Abstract
During co-translational protein translocation, the ribosome associates with a membrane channel, formed by the Sec61 complex, and recruits the translocon-associated protein complex (TRAP). Here, we report the structure of a ribosome-channel complex from mammalian endoplasmic reticulum in which the channel has been visualized at 11Å resolution. In this complex, single copies of Sec61 and TRAP associate with a non-translating ribosome and this stoichiometry was verified by quantitative mass spectrometry. A bilayer-like density surrounds the channel and can be attributed to lipid and detergent. The crystal structure of an archaeal homolog of the Sec61 complex was then docked into the map. In this model, two cytoplasmic loops of Sec61 may interact with RNA helices H6, H7 and H50, while the central pore is located below the ribosome tunnel exit. Hence, this copy of Sec61 is positioned to capture and translocate the nascent chain. Finally, we show that mammalian and bacterial ribosome-channel complexes have similar architectures.
Keywords: ribosome-channel complex, Sec61 complex, co-translational translocation, translocon associated protein complex (TRAP)
Introduction
Many proteins are translocated across the endoplasmic reticulum (ER) membrane as they are being translated by the ribosome (Rapoport, 2007). During translocation, the ribosome binds to a membrane channel that is formed by the heterotrimeric Sec61 complex which consists of the α-, β and γ-subunits. Secretory and other soluble proteins are transported completely through the channel, while hydrophobic segments exit the channel through a lateral gate and become trans-membrane (TM) segments. Protein translocation in bacteria and archaea uses a homolog of Sec61, the SecY complex, to form the channel.
The crystal structure of an archaeal-subunit is comprised of two helix bundles consisting of TMs 1-5 and 6-10 (van den Berg et al., 2004). The helix bundles form an hourglass-shaped pore that is plugged at the extra-cellular side by a short helix (TM2a). The constriction of the pore is formed by a ring of hydrophobic residues, whose side chains surround the translocating polypeptide chain (Cannon et al., 2005). During initiation of translocation, a signal sequence or TM segment of a nascent polypeptide chain intercalates into the walls of the channel between TMs 2b and 7 (Plath et al., 1998). These helices are part of the lateral gate and their separation likely destabilizes the interactions of TM2a, causing it to move towards the back of the channel to open the pore (Tam et al., 2005). The SecY crystal structure and other data indicate that the translocation pore is formed from a single copy of the SecY complex (van den Berg et al., 2004; Osborne and Rapoport, 2007).
A central, unresolved issue is how ribosomes interact with SecY or Sec61 during co-translational translocation. The structure of an E. coli ribosome with an associated nascent chain and SecY channel has been determined by electron cryo-microscopy at ∼15Å resolution (Mitra et al., 2005). Based on this structure, a model was proposed in which two copies of the SecY complex are bound to the ribosome in a near front-to-front orientation. It was further postulated that the pores of the two SecY molecules may fuse during translocation (Mitra et al., 2006). However, a recent structure shows that a non-translating ribosome binds a single copy of the SecY complex with the pore of SecY located beneath the ribosome tunnel exit (Ménétret et al., 2007). This SecY copy could thus form the channel. In addition, the location and the orientation of SecY in this model are not similar to either copy of SecY in the dimer model (Ménétret et al., 2007; Mitra et al., 2005).
In eukaryotes, ribosome-Sec61 complexes have a donut-like structure beneath the ribosome (Hanein et al., 1996; Beckmann et al., 1997; Ménétret et al., 2000). Based on the volume of the electron density, it was suggested that this feature may contain 3 or 4 copies of the Sec61 complex (Beckmann et al., 2001; Ménétret et al., 2005). In addition, 3-4 connections were seen between the ribosome and channel, consistent with the idea that multiple Sec61 molecules are present in the complex. However, at low resolution it may be difficult to distinguish between density contributed by protein, detergent or lipid. In addition, the choice of an appropriate threshold is problematic at lower resolution, as small errors can result in the appearance of spurious, yet reproducible connections between closely opposed objects. Thus, a clear picture of how Sec61 binds to the ribosome is not yet available.
In mammals, the active translocation complex contains additional membrane proteins. These include the signal peptidase, the oligosacharyl transferase, the translocating chain-associated membrane protein (TRAM) and the translocon-associated protein complex (TRAP) (Osborne et al., 2005; Johnson and Waes, 1999). The TRAP complex remains stably associated with detergent-solubilized, ribosome-Sec61 complexes and has a prominent lumenal domain that is located beneath the channel (Ménétret et al., 2005). The TRAP complex is comprised of four membrane protein subunits. The α-, β-, and δ-subunits are single-spanning membrane proteins, while the γ-subunit the crosses the membrane four times (Hartmann et al., 1993). TRAP can be crosslinked to nascent chains (Wiedmann et al., 1989; Görlich et al., 1992, Mothes et al., 1994) and may help translocate proteins that have prolonged access to the cytoplasm (Fons et al., 2003). However, the exact function of TRAP remains to be clarified.
Recently, we reported the structure of the mammalian ribosome at ∼8.7Å resolution in a ribosome-channel complex (Chandramouli et al., 2008). We have now performed a detailed study of the channel in this improved map. The new structure indicates that a single copy of Sec61 is bound to the non-translating ribosome. In particular, we were able to use a crystal structure of the archaeal SecY complex, as a model for Sec61, to dock two cytoplasmic loops into a central connection at the tunnel exit. This placed a single copy of Sec61 in the center of a membrane-like disk with a single copy of TRAP located next to Sec61. Quantitative mass spectrometry verified the 1:1 stoichiometry of Sec61 and TRAP in the complex. In the density map, we find that Sec61 is positioned below the ribosome tunnel exit, where it may capture and translocate the nascent chain. In addition, our data show that interactions of Sec61 with the mammalian ribosome are fundamentally similar to those observed in a bacterial ribosome-SecY complex (Ménétret et al., 2007).
Results
Single copies of Sec61 and TRAP in the native channel
We recently determined the structure of a mammalian ribosome at 8.7 Å resolution by analyzing images of frozen-hydrated, ribosome-channel complexes (RCCs) (Chandramouli et al., 2008). We have now used this electron density map to perform a detailed study of the channel. The samples were prepared from pancreatic rough microsomes treated with puromycin and 500 mM potassium acetate (PKRMs) (Morgan et al., 2002; Ménétret et al., 2005). This treatment strips ribosomes from the membranes and also moves the P-site tRNA into the E-site. The ribosomes were pelleted and then added back to an excess of stripped microsomes, so that all the ribosomes would associate with a channel complex. The membranes were floated in a sucrose gradient and solubilized in digitonin. The resulting RCCs were sedimented, resuspended and then analyzed by electron cryo-microscopy.
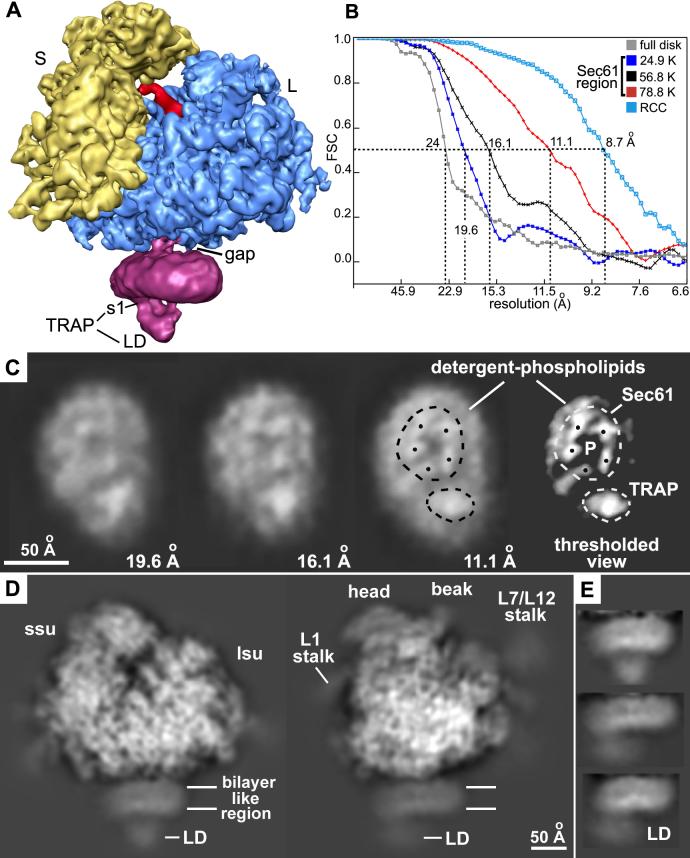
A front view of the final map derived from ∼79,000 particles is shown in Figure 1A. In the map, the channel (shown in magenta) is separated from the ribosome by a gap of 10-12Å and is linked to the large subunit by a central connection. In addition, the TRAP complex is present at the back of the channel and has a prominent lumenal domain. The region that would normally be contained in the ER membrane is preserved at ∼24Å resolution, based on the 0.5 value from a Fourier shell correlation (FSC) curve (Chandramouli et al., 2008, Figure 1B, grey curve). However, a calculated projection of the disk-like membrane region revealed two high density features (shown in white). The larger one is similar in size to the Sec61 complex and the smaller one may contain the TRAP complex (Figure 1C, see the third and fourth panels from the left). In this projection, the high-density area corresponding to Sec61 has five features arranged in a ring around a central pore. The resolution of this region was estimated to be ∼11.1Å based on the FSC0.5 with a suitable mask (Figure 1B, red curve). When the crystal structure of the archaeal SecY complex is low-pass filtered at 11Å resolution and viewed in projection, a similar density distribution can be seen for the membrane-embedded region (data not shown). Together, these data suggest that single molecules of Sec61 and TRAP are bound to the ribosome and that the additional density surrounding the proteins may originate from lipid and detergent.
Figure 1.
Single copies of Sec61 and TRAP are bound to a non-translating mammalian ribosome.
A. The 3D map of the RCC is shown as a rendered surface with the ribosome at 8.7Å resolution and the channel region (in magenta) filtered to ∼16Å resolution. The small (S) and large (L) subunits are shown in yellow and blue.
B. Resolution curves are shown for the complete RCC (light blue line), the membrane-like disk (grey) and for the Sec61 region in three maps calculated with a total of ∼24,900 (blue), 56,800 (black) and 78,800 (red) particles.
C. Projections of the membrane-like disk are shown for three maps in which the ribosome was determined at resolutions of 16, 14 and 8.7Å, respectively while the Sec61 regions were imaged at 19.6, 16.1 and 11.1Å resolution (left to right, panels 1-3). Higher density is shown in white. On the far right, the denisty of the projection of the membrane-like disk has been thresholded to show that the brightest features that arise from Sec61 and TRAP in the final map (panel 4).
D. Two projections are shown of the final map with the channel oriented at the bottom. The map has been filtered to ∼12Å resolution and strongly scattering material is shown in white. Small and large subunits are indicated (ssu and lsu), along with some flexible regions (head, beak of ssu and L7/L12 stalk). The density in the channel region is viewed edge-on and resembles a bilayer. The position of the TRAP lumenal domain (LD) is marked.
E. Close-ups are shown of edge-on projections of the membrane-like disk in which the contrast has been adjusted to more clearly show the bilayer-like appearance.
This data implies that previous structures were not able to resolve the membrane-embedded proteins, perhaps due to contrast matching (Ménétret et al., 2005; Morgan et al., 2002). We analyzed subsets of the final dataset to test this idea. Two smaller datasets were independently refined with EMAN and 3D maps were calculated with ∼25,000 and ∼57,000 particles, respectively. The resolution of the membrane-embedded region in these two maps was estimated to be ∼19.6Å and ∼16.8Å (Figure 1B; dark blue and black curves). Because the same mask was used for the Sec61 region in all the maps, the masking itself was not responsible for the increase in resolution. Projections of the membrane-embedded regions were calculated from these maps. In these projections, the protein features became more distinct as the resolution was improved (Figure 1C, compare the first 3 panels). Thus, higher resolution is required to overcome contrast matching between protein, lipid and detergent.
Further evidence that the membrane proteins are surrounded by lipid and detergent comes from projections of side views of the disk-like density beneath the ribosome (Figures 1D, 1E). These projections show two parallel, high density stripes separated by a low density region. This density profile and its thickness of ∼40Å are suggestive of a lipid bilayer-like structure. Thus, ER membrane solubilization with digitonin resulted in single copies of Sec61 and TRAP being surrounded by a disk in which the lipids may be organized in a bilayer-like arrangement. The steroidal detergent molecules might cap and stabilize the edges of this “mini-membrane”.
Quantitation of Sec61 and TRAP in the native channel
We then carried out a quantitative analysis with mass spectrometry to verify that single copies of Sec61 and TRAP are present in the RCCs. In these experiments, we used the AQUA method (Gerber et al. 2003), in which labeled peptides are added in known amounts to trypsin-digested samples to serve as internal standards. A similar method was used to obtain the ratio of components in ribosome-SecY complexes (Ménétret et al., 2007). In total we used 6 peptides, with two from ribosomal proteins (S5e, SA/p40). In addition, we chose one peptide each from the α- and β-subunits of the Sec61 complex and from the α- and β-subunits of the TRAP complex (Table 1). The RCCs were prepared as before (Morgan et al., 2002; Ménétret et al., 2005), except that the particles were purified on a sucrose gradient. Peak fractions from the gradient contained all of the expected components in the RCCs, as shown by blots and Coomassie stained gels (not shown). Data from the quantitative analysis is summarized in Table 1.
Table 1.
Quantitative mass spectrometry of purified ribosome-channel complexes
| Component | Proteins | Quantitation (pmoles) |
|---|---|---|
| Ribosomal small subunit | S5e | 1.11* (1.03-1.21) |
| SA/p40 1.18 | (1.05-1.31) | |
| Sec61 complex | Sec61α | 0.76 (0.72-0.82) |
| Sec61β | 1.08 (1.05-1.16) | |
| TRAP complex | TRAPα | 0.73 (0.65-0.83) |
| TRAPβ | 0.69* (0.64-0.80) | |
| Normalized ratios | 1.0 : 0.8 : 0.62 |
The samples were divided into 6 aliquots which were analyzed and averaged. The appropriate tryptic peptide fragment for proteins marked with an asterisk (*) could only be detected reliably in 5 of the runs.The range of values for each component in the experiments is shown on the right in parenthesis.
We found that the stoichiometry of ribosomes, Sec61, and TRAP was about 1.0 to 0.8 to 0.62 in these complexes. As an internal control, two ribosomal proteins from the small subunit were present in a 1:1 stoichiometry. In addition, the α- and β-subunits of sec61 and the α- and β-subunits of TRAP each had an approximate 1:1 ratio in their respective complexes. Although our data suggest that some ribosomes may not carry a channel, the occupancy estimated from our electron microscopy data appears to be higher. Indeed, about 95% of the particles contain a channel based on 3D classification with the multirefine option in EMAN (see Experimental Procedures). In addition, the observed differences between Sec61 (∼0.8 pmole) and TRAP (∼0.62 pmole) may be due to experimental errors. For example, trypsin digestion of TRAP subunits may have been incomplete. When taken together, the mass spectrometry and structural data both support the idea that single copies of Sec61 and TRAP are present in most of the purified RCCs.
Sec61 connections to the large ribosomal subunit
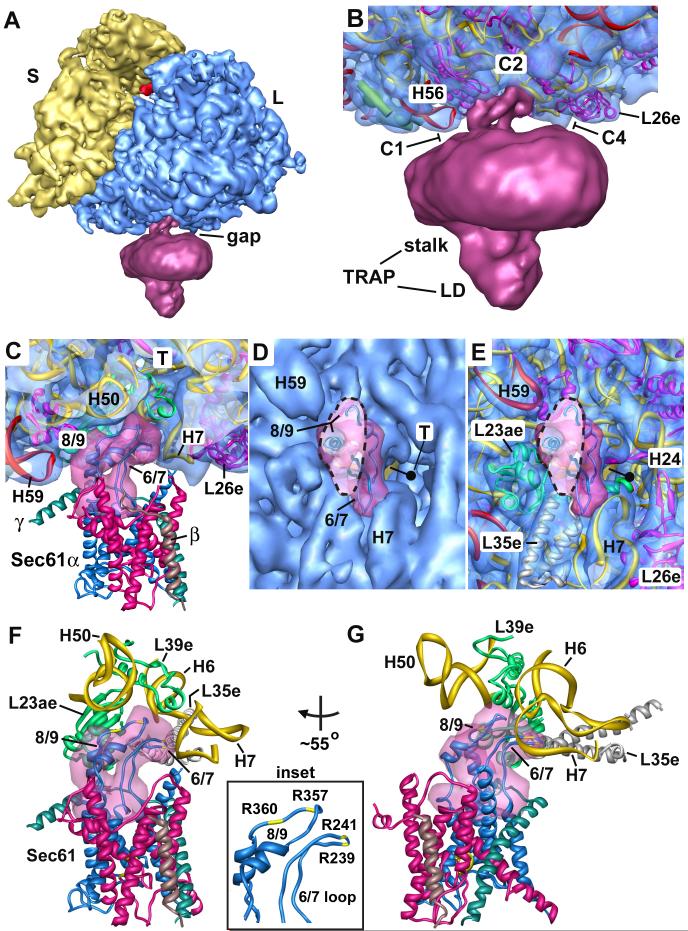
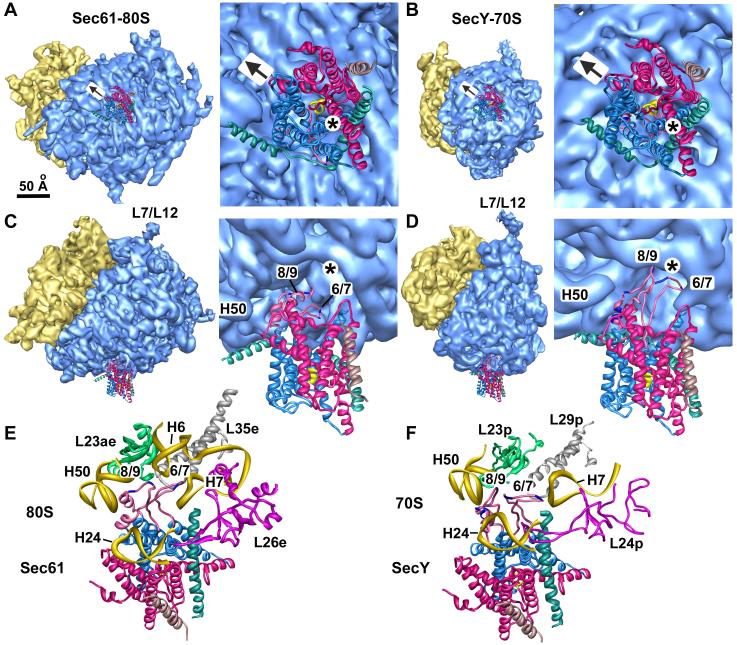
We used our improved map to evaluate the connections between the channel and the ribosome. This analysis was aided by a detailed model of the canine ribosome (Chandramouli et al., 2008), which allowed us to choose a reasonable threshold for the complex. As shown previously, a gap is present between the channel and the large ribosomal subunit (Figure 2A). However, only a single connecting region is seen, in contrast to previous studies in which three major connections were identified as C1, C2 and C4 (Morgan et al., 2002; Ménétret et al., 2000; 2005; Beckmann et al., 2001). The prominent connection in the new map corresponds to the C2 connection and is located near the exit tunnel (Figure 2B). Connections C1 and C4 observed previously correspond to close approaches of H59 and H7, respectively, to the surface of the membrane-like disk. While this map is qualitatively similar to those published previously, the improved resolution allowed us to identify C2 as the major, well ordered link.
Figure 2.
The 6/7 and 8/9 loops of Sec61 form the major connection with the ribosome.
A. An oblique front view of the RCC is shown. The RCC is color coded as described in Figure 1A. The small (S) and large (L) subunits are labeled. A single major connection spans the gap between the ribosome and the channel.
B. A close-up is shown of the junction between the ribosome and the membrane-like disk. The positions of connections observed in previous maps at a lower threshold (C1, C2 and C4) are indicated. Also shown are the regions of TRAP (stalk, lumenal domain, (LD)).
C. A thin slab containing the interface between the ribosome and the channel is shown. Helices 50 and 7 in the large subunit interact with the loops of sec61α near the tunnel exit (T). Density for the connection is shown as a transparent surface over-layed on the modeled loops (shown as ribbons).
D. A bottom view shows the insertion of the 6/7 and 8/9 loops into a pocket at the exit tunnel. The loop density is shown in magenta and the large subunit is shown in blue. The tunnel is marked with a dot and a line that points into the large subunit, towards the small subunit (yellow surface).
E. This view is similar to panel 2D but the surface of the large subunit is semi-transparent, to show the atomic model of the ribosome in this region.
F. The 6/7 and 8/9 loops are shown within a binding pocket which is formed by H6, H7 and H50, along with proteins L23ae, L35e and L39e. Basic residues in the Sec61 loops are shown in yellow and are labeled in the inset on the right.
G. A rotated view of panel 2F is shown. A small helix of L39e is close to the 8/9 loop and L35e helps to form the back of the binding pocket.
Cytoplasmic loops of Sec61α have been implicated in ribosome binding (Raden et al., 2000), including the loops between TMs 6 and 7 (the 6/7 loop) and between TMs 8 and 9 (the 8/9 loop; Cheng et al., 2005). In addition, these loops have been shown to mediate the binding of SecY to the bacterial ribosome (Ménétret et al., 2007). Thus, we docked the cytoplasmic loops from the crystal structure of archaeal SecY into connection 2. Only small adjustments were required to obtain a good fit of the loops within the density, after taking into account the fact that the mammalian 8/9 loop lacks two residues near its tip (Figure 2C, 2F, 2G). In particular, the two small α-helices of the 8/9 loop fit within short rod-like features in the connection density. The 6/7 loop is comprised of two strands with an extended β-hairpin like structure and this feature also fits well within the density (Figures 2C-2G, 4A, 4B and Supplemental Figure 1). Since the resolution of the membrane-embedded region of Sec61 and the ribosome are ∼11Å and 8.7Å, respectively, the connecting loops of Sec61 may be visualized at a resolution of ∼9-10Å. This explains why we could dock the loops into their respective density.
In the resulting model, the 6/7 and 8/9 loops of Sec61 are located in a pocket at the tunnel exit, as shown in a bottom view (Figures 2D, 2E). The 6/7 and 8/9 loops appear to interact with RNA helices in the large subunit that include H50, H6 and H7 (Figures 2F, 2G). In particular, the 6/7 loop may interact with both H6 and H7 as it is located between them. The 8/9 loop may interact with both H6 and H50. Additional contacts with surrounding large subunit proteins are also possible. These proteins help to form the binding pocket and include L23ae (L23p), L35e (L29p) and L39e. With the exception of L39e, these proteins are also found in bacteria. Basic residues in the 6/7 loop (Arg239 and Arg241) and the 8/9 loop (Arg357 and Arg360) may mediate binding to the phosphate backbone of rRNA (Figure 2F, inset). Indeed, mutations in residues equivalent to Arg239, Arg241 and Arg360 in yeast Sec61α led to a severe growth defect (Cheng et al., 2005). Overall, it appears that the cytoplasmic loops of Sec61 fit into the ribosome like a key into a lock. The extensive van der Waals interactions are consistent with the high affinity binding reported previously (∼12 nM; Prinz et al., 2000).
The membrane-embedded region of Sec61
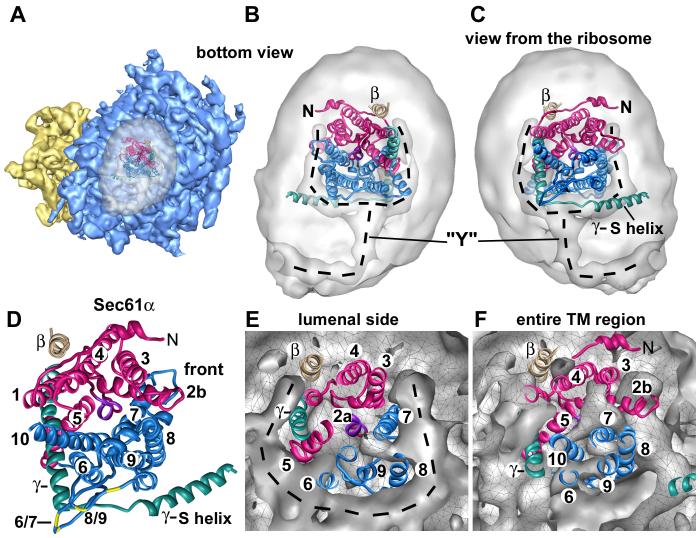
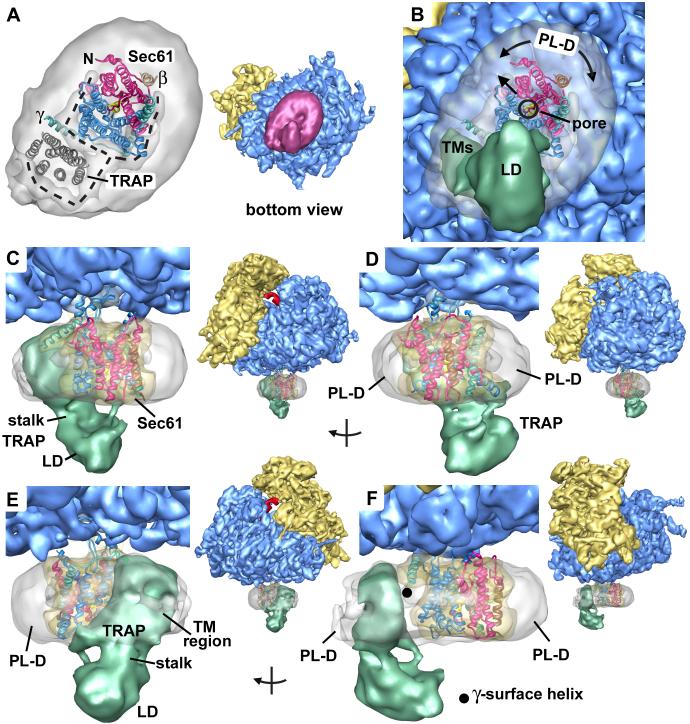
Next, we evaluated the fit of the archaeal SecY crystal structure within the membrane region that contains Sec61 in the complete map. Remarkably, we found that the initial fit of the SecY crystal structure was quite good, because the horizontal (in-plane) positioning within the membrane-like disk was dictated by TMs at the base of the 6/7 and 8/9 loops. We then improved the docking using the “fit to map” option in CHIMERA, which uses a 6D search to optimize the fit of a pdb file within the map (Goddard et al., 2005). We also repositioned the surface helix of the SecE/Sec61γ subunit within the map by tilting it slightly downwards.
The docking within the channel region is shown at ∼17Å resolution, viewed from either the ribosome (Figures 3A, 3B) or the ER lumen (Figure 3C). Note that Sec61 is nearly encircled by a low density feature in the center of the membrane-like disk. This low density feature may arise from disordered phospholipids tails. The overall fit of the SecY model into the map at ∼11Å resolution is shown in Figures 3E and 3F. A view of the SecY model on its own is shown in Figure 3D in the same orientation. The overall fit of the SecY model in the high density features suggests that we were able to position groups of α-helices accurately, commensurate with the estimated resolution of ∼11Å for this region. For example, we could easily follow the tilted trajectory of the TM helix of SecE/Sec61γ as it crosses a the membrane region, as shown in a mini-map which contains the high density channel features (see Experimental Procedures and Supplemental Figure 1B, Figure 4A). In addition, the TM helix of Sec61γ could be identified, as shown in a stereo view in Supplemental Figures 2A, 2B, and we were able to resolve the surface helix of Sec61γ (Supplemental Figures 1A, 1C and 2C). In fact, most of the α-helices do not cross significant low density regions in the map with the exception of TM6 (Figures 3E, 3F). Stereo views further demonstrate the overall fit of Sec61, as shown in a thin slab near the ER lumen and in a thicker slab that encompasses most of the membrane-embedded region (Supplemental Figures 2D and 2E).
Figure 3.
Docking the Sec61 complex into the electron density map.
A. A bottom view is shown of the RCC with the membrane-embedded region rendered semi-transparent to show the docked SecY model.
B. A low density Y-shaped region is present within the membrane-like disk and nearly encircles the embedded region of Sec61. The Y-like region is indicated by a dashed line. The electron density map was truncated to 17Å resolution for panels 3B and 3C. The Sec61 complex was modeled with a crystal structure of SecY and is color coded as follows. The N-terminal half of SecY/Sec61α is is colored in red, while the C-terminal half is shown in blue. The SecE/γ subunit is shown in green and the Secβ subunit is shown in tan.
C. In this panel, the docked SecY in the channel region is viewed from the ribosome.
D. A ribbon model of the SecY complex is shown. This view is from the ribosome and is similar to that in panels 3E and 3F. The surface helix of the SecE/Sec61γ is labeled (γ-S helix).
E. A cross-section is shown of the Sec61 region from the full 3D map truncated at 11Å resolution. A thin slab encompasses the lumenal side of the channel and the SecY model fits within a low density feature (marked with dashed line). Helices of the docked model are numbered (van den Berg et al., 2004).
F. A thicker slab is shown which contains the entire membrane-embedded region of Sec61.
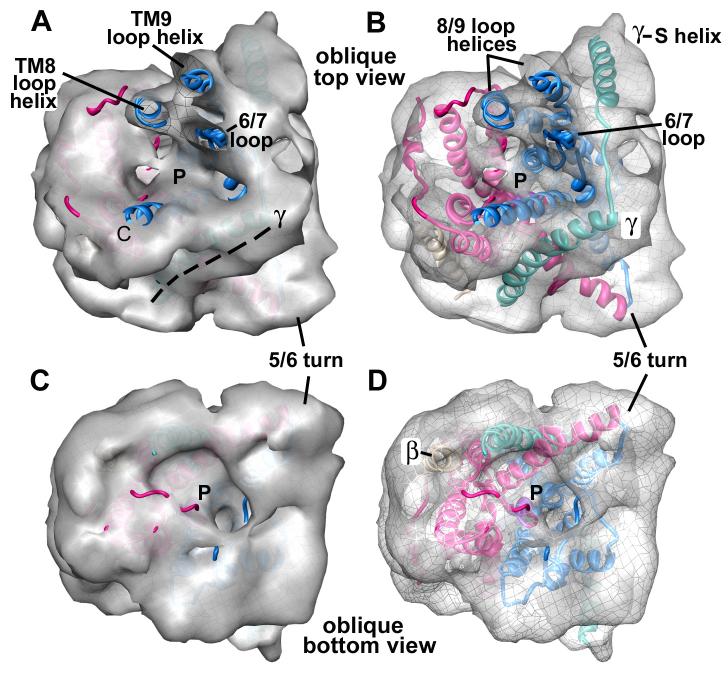
Figure 4.
The entrance and exit vestibules of the hour-glass shaped pore in Sec61.
A. The top surface of the mini-map is shown with the docked SecY model. The map has been rendered as a solid surface to show the depression leading into the pore. The 6/7 and 8/9 loops have been cut by a clipping plane to show their fit in the map. The TM of SecE/Sec61γ forms a ridge (see dashed lines). Note that the entrance and exit vestibules are similar in the complete map of the channel region.
B. The same view is shown as in panel 4A, except that the surface is semi-transparent to show the helices that form the surface depression.
C. The bottom surface of the mini-map is shown with the docked SecY model. The map has been rendered as a solid surface to show the depression that forms the pore exit.
D. The structure in panel 4C is shown with the mini-map rendered as a semi-transparent surface. The helices which form the exit vestibule are shown.
The cytoplasmic entrance of the pore is clearly visible at the center of Sec61 in the mini-map, adjacent to the 6/7 and 8/9 loops (Figures 4A, 4B). The exit vestibule of the pore is also visible (Figures 4C, 4D, Supplemental Figures 2A-2C), while the pore itself appears to be closed since there is density in this region. Our model positions the central pore of Sec61 below the ribosome tunnel exit, such that a nascent chain could cross the gap and insert into the channel (see next section). This analysis provides further evidence that a single copy of Sec61 is present in the RCC.
A comparison of Sec61- and SecY-ribosome complexes
Given the sequence homology between Sec61 and SecY (van den Berg et al., 2004), one might expect structural similarities between mammalian and bacterial RCCs. We therefore aligned the structure of the E. coli RCC (Ménétret et al., 2007) with the new mammalian structure. The independent docking of Sec61 and SecY in their respective maps showed that the channels are positioned similarly beneath the ribosome. In both cases, the lateral gate between TMs 2b and 7 is pointed towards the small ribosomal subunit and the tunnel exit (asterisks) is only slightly offset from the central pore (Figures 5A, 5B).
Figure 5.
A comparison of Sec61- and SecY-ribosome complexes.
For panels A-D, the complete ribosome with docked Sec61 or SecY is shown on the left and a close-up is shown on the right. The color coding is yellow for the small subunit and blue for the large subunit. The N- and C-terminal halves of the Sec61α/SecY subunits are shown in blue and red ribbons respectively, while the Sec61γ/SecE and Sec61β/β-subunits are shown in green and tan.
A. A bottom view is shown of the ribosome-Sec61 complex. The positions of the lateral gate (arrow) and the tunnel (*) are marked.
B. A bottom view is shown of the ribosome-SecY complex with the lateral gate and tunnel marked as in panel A.
C. A tilted view is shown of the ribosome-Sec61 complex in which the 6/7 and 8/9 loops are clearly visible near the tunnel exit (marked with an asterisk).
D. A tilted view is shown of the ribosome-SecY complex in which the cytoplasmic loops and their insertion into the tunnel can be seen.
E. The 6/7 and 8/9 loops are viewed from the exit tunnel for the Sec61-ribosome complex. The 6/7 loop is inserted between H6 and H7 while the 8/9 loop may interact with both H6 and H50. Three conserved ribosomal large subunit proteins are also shown.
F. A similar view to panel E is shown of the longer 6/7 and 8/9 loops for the SecY-ribosome complex (Ménétret et al., 2007). The 6/7 loop interacts with H7 while the 8/9 loop may bind to H50 and H24.
In both structures, the 6/7 and 8/9 loops tether the respective channels at the tunnel exit (Figures 5C, 5D). We note, however, that the 6/7 and 8/9 loops in bacterial SecY are longer than the corresponding loops in Sec61. Hence, the SecY loops extend further into the ribosome tunnel than the shorter loops of Sec61. The 6/7 loop in bacterial SecY points towards H7 while it is located between H6 and H7 in the canine ribosome (compare Figures 5F and 5E). The longer bacterial 6/7 loop partially blocks the tunnel exit but might adopt an alternate conformation when a nascent chain is present (Ménétret et al., 2007). The 8/9 loop in the canine complex is located between H6 and H50, while this loop in bacteria is located near H50 and could interact with H24. This difference can be ascribed to the shorter 8/9 loop that is present in archaea and mammals.
Finally, the Sec61 and SecY monomers are surrounded by an annulus of extra density in both specimens. This larger region in the mammalian complex may contain phospholipids and digitonin (Figure 1C). In the bacterial complex, the annulus may contain detergent, because the SecY complex was probably delipidated during its purification (Ménétret et al., 2007).
The TRAP complex
We evaluated the density contributed by the TRAP complex within the channel region. At the back of the membrane-like disk, a high-density feature is almost completely encircled by a low-density “tail” (Figure 6A). This region is large enough to contain seven TM segments, the predicted number of TMs in the TRAP complex and was modeled with bacteriorhodopsin. We then created a soft mask that included the membrane-embedded region, the stalk and the lumenal domain of TRAP. We used the mask to isolate this density and calculated an FSC curve in the usual way. This analysis suggested that TRAP has been visualized at ∼15Å resolution in the map (not shown).
Figure 6.
A model for TRAP in the mammalian ribosome-channel complex.
A. A bottom view is shown of the membrane-like disk from the RCC (see central icon view) with the lumenal domain of TRAP removed. The disk is rendered semi-transparent and shows the docked SecY/Sec61 model within the internal low density features attributed to lipid tails. The low density region (see dashed lines) extends to the back of the disk and nearly encircles a high density region that is the correct size to contain the 7 TMs of the TRAP complex (modeled with bR, see grey ribbons). Note that Sec61 and TRAP are separated by a low density feature; hence, there is room for a row of phospholipids between them.
B. This view is similar to panel A, except that TRAP is shown as a solid surface (green) and the ribosome is present. The lumenal domain is almost directly in line with the central pore of Sec61 (marked with a circle). The empty region of the disk probably contains phospholipids with digitonin at the edge (marked PL-D with curved arrows). A single arrow points from the central pore of Sec61 in the direction of the small subunit and marks the position of the proposed lateral gate.
C. Modeled Sec61 and TRAP are shown in a frontal view (see icon on the right). The outline of the Sec61 molecule is shown as transparent yellow surface using the mini-map.
D. A rotated view shows that empty areas in the disk on either side of Sec61 and TRAP that may contain phospholipids and detergent.
E. The TRAP complex is shown from the back and has a slanted orientation.
F. In a side view, there is a gap between TRAP and Sec61. The surface helix of Sec61γ may contact the membrane-embedded region of TRAP (black dot).
Upon closer inspection, we find that the TRAP and Sec61 complexes in the RCC are laterally offset relative to one another and separated by a low-density region (Figure 6A). Thus, a row of phospholipids is probably located between the two proteins. In addition, the surface helix of Sec61γ is located in close proximity to TRAP and may make a bridging contact (Figures 6B and 6F). There is no direct connection between TRAP and the ribosome, which suggests that this protein is recruited to the RCC by its association with Sec61. The small number of interaction points between TRAP and Sec61 may account for the measured difference in resolution of the two membrane proteins in the map (15.1 versus 11.1 Å). The lumenal domain of TRAP is connected to the transmembrane region by a large stalk (Figures 6C-6F) and the tip of the lumenal domain is located directly below the channel pore (Figure 6B, 6F). This suggests that TRAP may interact with the nascent chain or recruit lumenal chaperones to bind to the emerging polypeptide chain. Finally, the TRAP complex has a tilted appearance due to the offset between the membrane-embedded region and lumenal domain (Figure 6E).
Discussion
We have shown that a non-translating ribosome binds to single copies of Sec61 and TRAP in ribosome-channel complexes derived from mammalian ER membranes. The major connection is made between the ribosome and the 6/7 and 8/9 loops of Sec61. This connection is close to the ribosome tunnel exit, which positions the Sec61 complex so that an emerging nascent chain could move directly into the channel. Consistent with previous suggestions, this implies that a single copy of Sec61 would form the channel (van den Berg et al., 2004; Ménétret et al., 2007). In addition, the lumenal domain of the TRAP complex is positioned below the Sec61 channel so that it would be in close proximity to the nascent chain. Finally, our results demonstrate a fundamental similarity between mammalian and bacterial ribosome-channel complexes.
Architecture of the channel
The present structure gives the most detailed picture of a ribosome-bound channel that has been obtained to date. In this map, the resolution of the membrane-embedded region of Sec61 is estimated to be ∼11Å. Most of the α-helices of Sec61 reside within high density features in the map (with the exception of TM6), but are not fully resolved as individual rods. However, prominent features such as the TM and surface helix of Sec61γ, the 5/6 turn, and the entry and exit vestibules to the pore were visualized. In addition, the overall packing of the TM helices in the 11Å map was reasonable, based on our docking of a crystal structure of the archaeal SecY complex. When combined with the placement of the 6/7 and 8/9 loops, the fit of the membrane-embedded region allowed us to accurately dock the crystal structure. In the resulting model, the lateral gate of Sec61 points towards the small subunit. This gate may allow TM segments of nascent membrane proteins to move into the lipid bilayer. Even with the improved resolution, a gap is still present between the ribosome and channel (Ménétret et al., 2000, 2005; Morgan et al., 2002; Beckmann et al. 2001). This gap would allow loop segments of membrane proteins to emerge into the cytoplasm.
The present structure is remarkably similar to that of the non-translating ribosome-SecY complex (Ménétret et al., 2007). In both cases, a single copy of the channel is bound and the 6/7 and 8/9 loops interact with the ribosome at the tunnel exit. Despite some differences, the loops fit into the ribosome like a key in a lock and conserved basic loop residues in both channels may interact with conserved RNA helices at the tunnel exit. These cytoplasmic loops may also contact proteins in the vicinity of H7 and H50. Moreover, the general orientation of SecY is similar to that observed for Sec61 in the mammalian RCC. This similarity reflects a high degree of conservation in co-translational protein translocation. Hence, we propose that the copy of Sec61 or SecY which binds at the ribosome tunnel exit may form the active channel that captures and translocates the nascent polypeptide chain.
Previous structures of ribosome-Sec61 complexes had suggested that 3 or 4 copies of Sec61 may associate with the ribosome (Beckmann et al., 2001; Ménétret et al., 2005). Our current data show that only one copy is bound to the ribosome and thus, much of the additional density surrounding the Sec61 complex can be attributed to lipid and detergent. This is supported by the observation that the disk has a bilayer-like density distribution. The formation of a membrane-like disk presumably reflects the ability of steroidal detergents, such as digitonin, to cap the exposed hydrophobic edges of phospholipid bilayers. This may be similar in some respects to the stabilization of membrane disks by apolipoproteins (Zhu and Atkinson, 2007). Consistent with this idea, two ribosome-SecY structures contained a smaller annulus of density around the channel, presumably because extensively delipidated SecY was used to form the complexes (Ménétret et al., 2007). The association of Sec61 or SecY with lipid and/or detergent disks may also explain the size of ring-like particles observed previously with purified proteins (Hanein et al., 1996; Meyer et al., 1999; Manting et al., 2000).
In a recent structure, an E. coli ribosome that carried a nascent polypeptide chain was proposed to bind to two copies of SecY (Mitra et al., 2005), but the channel region was not much larger than that seen in a non-translating ribosome-SecY complex (Ménétret et al., 2007). Perhaps the translating ribosome-SecY complexes contained a single copy of SecY along with additional lipid and detergent. In any case, neither of the SecY complexes in the proposed dimer model are positioned beneath the ribosome like single copies of SecY or Sec61 observed in our structures with a non-translating ribosome (Ménétret et al., 2007; and this study).
Our data do not exclude the idea that oligomers of the SecY or Sec61 complex may form in a membrane during translocation. In fact, SecY dimers are likely required for bacterial post-translational translocation driven by the SecA ATPase (Osborne and Rapoport, 2007; Duong, 2003). In addition, SecYEG dimers have been observed in 2D membrane crystals (Breyton et al., 2002), large intra-membrane particles were seen in freeze-fracture experiments (Hanein et al., 1996, Meyer et al., 1999; Scheuring et al., 2005), and oligomers may be required for the tight binding of ribosomes to ER membranes (Schaletzky and Rapoport, 2006). Hence, solubilization of SecY and Sec61 in detergent may result in the disassembly of these oligomers. This is supported by the observation that only the active copy of SecY remains associated with SecA after solubilization (Duong, 2003). A similar situation may pertain to SecY or Sec61 complexes bound to ribosomes.
The TRAP complex
Our new structure shows that one copy of the TRAP complex is associated with the mammalian RCC. Together, the electron microscopy and mass spectrometry data suggest that nearly every translocon may contain a TRAP complex. In addition, the size of the TRAP lumenal domain is consistent with one copy of TRAP being present in the RCCs. Thus, TRAP is an integral part of the translocon (Ménétret et al., 2005). Intriguingly, we also find that membrane-embedded regions of TRAP and Sec61 are separated by a low density feature, which may correspond to a row of phospholipids. These phospholipids may form bridges between adjacent TM regions, as they do in some 2D membrane protein crystals (Gonen et al., 2005; Hite et al., 2007). Intervening lipids between the TM regions of Sec61 and TRAP may allow some flexibility of the proteins, while still maintaining their association. In addition, the interaction between membrane-embedded regions of TRAP and Sec61 does not block the lateral gate of Sec61.
We also verified that a stalk links the transmembrane region of TRAP to a prominent lumenal domain. Based on sequence analysis, the lumenal domain and stalk are likely comprised of the N-terminal regions of the α, β and δ subunits (Liu et al., submitted), and the size of this domain in the 3D map is consistent with this idea. The lumenal domain of TRAP is positioned so that it could interact with a nascent chain emerging from the channel, which would explain the crosslinking data (Wiedmann et al., 1989; Görlich et al., 1992). Alternatively, the lumenal domain of TRAP may direct chaperones to the nascent chain.
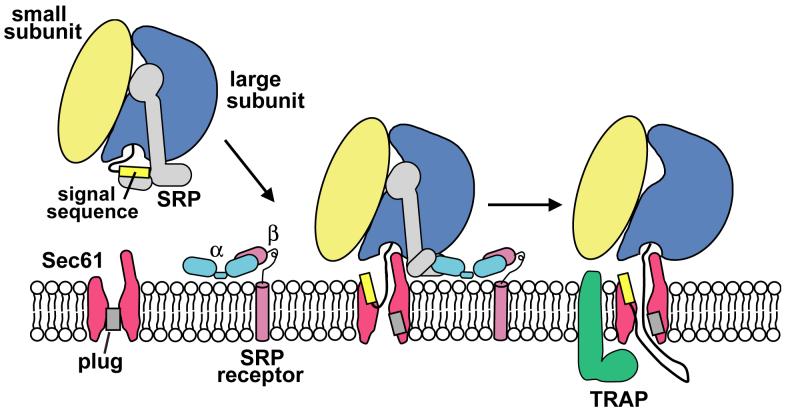
The initiation of protein translocation
We postulate that the observed ribosome-Sec61 structure resembles an early stage in co-translational protein translocation. When a ribosome synthesizes a nascent secretory protein, the signal sequence first binds to the M-domain of SRP to form a stalled, ribosome-SRP complex (Halic et al., 2004, 2006a; Figure 7, left). This stalled complex is then targeted to the ER membrane through reciprocal interactions between SRP and the α-subunit of its receptor (SR) (Egea et al., 2004). When the SRP-SR complex is formed, the M-domain is partially displaced from its position over the tunnel exit (Halic et al., 2006b). This may allow the cytoplasmic loops of Sec61 to bind to the ribosome as observed in our structure. This interaction with Sec61 may help to further displace the M-domain. At this point, the signal sequence may be transferred to a binding site within Sec61α, which sits below the tunnel exit (Figure 7, middle). Subsequent disassociation of the SRP-SR complex from the ribosome would precede the formation of a fully-active, ribosome-channel complex (Figure 7, right). The RCC may then recruit a second copy of Sec61 (not shown) to help stabilize the junction between the ribosome and the channel. At some point during this process, TRAP may associate with Sec61 to form a stable membrane-protein complex. With the exception of TRAP association, this sequence of events may be the same in bacteria (Ménétret et al., 2007). The structure of a translating mammalian RCC is now needed to provide additional insights into co-translational protein translocation.
Figure 7.
The initiation of protein translocation at the ER in higher eukaryotes.
A three step model is shown for the docking of a ribosome-nascent chain-SRP complex to the ER membrane and subsequent formation of an active ribosome-channel complex (see Discussion for details).
Experimental Procedures
Sample preparation and quantitative mass spectrometry
Ribosome-channel complexes for electron cryo-microscopy were prepared from canine ribosomes with a bound E-site tRNA and ribosome-stripped microsomes (PKRMs), as described (Ménétret et al., 2005; Morgan et al., 2002). For mass spectrometry, an excess of PKRMs was added to the ribosomes (∼22 pmol), the complexes were solubilized in 0.8% DBC and then separated on a 10-40% sucrose gradient in 50 mM Hepes-KOH buffer (pH 7.5) in 500 mM KAc, 10 mM MgCl2, 0.8% DBC. Solubilized membrane proteins ran at the top of the gradient. Peak fractions containing the RCCs (fractions 6-9) were identified by their absorbance at 260 nm. SDS-PAGE and immuno-blotting showed that they contained ribosomes (at ∼25 fmol/ul), Sec61 and TRAP. Appropriate fractions were quick-frozen in liquid nitrogen and stored at -80°C until they were analyzed. We chose 6 tryptic peptides for the quantitative analysis of Sec61 and TRAP in the RCC. The ribosomal proteins were (S5e, SA/p40), along with α- and β-subunits of Sec61 and the α- and β-subunits of TRAP. Labeled peptides for the AQUA method (Gerber et al. 2003) were obtained from Cell Signaling Technology, Inc. The general approach for precipitating the RCCs from sucrose density gradient fractions and the quantitative analysis have been described (Ménétret et al., 2007).
Image processing and modeling
The processing of ∼101,000 particles to obtain an improved 3D map of the RCC was described previously (Chandramouli et al., 2008). In the end, we kept ∼78,800 of the best particles for the final map. We also used the multirefine option in EMAN to classify particles into groups that contained or lacked a channel using appropriate 3D reference volumes. This study showed that the overall occupancy of the channel in the particle dataset is ∼95%. We also processed two subsets of the final dataset separately with EMAN (v1.8) running on a Linux cluster with ∼28 nodes (Ludtke et al., 1999), to create lower resolution maps of the RCC. The final 3D maps for these two datasets contained ∼24,900 and 56,800 particles, respectively. Fourier shell correlation curves for each of the 3D structures were calculated with the eotest in EMAN, using volumes calculated from even and odd numbered particles and an appropriate mask for the various regions (see below; Ménétret et al., 2005). Projections of the respective transmembrane regions were created in EMAN (Ludtke et al., 1999).
The 6/7 and 8/9 loops of the crystal structure of archaeal SecY (1rhz) were docked into the final 3D map manually in ‘O’ (Jones et al., 1991) after converting the file to brix with SPIDER (Frank et al., 1996). The fit was checked in Chimera (Goddard et al., 2005) and the loop geometry was refined and regularized with Coot (Emsley and Cowtan, 2004). The transmembrane region of the SecY model was docked into the final map by calculating rotational and translational fits in Chimera using the “fit in map” option. In this option, a 6D search was made between the pdb file and the 3D map. For each orientation, an average map value is tabulated from the intersection of the atoms in the pdb model with the map. The maximal value gives the best local fit. In a second approach, the SecY model was converted into a density map with EMAN. The sum of the product of the two map values for each (x,y,z) in the SecY map and the channel region of the RCC map was also calculated in Chimera with a 6D search to find the best fit, again using the “fit in map” option (T. Goddard, personal communication). The surface helix of SecE was also moved downwards a bit to better fit into the density.
The docked SecY model was used to create a soft mask in EMAN, which was used to isolate the channel region for the FSC calculation in the three maps. We also low pass filtered the mask to create a nearly featureless “blob” mask that was used to cut out the central high density features of the membrane-embedded region for further visualization in Chimera. The borders of this “mini-map” were formed in part by a central, low density region that nearly encircles Sec61. This low density region may be due to phospholipid hydrocarbon tails. However, higher density regions at the top and bottom surface of the channel are not as clearly demarcated due to local contrast matching. Hence, the blob mask does cut through some higher density in these regions. Thus, the mini-map was used mainly for display purposes and to show features in the membrane-like disk that are not affected by the masking.
Ribosomes in the 3D maps were aligned as follows to compare bacterial and mammalian channel complexes. We started with the pseudo-atomic model of the canine ribosome which was docked within the appropriate 3D map in previous work (Chandramouli et al., 2008). We also used an atomic model of the E. coli ribosome (Berk et. al., 2006) docked within a map of the ribosome-SecY complex at ∼9.6Å resolution (Ménétret et al., 2007). Core regions of the two large subunits were super-imposed in Chimera with the fit-model-to-model option and then the appropriate 3D maps were transformed into the same orientation. This docking was then checked with the fit in map option using the 3D volumes. Figures were made using Chimera (Goddard et al., 2005), GIMP and Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank Andrea Neuhof for preparation of the ribosome-channel complexes that were used for data collection. We are also indebted to Preethi Chandramouli and Steve Ludtke for helpful discussions. Work in the T.A.R. and C.W.A. laboratories was supported by NIH grants. T.A.R. is a member of the Howard Hughes Medical Institute. The modeled ribosome-Sec61 complex has been deposited in the PDB (XXXX) and the EM map has been deposited in the EMD (ZZZZZZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. U S A. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of SecYEG, the bacterial protein-translocation complex. Nature. 2002;418:662–664. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM, Jr., Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli P, Topf M, Ménétret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7Å resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS. The translocon-associated protein complex facilitates the initiation of substrate translocation across the ER membrane. J. Cell Biol. 2003;129(160):529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure (Camb) 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–68. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006a;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006b;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KES, Jungnickel B, Plath K, Kalies K-U, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Görlich D, Kostka S, Otto A, Kraft R, Knespel S, Burger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur. J. Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- Hite RK, Gonen T, Harrison SC, Walz T. Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Arch. 2007 doi: 10.1007/s00424-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE, Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sommer N, Hegde RS, Ménétret JF, Hartmann E, Akey CW. Subunit interactions of the translocon-associated protein complex. J. Biol. Chem. submitted. [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Manting EH, van Der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétret JF, Schaletsky J, Clemons WM, Jr., Osborne AR, Skanland S, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, Rapoport TA, Akey CW. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Molec. Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Hegde RS, Heinrich S, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Molec. Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Meyer TH, Ménétret JF, Breitling R, Miller KR, Akey CW, Rapoport TA. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J. Mol. Biol. 1999;285:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, 3rd, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Frank J. A model for co-translational translocation: ribosome-regulated nascent polypeptide translocation at the protein-conducting channel. FEBS Lett. 2006;580:3353–3360. doi: 10.1016/j.febslet.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Ménétret JF, Neuhof A, Rapoport TA, Akey CW. Structure of the mammalian ribosome-channel complex at 17Å resolution. J. Mol. Biol. 2002;324:871–886. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raden D, Song W, Gilmore R. Role of the cytoplasmic segments of Sec61alpha in the ribosome-binding and translocation-promoting activities of the Sec61 complex. J. Cell Biol. 2000;150:53–64. doi: 10.1083/jcb.150.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic ER and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Schaletzky J, Rapoport TA. Ribosome binding to and dissociation from translocation sites of the ER membrane. Mol. Biol. Cell. 2006;17:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring J, Braun N, Nothdurft L, Stumpf M, Veenendaal AK, Kol S, van der Does C, Driessen AJ, Weinkaufs S. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J. Mol. Biol. 2005;354:258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Crystal structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Görlich D, Hartmann E, Kurzchalia TV, Rapoport TA. Photocrosslinking demonstrates proximity of a 34 kDa membrane protein to different portions of preprolactin during translocation through the endoplasmic reticulum. FEBS Lett. 1989;257:263–268. doi: 10.1016/0014-5793(89)81549-2. [DOI] [PubMed] [Google Scholar]
- Zhu HL, Atkinson D. Conformation and lipid binding of a C-terminal (198-243) peptide of human apolipoprotein A-I. Biochemi. 2007;46:1624–1634. doi: 10.1021/bi061721z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.