Abstract
Salmonella enterica serovar Typhimurium has at least nine multidrug efflux pumps. Among these pumps, AcrAB is effective in generating drug resistance and has wide substrate specificity. Here we report that indole, bile, and an Escherichia coli conditioned medium induced the AcrAB pump in Salmonella through a specific regulator, RamA. The RamA-binding sites were located in the upstream regions of acrAB and tolC. RamA was required for indole induction of acrAB. Other regulators of acrAB such as MarA, SoxS, Rob, SdiA, and AcrR did not contribute to acrAB induction by indole in Salmonella. Indole activated ramA transcription, and overproduction of RamA caused increased acrAB expression. In contrast, induction of ramA was not required for induction of acrAB by bile. Cholic acid binds to RamA, and we suggest that bile acts by altering pre-existing RamA. This points to two different AcrAB regulatory modes through RamA. Our results suggest that RamA controls the Salmonella AcrAB-TolC multidrug efflux system through dual regulatory modes in response to environmental signals.
Salmonella enterica is a bacterial pathogen that causes a variety of diseases in humans, including gastroenteritis, bacteremia, and typhoid fever (1). In the 1990s, the prevalence of multidrug-resistant Salmonella increased in the United Kingdom (2, 3), the United States (4, 5), and Canada (6). Many countries documented outbreaks associated with drug-resistant Salmonella in poultry, cattle, and swine (4, 7-10). Emerging resistance to antibiotics in Salmonella has been found in both humans and animals and is a potentially serious public health problem (11, 12). High level fluoroquinolone resistance in S. enterica serovar Typhimurium phage type DT204 has been reported to result from multiple target gene mutations and active efflux by the AcrAB-TolC multidrug efflux pump (13, 14).
Multidrug efflux pumps have important physiological functions, including transport of drugs, bile salts, toxins, and environmental compounds (15, 16). In bacteria, drug resistance is often associated with multidrug efflux pumps that decrease cellular drug accumulation (17, 18). In bacteria, such pumps have been classified into five families on the basis of sequence similarity as follows: the major facilitator, resistance-nodulation-cell division, small multidrug resistance, multidrug and toxic compound extrusion, and ATP-binding cassette families (19-21). In Gram-negative bacteria, resistance-nodulation-cell division pumps are especially effective in generating resistance (17, 22-24). Recent studies have shown that Gram-negative S. enterica serovar Typhimurium has nine functional drug efflux pumps (25). Many multidrug pumps have overlapping substrate spectra, and it is intriguing that bacteria, with their economically organized genomes, harbor large sets of multidrug efflux genes (17).
The key to understanding how bacteria utilize these pumps lies in the regulation of their expression (19, 26-28). Currently available data indicate that multidrug efflux pumps are often expressed under precise and elaborate transcriptional control. For example, expression of acrAB, which encodes the AcrAB pump, may be subject to multiple levels of regulation. In Escherichia coli, it is modulated locally by the repressor AcrR (29). At a more global level, it is modulated by stress conditions and by regulators such as MarA, SoxS, and Rob (30, 31). Olliver et al. (32) reported that mutation in acrR contributes to overexpression of acrAB in Salmonella and increases resistance to multiple drugs. Eaves et al. (33) reported that acrB, acrF, and acrD are coordinately regulated and that their expression influences expression of transcriptional activators marA and soxS. Furthermore, integration of IS1 and IS10 elements into the upstream region of the acrEF operon has been reported to cause increased expression of acrEF (34). These examples illustrate the complexity and diversity of the mechanisms regulating bacterial multidrug efflux pumps. However, few data are available on signals that induce multidrug efflux genes in Salmonella.
Previously, it was reported that indole induces the acrD, acrE, cusB, emrK, mdtA, mdtE, and mdtH multidrug efflux pump genes in E. coli (35). They also reported that indole induction of acrD and mdtA is mediated by the BaeSR and CpxAR systems. However, the effect of indole on the AcrAB-TolC multidrug efflux pump, which plays a major role in antibiotic resistance, remains unknown. Very few signals inducing multidrug efflux pumps in Salmonella have been identified so far. Here we report on induction of acrAB in Salmonella via the specific regulator RamA in response to indole, bile, and an E. coli conditioned medium. This study describes the dual regulatory mode of acrAB via RamA in response to environmental signals.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions—The bacterial strains and plasmids used in this study are listed in Table 1. The S. enterica serovar Typhimurium strains were derived from the wild-type (WT)4 strain ATCC14028s (36). P22-mediated transductions were performed as described by Davis et al., (37). Bacterial strains were grown at 37 °C in LuriaBertani (LB) broth or plates. Antibiotics such as ampicillin (100 μg/ml), kanamycin (25 μg/ml), or chloramphenicol (25 μg/ml) were added when required.
TABLE 1.
S. enterica strains and plasmids used in this study
| Strain or plasmid | Original name | Characteristics | Source or Ref. |
|---|---|---|---|
| Strains as in text | |||
| WT | ATCC14028s | S. enterica serovar Typhimurium wild-type | 36 |
| tolC-lac | EG15109 | ΔtolC-lacZY+ KmR | 25 |
| acrAB-lac | NKS505 | ΔacrAB-lacZY+ KmR | This study |
| acrEF-lac | EG15114 | ΔacrEF-lacZY+ KmR | 25 |
| acrD-lac | EG15120 | ΔacrD-lacZY+ KmR | 25 |
| mdtABC-lac | EG15124 | ΔmdtABC-lacZY+ KmR | 25 |
| mdsABC-lac | NKS517 | ΔmdsABC-lacZY+ KmR | This study |
| emrAB-lac | NKS522 | ΔemrAB-lacZY+ KmR | This study |
| mdfA-lac | NKS524 | ΔmdfA-lacZY+ KmR | This study |
| mdtK-lac | EG15132 | ΔmdtK-lacZY+ KmR | 25 |
| macAB-lac | NKS530 | ΔmacAB-lacZY+ KmR | This study |
| ΔbaeSR cpxAR/acrAB-lac | NES56 | ΔbaeSR ΔcpxAR::CmR ΔacrAB-lacZY+ KmR | This study |
| ΔbaeSR cpxAR/acrD-lac | NES26 | ΔbaeSR ΔcpxAR::CmR ΔacrD-lacZY+ KmR | This study |
| ΔmarA/acrAB-lac | NES20 | ΔmarA::CmR ΔacrAB-lacZY+ KmR | This study |
| ΔsoxS/acrAB-lac | NES28 | ΔsoxS::CmR ΔacrAB-lacZY+ KmR | This study |
| Δrob/acrAB-lac | NES29 | Δrob::CmR ΔacrAB-lacZY+ KmR | This study |
| ΔsdiA/acrAB-lac | NES35 | ΔsdiA::CmR ΔacrAB-lacZY+ KmR | This study |
| ΔacrR/acrAB-lac | NES48 | ΔacrR::CmR ΔacrAB-lacZY+ KmR | This study |
| ΔmarA rob soxS sdiA acrR/acrAB-lac | NES55 | ΔmarA Δrob ΔsoxS ΔsdiA ΔacrR::CmR ΔacrAB-lacZY+ KmR | This study |
| ΔramA/acrAB-lac | NES58 | ΔramA::CmR ΔacrAB-lacZY+ KmR | This study |
|
ΔramA/tolC-lac |
NES65
|
ΔramA::CmR ΔtolC-lacZY+
KmR |
This study
|
| Plasmids | |||
| pKD3 | repR6Kg[r]ApRFRT CmRFRT | 39 | |
| pKD4 | repR6Kg[r]tsApRFRT KmRFRT | 39 | |
| pCP20 | reppSC101 ApR CmRcI857lPRflp | 39 | |
| pMALc2X | Vector, AmpR | New England Biolabs | |
| pMALc2X ramA-His6 | ramA-His6 gene cloned into pMALc2X, AmpR | This study | |
| pMALc2X ramA-His6 (truncated) | 5 -Terminal (69 bp) deleted ramA-His6 gene cloned into pMALc2X, AmpR | This study | |
| pNN387 | Single copy vector, CmR, NotI-HindIII cloning site upstream of promoter-less lacZ | 38 | |
| pNNramA | pNN387 (ramA gene promoter-lacZ) | This study |
Plasmid Construction—ramA was amplified by PCR from the genomic DNA of strain ATCC14028s (36) using LA-Taq polymerase (Takara Bio Inc., Otsu, Japan) and the primers listed in Table 2. This process introduced the EcoRI and HindIII restriction sites. The PCR fragment was cloned between the EcoRI and HindIII sites of the pMAL-c2X vector (New England Biolabs Inc., Ipswich, MA). The ramA promoter was amplified by PCR, and the PCR fragment was cloned between the NotI and HindIII sites of the pNN387 vector (38). The nucleotide sequences of the recombinant plasmids were determined using an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems Foster City, CA).
TABLE 2.
Primers used in this study
| Primer | Sequences (5′ to 3′) |
|---|---|
| For gene deletion | |
| marA-P1 | TATTTGCTCAAGAAAATTCTGCCGTAGACAAAAAAGACGTGTGTAGGCTGGAGCTGCTTC |
| marA-P2 | CAGCATTTTCATGGTGCTCTTCGCGTGGCGCATAAACAAACATATGAATATCCTCCTTAG |
| soxS-P1 | GACAAAACGACGAATCGAATACTGTTTAAGAGGCAACAATGTGTAGGCTGGAGCTGCTTC |
| soxS-P2 | GCCTCTGACGATACGCGGGCAGACGCCCTACAGGCGGTGACATATGAATATCCTCCTTAG |
| rob-P1 | TGATATCGGGTGCTCGTTGTTGAAAGGATGGGGATATTTTGTGTAGGCTGGAGCTGCTTC |
| rob-P2 | GCATCTGGACGCCCCTGCATTGGATGAGCTACAGCGTTAACATATGAATATCCTCCTTAG |
| sdiA-P1 | ATAATAATGATTATCAATATCAAAGGCGTGACCATAAAATGTGTAGGCTGGAGCTGCTTC |
| sdiA-P2 | TCAGCCGGTTTCGCGTCTGGCGCGAAGCATCGTCAGCACGCATATGAATATCCTCCTTAG |
| acrR-P1 | AGCGACACAGAAAATGTCCAGGAGTTTTCCTGGAATATTAGTGTAGGCTGGAGCTGCTTC |
| acrR-P2 | TAAATGTATGTAAATCTAACGCCTGTAAATTCACCGACATCATATGAATATCCTCCTTAG |
| ramA-P1 | CCATGCGATAAGCTGTCTCACAATTTGTGTGGTTATTACTGTGTAGGCTGGAGCTGCTTC |
|
ramA-P2
|
CGATTAAACATTTCAATGCGTACGGCCATGCTTTTCTTTACATATGAATATCCTCCTTAG
|
| For plasmid construction | |
| ramA-F-EcoRI (for histidine-tagged RamA) | GCGAATTCATGACCATTTCCGCTCAGG |
| ramA-F-EcoRI (for N-terminal truncated RamA) | GCGAATTCCGTATTGATGATATTGCC |
| ramA-R-HindIII | GCAAGCTTTCAATGATGATGATGATGATGCGTACGGCCATGCTT |
| ramA-F-NotI (for pNN387ramA) | CGCGCGGCCGCGTTGTTTCAGCCAGCGCC |
|
ramA-R-HindIII (for pNN387ramA)
|
GAGAAGCTTCGTGCTCTCCCCTCTATCAA
|
| For EMSA | |
| acrA-PF | CCATATGTCGGTGAATTTACAGGCG |
| acrA-PR1 (for pAcrA1) | TCAGGGGGAGCCGTTGACCGTC |
| acrA-PR2 (for pAcrA2) | CCTCTGTTTTTGTTCATATGTAAACCTCG |
| acrA-PR3 (for pAcrA3) | TGTTTGGTTTTTCGTGCCATATGTCG |
| acrA-PR4 (for pAcrA4) | TCTGATAGCTCCCAGATCTCACTG |
| acrA-PR5 (for pAcrA5) | AGTAGCTTCAAGAATATGAACTAAAATTTC |
| acrA-PR6 (for pAcrA6) | AAGGCTACGTTGCGCCTGTTGAACC |
| acrA-PR7 (for pAcrA7) | ATCAGTATCGCCGCACGACGG |
| acrA-PR8 (for pAcrA8) | AGGATGTGTTGTCGTGTCTCCAGCGC |
| acrA-PR9 (for pAcrA9) | TTCGTGCCATATGTCGGTGAATTTACAG |
| acrA-PR10 (for pAcrA10) | TGTTTGGTTTTTCGTGCCATATGTCG |
| acrA-PR11 (for pAcrA11) | CAGCGCTTGTTGTTTGGTTTTTCG |
| acrA-PR12 (for pAcrA12) | GTCGTGTCTCCAGCGCTTGTTGTTTGG |
| tolC-PF1 | CTTTACCTCGCCACTCATTTCTCCG |
| tolC-PF2 (for pTolC1) | ATGGCACGTAACGCCAACTTTTTGC |
| tolC-PF3 (for pTolC2) | GTGCTGCCCTGCTAGCAATAATG |
| tolC-PR | TCCTTGTTGTGAAGCAGTATTTAGCG |
Construction of Gene Deletion Mutants—Genes were disrupted as described by Datsenko and Wanner (39). The chloramphenicol resistance cat gene, flanked by Flp recognition sites, was amplified by PCR using the primers listed in Table 2. The resulting PCR products were used to transform the recipient ATCC14028s strain harboring the pKD46 plasmid that expresses Red recombinase. Chromosomal structure of the mutated loci was verified by PCR (39). The deletions were transferred to strains by P22 transduction. The cat gene was eliminated using plasmid pCP20 (39).
β-Galactosidase Assays—Single colonies of each bacterial strain to be assayed were inoculated into 2 ml of LB containing the appropriate selected antibiotics. After overnight growth at 37 °C, the cultures were diluted to 1:50 into E. coli conditioned medium or LB media. The cells were then grown at 37 °C until the optical density of 0.8 at 600 nm. To test the effect of indole or bile on gene expression, 2 mm indole, 0.25 mm bile salts, 0.25 mm cholic acid, or 0.25 mm deoxycholic acid were added to secondary cultures. β-Galactosidase activities were determined as described by Miller (40). All assays were performed in triplicate.
Survival Assay—Salmonella WT strain was grown at 37 °C in LB medium, with or without indole, for 7 h. Benzalkonium was added to create a concentration of 150 mg/ml in LB. After incubation for 10 min, the number of colony-forming units was determined by serial dilutions in phosphate-buffered saline on LB agar. The percentage of cells surviving under benzalkonium was the number of colony-forming units per ml remaining after the benzalkonium treatment divided by the initial number of colony-forming units per ml. Survival levels of the indole-treated cells were standardized to 100%, and untreated cell values were displayed relative to those of the indole-treated cells. All assays were performed in triplicate.
Preparation of Conditioned Medium—Conditioned medium was prepared by inoculating 30 ml of LB broth with 300 μl of a 10-3 dilution of an overnight culture of E. coli MG1655, followed by shaking (170 rpm) at 37 °C for 24 h (A600 of 5.0) (41). The cells were pelleted by centrifugation, and the resulting supernatants supplemented with 20× LB broth to create a final concentration of 0.5× and adjusted to a pH of 7.5. The conditioned medium was then filter-sterilized through a 0.2-μm pore filter.
Purification of Histidine-tagged RamA and Truncated RamA Protein—Full-length ramA gene or truncated ramA gene (69 bases at 5-terminal was deleted) were amplified from genomic DNA of ATCC14028s by PCR with the primers listed in Table 2. The DNA fragments were cloned into pMALc2x vector (New England Biolabs). Constructed plasmids were transformed into BL21(DE3) to produce histidine-tagged RamA or N-terminal truncated (23 amino acids) His-RamA. For purification of RamA protein, E. coli was grown at 37 °C to an A600 of 0.5. RamA production was induced by addition of 0.3 mm isopropyl 1-thio-β-d-galactoside. Cultures were incubated for 3 h, and bacterial cells were then disrupted by French press (SLM Instruments, Inc., Urbana, IL). The protein was purified using TALON metal affinity resin (Clontech).
DNA Mobility Shift Assay—Upstream regions of acrA and tolC were amplified by PCR. The PCR products were purified for a DNA mobility shift assay. Ten microliters of reaction mixture for the DNA mobility shift assay contained 0.15 pmol of DNA and RamA protein. The reaction buffer contained 10 mm Tris-HCl (pH 7.5), 50 mm KCl, and 1 mm dithiothreitol. Reaction mixtures were incubated for 30 min at room temperature and separated on a 5% native polyacrylamide gel at 4 °C. The gel was soaked in 10,000× diluted SYBR Green I nucleic acid stain (Cambrex Corp., East Rutherford, NJ). DNA was visualized under blue incident light at 460 nm (Luminescent Image Analyzer LAS-3000, Fujifilm Life Science, Stamford, CT).
Intrinsic Fluorescence Spectrum of RamA in the Presence of Cholic Acid—Fluorescence spectra of RamA and truncated RamA were measured as described by Rosenberg et al. (31). The fluorescence emission spectra were recorded using a LS 55 fluorescence spectrometer, 120 V (PerkinElmer Life Sciences).
RESULTS
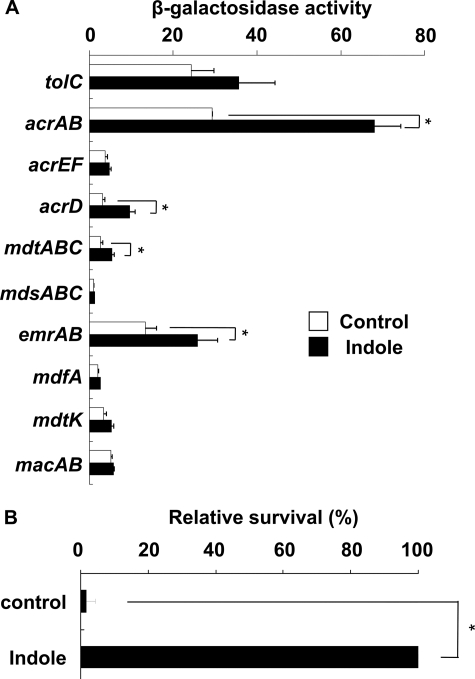
Indole Induces Four Multidrug Efflux Pumps and Drug Tolerance of Salmonella—In E. coli, indole is produced from tryptophan by tryptophanase and is excreted from the cell (42). However, Salmonella does not produce indole because it lacks the tnaA gene encoding tryptophanase (43). Indole has also been reported to auto-regulate multidrug efflux genes in E. coli (35). We postulated that Salmonella multidrug efflux genes may respond to indole. To investigate the effect of indole on the expression of multidrug efflux pumps, Salmonella strains, in which the efflux genes were replaced with a reporter gene (lacZ), were inoculated into cultures, with or without indole. Expression levels of drug efflux pumps were measured by a β-galactosidase reporter assay. Indole significantly induced expression of the acrAB, emrAB, acrD, and mdtABC efflux genes in Salmonella (Fig. 1A). A survival assay using benzalkonium showed that indole enhanced drug tolerance of Salmonella (Fig. 1B).
FIGURE 1.
Indole induction of multidrug efflux genes and drug tolerance of Salmonella enterica serovar Typhimurium. The data correspond to mean values from three independent experiments. Bars correspond to the standard deviation. Asterisks indicate statistically significant differences (*, p < 0.01) in the paired Student's t test. A, differences in β-galactosidase activity in tolC-lac (EG15109), acrAB-lac (NKS505), acrEF-lac (EG15114), acrD-lac (EG15120), mdtABC-lac (EG15124), mdsABC-lac (NKS517), emrAB-lac (NKS522), mdfA-lac (NKS524), mdtK-lac (EG15132), and macAB-lac (NKS530) strains grown in LB medium with (solid bars) or without (open bars) 2 mm indole. B, drug tolerance of S. enterica serovar Typhimurium induced by indole. WT strain (ATCC14028s) was incubated with or without 2 mm indole. Cells were then challenged to benzalkonium (500 mg/ml) for 10 min. The survival levels of the indole-treated cells were normalized to 100%, and untreated cells are displayed relative to those of the indole-treated cells. The actual survival of indole-treated cells was 0.025%.
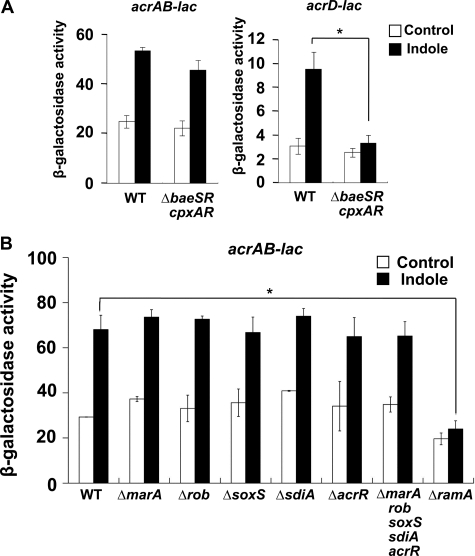
Indole Induces acrAB Expression via the RamA Regulator—Among the multidrug efflux pumps, AcrAB plays a major role in the intrinsic resistance of Salmonella (25). Also, Hirakawa et al. (35) reported that the baeSR and cpxAR signal transduction system genes are required for indole induction of multidrug efflux pumps in E. coli. To identify the regulatory elements that induce acrAB in response to indole in Salmonella, we constructed a mutant that lacked baeSR and cpxAR. In the ΔbaeSR cpxAR mutant, the expression of acrAB was not significantly different from that in the wild-type (WT) strain; however, indole induction of acrD was significantly lower in the mutant compared with the WT strain (Fig. 2A). The result indicates that the BaeSR and CpxAR signal transduction systems are not involved in indole induction of acrAB, whereas they are required for acrD induction. Other regulators, marA, soxS, rob, sdiA, and acrR, have been previously reported to control acrAB expression in E. coli (27). With the exception of ramA, none significantly altered the indole induction of acrAB in Salmonella (Fig. 2B). The stimulatory effect of indole on acrAB expression was completely eliminated in the ΔramA mutant (Fig. 2B). The results indicate that the RamA regulator is required for indole induction of acrAB in Salmonella.
FIGURE 2.
Indole activation of acrAB expression through the RamA regulator. The data correspond to mean values from three independent experiments. Bars correspond to the standard deviation. Asterisks indicate statistically significant differences (*, p < 0.01) in the paired Student's t test. A, β-galactosidase levels in WT or ΔbaeSR cpxAR strains, carrying acrAB-lac and acrD-lac transcriptional fusions, grown in LB medium with (solid bars) or without (open bars) 2 mm indole. B, β-galactosidase levels were assayed in strains carrying the acrAB-lac transcriptional fusion in the WT (NKS505), ΔmarA (NES20), Δrob (NES29), ΔsoxS (NES28), ΔsdiA (NES35), ΔacrR (NES48), ΔmarA rob soxS sdiA acrR (NES55), and ΔramA (NES58) strains. Strains were grown in LB medium with (solid bars) or without (open bars) 2 mm indole.
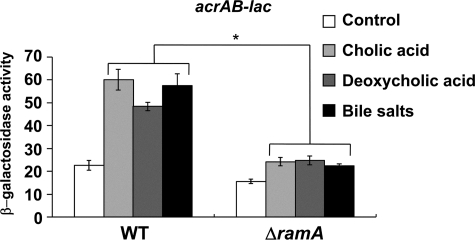
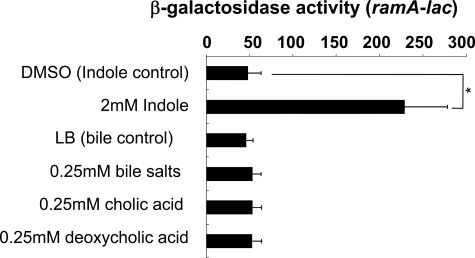
acrAB Activation by Bile Is Dependent on the RamA Regulator—The AcrAB pump is reported to export bile salts and play a role in bile resistance in E. coli and Salmonella (44-46). Also, acrAB is reportedly induced by bile in a Robdependent manner in E. coli (31). Although acrAB is also induced by bile in Salmonella, the induction mediating regulator is unknown (47). Prouty et al. (47) further reported that acrAB activation by bile is independent of MarA, Rob, PhoP/PhoQ, and RpoS. We investigated the possibility that RamA controls acrAB expression in response to bile. In agreement with Prouty et al. (47), bile salts, cholic acid, and deoxycholic acid significantly induced acrAB expression in Salmonella (Fig. 3). When ramA was deleted, acrAB induction was eliminated (Fig. 3). These findings indicate a novel RamA-dependent pathway for bile-mediated regulation of the AcrAB efflux pump in Salmonella, different from that observed in E. coli.
FIGURE 3.
Requirement of RamA for induction of acrAB by bile. β-Galactosidase levels were assayed in WT (NKS505) or ΔramA (NES58) strains carrying the acrAB-lac transcriptional fusion. Cells were grown in LB medium (control) or LB medium supplemented with 0.25 mm cholic acid, 0.25 mm deoxycholic acid, or 0.25 mm bile salts. The data correspond to mean values from three independent experiments. Bars correspond to the standard deviation. Student's t test; *, p < 0.01 versus WT.
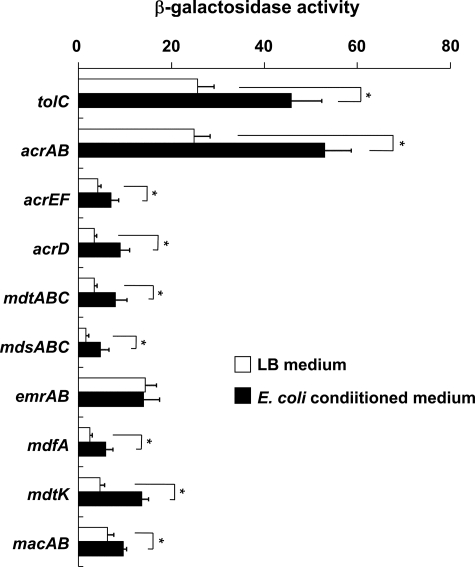
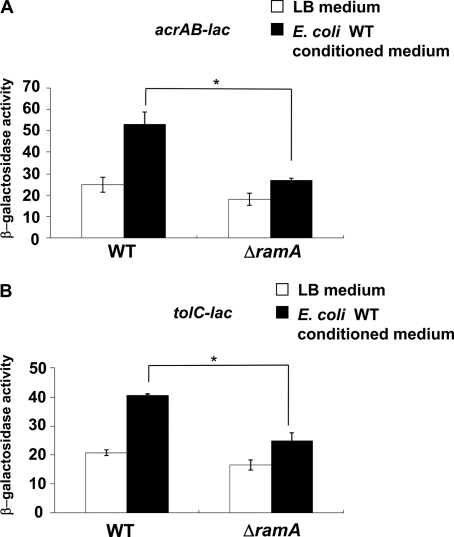
Conditioned Medium from E. coli Induces Salmonella acrAB and tolC Genes via the RamA Regulator—Indole accumulates and MdtEF is induced in stationary phase cultures of E. coli, but experiments with a tnaAB mutant showed that indole partially contributes to this induction (48). These results indicate that E. coli produces indole as well as other efflux pump inducers. Therefore, we investigated whether an E. coli conditioned medium would induce multidrug efflux pumps in Salmonella. Conditioned medium, prepared from E. coli MG1655, significantly induced eight Salmonella multidrug efflux pumps, including acrAB and the outer membrane protein gene tolC (Fig. 4). Inductions of acrAB and tolC were significantly decreased in ΔramA (Fig. 5, A and B), indicating that the E. coli conditioned medium induced acrAB and tolC via the RamA regulator.
FIGURE 4.
Multidrug efflux genes in Salmonella enterica serovar Typhimurium induced by an E. coli conditioned medium. The tolC-lac (EG15109), acrAB-lac (NKS505), acrEF-lac (EG15114), acrD-lac (EG15120), mdtABC-lac (EG15124), mdsABC-lac (NKS517), emrAB-lac (NKS522), mdfA-lac (NKS524), mdtK-lac (EG15132), and macAB-lac (NKS530) strains were grown in conditioned medium prepared from E. coli culture. Expression levels of multidrug efflux genes were determined by β-galactosidase assay. The data correspond to mean values of three independent experiments. Error bars correspond to the standard deviation. Asterisks indicate statistically significant differences (*, p < 0.01) in the paired Student's t test.
FIGURE 5.
RamA induction of acrAB and tolC by conditioned medium of E. coli. The expression of acrAB (A) and tolC (B) determined by β-galactosidase assay using strains acrAB-lac (NKS505), ΔramA/acrAB-lac (NES58), tolC-lac (EG15109), or ΔramA/tolC-lac (NES65) grown in LB medium or conditioned medium from E. coli MG1655. The data correspond to mean values from three independent experiments. Bars correspond to the standard deviation. Asterisks indicate statistically significant differences (*, p < 0.01) in the paired Student's t test.
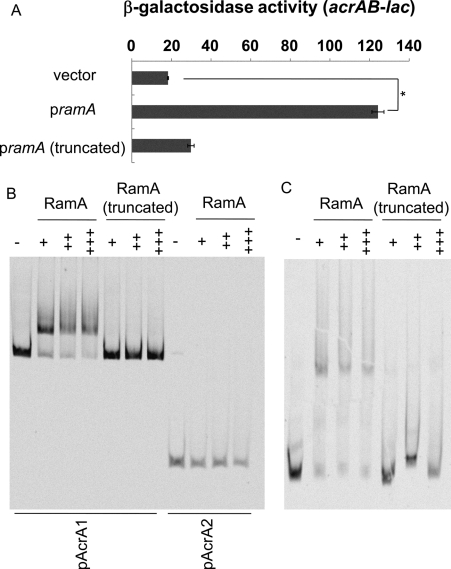
RamA Binds to the Upstream Regions of acrA and tolC—The aforementioned results indicate that RamA plays a major role in inducing acrAB in response to environmental signals such as indole and bile. To understand the regulation of acrAB by RamA, electrophoretic mobility shift assays (EMSA) with the RamA protein were performed. Plasmids encoding the histidine-tagged RamA or the N terminus truncated RamA proteins were constructed (Table 2). Because it was reported that RamA overproduction was related to increased AcrAB expression in clinical Klebsiella pneumoniae isolates (49), we investigated the effect of histidine-tagged RamA on acrAB expression. Overproduction of histidine-tagged RamA significantly induced the expression of acrAB (Fig. 6A); however, the truncated RamA did not induce acrAB (Fig. 6A) and was used as a negative control in subsequent EMSA. Upstream regions of acrA were amplified by PCR, and the fragments were incubated with RamA or truncated RamA protein. RamA bound to pAcrA1, whereas truncated RamA did not (Fig. 6B). However, RamA did not bind to pAcrA2, indicating that the RamA-binding site is located between -795 and -142 upstream of acrA (Fig. 6B). RamA did bind to the upstream region of tolC, whereas the truncated RamA did not (Fig. 6C). These results indicate that RamA directly controls the expression of acrAB and tolC.
FIGURE 6.
RamA binds to the upstream region of acrA and tolC. A, β-galactosidase activity measured with acrAB-lac (NKS505) harboring a plasmid expressing ramA, truncated ramA, or the vector control (pMALc2X). The data correspond to mean values from three independent experiments. Error bars correspond to the standard deviation. Student's t test; *, p < 0.01 versus control. B and C, EMSA images for RamA binding to the upstream regions of acrA (B) and tolC (C). Upstream regions of acrA (pAcrA1, -795 to +16 region relative to the start codon of acrA; pAcrA2, -141 to +16) (A) and tolC (-250 to -1 region relative to the start codon of tolC)(C) were incubated with various concentrations of RamA or N-terminal truncated RamA. Protein concentrations are as follows: -, without protein; +, 1.0 μm; ++, 1.5 μm; +++, 2.0 μm.
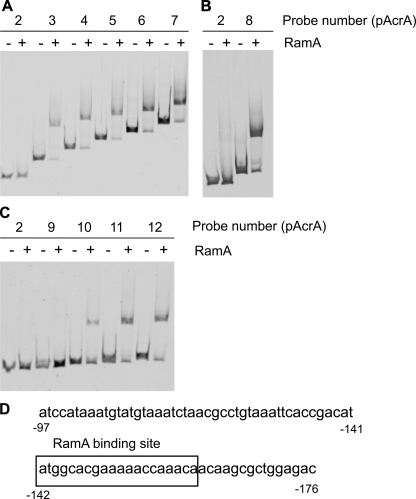
Determination of RamA-binding Sites for acrA and tolC—To determine the RamA-binding site for acrA, we prepared different lengths of DNA fragments for EMSA. The fragments used were as follows: pAcrA3 (-241 to +16, the numbering is relative to the start codon of acrA), pAcrA4 (-341 to +16), pAcrA5 (-441 to +16), pAcrA6 (-541 to +16), and pAcrA7 (-641 to +16). RamA bound to pAcrA3-7, but it did not bind to pAcr2 (Fig. 7A). These results indicate that the RamA-binding site was between -241 and -142. We then examined fragment pAcrA8 (-191 to +16); RamA bound to this fragment, indicating a binding site between -191 and -142 (Fig. 7B). Further examination with pAcrA9 (-151 to +16), pAcrA10 (-161 to +16), pAcrA11 (-171 to +16), and pAcrA12 (-181 to +16) revealed that RamA bound to pAcrA10-12 but not to pAcrA9 (Fig. 7C). These results indicate that the -161 to -152-bp region is required for RamA binding. It was previously reported that RamA bound to a 20-bp asymmetric sequence with a degenerate consensus soxbox of AYNGCAC-NNWNNRYYAAAYN (N = any base; R = A/G; W = A/T; Y = C/T) (50, 51). A DNA sequence resembling this consensus soxbox sequence, ATGGCACGAAAAACCAAACA, was located at -161 to -142 (Fig. 7D).
FIGURE 7.
A-C, determination of RamA-binding site for acrA. EMSA of RamA binding to the upstream regions of acrA is shown. DNA fragments, including upstream regions of acrA, were incubated without (-) or with (+) purified RamA (1.0 μm). DNA fragments are as follows: pAcr2 (-141 to +16, the numbering is relative to the start codon of acrA), pAcrA3 (-241 to +16), pAcrA4 (-341 to +16), pAcrA5 (-441 to +16), pAcrA6 (-541 to +16), pAcrA7 (-641 to +16) (A), pAcrA8 (-191 to +16) (B), pAcrA9 (-151 to +16), pAcrA10 (-161 to +16), pAcrA11 (-171 to +16), and pAcrA12 (-181 to +16) (C). D, nucleotide sequence in the upstream region of acrA. Boxed sequence corresponds to the RamA-binding site. The numbers indicate the positions from the start codon of acrA.
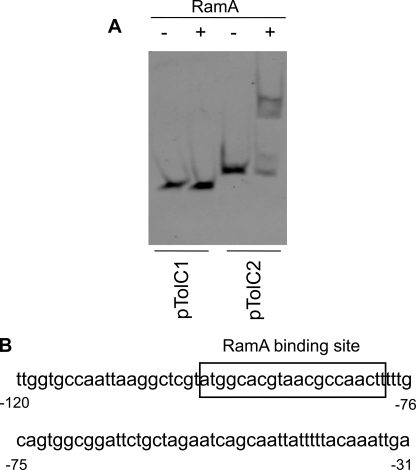
We also determined a RamA-binding site located upstream of tolC. In a tolC promoter, we found a soxbox sequence between -99 and -80 (the numbering is relative to the start codon of tolC). Therefore, we prepared fragments of pTolC1 (-79 to -1) and pTolC2 (-99 to -1) to determine binding location. RamA bound to pTolC2 but did not bind to pTolC1 (Fig. 8A) indicating that RamA binds between -99 to -80 and contains the ATGGCACGTAACGCCAACTT consensus sequence (Fig. 8B).
FIGURE 8.
RamA-binding site for tolC. A, DNA fragments, including tolC promoter regions pTolC1 (-80 to -1; the numbering is relative to the start codon of tolC) and pTolC2 (-100 to +1), were incubated without (-) or with (+) purified RamA (1.0 μm). B, nucleotide sequence upstream region of tolC. Boxed sequence corresponds to the RamA-binding site. The numbers indicate the positions from the start codon of tolC.
Indole Induces ramA Expression but Bile Does Not—The effects of indole and bile on ramA expression levels were investigated because increased ramA expression has been reported to cause increased production of the AcrAB-TolC efflux system (49). Using a reporter plasmid of ramA, a β-galactosidase assay showed that indole enhanced the promoter activity of ramA (Fig. 9). This suggests that indole induces acrAB through increased expression of ramA. Bile salts, cholic acid, and deoxycholic acid did not affect the expression level of ramA despite its requirement for induction of acrAB. This indicates an acrAB regulatory mode other than through increased production of RamA.
FIGURE 9.
Effect of indole and bile on ramA transcription. β-Galactosidase levels were assayed in the WT strain carrying the ramA reporter plasmid (pNNramA) (NES84). Cells were grown in LB medium supplemented with 2 mm indole, 0.25 mm cholic acid, 0.25 mm deoxycholic acid, or 0.25 mm bile salts. The data correspond to mean values from three independent experiments. Bars correspond to the standard deviation. Student's t test; *, p < 0.01 versus control.
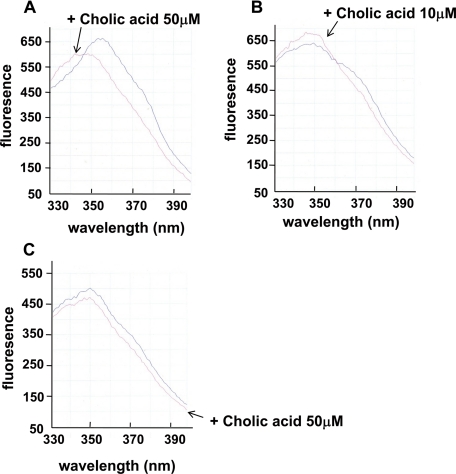
Binding of Bile to RamA Protein—The failure of bile to affect the expression level of ramA suggests that RamA may detect the presence of a bile acid component such as cholic acid. This was explored using the intrinsic (tryptophan) spectrum of RamA, as described by Rosenberg et al. (31). When 50 μm cholic acid was added to 75 nm RamA, there was a strong blue shift in the emission spectrum (Fig. 10A). The blue shift was also slightly seen with 10 μm cholic acid and 75 nm RamA (Fig. 10B). In contrast, a blue shift was not observed when 50 μm cholic acid was added to 75 nm truncated RamA (Fig. 10C). These results indicate that bile binds to RamA to induce acrAB expression in Salmonella.
FIGURE 10.
Binding of bile to the RamA protein. A, cholic acid (50 μm) produces a blue shift in the intrinsic spectrum of RamA. Intrinsic fluorescence of RamA (75 nm) was measured in the absence or presence of cholic acid. B, cholic acid (10 μm) also produces a blue shift in the intrinsic spectrum of RamA. C, blue shift in the intrinsic spectrum of truncated RamA was not produced by 50 μm cholic acid.
DISCUSSION
Very few signals inducing multidrug efflux pumps in Salmonella have been identified so far (47, 52). In this study, we found that indole, bile, and an E. coli conditioned medium induced several multidrug efflux genes in Salmonella. We found that acrAB induction by these three signal sources is completely dependent on the Salmonella-specific regulator RamA, indicating that RamA plays a major role in inducing acrAB. RamA belongs to the AraC transcriptional activator family, and activation of RamA is reported to confer drug resistance on Salmonella (53). In Salmonella, RamA is also involved in resistance to superoxide (54) and in paraquat induction of the flavohemoglobin gene (50).
The ramA gene appears to be specific for Salmonella serovars and is absent in many other Gram-negative microorganisms; notable exceptions are K. pneumoniae and Enterobacter spp. (54-56). The results of genomic comparison indicate that the gene organization surrounding ramA gene and the corresponding region in E. coli are similar, with two exceptions as follows: the absence of ramA and the presence of Yi81-2 in E. coli (57). We suggest that the AcrAB induction pathway in Salmonella is different from that in E. coli. Bile induces AcrAB in both Salmonella and E. coli. In E. coli, the transcriptional factor Rob plays a major role in inducing acrAB expression in response to bile (31). However, our data indicate that bile induction of acrAB in Salmonella is completely dependent on RamA, not Rob. Other regulators, including MarA, SoxS, SdiA, and AcrR, are not involved in AcrAB induction by indole and bile. These results suggest that RamA is a master regulator of Salmonella acrAB and may mask the contributions of any other acrAB regulators. In E. coli, it was reported that multiple regulators, including MarA, Rob, SoxS, and SdiA, work coordinately in controlling acrAB expression in response to acrAB inducers. This may be related to the lack of RamA in E. coli. Indeed, overproduction of RamA has induced the drug resistance level of E. coli (53, 57). A recent report suggests that RamA and RamR, not SoxS and MarA, are involved in AcrAB-mediated multidrug resistance in Salmonella (58). Based on our results and these other studies, RamA appears to be the master regulator of acrAB in Salmonella.
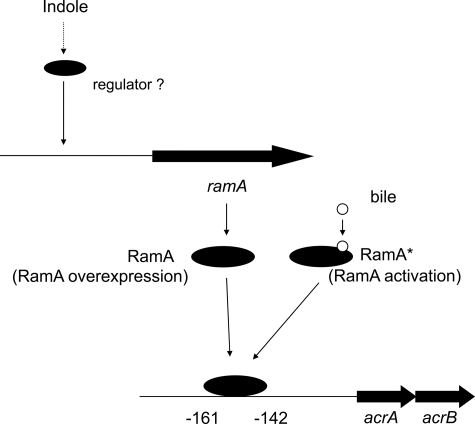
We also suggest the existence of a different induction mechanism for acrAB via the RamA regulator (Fig. 11). Indole was shown to induce ramA expression (Fig. 9), and such an increased expression of ramA can induce acrAB (Fig. 6A). On the other hand, bile did not affect expression of ramA (Fig. 9), but it did bind to RamA (Fig. 11). This is reminiscent of the binding of bile to the Rob protein involved in regulation of acrAB in E. coli (31). We also suggest that the N-terminal domain of RamA may be required for binding of bile because cholic acid did not bind to the truncated RamA (Fig. 10). Our results suggest a mechanism in which RamA can change between an “activated state” and an “overexpressed state” in response to environmental signals, thereby inducing the AcrAB-TolC system (Fig. 11). Thus, RamA can be converted from a low activity state to a high activity state in response to bile. We also suggest that Salmonella may have an additional sensor for indole that controls ramA expression (Fig. 11).
FIGURE 11.
Proposed model for the expression of acrA and acrB multidrug efflux genes by RamA. In one pathway, bile may bind to the RamA protein, which is then converted from a low to a high activity state. In the other path, indole may activate ramA transcription to directly induce acrA and acrB.
Indole and bile are found in various internal human environments, especially in the intestine (59, 60). Indole is produced by many enteric bacterial species (60), and bile is often present at high concentration in the intestinal tract (59). Therefore, RamA may be required for Salmonella to detect environmental signals and for subsequent induction of the AcrAB-TolC system, resulting in excretion of toxic compounds by Salmonella into surrounding environments, such as the intestine.
Acknowledgments
We thank Eduardo A. Groisman, Barry L. Wanner, and Ronald W. Davis for providing strains and plasmids and Hiroki Sakata for communicating unpublished results.
Author's Choice—Final version full access.
This work was supported in part by Takeda Science Foundation, Inamori Foundation, the PRESTO program of the Japan Science and Technology Agency (to K. N.), the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, and by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Society for the Promotion of Science (to A. Y. and K. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: WT, wild type; LB, Luria-Bertani; EMSA, electrophoretic mobility shift assay.
References
- 1.Scherer, C. A., and Miller, S. I. (2001) in Principles of Bacterial Pathogenesis (Groisman, E. A., ed), pp. 266-333, Academic Press, New York
- 2.Threlfall, E. J., Frost, J. A., Ward, L. R., and Rowe, B. (1996) Lancet 347 1053-1054 [DOI] [PubMed] [Google Scholar]
- 3.Threlfall, E. J., Ward, L. R., Skinner, J. A., and Rowe, B. (1997) Microb. Drug Resist. 3 263-266 [DOI] [PubMed] [Google Scholar]
- 4.Grein, T., O'Flanagan, D., McCarthy, T., and Bauer, D. (1999) Ir. Med. J. 92 238-241 [PubMed] [Google Scholar]
- 5.Hosek, G., Leschinsky, D. D., Irons, S., and Safranek, T. J. (1997) Morb. Mortal. Wkly. Rep. 46 308-310 [Google Scholar]
- 6.Ng, L. K., Mulvey, M. R., Martin, I., Peters, G. A., and Johnson, W. (1999) Antimicrob. Agents Chemother. 43 3018-3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cody, S. H., Abbott, S. L., Marfin, A. A., Schulz, B., Wagner, P., Robbins, K., Mohle-Boetani, J. C., and Vugia, D. J. (1999) J. Am. Med. Assoc. 281 1805-1810 [DOI] [PubMed] [Google Scholar]
- 8.Davies, A., O'Neill, P., Towers, L., and Cooke, M. (1996) Commun. Dis. Rep. CDR Rev. 6 R159-R162 [PubMed] [Google Scholar]
- 9.Molbak, K., Baggesen, D. L., Aarestrup, F. M., Ebbesen, J. M., Engberg, J., Frydendahl, K., Gerner-Smidt, P., Petersen, A. M., and Wegener, H. C. (1999) N. Engl. J. Med. 341 1420-1425 [DOI] [PubMed] [Google Scholar]
- 10.Villar, R. G., Macek, M. D., Simons, S., Hayes, P. S., Goldoft, M. J., Lewis, J. H., Rowan, L. L., Hursh, D., Patnode, M., and Mead, P. S. (1999) J. Am. Med. Assoc. 281 1811-1816 [DOI] [PubMed] [Google Scholar]
- 11.Cloeckaert, A., and Chaslus-Dancla, E. (2001) Vet. Res. 32 291-300 [DOI] [PubMed] [Google Scholar]
- 12.Piddock, L. J. (2002) FEMS Microbiol. Rev. 26 3-16 [DOI] [PubMed] [Google Scholar]
- 13.Baucheron, S., Chaslus-Dancla, E., and Cloeckaert, A. (2004) J. Antimicrob. Chemother. 53 657-659 [DOI] [PubMed] [Google Scholar]
- 14.Baucheron, S., Imberechts, H., Chaslus-Dancla, E., and Cloeckaert, A. (2002) Microb. Drug Resist. 8 281-289 [DOI] [PubMed] [Google Scholar]
- 15.Li, X. Z., and Nikaido, H. (2004) Drugs 64 159-204 [DOI] [PubMed] [Google Scholar]
- 16.Schuldiner, S. (2006) Nature 443 156-157 [DOI] [PubMed] [Google Scholar]
- 17.Nikaido, H. (1996) J. Bacteriol. 178 5853-5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zgurskaya, H. I., and Nikaido, H. (2000) Mol. Microbiol. 37 219-225 [DOI] [PubMed] [Google Scholar]
- 19.Brown, M. H., Paulsen, I. T., and Skurray, R. A. (1999) Mol. Microbiol. 31 394-395 [DOI] [PubMed] [Google Scholar]
- 20.Paulsen, I. T., Chen, J., Nelson, K. E., and Saier, M. H., Jr. (2001) J. Mol. Microbiol. Biotechnol. 3 145-150 [PubMed] [Google Scholar]
- 21.Putman, M., van Veen, H. W., and Konings, W. N. (2000) Microbiol. Mol. Biol. Rev. 64 672-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., and Yamaguchi, A. (2006) Nature 443 173-179 [DOI] [PubMed] [Google Scholar]
- 23.Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002) Nature 419 587-593 [DOI] [PubMed] [Google Scholar]
- 24.Yu, E. W., Aires, J. R., and Nikaido, H. (2003) J. Bacteriol. 185 5657-5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishino, K., Latifi, T., and Groisman, E. A. (2006) Mol. Microbiol. 59 126-141 [DOI] [PubMed] [Google Scholar]
- 26.Ahmed, M., Borsch, C. M., Taylor, S. S., Vazquez-Laslop, N., and Neyfakh, A. A. (1994) J. Biol. Chem. 269 28506-28513 [PubMed] [Google Scholar]
- 27.Grkovic, S., Brown, M. H., and Skurray, R. A. (2002) Microbiol. Mol. Biol. Rev. 66 671-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomovskaya, O., Lewis, K., and Matin, A. (1995) J. Bacteriol. 177 2328-2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, D., Alberti, M., Lynch, C., Nikaido, H., and Hearst, J. E. (1996) Mol. Microbiol. 19 101-112 [DOI] [PubMed] [Google Scholar]
- 30.Randall, L. P., and Woodward, M. J. (2002) Res. Vet. Sci. 72 87-93 [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg, E. Y., Bertenthal, D., Nilles, M. L., Bertrand, K. P., and Nikaido, H. (2003) Mol. Microbiol. 48 1609-1619 [DOI] [PubMed] [Google Scholar]
- 32.Olliver, A., Valle, M., Chaslus-Dancla, E., and Cloeckaert, A. (2004) FEMS Microbiol. Lett. 238 267-272 [DOI] [PubMed] [Google Scholar]
- 33.Eaves, D. J., Ricci, V., and Piddock, L. J. (2004) Antimicrob. Agents Chemother. 48 1145-1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olliver, A., Valle, M., Chaslus-Dancla, E., and Cloeckaert, A. (2005) Antimicrob. Agents Chemother. 49 289-301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T., and Yamaguchi, A. (2005) Mol. Microbiol. 55 1113-1126 [DOI] [PubMed] [Google Scholar]
- 36.Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 5189-5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis, R. W., Bolstein, D., and Roth, J. R. (1980) Advanced Bacterial Genetics, pp. 100-105, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 38.Elledge, S. J., and Davis, R. W. (1989) Genes Dev. 3 185-197 [DOI] [PubMed] [Google Scholar]
- 39.Datsenko, K. A., and Wanner, B. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6640-6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. (1972) Experiments in Molecular Genetics, pp. 352-355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 41.Baca-DeLancey, R. R., South, M. M., Ding, X., and Rather, P. N. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4610-4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanofsky, C., Horn, V., and Gollnick, P. (1991) J. Bacteriol. 173 6009-6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., Hou, S., Layman, D., Leonard, S., Nguyen, C., Scott, K., Holmes, A., Grewal, N., Mulvaney, E., Ryan, E., Sun, H., Florea, L., Miller, W., Stoneking, T., Nhan, M., Waterston, R., and Wilson, R. K. (2001) Nature 413 852-856 [DOI] [PubMed] [Google Scholar]
- 44.Lacroix, F. J., Cloeckaert, A., Grepinet, O., Pinault, C., Popoff, M. Y., Waxin, H., and Pardon, P. (1996) FEMS Microbiol. Lett. 135 161-167 [DOI] [PubMed] [Google Scholar]
- 45.Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H., and Hearst, J. E. (1995) Mol. Microbiol. 16 45-55 [DOI] [PubMed] [Google Scholar]
- 46.Thanassi, D. G., Cheng, L. W., and Nikaido, H. (1997) J. Bacteriol. 179 2512-2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prouty, A. M., Brodsky, I. E., Falkow, S., and Gunn, J. S. (2004) Microbiology 150 775-783 [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi, A., Hirakawa, H., Hirata, T., Nishino, K., and Yamaguchi, A. (2006) J. Bacteriol. 188 5693-5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneiders, T., Amyes, S. G., and Levy, S. B. (2003) Antimicrob. Agents Chemother. 47 2831-2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez-Urzua, E., Zamorano-Sanchez, D. S., Ponce-Coria, J., Morett, E., Grogan, S., Poole, R. K., and Membrillo-Hernandez, J. (2007) Arch. Microbiol. 187 67-77 [DOI] [PubMed] [Google Scholar]
- 51.Martin, R. G., and Rosner, J. L. (2002) Mol. Microbiol. 44 1611-1624 [DOI] [PubMed] [Google Scholar]
- 52.Nishino, K., Nikaido, E., and Yamaguchi, A. (2007) J. Bacteriol. 189 9066-9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Straaten, T., Janssen, R., Mevius, D. J., and van Dissel, J. T. (2004) Antimicrob. Agents Chemother. 48 2292-2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Straaten, T., Zulianello, L., van Diepen, A., Granger, D. L., Janssen, R., and van Dissel, J. T. (2004) Infect. Immun. 72 996-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chollet, R., Chevalier, J., Bollet, C., Pages, J. M., and Davin-Regli, A. (2004) Antimicrob. Agents Chemother. 48 2518-2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komatsu, T., Ohta, M., Kido, N., Arakawa, Y., Ito, H., Mizuno, T., and Kato, N. (1990) J. Bacteriol. 172 4082-4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yassien, M. A., Ewis, H. E., Lu, C. D., and Abdelal, A. T. (2002) Antimicrob. Agents Chemother. 46 360-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abouzeed, Y. M., Baucheron, S., and Cloeckaert, A. (2008) Antimicrob. Agents Chemother. 52 2428-2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batta, A. K., Salen, G., Batta, P., Tint, G. S., Alberts, D. S., and Earnest, D. L. (2002) J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 775 153-161 [DOI] [PubMed] [Google Scholar]
- 60.Sonnenwirth, A. C. (1980) in The Enteric Bacteria and Bacteroides (Davis, B. D., Dulbecco, R., Eisen, H. N., and Ginsberg, H. S., eds), pp. 645-652, Harper & Row, Publishers, Inc., Philadelphia, PA