schizosaccharomyces pombe (ter1+) has opened the door for structure-function analyses in a model that shares many features with the telomere maintenance machinery of higher eukaryotes. Here we describe a structural element in TER1 that defines the 5′ boundary of the template. Disruption of a predicted long range base pairing interaction between template-adjacent nucleotides and a sequence further upstream resulted in reverse transcription beyond the template region and caused telomere shortening. Normal telomere length was restored by combining complementary nucleotide substitutions in both elements, showing that base pairing, not a specific sequence, limits reverse transcription beyond the template. The template boundary described here resembles that of budding yeasts and some mammalian telomerases. However, unlike any previously characterized boundary element, part of the paired region overlaps with the template itself, thus necessitating disruption of these interactions during most reverse transcription cycles. We show that changes in the paired region directly affect the length of individual telomeric repeat units. Our data further illustrate that marginal alignment of the telomeric 3′ end with RNA sequences downstream of the template is responsible for primer slippage, causing incorporation of strings of guanosines at the start of a subset of repeats.
The reverse transcriptase telomerase replenishes terminal DNA sequences lost through incomplete DNA replication and nucleolytic degradation (1). Without functional telomerase, telomeres undergo progressive shortening with each cell division, a process that eventually limits the proliferative potential of affected cells. Mutations in human telomerase subunits cause dyskeratosis congenita, aplastic anemia, and idiopathic pulmonary fibrosis, a group of disorders characterized by insufficient renewal capacity of cells caused by shortening of telomeres (2-6). Conversely, activation of telomerase is critical for continued proliferation of most cancer cells, making the enzyme a promising target for anti-cancer drugs.
At its core, telomerase is comprised of the catalytic protein subunit TERT (telomerase reverse transcriptase) and a TER (telomerase RNA) component (7). Together, TERT and TER are sufficient to reconstitute activity in vitro, but additional factors are required for telomere maintenance in vivo. Whereas TERT has a highly conserved catalytic domain, telomerase RNA subunits vary widely in size from ∼150 nucleotides in ciliates and ∼450 nucleotides in vertebrates to over 1,500 nucleotides in some yeasts (8, 9). Even among closely related species, substantial variation in telomerase RNA sequence exists. For example, the telomerase RNA subunits of four sensu stricto Saccharomyces species share only 43% sequence identity (10). Despite widely divergent primary sequences, conserved structural elements have been identified in ciliates (11, 12), vertebrates (13), and budding yeasts (10, 14), and a universal RNA core has been proposed (15).
Whereas viral reverse transcriptases make DNA copies of large RNA molecules, telomerase repeatedly copies only a few nucleotides of its RNA subunit to generate telomeric repeats. In most eukaryotes, the template region consists of one and a half telomeric repeats. To ensure that only the template sequence is copied onto the ends of chromosomes, the boundaries must be precisely defined. This challenge has been met with surprisingly diverse solutions over the course of eukaryotic evolution. In ciliates, the conserved sequence motif 5′-(U)GUCA-3′ is located two nucleotides upstream of the template (12, 16) and defines the template boundary (17). Closer examination revealed that the template boundary sequence overlaps with a high affinity binding site recognized by the RNA binding domain of TERT, suggesting that a TERT-RNA interaction blocks copying of the template past the boundary (18). In Kluyveromyces and Saccharomyces yeast species, nucleotides directly adjacent to the template are base-paired with complementary sequences several hundred nucleotides upstream. This bulged stem structure, rather than a specific sequence, determines the 5′ end of the template and prevents read-through into paired sequences (19, 20). In the secondary structure of human telomerase, the template-proximal element of the P1 helix starts eight nucleotides upstream of the template. Mutational analysis demonstrated that the P1 helix and its distance from the template are critical for template boundary definition in human telomerase (21). The 5′ end of the RNA subunit in mouse is located only two nucleotides upstream of the template, and the distance to the end defines the template boundary (21).
In all vertebrates, telomerase adds the hexameric repeat sequence GGTTAG, but substantial variability in repeat length and sequence is found in lower eukaryotes. For example, in Kluyveromyces and Candida yeast species, perfect copies of up to 26-nucleotide template sequences are repeatedly copied onto chromosome ends. In other fungi, some protozoa, and slime molds, shorter but heterogeneous repeats are found at chromosome ends (reviewed in Ref. 22). Multiple possible alignment registers (23), slippage during repeat synthesis (24), and nucleotide misincorporation (25) have all been found to account for the addition of variable repeat sequences from a single RNA template.
Among well studied species, repeat heterogeneity is most prominent in the two evolutionarily distant fungi, Saccharomyces cerevisiae and Schizosaccharomyces pombe. Careful examination of telomeric sequences added by telomerase in S. cerevisiae revealed that heterogeneity arises through a combination of two mechanisms: abortive reverse transcription partway through the template and multiple alignment possibilities of the telomeric 3′ overhang with the RNA (26). Early analysis of S. pombe telomeric sequences suggested a high degree of sequence heterogeneity with a consensus of T1-2ACA0-1C0-1 G1-6 or T1-3ACA0-2C0-1G1-8 (27, 28). It was later pointed out that this consensus included uncommon variations and that 5′-GGTTACA-3′ describes the majority of telomeric repeats (29). Subsequent analysis of a much larger sample of cloned telomeres revealed that S. pombe telomeres are comprised of constant GGTTAC core repeats separated by up to 10 nucleotides of spacer sequence (30). How this unusual sequence pattern is generated has remained enigmatic largely because the gene encoding the telomerase RNA subunit in fission yeast was unknown until earlier this year (31, 32).
As expected for a telomerase core component, deletion of S. pombe ter1+ results in progressive telomere attrition, followed by widespread cell death and the emergence of survivors with circular chromosomes. A series of point mutations helped define which nucleotides in the RNA sequence are copied into telomeric repeats and thus constitute the template region (31). In an effort to identify the boundary element and to understand the mechanism underlying telomere repeat heterogeneity, we have now examined the sequence and structure adjacent to the template. We show that long range base pairing interactions create a boundary element that defines the 5′ end of the template. Surprisingly, the boundary element partially overlaps the template, and synthesis of most repeats found in natural telomere sequences involves partial opening of the paired region.
EXPERIMENTAL PROCEDURES
Constructs and Strains—A knock-out for ter1+ was generated by replacing nucleotides 23-1422 of the RNA-encoding region with the kanamycin resistance gene in a diploid strain as described (33). The resulting strain is referred to as PP407 (h+/- ade6-M210/ade6-M216 his3-D1/his3-D1 leu1-32/leu1-32 ura4-D18/ura4-D18 ter1+ ter1:kanr). Mutations in ter1 were introduced using the QuikChange II XL site-directed mutagenesis kit (Stratagene) on pJW10, a plasmid containing a genomic DNA fragment of the ter1+ locus (31). DNA constructs were sequence verified and introduced into PP407 by electroporation. Diploid transformants were selected on Edinburgh minimal media (EMM) supplemented with adenine, histidine, and leucine (EMM AHL) and sporulated on malt extract agar plates. Spores harboring a mutant ter1 plasmid were germinated on pombe minimal glutamate (PMG) supplemented with adenine, histidine, and leucine, and haploid strains deleted for the genomic copy of ter1+ were identified by growth on YEA geneticin. At least four isolates of each mutant were subjected to four serial restreaks on EMM AHL before cells were transferred into 20 ml of liquid EMM AHL for 18 h followed by genomic DNA isolation.
Primer Extension—Total or poly(A) enriched RNA (5 μg) and RNA isolated from Trt1-Myc immunoprecipitations were incubated with 32P-labeled oligonucleotide BLoli1116 (tatacttaaggcctatgaatc; 2 pmol) and dNTPs (10 nmol) in 13 μl of double distilled H2O at 65 °C for 5 min. The reaction volume was increased to 20 μl by the addition of RNase inhibitor (RNasin Plus, 40 units), dithiothreitol (final concentration, 5 mm), first strand buffer (Invitrogen), and Superscript III reverse transcriptase (200 units; Invitrogen), and reactions were incubated at 55 °C for 60 min. The reactions were terminated by the addition of 5 μl of stop buffer (100 mm Tris-HCl, pH 7.5, 0.2 m EDTA, 2.5% (w/v) SDS, and 1% (w/v) proteinase K) and incubation at 42 °C for 10 min. Nucleic acids were purified by phenol/chloroform extraction and ethanol precipitation and solubilized in 3 μl of NaOH (0.1 m) and 7 μl of formamide. Radiolabeled primer extension products were separated on 8% Tris borate-EDTA, 8 m urea gels next to dideoxynucleotide chain termination sequencing reactions using cloned ter1+ as a template.
Telomere Length and Capture Assay—Genomic DNA isolation and telomeric Southern blots were performed as described (34). S. pombe telomeres were cloned from genomic DNA samples using the G overhang capture assay (31). In brief, a partial duplex (0.5 pmol) comprised of DNA oligonucleotides PBoli733 (gcgtacgactcactgtagatnnnnn-3′-O(CH2)2CH2OH) and PBoli749 (5′-phosphate-atctacagtgagtcgtacgcaa-3′ biotin) was incubated with 1 μg of S. pombe genomic DNA in a “Quick Ligation” reaction (New England Biolabs). Products were digested with EcoRI (40 units) for 3 h at 37 °C, and terminal DNA fragments ligated to the biotinylated tag were recovered on magnetic streptavidin beads (Dynal). After two washes in 10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.3 m NaCl and two washes in 10 mm Tris-HCl, pH 8.0, 1 mm EDTA, chromosome end fragments were amplified by polymerase chain reaction (PCR) with PBoli434 (gtgtggaattgagtatggtga) and PBoli745 (gcgtacgactcactgtagat). PCR products were cloned into the pCR4blunt-TOPO vector (Invitrogen) for sequence analysis of individual telomeres.
Telomere Sequence Analysis—Telomeric sequences from each strain were compiled in FASTA format, and the relative abundance of different repeat sequences in each sample was determined using TweenMotif (30) and Excel. The invariant sequence gggttacaaggttacgtggttacacggttaca found at the beginning of all telomeres was excluded from the analysis. Tween-Motif is an interactive Windows program for visualizing the locations of a specified target motif within a set of nucleotide sequences, as well as the gaps between the repeating motifs. The program creates a summary table of the frequency counts of the gap sequences that can be pasted into an Excel spread-sheet for further analysis.
The TweenMotif program is available for download from the Baumann Lab web site, along with source code and sample analysis files.
RESULTS
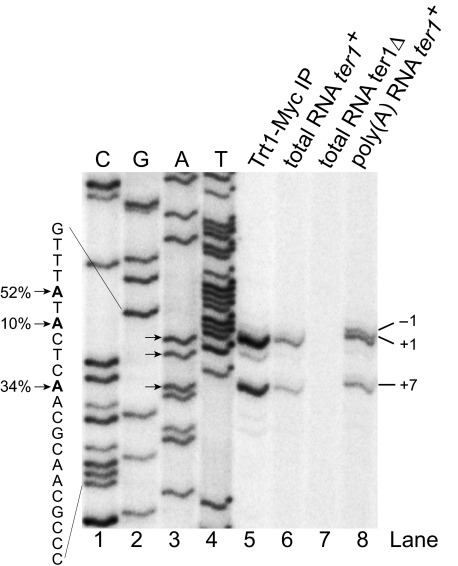
Determining the 5′ End of TER1—To appropriately characterize structure and function of an RNA, the 5′ and 3′ ends need to be accurately determined. The 5′ end of TER1 had previously been mapped by reverse transcribing this part of the RNA, followed by terminal transferase-mediated addition of a homooligomeric nucleotide tail for PCR amplification of the cDNA product (31). The reported 5′ end was based on four independently derived clones. Using a similar experimental approach, Webb and Zakian (32) identified two alternative starting positions: one 6 nucleotides further upstream and another one 1 nucleotide downstream. Shorter clones could be the product of degradation or may reflect naturally occurring 5′ end heterogeneity, as reported for budding yeast (14). We have now used reverse transcription from a radiolabeled primer to quantitatively assess the abundance of different 5′ ends in multiple RNA samples (Fig. 1). The dominant end (52%) was observed at position -6 relative to that of our original clones isolated from total cellular RNA preparations and from affinity-purified telomerase. We will therefore refer to this position as +1 from here onwards. Interestingly, a band at position -1 was observed in oligo(dT)-purified RNA (Fig. 1, lane 8), suggesting that longer transcripts may be processed to the +1 position prior to capping.
FIGURE 1.
Determination of the 5′ end of TER1 by primer extension. Oligonucleotide BLoli1116 (complementary to nucleotides 98-118) was radiolabeled and used for primer extension with total RNA, poly(A) enriched fraction, and RNA isolated from Trt1-Myc immunopurifications. No signal was observed with total RNA from a ter1Δ strain (lane 7). The three major 5′ ends are marked with arrows. The percentages next to the sequence indicate relative amount of RNA isolated from the Trt1-Myc sample starting at this position. Dideoxynucleotide sequencing (lanes 1-4) was performed with BLoli1116 and a cloned fragment of the ter1+ gene. Sequencing lanes are labeled for the strand representing the TER1 RNA.
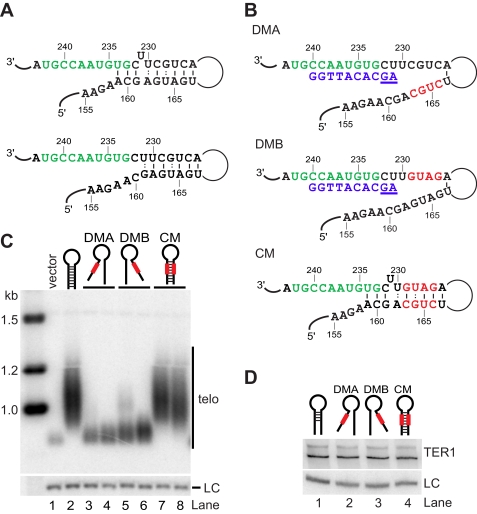
An Essential Paired Element Borders the Template Region—We previously reported that a nucleotide substitution in TER1 at position 233 (formerly 227) results in corresponding changes in telomeric DNA, suggesting that G233 is part of the template (31). In contrast, a C232A mutation was not incorporated into telomeres, consistent with this residue being outside of the template. To elucidate what defines the 5′ boundary element, we examined the sequence and putative structure upstream of the template. In the absence of mutational studies or co-variation analysis, secondary structure elements are inherently difficult to predict for large RNAs. Despite this caveat, we were intrigued to find that Mfold software predicts RNA sequences upstream of the template to be base-paired. The two most commonly observed folds are shown in Fig. 2A. The structure shown on top forms a longer, energetically favored helix including a bulged U at position 231. However, two nucleotides that are part of the template (U234 and G233) are engaged in base pairing interactions. If this conformation exists in vivo, the pairing at the base of the stem would have to be disrupted in most rounds of extension because at least U234 is copied 80% of the time (31). In an alternative local structure, the bulged U231 forms a G:U wobble, with G161 placing the start of the helix two nucleotides away from the 5′ end of the template (Fig. 2A, bottom panel).
FIGURE 2.
Disruption of the template boundary element causes telomere shortening. A, schematic of template (green) and boundary element. Structure predictions on nucleotides 1-1213 were performed with Mfold. Two alternative local structures are shown. B, schematic of mutations designed to test the predicted helix adjacent to the template. Mutated nucleotides are in red in DMA, DMB, and the CM. Telomeric DNA sequence complementary to the template is shown in blue, and two nucleotides of read-through product are underlined. C, telomeric Southern blot for ter1- cells harboring vector only (lane 1) or plasmids with wild type ter1+ (lane 2) or mutants as indicated (lanes 3-8). Genomic DNA was digested with EcoRI, separated on a 1% agarose gel, transferred to a nylon membrane, and probed with the nick translation products from a 300-bp telomeric DNA fragment. A probe against the rad16+ gene was used as loading control (LC). D, TER1 levels from wild type (lane 1), DMA (lane 2), DMB (lane 3), and CM (lane 4) were analyzed by Northern blotting of total RNA samples (15 μg/lane). The small nucleolar RNA snR101 was used as loading control (LC).
To test whether base pairing upstream of the template is critical for telomerase function, we substituted four nucleotides with their Watson-Crick complementary base in the two regions predicted to form the paired element. Disruption mutant A (DMA)3 replaces nucleotides 163-166 in the distal paired element with the complementary sequence of the template-proximal paired element, thereby disrupting the predicted pairing interactions (Fig. 2B). Reciprocally, disruption mutant B (DMB) replaces nucleotides 226-229 in the template-proximal paired element. We also generated a compensatory mutant (CM) that combines the mutant sequences of DMA and DMB, thereby restoring the potential for base pairing while changing the sequence of both paired elements (Fig. 2B). Computational analysis of DMA and DMB using Mfold predicted that these sequence changes disrupted the putative helix and the central domain of the predicted structure without affecting the global fold of the RNA. Similarly, in silico folding supported that CM would restore the predicted pairing interactions while maintaining the same global fold (data not shown).
The three mutant RNA subunits, as well as wild type TER1 and empty vector controls were introduced into a ter1+/- diploid strain. Transformants were selected and sporulated to derive haploid strains containing the respective plasmid but lacking the genomic copy of ter1+. These strains were propagated for ∼70 generations prior to analyzing telomere length. At this point, cells containing the vector control had lost most telomeric repeats (Fig. 2C, lane 1). In contrast, cells harboring a copy of the wild type ter1+ gene maintained normal telomeres (lane 2). The DMA and DMB cells had very short telomeres, indicating that telomerase activity was compromised (lanes 3-6). Combining the deleterious DMA and DMB mutations in the CM restored normal telomere length (lanes 7 and 8). Northern blotting confirmed that telomere shortening in the DMA and DMB mutants is not a consequence of reduced TER1 RNA levels (Fig. 2D). These results support that the computationally predicted helix downstream of the template exists in vivo. Disruption of the paired region compromises telomerase function, whereas restoring base pairing with a different sequence was sufficient to rescue telomerase function to wild type levels.
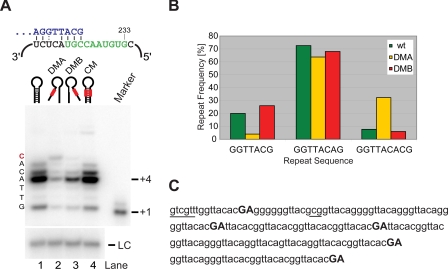
The Paired Region Functions as a Boundary Element in Vivo and in Vitro—To characterize the effects of the paired element mutations more directly, we introduced them into a pre-senescent ter1- strain containing Myc epitope-tagged Trt1. Telomerase was immunopurified from extracts, and activity was assayed on beads (35). Using an S. pombe telomeric primer ending in the sequence... GGTTACG-3′, wild type telomerase added up to six nucleotides with a dominant pausing site at position +4 (Fig. 3A). Previous work using the same primer and assay conditions showed that nucleotide incorporation was consistent with addition of the sequence 5′-GTTACA-3′ (31). Telomerase activity was 3.3- and 2.5-fold reduced for the DMA and DMB mutants, respectively. Despite the reduction in overall activity, incorporation of an additional nucleotide was observed for DMA and, to a lesser extent, for DMB (Fig. 3A). Reverse transcription did not technically extend beyond the last nucleotide of the flexible template boundary, but this last nucleotide, G233, is only used ≤8% of the time in vivo.
FIGURE 3.
Boundary element disruption mutants result in extended reverse transcription products in vitro and in vivo. A, in vitro activity assay for wild type (lane 1), DMA (lane 2), DMB (lane 3), and CM (lane 4). Telomerase assays were carried out as described in Ref. 31. A 100-mer oligonucleotide was used as loading control (LC). A schematic for the alignment of the telomeric primer (blue) upstream of the template (green) is shown above the gel. Nucleotides added by telomerase are shown to the left of the gel. B, analysis of cloned telomere sequences from wild type (wt, n = 141), DMA (n = 83), and DMB (n = 79). Telomeres were isolated after 80 generations in the presence of the ter1 mutant, cloned by G overhang capture assay and sequenced. After trimming of the invariant proximal part of each telomere, the relative abundance of the three most common repeats was determined. C, sequences for the distal part of four telomeres isolated from DMA mutant cells. Read-through products are highlighted in bold, capital letters. Aberrant sequences found in only one telomere are underlined.
To assess whether the DMA and DMB mutations caused a shift toward longer repeat units in vivo, we cloned telomeres from cells expressing each telomerase RNA and compared the frequencies of the three most common repeats with wild type. The sequence GGTTACA(G), describing the most common repeat for wild type telomeres, was only modestly reduced in DMA and DMB strains (Fig. 3B). However, the extended repeat described by the sequence GGTTACAC(G) was over 4-fold more abundant in telomeres generated in the presence of DMA. Conversely, the frequency of short GGTTAC(G) repeats was proportionally reduced in this mutant (Fig. 3B). Despite the presence of similarly short telomeres in DMB, the shift toward longer repeat units was not apparent for this mutant (Fig. 3B).
Closer examination of 83 telomeres cloned from the DMA mutant revealed direct evidence for reverse transcription of nontemplate nucleotides, with the sequence 5′-ACACGA-3′ (bold nucleotides are encoded by C232 and U231 5′ of the template; see Fig. 2, A and B) being identified (Fig. 3C). On two occasions telomeres terminated in this sequence, and in one instance the sequence was followed by a stutter of six guanosines at the start of the next repeat. In the remaining two cases, found within the same telomere, the sequence ACACGA was followed by TTA, suggesting that reverse transcription beyond the template had resulted in the use of an alternative register of alignment in the following cycle. Evidence for reverse transcription beyond the template was also observed in the DMB strain, whereas telomere sequences from the CM strain were indistinguishable from wild type (data not shown). The presence of the ACACGA sequence at the ends of telomeres and preceding aberrant repeats indicates that additional rounds of telomerase action are compromised once read-through into telomere-adjacent sequences has occurred. Read-through products may also be subject to exonucleolytic degradation because protection by telomere-binding proteins is compromised by the nontelomeric sequence. This may explain why read-through products were detected in <10% of telomeres from the mutant strains. Taken together, these results provide strong support for a paired region 5′ of the template acting as a boundary element in vivo.
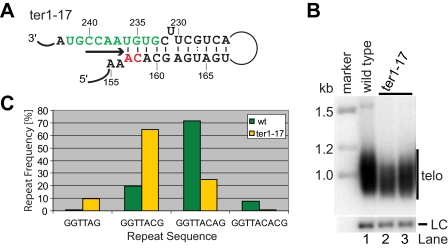
An Extended Helix Shortens Repeat Length and Compromises Telomere Maintenance—Pairing of nucleotides immediately 5′ of the template with a distal element in the RNA appears to be the most widespread solution for defining the template boundary of telomerase in different species. However, unlike other species where the paired element is either directly adjacent or several nucleotides away from the template itself, the paired region and the template overlap by two nucleotides in the energetically favored conformation of fission yeast TER1. We hypothesized that telomere repeat heterogeneity in fission yeast may be a direct consequence of this overlap. To test whether a longer, more stable boundary element would result in a greater abundance of short repeats, we extended the potential for base pairing by replacing two nucleotides upstream of the distal paired element (Fig. 4A). If a telomeric DNA end aligns with G241, only the first four of the eight bases that normally constitute the template are unpaired in this ter1-17 mutant.
FIGURE 4.
An extended boundary helix causes shortening of telomeric repeat units. A, schematic of the template and boundary element region in the ter1-17 mutant. The template is in green, and mutated nucleotides are shown in red. An arrow underneath the unpaired core template nucleotides denotes the direction of reverse transcription. The energetically favored structure of this region was determined by Mfold. B, Southern blot of EcoRI-digested genomic DNA from wild type (wt) and ter1-17 mutant cells probed with a telomeric fragment. A probe against the rad16+ gene was used as loading control (LC). C, a graphical representation of the distribution of different telomeric repeats in wild type (n = 141) and ter1-17 (n = 72) cells 80 generations after the introduction of mutant telomerase.
Cells expressing ter-17 in place of wild type ter1+ had only slightly shortened telomeres (Fig. 4B). The overall reduction in telomere hybridization signal relative to the loading control may be due to the reduced ability of a wild type telomere probe to hybridize to the telomeric repeats generated in this mutant. Alternatively, or in addition, a fraction of telomeres may have been lost altogether as a consequence of the mutation. Although we did not further investigate the latter possibility, telomeric hybridization of similar intensity was observed after 40 and 70 generations, arguing against the possibility that the cultures were being taken over by survivors with circular chromosomes (data not shown).
Sequence analysis of 70 cloned telomeres from ter1-17 cells revealed a clear shift toward shorter repeats with GGTTAC being 3.3-fold more abundant in ter1-17 mutant cells than in wild type (Fig. 4C). Our telomere sequence analysis of this and other mutants is conservative in that only the four most proximal and invariant telomeric repeats have been excluded from the analysis. It is therefore expected that not all scored repeats have been newly synthesized since introduction of the mutant template. GGTTA(G) repeats, which make up less than 1% in wild type cells, were enriched by 9.7-fold in the ter1-17 mutant. Conversely, GGTTACA(G), the most abundant repeat in wild type cells, was reduced 2.9-fold, and repeats ending in -ACAC were reduced by 10.7-fold in ter1-17. In summary, extending the boundary element helix further into the template region resulted in the addition of shorter repeats in vivo but did not eliminate telomere repeat heterogeneity.
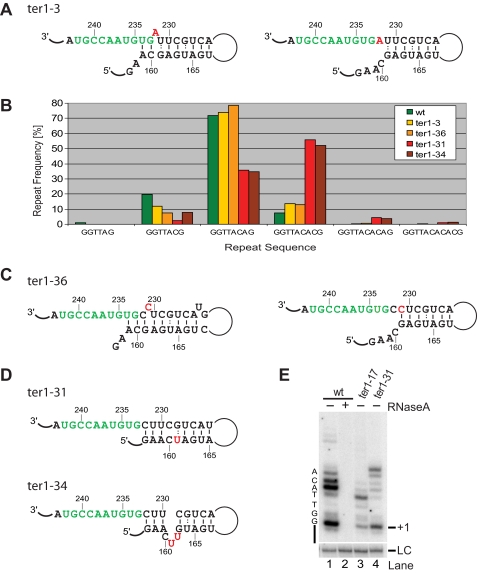
A Destabilized Boundary Element Favors Longer Repeats—The results obtained with wild type and the ter1-17 mutant suggested that fission yeast telomerase is unique in that pairing interactions at the base of the boundary element are disrupted during most extension cycles. To test whether destabilizing the boundary element helix near the template would result in longer repeats, we examined telomere sequences from the ter1-3 mutant strain (Fig. 5A). This mutant had previously been used together with others in mapping the template (31). No incorporation of the altered nucleotide into telomeres had been observed, and telomere length was normal. However, when we now compared the relative abundance of the different repeat units ending in A, AC, ACA, etc., we noticed a 40% drop in GGTTAC repeats and a concomitant increase in GGTTACAC repeats (Fig. 5B). This observation is consistent with the C232A mutation destabilizing the boundary element by eliminating the G161-C232 interaction. The energetically favored fold for this mutant pairs the otherwise bulged U231 with G161 and leaves a bulged A232 and only two paired nucleotides adjacent to the template. Weakening the boundary element in such a manner resulted in more frequent synthesis of longer repeat units. These observations argue against the alternative local structure depicted in Fig. 5A (right panel), in which the C232A mutation has no effect on base pairing interactions.
FIGURE 5.
Longer repeat units in the presence of a destabilized boundary element. A, schematic of template and boundary element of TER1 with boundary element mutation C232A (ter1-3). The mutated nucleotide is shown in red. Structures were determined by Mfold, and two alternative folds are shown. B, graphical representation of the relative abundance of telomeric repeat units within the variable part of telomeres after 80 generations. The number of telomeres (n) in each data set was 141 (wild type), 72 (ter1-3), 77 (ter1-36), 72 (ter1-31), and 71 (ter1-34). C, template and boundary element structures for U231C (ter1-36). D, schematics of local structures for ter1-31 and ter1-34. E, telomerase activity assay for wild type and ter1 mutants. For lane 2, telomerase was incubated with RNase A (5 ng/μl) for 10 min prior to the addition of primer and nucleotides. The +1 position is indicated on the right, and nucleotides added by telomerase are on the left of the gel. wt, wild type.
However, telomere sequence analysis for the ter1-36 mutant indicated that the two alternative structures may exist in equilibrium (Fig. 5C). Changing the bulged U to C carries no significant energetic penalty, whereas replacing the terminal G:U wobble with a more stable G-C base pair is energetically favorable. Consistent with this idea, longer repeat units were observed for the ter1-36 mutant (Fig. 5B).
Two additional mutants containing nucleotide substitutions that affect pairing at the base of the boundary element further supported the results described above (Fig. 5D). Whereas ter1-3 and ter1-36 show a modest change toward longer repeat units, further destabilization of the boundary element in ter1-31 and ter1-34 resulted in a 7.5-fold increase in GGTTACAC sequences at the expense of shorter repeats (Fig. 5B). Notably, GGTTACACA and GGTTACACAC repeats, not normally seen in telomeres, were being synthesized in these mutants. The presence of such long repeats cannot be explained by simple reverse transcription proceeding into template adjacent sequences. Rather the primer has to realign prior to the addition of the last A or AC at the end of the repeat. How this realignment occurs is presently unclear. Despite these obvious changes in favor of longer repeat units, no change in telomere length was detected in any of these mutant strains (supplemental Fig. S1).
Nucleotide Addition Processivity Is Controlled by the Boundary Element in Vitro and in Vivo—The data discussed above supported the conclusion that heterogeneity at the end of telomeric repeats is a direct consequence of a variable degree of invasion into the boundary element during each cycle of reverse transcription. To test this hypothesis more directly, two boundary element mutants were introduced into a strain with Myc-tagged Trt1 to allow direct analysis of telomerase activity in vitro. A preference for shorter repeat units had been observed in telomeres from ter1-17. Consistent with the extended boundary element limiting extension by telomerase, nucleotide addition processivity was notably reduced in this mutant compared with wild type (Fig. 5E, compare lanes 1 and 3). Conversely, ter1-31, a mutation that opened the base of the boundary element resulting in longer repeat units in vivo, had increased nucleotide addition processivity (lane 4). These results support the notion that the boundary element is directly responsible for the changes in telomere sequence observed in vivo.
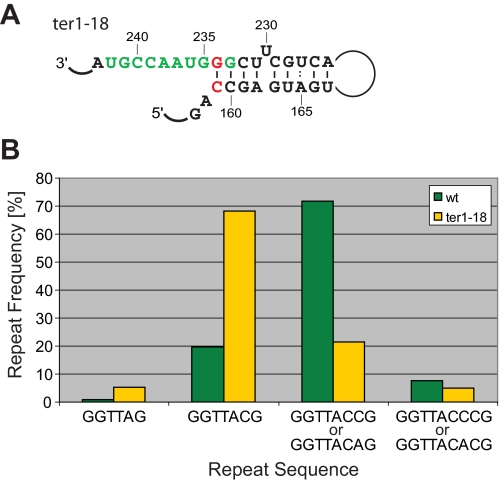
The fact that U234 is copied into 80% of telomeric repeats supports the notion that the A159-U234 base pair constitutes a weak block. We replaced the A-U with a C-G base pair to generate a more stable boundary element in the mutant ter1-18 (Fig. 6A). In this mutant, three consecutive G-C base pairs are located at the template-proximal end of the boundary element. If reverse transcription is blocked by the first G-C base pair, only GGTTAC repeats should be observed. In contrast, if part of the boundary element is disrupted during reverse transcription, the U234G mutation will result in the synthesis of GGTTACC or even GGTTACCC repeats. We have previously shown that such aberrant repeats are generated in a U234G mutant (ter1-1) in the absence of the compensatory A159C change (31). A comparison of 74 telomeres from the ter1-18 mutant with our wild type data set provided experimental support for a stabilized boundary. Whereas GGTTA and GGTTAC repeats make up about 21% of all repeats in wild type cells, replacing the A-U with a C-G base pair in ter1-18 raised this number to 74% (Fig. 6B). It thus appears that stabilizing the boundary element by replacing the terminal A-U with a C-G base pair generated a more stringent boundary element. However, read-through into the paired region was not entirely blocked, because some GGTTACC and GGTTACCC repeats were still observed.
FIGURE 6.
Effect of replacing the template-proximal A-U base pair with G-C. A, schematic of ter1-18 mutation. B, graph depicting the relative abundance of different repeat units. GGTTACCG and GGTTACCCG repeats are not present in wild type telomeres. As the part of ter1-18 telomeres included in the analysis contained wild type GGTTACA and mutant GGTTACC repeats, the sum of both was plotted. GGTTACCC and GGTTACAC sequences were treated in the same manner. wt, wild type.
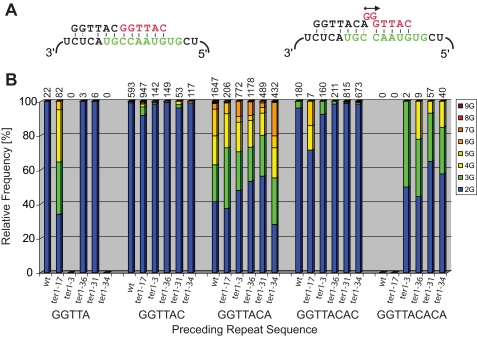
Translocation and the Origin of Stuttering—Intermittent reverse transcription of nucleotides encoded at the 5′ end of the template accounts for a second facet of telomere repeat heterogeneity in fission yeast. Telomeric repeats also vary in the number of G residues found at the start of each repeat. Because the template contains only one instance of two consecutive cytosines, the addition of more than two guanosines requires that the telomeric 3′ end slip back repeatedly to generate longer runs of G. It has previously been noted that such stuttering occurs frequently if the preceding repeat is GGTTACA, but not following a GGTTAC repeat (i.e. without the 3′-most A) (36). A telomeric... GGTTACA end cannot pair with the alignment region of the RNA template in such a manner that another GGTTAC(A) repeat can be added processively. Instead, the alignment register is thought to shift by one nucleotide to allow formation of three consecutive base pairs and a noncanonical A-C interaction at the 3′ end of the telomere (31, 32). This arrangement allows templated addition of a single G followed by presumed template slip-page to account for the addition of more guanosines (Fig. 7A).
FIGURE 7.
G stuttering in relation to the previous repeat sequence. A, schematic of alignment for telomeric DNA ending in AC (left) or ACA (right). The template is shown in green, and the newly added telomeric sequence is in red. B, telomere data sets for wild type and mutants were processed with TweenMotif to reveal the relative abundance of two to nine guanosines following each of the five repeats shown below. The total number of repeats analyzed in each category is shown above the columns.
Drawing on telomere sequencing data from wild type and the mutants described here, we have analyzed the number of consecutive guanosines after all repeat variations. With few exceptions, the addition of more than two guanosines correlated with the preceding repeat ending in adenosine (Fig. 7B). For wild type and all five template boundary mutants, between 45 and 70% of repeats 3′ of a GGTTACA sequence started with three to nine guanosines. In contrast, GGTTAC and GGTTACAC repeats were almost always followed by only two Gs. Interestingly, this correlation also existed for shorter and longer repeats that are rarely, if ever, seen in wild type telomeres. For example, GGTTACACA repeats are absent from wild type telomeres but are generated in ter1 mutants with a destabilized boundary element. In these, 35-65% of GGTTACACA repeats were followed by three or more guanosines. Similarly, GGTTA repeats are uncommon in wild type cells (<1%) but make up 10% of all repeats in the ter1-17 mutant with an extended boundary element. Consistent with a correlation between a terminal adenosine and G stuttering, 66% of repeats following GGTTA initiate with runs of three to five guanosines. In the few cases where terminal adenosines did not correlate with a high incidence of three or more guanosines in the adjacent repeat, sample numbers were generally small, and the results are statistically insignificant (Fig. 7B).
DISCUSSION
Here we have shown that disruption of a predicted long range base pairing interaction between template-adjacent nucleotides and sequences upstream results in reverse transcription beyond the template. Normal telomere length was restored by combining complementary mutant sequences in both paired elements, confirming that base pairing rather than a specific sequence defines the template boundary. Analysis of a series of template boundary element mutants established a correlation between length and stability of the boundary element and telomere repeat sequences. While this manuscript was in preparation, the existence of the template boundary element described here was proposed based on another set of mutations (32).
Paired regions define template boundaries in a variety of organisms, but the relative location of the template-distal paired element varies widely. In budding yeasts the boundary helix is formed by nucleotides between the 5′ edge of the template and the 5′ end of the RNA (19, 20), whereas the functionally equivalent sequence in the human RNA is formed by the 5′ template-adjacent nucleotides pairing with a stretch of sequence 3′ of the template and pseudoknot (21). The boundary element in fission yeast resembles budding yeasts in that both paired elements are located upstream of the template. However, unlike other characterized boundary elements, the paired region partially over-laps with the template, suggesting that synthesis of most repeats involves dissociation of pairing interactions at the base of the boundary element. Alternatively, the pairing interactions that are energetically favored when the RNA is folded in isolation may not constitute the most stable structure in the presence of the catalytic protein subunit. If protein-RNA interactions stabilize the structure shown in the lower panel in Fig. 2A, the 5′ end of the template would be two nucleotides away from the first base pair of the boundary element. Such spacing would be similar to the distance between the 5′ end of the template and the boundary element in S. cerevisiae (20). Although our experiments did not address whether protein components alter local structural elements, several of the mutants presented here argue against this model. If a structure that excludes all templating nucleotides from pairing was favored, the ter1-3, ter1-17, and ter1-18 mutations would not be expected to affect the boundary of reverse transcription because the affected nucleotides are unpaired in wild type and mutants. In reality, each of these mutations shifted the distribution toward shorter or longer repeats in a manner consistent with the structures drawn in Figs. 4, 5 and 6. A third possibility is that the boundary element may switch between both conformations, a structural flexibility that could be instrumental in generating repeat heterogeneity.
It is presently unclear why some species have heterogeneous telomeric repeats, whereas others have precisely defined tandem repeats of a specific sequence. Telomeres are bound by at least two sequence-specific telomere-binding proteins, and the requirement for co-evolution between the RNA template and the DNA-binding domains of two or more proteins would be expected to severely restrict the freedom of telomeric sequences to diverge over the course of evolution. Consistent with this notion, the sequence GGTTAG describes telomeric repeats in all vertebrates. However, a surprising degree of sequence divergence is observed among yeasts ranging from perfect 26-nucleotide repeats in Saccharomyces kluyveri (37) to the variable (TG)0-6TGGGTGTG(G) repeats in S. cerevisiae (26). It appears that the telomere maintenance machinery is far less constrained in these single-celled organisms, allowing for substantial divergence in telomeric sequences without compromising telomere function.
S. pombe repeats have previously been described as a composite of GGTTAC cores, which are important for Pot1 binding, and variable spacer sequences (30). Here we have shown that variability in the spacer arises by two mechanisms: a flexible boundary element that permits intermittent addition of A or AC at the end of repeat synthesis and G stuttering caused by unstable primer alignment when the previous repeat terminated in A rather than C. At least under laboratory conditions, changes in the spacer sequence appear to be tolerated very well. Even mutations that caused a dramatic shift toward longer repeats had no apparent effect on telomere length or the incidence of telomere loss. It is important to note, however, that replacement of wild type TER1 with a mutant form does not result in the exchange of proximal repeats for many hundreds of generations. We can therefore not exclude the possibility that a shift toward shorter or longer repeats would have a more dramatic effect once proximal repeats have been exchanged. Some template mutations in the telomerase RNA from Kluyveromyces lactis have no effect on telomere length for 400-500 generations but cause a 100-fold increase in telomere length when a threshold number of internal repeats has been replaced with mutant sequences (38). In fission yeast, half to two-thirds of telomeric sequence is replaced by mutant repeats during the first 50-100 generations, but proximal repeats appear to be sheltered from exchange for many generations thereafter.4
On the other hand, S. pombe telomere length is highly sensitive to perturbation of Pot1-binding, because minor changes in the amount of telomere-bound Pot1 have a dramatic effect on telomere length (34). Mutations in the Pot1 DNA-binding domain predicted to lower its affinity for telomeric DNA cause dramatic telomere lengthening in vivo.5 The same phenotype would be expected for mutant telomeric repeats that are bound with reduced affinity by Pot1. Perhaps surprisingly, not one mutation that affects telomere sequence was associated with telomere elongation in our studies. A possible explanation comes from in vitro binding experiments indicating that fission yeast Pot1 is well adapted to accommodating telomere repeat heterogeneity (30). It appears that the two OB-folds in the DNA-binding domain interact independently with two GGTTAC repeats, allowing for intervening spacer sequences to be looped out (39). In this manner, Pot1 binding may be largely unaffected by the kind of telomere repeat heterogeneity observed here. Although flexible template boundaries and variable repeats may simply be accommodated by adaptable proteins, it is too early to refute the idea that repeat divergence may hold some selective advantage in certain organisms. It will thus be interesting to engineer S. pombe strains with perfect telomeric repeats as well as examine the effects of subtle changes in repeat composition in competitive growth assays.
Supplementary Material
Acknowledgments
We thank Elodie Brun for generating the ter1 deletion strain used in this study, Kym Delventhal, Eric Jessen, and the Stowers Institute Molecular Biology Facility for site-directed mutagenesis and sequencing services, and Diana Baumann, Rachel Helston, and Carla Anderson for proofreading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant K99GM80400 (to D. C. Z.). This work was also supported by the Stowers Institute for Medical Research and a Pew Scholars in the Biomedical Sciences Award (to P. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: DMA, disruption mutant A; DMB, disruption mutant B; CM, compensatory mutant.
J. T. Bunch and P. Baumann, unpublished data.
P. Baumann, unpublished data.
References
- 1.Collins, K. (2006) Nat. Rev. Mol. Cell. Biol. 7484 -494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell, J. R., Wood, E., and Collins, K. (1999) Nature 402551 -555 [DOI] [PubMed] [Google Scholar]
- 3.Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P. J., and Dokal, I. (2001) Nature 413432 -435 [DOI] [PubMed] [Google Scholar]
- 4.Vulliamy, T., Marrone, A., Dokal, I., and Mason, P. J. (2002) Lancet 3592168 -2170 [DOI] [PubMed] [Google Scholar]
- 5.Armanios, M., Chen, J. L., Chang, Y. P., Brodsky, R. A., Hawkins, A., Griffin, C. A., Eshleman, J. R., Cohen, A. R., Chakravarti, A., Hamosh, A., and Greider, C. W. (2005) Proc. Natl. Acad. Sci. U. S. A. 10215960 -15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi, H., Calado, R. T., Ly, H., Kajigaya, S., Baerlocher, G. M., Chanock, S. J., Lansdorp, P. M., and Young, N. S. (2005) New Engl. J. Med. 3521413 -1424 [DOI] [PubMed] [Google Scholar]
- 7.Cech, T. R. (2004) Cell 116273 -279 [DOI] [PubMed] [Google Scholar]
- 8.Theimer, C. A., and Feigon, J. (2006) Curr. Opin. Struct. Biol. 16307 -318 [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. L., and Greider, C. W. (2004) Trends Biochem. Sci. 29183 -192 [DOI] [PubMed] [Google Scholar]
- 10.Zappulla, D. C., and Cech, T. R. (2004) Proc. Natl. Acad. Sci. U. S. A. 10110024 -10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero, D. P., and Blackburn, E. H. (1991) Cell 67343 -353 [DOI] [PubMed] [Google Scholar]
- 12.Lingner, J., Hendrick, L. L., and Cech, T. R. (1994) Genes Dev. 81984 -1998 [DOI] [PubMed] [Google Scholar]
- 13.Chen, J. L., Blasco, M. A., and Greider, C. W. (2000) Cell 100503 -514 [DOI] [PubMed] [Google Scholar]
- 14.Dandjinou, A. T., Levesque, N., Larose, S., Lucier, J. F., Abou Elela, S., and Wellinger, R. J. (2004) Curr. Biol. 141148 -1158 [DOI] [PubMed] [Google Scholar]
- 15.Lin, J., Ly, H., Hussain, A., Abraham, M., Pearl, S., Tzfati, Y., Parslow, T. G., and Blackburn, E. H. (2004) Proc. Natl. Acad. Sci. U. S. A. 10114713 -14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormick-Graham, M., and Romero, D. P. (1995) Nucleic Acids Res. 231091 -1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Autexier, C., and Greider, C. W. (1995) Genes Dev. 92227 -2239 [DOI] [PubMed] [Google Scholar]
- 18.Lai, C. K., Miller, M. C., and Collins, K. (2002) Genes Dev. 16415 -420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzfati, Y., Fulton, T. B., Roy, J., and Blackburn, E. H. (2000) Science 288863 -867 [DOI] [PubMed] [Google Scholar]
- 20.Seto, A. G., Umansky, K., Tzfati, Y., Zaug, A. J., Blackburn, E. H., and Cech, T. R. (2003) RNA 91323 -1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, J. L., and Greider, C. W. (2003) Genes Dev. 172747 -2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellinger, R. J., and Sen, D. (1997) Eur. J. Cancer 33735 -749 [DOI] [PubMed] [Google Scholar]
- 23.Singer, M. S., and Gottschling, D. E. (1994) Science 266404 -409 [DOI] [PubMed] [Google Scholar]
- 24.Collins, K. (1999) Annu. Rev. Biochem. 68187 -218 [DOI] [PubMed] [Google Scholar]
- 25.McCormick-Graham, M., Haynes, W. J., and Romero, D. P. (1997) EMBO J. 163233 -3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forstemann, K., and Lingner, J. (2001) Mol. Cell. Biol. 217277 -7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konig, P., and Rhodes, D. (1997) Trends Biochem. Sci 2243 -47 [DOI] [PubMed] [Google Scholar]
- 28.Zakian, V. A. (1995) Science 2701601 -1607 [DOI] [PubMed] [Google Scholar]
- 29.Hiraoka, Y., Henderson, E., and Blackburn, E. H. (1998) Trends Biochem. Sci. 23 126. [DOI] [PubMed] [Google Scholar]
- 30.Trujillo, K. M., Bunch, J. T., and Baumann, P. (2005) J. Biol. Chem. 2809119 -9128 [DOI] [PubMed] [Google Scholar]
- 31.Leonardi, J., Box, J. A., Bunch, J. T., and Baumann, P. (2008) Nat. Struct. Mol. Biol. 15 26-33 [DOI] [PubMed] [Google Scholar]
- 32.Webb, C. J., and Zakian, V. A. (2008) Nat. Struct. Mol. Biol. 1534 -42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baumann, P., and Cech, T. R. (2000) Mol. Biol. Cell 113265 -3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunch, J. T., Bae, N. S., Leonardi, J., and Baumann, P. (2005) Mol. Cell. Biol. 255567 -5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haering, C. H., Nakamura, T. M., Baumann, P., and Cech, T. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 976367 -6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ares, M., Jr., and Chakrabarti, K. (2008) Nat. Struct. Mol. Biol. 15 18-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohn, M., McEachern, M. J., and Blackburn, E. H. (1998) Curr. Genet. 33 83-91 [DOI] [PubMed] [Google Scholar]
- 38.McEachern, M. J., and Blackburn, E. H. (1995) Nature 376403 -409 [DOI] [PubMed] [Google Scholar]
- 39.Croy, J. E., Podell, E. R., and Wuttke, D. S. (2006) J. Mol. Biol. 36180 -93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.