Abstract
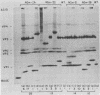
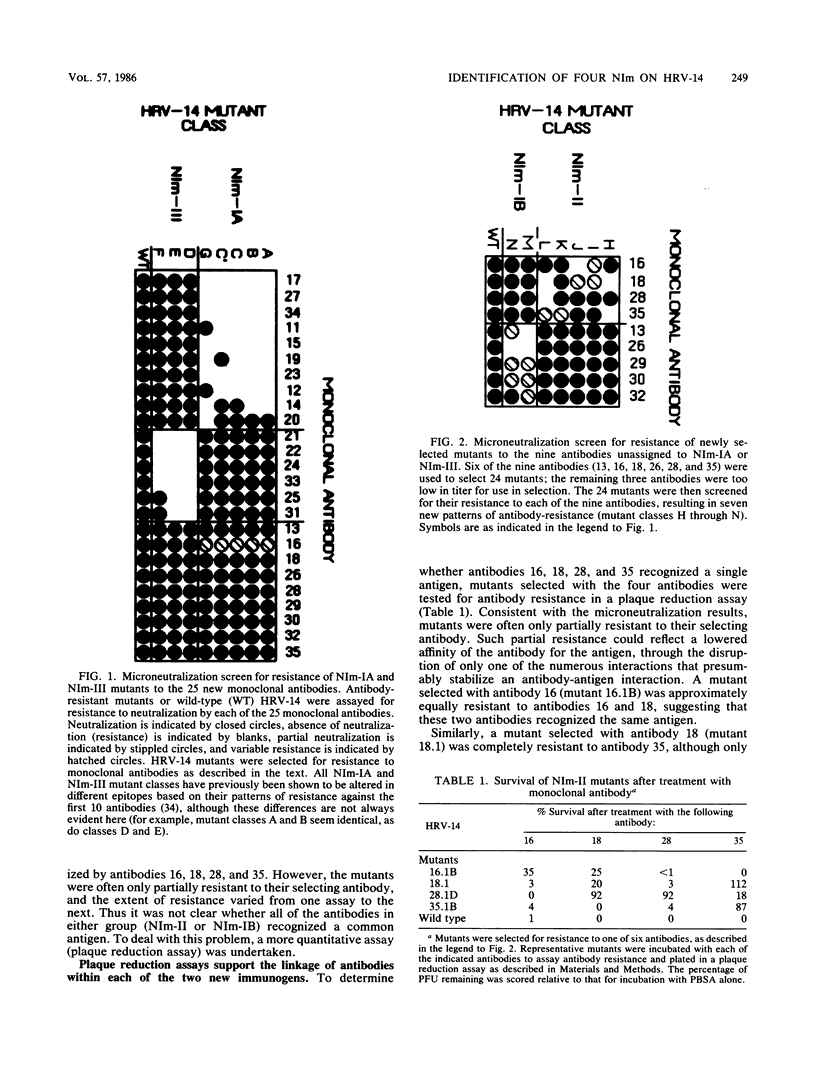
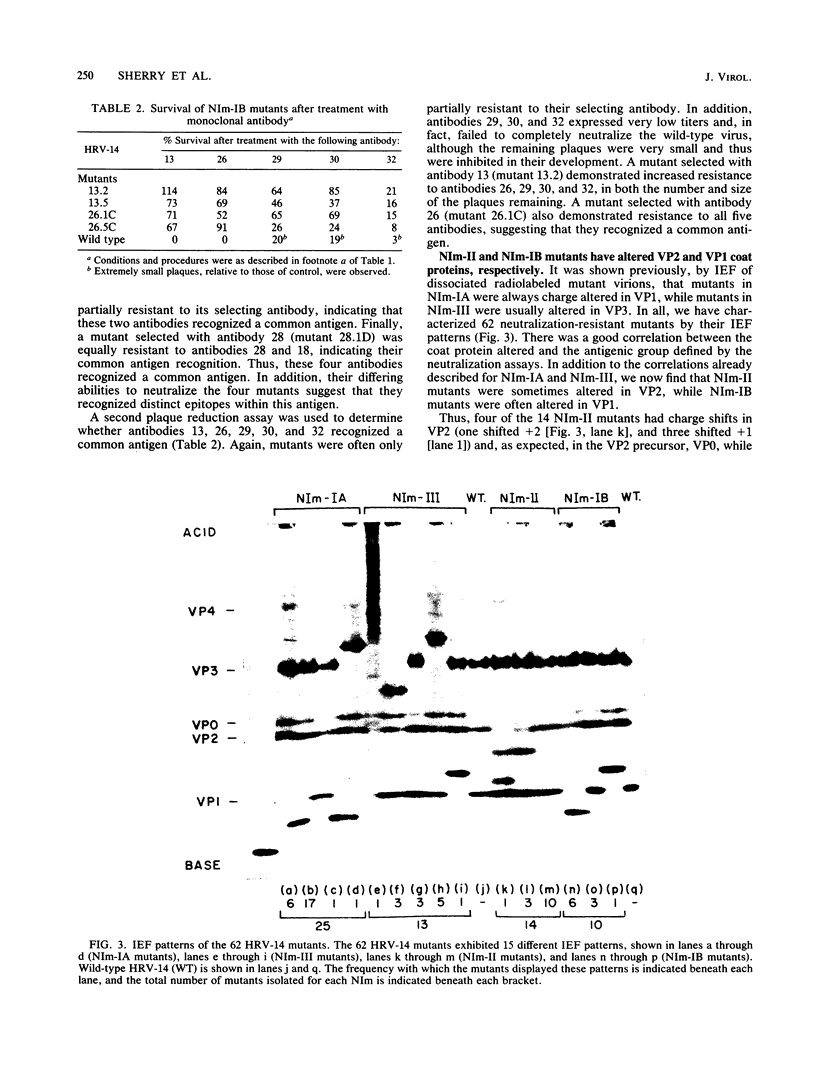
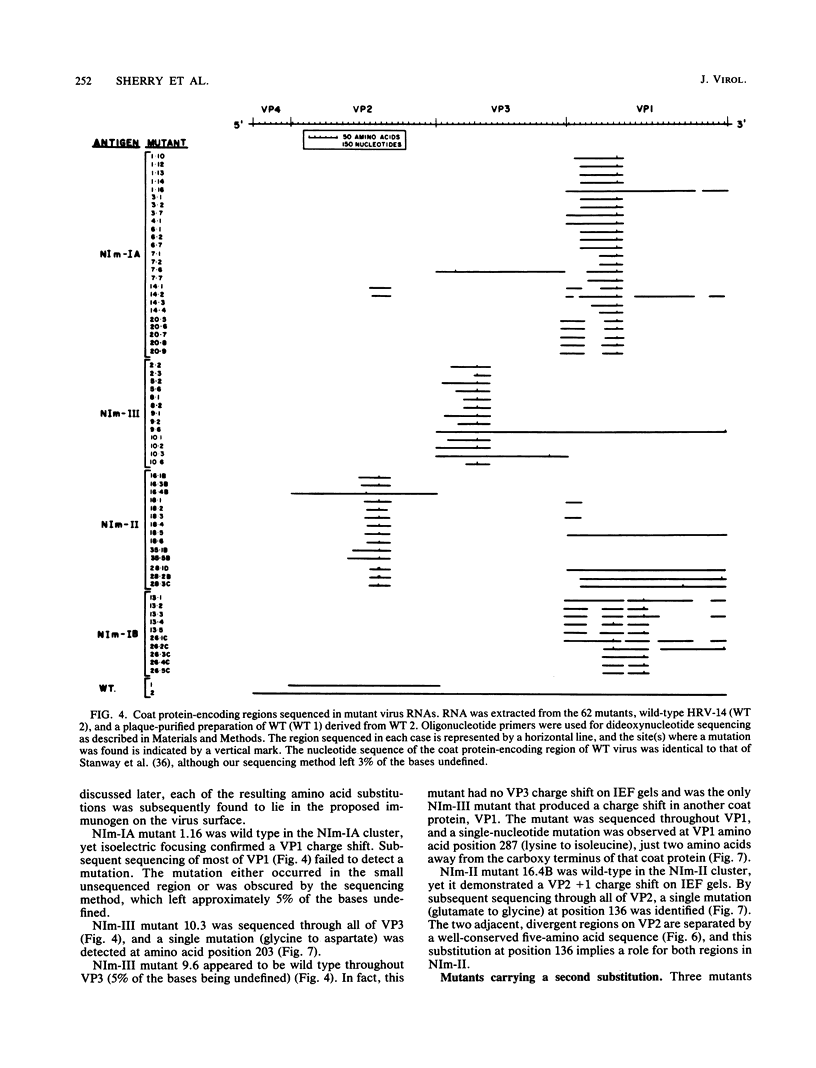
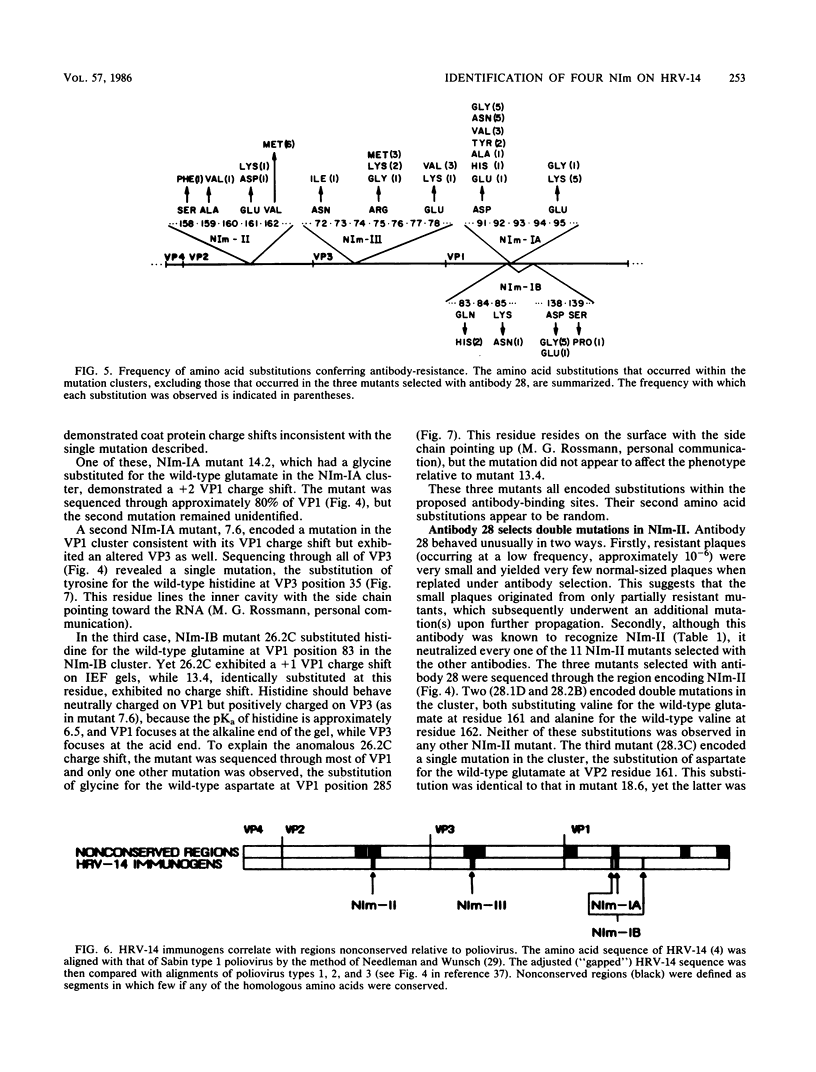
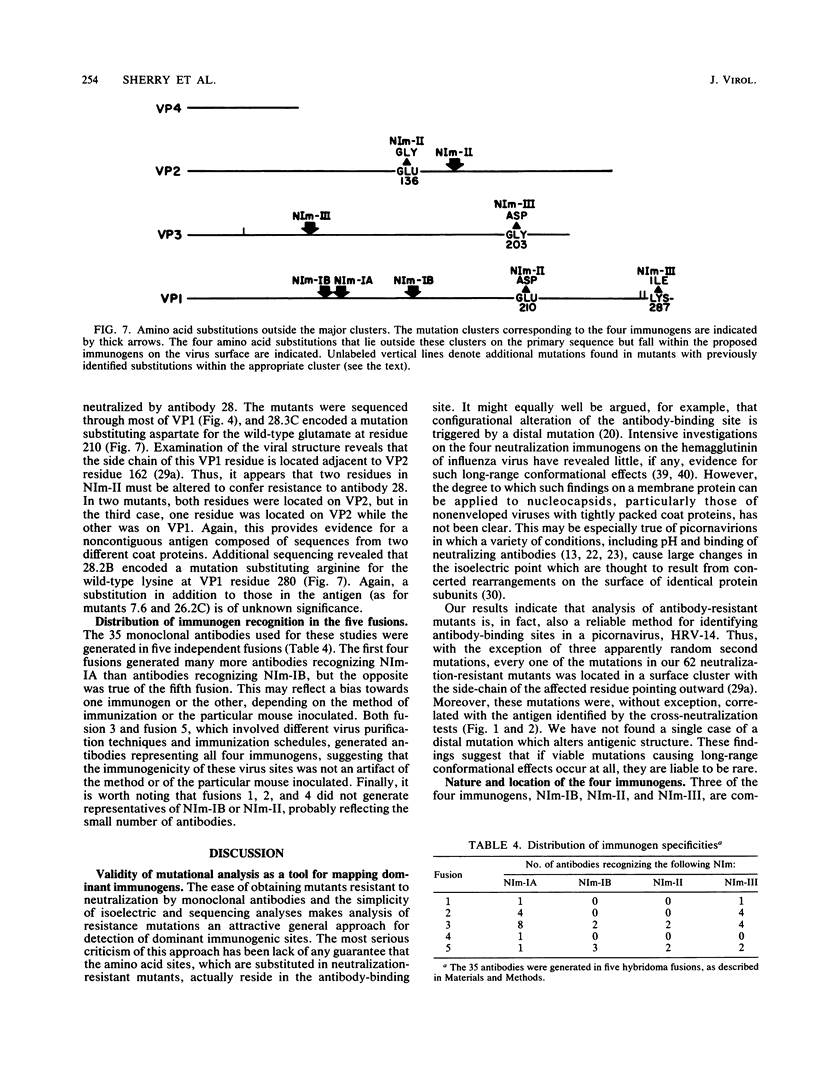
A collection of 35 mouse monoclonal antibodies, raised against human rhinovirus 14 (HRV-14), was used to isolate 62 neutralization-resistant mutants. When cross-tested against the antibodies in a neutralization assay, the mutants fell into four antigenic groups, here called neutralization immunogens: NIm-IA, -IB, -II, and -III. Sequencing the mutant RNA in segments corresponding to serotype-variable regions revealed that the amino acid substitutions segregated into clusters, which correlated exactly with the immunogenic groups (NIm-IA mutants at VP1 amino acid residue 91 or 95; NIm-II mutants at VP2 residue 158, 159, 161, or 162; NIm-III mutants at VP3 residue 72, 75, or 78; and NIm-IB mutants at two sites, either VP1 residue 83 or 85, or residue 138 or 139). Examination of the three-dimensional structure of the virus (M. G. Rossmann, E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H.-J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend, Nature [London], 317:145-153, 1985) revealed that each of the substitution clusters formed a protrusion from the virus surface, and the side chains of the substituted amino acids pointed outward. Moreover, four of the amino acid substitutions, which initially appeared to be anomalous because they were encoded well outside the cluster groups, could be traced to surface positions immediately adjacent to the appropriate viral protrusions. We conclude that three of the four antigens, NIm-IB, -II, and -III, are discontinuous. Thus, the amino acid substitutions in all 62 mutants fell within the proposed immunogenic sites; there was no evidence for alteration of any antigenic site by a distal mutation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A collaborative report: rhinoviruses--extension of the numbering system. Virology. 1971 Feb;43(2):524–526. [PubMed] [Google Scholar]
- Abraham G., Colonno R. J. Many rhinovirus serotypes share the same cellular receptor. J Virol. 1984 Aug;51(2):340–345. doi: 10.1128/jvi.51.2.340-345.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton M. T., Webster R. G. The antigenic structure of the influenza B virus hemagglutinin: operational and topological mapping with monoclonal antibodies. Virology. 1985 Jun;143(2):583–594. doi: 10.1016/0042-6822(85)90396-4. [DOI] [PubMed] [Google Scholar]
- Bittle J. L., Houghten R. A., Alexander H., Shinnick T. M., Sutcliffe J. G., Lerner R. A., Rowlands D. J., Brown F. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature. 1982 Jul 1;298(5869):30–33. doi: 10.1038/298030a0. [DOI] [PubMed] [Google Scholar]
- Callahan P. L., Mizutani S., Colonno R. J. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci U S A. 1985 Feb;82(3):732–736. doi: 10.1073/pnas.82.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow M., Yabrov R., Bittle J., Hogle J., Baltimore D. Synthetic peptides from four separate regions of the poliovirus type 1 capsid protein VP1 induce neutralizing antibodies. Proc Natl Acad Sci U S A. 1985 Feb;82(3):910–914. doi: 10.1073/pnas.82.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classification and nomenclature of viruses. Fourth report of the International Committee on Taxonomy of Viruses. Intervirology. 1982;17(1-3):1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- Cooney M. K., Fox J. P., Kenny G. E. Antigenic groupings of 90 rhinovirus serotypes. Infect Immun. 1982 Aug;37(2):642–647. doi: 10.1128/iai.37.2.642-647.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M. K., Wise J. A., Kenny G. E., Fox J. P. Broad antigenic relationships among rhinovirus serotypes revealed by cross-immunization of rabbits with different serotypes. J Immunol. 1975 Feb;114(2 Pt 1):635–639. [PubMed] [Google Scholar]
- Diamond D. C., Jameson B. A., Bonin J., Kohara M., Abe S., Itoh H., Komatsu T., Arita M., Kuge S., Nomoto A. Antigenic variation and resistance to neutralization in poliovirus type 1. Science. 1985 Sep 13;229(4718):1090–1093. doi: 10.1126/science.2412292. [DOI] [PubMed] [Google Scholar]
- Emini E. A., Jameson B. A., Wimmer E. Identification of a new neutralization antigenic site on poliovirus coat protein VP2. J Virol. 1984 Nov;52(2):719–721. doi: 10.1128/jvi.52.2.719-721.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini E. A., Jameson B. A., Wimmer E. Priming for and induction of anti-poliovirus neutralizing antibodies by synthetic peptides. Nature. 1983 Aug 25;304(5928):699–703. doi: 10.1038/304699a0. [DOI] [PubMed] [Google Scholar]
- Emini E. A., Ostapchuk P., Wimmer E. Bivalent attachment of antibody onto poliovirus leads to conformational alteration and neutralization. J Virol. 1983 Nov;48(2):547–550. doi: 10.1128/jvi.48.2.547-550.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. W., Frankenberger E. A., Rossmann M. G., Fout G. S., Medappa K. C., Rueckert R. R. Crystallization of a common cold virus, human rhinovirus 14: "isomorphism" with poliovirus crystals. Proc Natl Acad Sci U S A. 1983 Feb;80(4):931–934. doi: 10.1073/pnas.80.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. M., Minor P. D., Schild G. S., Almond J. W. Critical role of an eight-amino acid sequence of VP1 in neutralization of poliovirus type 3. Nature. 1983 Aug 4;304(5925):459–462. doi: 10.1038/304459a0. [DOI] [PubMed] [Google Scholar]
- Hogle J. M., Chow M., Filman D. J. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985 Sep 27;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Hogle J. M. Preliminary studies of crystals of poliovirus type I. J Mol Biol. 1982 Oct 5;160(4):663–668. doi: 10.1016/0022-2836(82)90322-9. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Hovanec D. L., Air G. M. Antigenic structure of the hemagglutinin of influenza virus B/Hong Kong/8/73 as determined from gene sequence analysis of variants selected with monoclonal antibodies. Virology. 1984 Dec;139(2):384–392. doi: 10.1016/0042-6822(84)90384-2. [DOI] [PubMed] [Google Scholar]
- Jameson B. A., Bonin J., Wimmer E., Kew O. M. Natural variants of the Sabin type 1 vaccine strain of poliovirus and correlation with a poliovirus neutralization site. Virology. 1985 May;143(1):337–341. doi: 10.1016/0042-6822(85)90121-7. [DOI] [PubMed] [Google Scholar]
- Mandel B. Characterization of type 1 poliovirus by electrophoretic analysis. Virology. 1971 Jun;44(3):554–568. doi: 10.1016/0042-6822(71)90369-2. [DOI] [PubMed] [Google Scholar]
- Medappa K. C., McLean C., Rueckert R. R. On the structure of rhinovirus 1A. Virology. 1971 May;44(2):259–270. doi: 10.1016/0042-6822(71)90258-3. [DOI] [PubMed] [Google Scholar]
- Melnick J. L. Taxonomy of viruses, 1980. Prog Med Virol. 1980;26:214–232. [PubMed] [Google Scholar]
- Minor P. D., Evans D. M., Ferguson M., Schild G. C., Westrop G., Almond J. W. Principal and subsidiary antigenic sites of VP1 involved in the neutralization of poliovirus type 3. J Gen Virol. 1985 May;66(Pt 5):1159–1165. doi: 10.1099/0022-1317-66-5-1159. [DOI] [PubMed] [Google Scholar]
- Minor P. D., Pipkin P. A., Hockley D., Schild G. C., Almond J. W. Monoclonal antibodies which block cellular receptors of poliovirus. Virus Res. 1984;1(3):203–212. doi: 10.1016/0168-1702(84)90039-x. [DOI] [PubMed] [Google Scholar]
- Minor P. D., Schild G. C., Bootman J., Evans D. M., Ferguson M., Reeve P., Spitz M., Stanway G., Cann A. J., Hauptmann R. Location and primary structure of a major antigenic site for poliovirus neutralization. Nature. 1983 Feb 24;301(5902):674–679. doi: 10.1038/301674a0. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Arnold E., Erickson J. W., Frankenberger E. A., Griffith J. P., Hecht H. J., Johnson J. E., Kamer G., Luo M., Mosser A. G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985 Sep 12;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Rueckert R. R., Pallansch M. A. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 1981;78(Pt A):315–325. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry B., Rueckert R. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J Virol. 1985 Jan;53(1):137–143. doi: 10.1128/jvi.53.1.137-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skern T., Sommergruber W., Blaas D., Gruendler P., Fraundorfer F., Pieler C., Fogy I., Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985 Mar 25;13(6):2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Minor P. D., Almond J. W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984 Oct 25;12(20):7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TYRRELL D. A., CHANOCK R. M. Rhinoviruses: a description. Science. 1963 Jul 12;141(3576):152–153. doi: 10.1126/science.141.3576.152. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Kohara M., Kataoka Y., Suganuma T., Omata T., Imura N., Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984 Apr 25;174(4):561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Zimmern D., Kaesberg P. 3'-terminal nucleotide sequence of encephalomyocarditis virus RNA determined by reverse transcriptase and chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4257–4261. doi: 10.1073/pnas.75.9.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]