Abstract
Fluorescence recovery after photobleaching has been widely used to study dynamic processes in the cell, but less frequently to analyze binding interactions and extract binding constants. Here we use it to analyze γ-tubulin binding to the mitotic spindle and centrosomes to determine the role of γ-tubulin in microtubule nucleation in the spindle. We find rapid γ-tubulin turnover in mitotic spindles of Drosophila early embryos, characterized by diffusional interactions and weak binding, differing from centrosomes with tight binding interactions. The diffusion coefficient of γ-tubulin is consistent with a major species existing in the cytoplasm as the less efficiently nucleating γ-tubulin small complex (γTuSC) or γ-tubulin, rather than γ-tubulin ring complex (γTuRC). The fluorescence recovery kinetics we observe implies that γ-tubulin functions by binding weakly to spindle microtubules. γ-Tubulin may interact transiently with the spindle, nucleating microtubules very rapidly, differing from centrosomes, where γ-tubulin binds tightly to nucleate microtubules.
INTRODUCTION
γ-Tubulin exists as a large complex in centrosomes, where it is essential for microtubule nucleation. It is also found at low density in spindles of animal cells (1–7) and anastral spindles of higher plants (8), where its role is largely unknown, although it is thought to nucleate microtubules from chromatin or preexisting microtubules (4,5). γ-Tubulin could play a potentially important role in the mitotic spindle by nucleating new microtubules from the sides of existing microtubules, as observed for cortical microtubules in plant cells (9) or interphase microtubules in yeast (10), and by bundling and organizing spindle microtubules (5,11).
Despite many reports that γ-tubulin is present in spindles, little data are available clarifying the role of γ-tubulin in the spindle or how it binds to spindles and nucleates microtubules, largely because of the dominant role centrosomes play in nucleating microtubules for spindle assembly and function. Although Drosophila lacking centrosomes have been reported to assemble mitotic spindles around chromatin (12), the requirement for γ-tubulin in microtubule nucleation in the spindle is uncertain. This information is needed to understand mitotic spindle assembly and spindle function. The recent identification of previously unknown proteins that recruit γ-tubulin to spindle microtubules may shed light on its role in the spindle (7), but information is needed regarding the interactions of γ-tubulin with spindle microtubules and differences or similarities in its interactions at centrosomes to understand how it functions in the spindle. In particular, binding on-rates and dissociation rates in cells would give a clearer idea of γ-tubulin interactions with the spindle and centrosomes, and its possible function.
Fluorescence photobleaching recovery kinetics can be used to analyze protein binding interactions in cells and has the advantage that it is minimally invasive. Fluorescence recovery after photobleaching (FRAP) is now widely used to study dynamic processes using confocal microscopy for bleaching and monitoring recovery (13), but it is less frequently used to analyze protein binding kinetics. This is largely because exact mathematical solutions have not been reported for either photobleaching or recovery using high numerical aperture (NA) objectives and small bleach spots like those typically used in confocal microscopy. Theoretical models have assumed that the bleached region is cylindrical and recovery occurs by two-dimensional (2D) diffusion (14), although recent models account for recovery by different contributions of diffusion and binding interactions (15). Models for three-dimensional (3D) diffusional recovery have been reported, e.g., the uniform disk model (16), but using objectives of low NA and large bleach spots, unlike those used in most current studies. Fluorescence recovery due to diffusion is greatly influenced by the bleach profile and 2D or 3D diffusion, but interactions of proteins in the cell often display binding-dominant, rather than diffusion-dominant, interactions (15). In these cases, when data are fit to models to obtain kinetic binding constants and diffusion coefficients, the binding parameters are much less affected by inaccuracies in the bleach profile assumed by the model and can be determined from the later binding-dominant phase of a curve fit, despite a poor fit to the early diffusion-dominant phase. We simulated diffusional fluorescence recovery in a bleach profile using a high NA objective and small region of interest (ROI) like those used in confocal microscopy, compared to bleach profiles assumed by two different models, and found that the recovery most closely approximated a cylinder profile.
We then analyzed γ-tubulin binding interactions with the mitotic spindle and centrosomes by fitting models for fluorescence recovery based on a cylinder bleach profile. The kinetic constants derived from the fits of the models to our FRAP data show rapid turnover of γ-tubulin in the spindle dominated by diffusion and weak binding interactions, unlike centrosomes, where turnover is dominated by binding interactions. The differences we observe imply different interactions of γ-tubulin with the spindle and centrosomes, and potentially different mechanisms of microtubule nucleation.
MATERIALS AND METHODS
γTub37C-gfp transgenes
Drosophila expressing γTub37C or γTub37C E116R fused to S65T gfp (17), regulated by the oocyte- and early embryo-specific ncd promoter (18), were recovered in γTub37C+ flies. Embryo viability was assayed with ncd-gfp No. 4121, a wild-type line expressing nonclaret disjunctional-green fluorescent protein (NCD-GFP) (19,20), as a control. The frequencies of viable embryos for γTub37C-gfp (0.816, n = 298, total = 365) and ncd-gfp (0.833, n = 219, total = 263) were similar to wild-type (Ore R) (0.815, n = 1,073, total = 1316) (21). Tests of the null hypothesis that γTub37C-gfp and ncd-gfp were from the same population gave a high p value (χ2 = 0.180, 1 degree of freedom, p = 0.67). Thus, the transgene does not cause dominant mutant effects resulting in embryo inviability.
Tests for rescue by γTub27C-gfp of the loss-of-function γTub37C APL10 (E117K) mutant (22) were performed in heterozygous mutant females carrying a deficiency, Df(2L)VA23, and one or two copies of the γTub37C-gfp F13F3 transgene. APL10/Df(2L)VA23 females without the transgene produced many eggs, but none hatched. The γTub37C-gfp transgene rescued APL10/Df(2L)VA23 female sterility: one copy gave 0.956 viable embryos (n = 65, total = 68) and two copies gave 0.868 (n = 231, total = 266). These frequencies are higher than ncd-gfp No. 4121 (0.833) or Ore R (0.815) (21), presumably due to out-crossing of deleterious genes during construction of the test females. The APL10 mutation (E117K) (23) was confirmed present in one test female by polymerase chain reaction using a primer that anneals to γTub37C intron 2 but not to the γTub37C-gfp cDNA transgene, followed by DNA sequence analysis.
Live imaging of embryos
Time-lapse images were acquired at 20°C using a confocal microscope (Bio-Rad Radiance2100; Carl Zeiss, Thornwood, NY) with a 40×/1.3 NA oil immersion objective and Bio-Rad LaserSharp 2000 software. Image analysis was performed using ImageJ 1.38× (National Institutes of Health, Bethesda, MD). Plot profiles of a line through the long axis of a metaphase or anaphase spindle were made in ImageJ and plotted in Kaleidagraph (Synergy Software, Reading, PA) after normalizing fluorescence to the highest value. γTub37C fluorescence in the spindle was estimated by modeling the spindle as two half-ellipsoids, each of radius, r, height, h, and volume, v = (2/3)πr2h. The half-spindle radius, height, and integrated and mean density of a ∼1 μm-thick optical section through the long axis were measured from images of cycle 10 embryos at metaphase (embryos, n = 5; spindles, n = 19). Total fluorescence was calculated by dividing the integrated density of an elliptical half-disk of thickness, w, by its volume, v = (πrhw)/2, and multiplying by the spindle volume. The centrosome was modeled as a sphere of radius, r, and volume, v = (4/3) πr3. Total fluorescence was obtained by dividing the integrated density of a disk through the centrosome of thickness, w, by the disk volume, v = πr2w, and multiplying by the centrosome volume. Mean fluorescence was measured for γTub37C-gfp (embryos, n = 5; half-spindles, centrosomes, or cytoplasmic regions, n = 19) or γTub37C E116R-gfp (embryos, n = 2; half-spindles, centrosomes, or cytoplasmic regions, n = 20) in metaphase spindles from time-lapse sequences.
Antibody-stained embryos
Methanol-fixed embryos were stained (24) with anti-γTub37C antibodies provided by Y. Zheng (Carnegie Institution, Baltimore, MD) and rhodamine-α-tubulin antibody from W. Sullivan (University of California, Santa Cruz, CA), followed by a DNA stain, DAPI.
γTub37C cytoplasmic concentration
The cytoplasmic concentration of γTub37C-GFP was estimated by serially diluting purified S65T GFP, preparing slides, and recording images by confocal microscopy using the same conditions as imaging embryos. The mean fluorescence of each dilution was determined and compared with the fluorescence of γTub37C-gfp or ncd-gfp embryo cytoplasm (n = 36, 3 measurements/embryo). GFP protein concentration was determined from the spectrum from 240 to 510 nm of the 1:1 dilution. The concentration was ∼2–4 μM from the OD490 and OD280 values, the S65T GFP extinction coefficient, λ489 = 56,000 M−1 cm−1, and λ489/λ280 = 2.25 (25), giving λ280 = 24,889 M−1 cm−1. γTub37C-gfp cytoplasmic fluorescence (27.4 ± 0.2) was similar to the 1:4 dilution (22.1 ± 1.2; 0.5–1 μM) and ncd-gfp cytoplasmic fluorescence (44.0 ± 1.1) was close to the 1:2 dilution (41.6 ± 0.4; 1–2 μM), consistent with the twofold higher copy number of the transgene in the four-dose ncd-gfp line used in this study. The GFP dilutions were analyzed by SDS-PAGE and quantitated relative to known concentrations of bovine serum albumin, giving values for the 1:1 and 1:2 dilution of 4 and 2 μM, respectively, within the range of concentrations estimated by protein absorbance.
Fluorescence recovery simulation
Fluorescence recovery was calculated for a bleach profile that resembles the actual profile for a confocal microscope with a high NA objective and small ROI, and two profiles assumed by different models, by simulating diffusion without binding using a 3D array of fluorophore concentration. Mean fluorescence values for a double cone resembling the profile of a small bead made by the high NA objective used in this study, a cylinder (14), and a disk (16) were calculated over a disk-shaped region corresponding to the region monitored in experimental assays using the following solution to the diffusion equation (derived by the Green's function method):
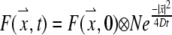 |
(1) |
where N is a normalization constant chosen such that the array used to approximate the time-dependent factor sums to 1 and ⊗ denotes convolution. The diffusion constant we used was 19.1 μm2/s (the value we derived for γ-tubulin in the cytoplasm). The first profile was modeled as two inverted photobleached cones with the angle determined by our 40×/1.3 NA objective, spreading outward axially and meeting in a circle at the focal disk. The bleaching above and below the circle was assumed to be inversely proportional to the cross-sectional area of the cone at each value of the vertical distance from the circle. This gives an initial profile, in cylindrical coordinates, of
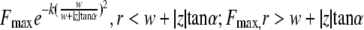 |
(2) |
where Fmax is the maximum fluorescence, w is the bleach spot radius (w = 1.3 μm for our small ROI), α is the cone angle, equal to the inverse sine of the NA divided by the index of refraction of the immersion medium (n = 1.518 for our oil), and k is a constant representing the bleaching depth, set so that the initial mean fluorescence value is 0.1 Fmax (this condition was used for all three models). Because the bleach spot is much larger than a diffraction-limited spot, the double cone effect arises only at the edges of the spot, whereas the interior is more uniform due to the overlap of many double cones, making the profile resemble the cylinder at the plane of focus. The second profile consisted of a bleached cylinder of radius r and 0.1 Fmax fluorescence, uniform along the z axis and surrounded by Fmax fluorophore; this profile underlies the 2D treatment by Axelrod et al. (14) and is assumed by the models (15) used in our analysis. Soumpasis (26) derived an exact solution for fluorescence recovery for the Axelrod photobleached cylinder. The third profile consisted of a disk with a Gaussian axial distribution of photobleaching; this is the 3D model used for much larger bleach spots and lower NA objectives (16). We set the initial fluorescence to Fmax outside the disk of radius w; inside the disk, we used their expression for the fluorescence value
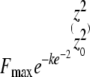 |
(3) |
The axial resolution, z0, is given by 2λ/NA2 for the half volumes used in our calculations (27), where λ is the light wavelength (488 nm in our assays). The constant k was calculated separately for this model and found to be greater than for the cylinder model. The small bleach spot and high NA objective used in our assays results in an elongated disk.
The mean fluorescence values for each of the three bleach profiles were calculated at different times during diffusional fluorescence recovery and plotted versus time. Values were also calculated using the Soumpasis equation for a cylinder as a control. We fit the cone values to the Soumpasis equation to estimate the error in D from use of the cylinder model. They gave D = 12.05 μm2/s compared to the 19.1 μm2/s that we used to calculate the cone values, suggesting that models assuming a cylinder bleach profile can give D within a factor of 2 or less when applied to confocal microscopy data.
Photobleaching analysis
FRAP assays were performed at 22–25°C on an LSM 510 confocal microscope (Carl Zeiss) using LSM 510 software, a 40×/1.3 NA oil immersion objective, and the 488 nm line of a 30 mW Ar laser operating at 75% power. Photobleaching spindles at prometaphase or metaphase did not disrupt cell division: the spindles recovered fluorescence and progressed to telophase, completing division with the same kinetics as unbleached spindles in the same field. Six prebleach images were recorded, followed by 3–4 bleach scans in ROIs of radius w = 2.66 or 1.3 μm and 494 recovery images at ∼165 ms time resolution. The mean pixel value of the photobleached structure during the recovery scans was recorded using LSM 510 software or tracked manually in ImageJ. Data were normalized to the fluorescence in the first prebleach image and corrected for loss during recovery imaging by adding back the fluorescence lost from an adjacent unbleached structure. Data from 7–14 assays were averaged and plotted versus time, then fit to kinetic models for fluorescence recovery representing pure diffusion, binding-dominant interactions, or different contributions of diffusion and binding (15,28). Where indicated in Table 1, normalized corrected data for the large and small ROIs were fit concurrently to a diffusion-binding model (15) with the first 120 data points weighted more heavily than the rest.
TABLE 1.
FRAP kinetic parameters
| Protein | Curve fit method* | D (μm2/s) | k*on (s−1) | koff (s−1) | t1/2 (s) | Feq | Ceq | Recovery† (%) |
|---|---|---|---|---|---|---|---|---|
| γTub37C-GFP | ||||||||
| Spindle | Concurrent weighted | 0.030 ± 0.004 | 0.16 ± 0.01 | 4 | 0.85 | 0.15 | 98 | |
| Centrosome‡ | Direct | 0.030 ± 0.001 | 0.0272 ± 0.0008 | 26 | 0.47 | 0.53 | 94.4 ± 0.5 | |
| Cytoplasm | Concurrent weighted | 19.1 ± 2.2 | 0.028 ± 0.004 | 0.14 ± 0.01 | 5 | 0.84 | 0.16 | 98 |
| NCD-GFP | ||||||||
| Cytoplasm | Concurrent weighted | 9.5 ± 0.8 | 0.026 ± 0.005 | 0.16 ± 0.02 | 4 | 0.86 | 0.14 | 98 |
| GFP-α-tubulin | ||||||||
| Spindle | Concurrent weighted | 0.051 ± 0.002 | 0.081 ± 0.002 | 9 | 0.61 | 0.39 | 103 | |
| Cytoplasm | Concurrent weighted | 14.6 ± 0.9 | 0.07 ± 0.01 | 0.44 ± 0.03 | 2 | 0.85 | 0.15 | 99 |
Value ±95% confidence interval from the curve fit.
Curve fit methods are given in Materials and Methods.
Average for large and small ROI.
Data for small ROI.
Fitting of FRAP data to kinetic models
FRAP data were fit to models for fluorescence recovery using Kaleidagraph and MATLAB (The MathWorks, Natick, MA). We first tried the single-state reaction-dominant model, which supposes that diffusion occurs very fast and recovery is dominated by the kinetics of a single binding state (15,28). In this case, recovery shows a much stronger dependence on the kinetics of protein binding than on the shape of the bleached region. Recovery is given by
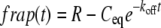 |
(4) |
where R is the fluorescence intensity at equilibrium after recovery, Ceq is the fraction of fluorescence at equilibrium due to binding, and koff is the dissociation rate constant. The pseudo first-order binding constant, k*on, was calculated by k*on = (Ceq koff)/(1 − Ceq) using koff from the curve fit. The turnover time was calculated by t1/2 = ln(2)/koff and % recovery by 100% Ceq/(1 − R + Ceq). The model fit the γ-tubulin centrosome data for the small bleach spot except for the first few points (see Fig. 3 D). The γ-tubulin centrosome data were also analyzed after bleaching with a large ROI, where the centrosome comprised ∼14% of the ROI surrounded by cytoplasm; recovery of only the centrosome was analyzed. The averaged data points for the large ROI overlapped with those for the small ROI (see Fig. 3 D), indicating that binding interactions, rather than diffusion, dominate during recovery. In contrast, the γ-tubulin spindle and cytoplasm data did not fit well to this model; they showed a rapid early phase that deviated significantly from the curve fit (see Fig. 3 B, inset, and Fig. 4 B). Furthermore, behavior in this early phase differed with bleach spot size; recovery was faster for the smaller bleach spot, indicating a significant diffusional component. Attempts to fit a pure diffusion model (Eq. 8 of Sprague et al. (15) or Eq. 16 of Soumpasis (26)) to the data gave poor fits due to slower fluorescence recovery than predicted by the model.
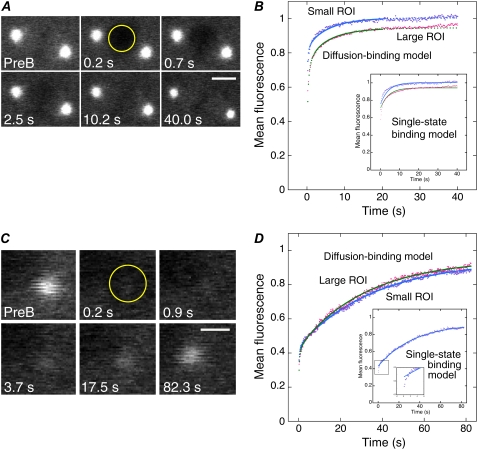
FIGURE 3.
FRAP analysis of γ-tubulin in the spindle and centrosome. (A and C) γTub37C-GFP fluorescence recovery at exponentially increasing time points in the spindle (ROI = 2.66 μm radius) (A) and centrosome (ROI = 1.3 μm radius) (C). PreB, prebleach. Bars, 5 μm (A) and 2 μm (C). (B and D) Mean data for small (1.3 μm radius) and large (2.66 μm radius) ROIs, normalized to the first prebleach value, showing fluorescence recovery at the spindle (B) or centrosome (D). Fits of the data (small ROI, purple; large ROI, pink) to the diffusion-binding model (small ROI, blue; large ROI, green) are shown. (Insets) Fits to the single-state binding model; the curve fit to the small ROI data points in D deviates at initial points (second inset).
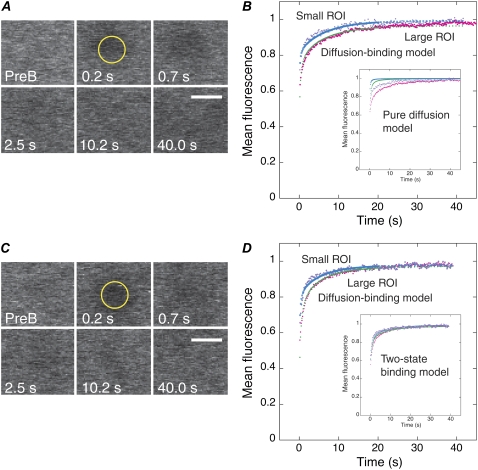
FIGURE 4.
FRAP analysis of γ-tubulin and NCD in the cytoplasm. (A and C) Fluorescence recovery at exponentially increasing time points in a small ROI (1.3 μm radius) of γTub37C-GFP (A) and NCD-GFP (C) in the cytoplasm. PreB, prebleach. Bars, 3 μm. (B and D) Mean data for small and large ROIs, normalized to the first prebleach value, showing fluorescence recovery for γTub37C-GFP (B) or NCD-GFP (D). Fits of the data to the diffusion-binding model are shown; colors are the same as Fig. 3. (Insets) Fits of the data to a pure diffusion model (recovery in the absence of a binding state) (B) and a two-state binding model (15), in which the protein binds to cellular structures in two states with different kinetics (D).
The two-state reaction-dominant model was then tried. Like the single binding state model, this model supposes that diffusion occurs very fast, but that recovery is dominated by the kinetics of two different binding states. Again, recovery depends more on the kinetics of binding than on the shape of the bleached region, and is given by
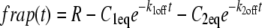 |
(5) |
where the two states are denoted as 1 and 2 (15). This model fit the γ-tubulin spindle and cytoplasm data well, but gave different rate constants for the two ROIs, indicating that the recovery behavior had a significant diffusional component unaccounted for by this model. However, both the two-state reaction-dominant and full model curve fits for the γ-tubulin and α-tubulin spindle and cytoplasm, and NCD cytoplasm FRAP data, gave k*onw2/D ≪ 1 for the slow binding state, using estimates of D based on the molecular weight of the proteins and w = 2.66 or 1.3 μm for the bleach spot radius. When the data exhibit these properties, they can be described by a two-term model with a fast phase corresponding to diffusion and a slower phase corresponding to binding, assuming a circular bleach spot, a cylindrical bleached region, and recovery by 2D diffusion in the plane of focus. Sprague et al. (15) derived a model (Eq. 41) with this behavior that we refer to here as the diffusion-binding model:
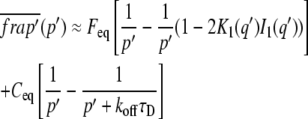 |
(6) |
where 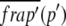 is the Laplace transform of the fluorescence scaled to the diffusion time constant
is the Laplace transform of the fluorescence scaled to the diffusion time constant 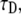 which equals w2/D; Feq is the fraction of protein free at equilibrium and Ceq is the bound fraction, q′ is the square root of p′, and I1(x) and K1(x) are modified Bessel functions. The inverse Laplace transform of this equation yields
which equals w2/D; Feq is the fraction of protein free at equilibrium and Ceq is the bound fraction, q′ is the square root of p′, and I1(x) and K1(x) are modified Bessel functions. The inverse Laplace transform of this equation yields
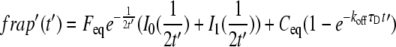 |
(7) |
Unscaling by substituting t/τD for t′ and letting Feq = koff/(k*on + koff) and Ceq = k*on/(k*on + koff), and multiplying by a normalization factor N so that fluorescence goes to 1 for large t, we obtained
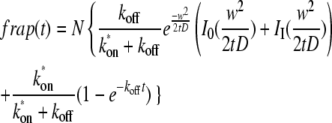 |
(8) |
which consists of a diffusional term, e.g., Eq. 16 (26), and a binding term. It can be fit to the data with the parameters N, D, k*on, and koff, allowing determination of the diffusion coefficient and kinetic constants for the protein in the spindle and cytoplasm. Fits of the data sets for the large and small ROIs were performed concurrently using the MATLAB routine leasqr.m (29) by appending the two data sets together in a vector as the dependent variable, entering the time points twice in a vector as the independent variable, and fitting these vectors to Eq. 8 with the same D, k*on, and koff for both data sets, but the radius, w, entered for each. A different normalization factor was fit for each data set.
The above method of concurrent fitting assumes that the binding constants, k*on and koff, for the weak binding phase do not change with bleach spot size, as predicted for interactions of a protein with a cellular structure, and that there is a single value for D in the region under study. It also assumes that the diffusion-binding model is appropriate for both bleach spot sizes.
The data for the cytoplasm and γ-tubulin in the spindle were truncated at 40 s (see Fig. 3 B, Fig. 4, and Fig. S3F in the Supplementary Material, Data S1), since the curves had already leveled off and the later data points added noise and made the fits worse. The first 120 points of the data, corresponding to the major part of the recovery, were weighted higher than the others: for every set of 5 points after the 120th, the time and fluorescence values were averaged and entered into the fitting routine as a single point (15). The NCD spindle data were not truncated because of the slower recovery and the α-tubulin spindle data were weighted but not truncated because the noise was more significant before 40 s than after (see Fig. S3, B and D, Data S1).
Data for the spindle and cytoplasm were analyzed with the diffusion-binding model using the concurrent fitting method described above. The kinetic parameters from the curve fits are shown in Table 1. Data for γ-tubulin at the centrosome were also analyzed using the diffusion-binding model, but by fitting the small and large ROI data separately, rather than concurrently. Here k*onw2/D ≪ 1, but k*on ≈ koff, indicating that the binding phase is dominant. Data for the large ROI fit to the diffusion-binding model are shown for comparison in Fig. 3 D; kinetic parameters for the curve fit were almost identical to the values shown in Table 1 for the small ROI. The Fig. 3 D inset shows that the single-state binding model fits well to the small ROI data, but deviates at early points.
The FRAP recovery of NCD in the spindle differs from the others studied in that it is not characterized by a rapid diffusion-dominated (or even effective-diffusion) phase, followed by a binding phase. The difference is visible qualitatively in the large discrepancy between the large and small ROI recovery curves; this discrepancy indicates that diffusion is a major influence throughout the recovery. This could be explained by a transient binding state that depletes the NCD that is free to diffuse, as in effective diffusion; the depletion is the same in both spot sizes, but in the case of NCD, unlike the other proteins, the slowdown is sufficient to make the large ROI take longer throughout recovery. Quantitatively, fitting the NCD spindle data to the diffusion-binding model yielded k*on = 6.2 ± 1.8 s−1 and D = 0.106 ± 0.004 μm2/s. Since the diffusion-binding model only applies when k*onw2/D ≪ 1 and here k*onw2/D is ∼100 for the small ROI and ∼400 for the large ROI, the model is not consistent with the data. NCD has microtubule-binding sites both in the head and the tail, which means there are at least three possible bound states—one each for the head and tail, and a third in which both the head and tail are bound—and it also binds cooperatively. Depending on the kinetics of the states (e.g., whether binding at one site induces rapid binding at the other), these properties may rule out all of the one- or two-binding state models.
As noted above, the assumption of a cylindrical bleached profile, which simplifies the mathematical treatment of recovery and allows the kinetic analysis, can introduce error into the estimated D values. Our analysis indicates that D can vary by a factor of ∼2 or more depending on the bleach profile that is assumed. The kinetic values can also be affected, although not to the same extent. For example, the values for γ-tubulin in the spindle are within 30% of those we report in Table 1 for the diffusion-binding model (k*on = 0.030 ± 0.004 s−1, koff = 0.16 ± 0.01 s−1) when a two-state reaction-dominant model is fit to the data (k*on = 0.02 s−1, koff = 0.12 s−1). The D values could also represent effective rather than free diffusion coefficients, as we believe to be the case for NCD and possibly α-tubulin in the cytoplasm. In this case, the data would fit a reduced two-state model in which diffusion slowed by binding interactions dominates in the early phase and weak binding dominates in the second phase, assuming a cylinder of bleaching and recovery by 2D diffusion in the plane of focus. This kinetic model,
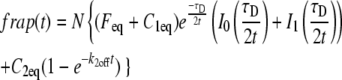 |
(9) |
where 1 and 2 represent the faster and slower states, respectively, holds when k*2onw2/D ≪ 1 (i.e., the second phase is significantly slower than diffusion; this is observed for NCD and γ-tubulin in the cytoplasm) and k*1onw2/D ≫ 1 (i.e., the initial faster phase is slowed by significant binding interactions). Deff = Dfree/(1 + k*1on/k1off), so the discrepancy between Deff and Dfree is large only if the fast binding state is relatively tight. The curves produced by this model are identical to the diffusion-binding model; when there is a fast second binding state of this nature, the diffusion-binding model will yield koff and Ceq (since these correspond to the unaltered, slower binding state), but will give Feq + C1eq instead of Feq, and Deff instead of Dfree.
Values for D were obtained from the curve fits to the diffusion-binding model and are given in Table 1 for γ-tubulin, NCD, and α-tubulin in the cytoplasm. These values are within the applicable range for this model. Since the diffusional phase of the recovery contains many fewer points than the binding phase, fits for D are more error-prone, so we used other methods to confirm D. One method involved separating the data points into two phases by approximating the two curves with a double-exponential fit and extracting a single-exponential curve corresponding to the fast phase:
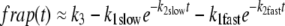 |
(10) |
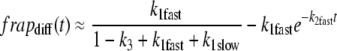 |
(11) |
The constant term in the fast phase curve is the fraction of recovery due to diffusion. Fitting this curve to Eq. 12,
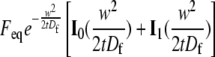 |
(12) |
assuming 2D diffusion during the recovery phase, with fitted parameters Feq and D yielding the D value. Another check was performed by fitting Eq. 8 to the data sets obtained only with the larger bleach spot, since they had more points in the rapidly recovering diffusion-dominant curve regions. The resulting D values for γTub37C and NCD are shown in Table 2.
TABLE 2.
Comparison of D (μm2/s) in the cytoplasm obtained by different fitting methods
| Protein | Concurrent fitting | Fast phase small ROI | Fast phase large ROI | Large ROI fit | Average |
|---|---|---|---|---|---|
| γTub37C | 19.1 | 6.7 | 12.3 | 32.6 | 17.7 |
| NCD | 9.5 | 4.3 | 9.1 | 13.8 | 9.2 |
The averages with or without the concurrent fitting values are close to the D values obtained with the concurrent fitting method, validating the use of this method. All of the methods gave a larger D for γTub37C than NCD.
Impact of noise on determination of k*on, koff, and D
The impact of noise in the data on determination of k*on, koff, and D can be quantified based on how much an error in a data point will affect the fit parameters. Thus, there will be an approximate inverse relationship between the error in D due to noise and
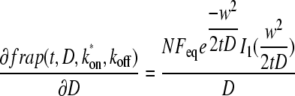 |
(13) |
using the diffusion-binding model. This value is on the order of 10−3 s/μm2 in the early part of recovery and drops rapidly to ∼10−5 s/μm2. This means that δD/δfrap increases from ∼50 to several thousand times the value of D. The large amplification of noise later in the run (after the first few seconds) reflects the fact that the later part of the curve is not significantly affected by diffusion, so changes in the later part of the curve will cause the fitting routine to alter the other fit parameters instead of D. This strongly exacerbates the effect of noise in the first few points on D.
By contrast,
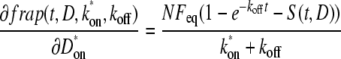 |
(14) |
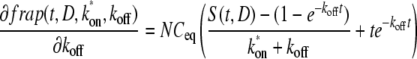 |
(15) |
where S(t,D) is the Soumpasis model recovery. Both of these expressions are approximately a second: δk*on/δfrap ≈ δkoff/δfrap ≈ 1 s−1, which is ∼1 order of magnitude greater than the values of k*on and koff obtained by fitting, rather than 2–4 orders of magnitude as for D.
Thus, over the major part of the FRAP recovery curve, the effect of noise on D is expected to exceed that on k*on and koff by at least an order of magnitude; as a result, the overall effect on D of noise in the data is much greater than on the kinetic parameters k*on and koff.
Confidence intervals
Ninety-five percent confidence intervals for each parameter were calculated from the variances given by the MATLAB leasqr.m (29) routine as part of its parameter covariance matrix and are shown in Table 1. The confidence interval was taken as 1.96 times the square root of the variance, since a normally distributed random variable lies within 1.96 standard deviations of the mean with 95% probability. The confidence intervals do not include errors associated with covariances of the parameters in the curve fit, or errors that uniformly alter the recovery curve.
RESULTS AND DISCUSSION
γ-Tubulin in Drosophila mitotic spindles
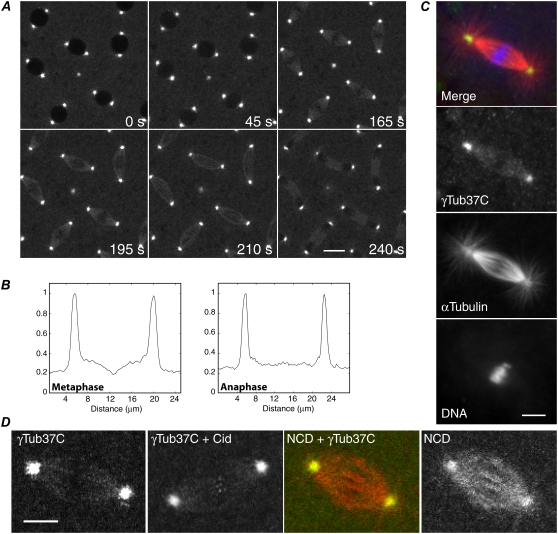
To study γ-tubulin in the spindle, we recovered flies expressing γTub37C, an oocyte- and early embryo-specific γ-tubulin, fused to GFP. The γTub37C-gfp transgene did not cause dominant negative mutant effects in γTub37C+ flies in genetic tests, and one or two copies fully rescued the γTub37C female-sterile mutant phenotype. The division time of γTub37C-gfp embryos by time-lapse imaging was comparable to wild-type, indicating that the line is wild-type with respect to cell division. γTub37C-gfp embryos displayed faintly labeled mitotic spindles with bright fluorescence at centrosomes, but little or no astral microtubule decoration (Fig. 1 A; Movie S1 in the Supplementary Material). Spindles showed a break in fluorescence at the metaphase plate, but it was interzonal in early anaphase (Fig. 1 B) and present in the midzone in late anaphase and telophase. γTub37C localization was confirmed by antibody-staining wild-type Ore R embryos (Fig. 1 C). γTub37C antibody staining closely resembled the GFP localization, but differed from α-tubulin antibody labeling of the spindle, centrosomes, and astral microtubules, indicating that γTub37C binds only to a subset of spindle microtubules. γTub37C-gfp embryos expressing Cid-GFP, a centromere-specific histone (30), showed labeled spindle fibers with ends close to chromosome centromeres, and embryos expressing a kinesin-14 spindle motor fused to a red fluorescent protein, NCD-RFP, showed faint γ-tubulin labeling of spindle fibers with bright fluorescence at centrosomes, differing from the bright labeling by NCD of spindles, centrosomes, and asters (Fig. 1 D).
FIGURE 1.
γ-Tubulin in Drosophila mitotic spindles. (A) γTub37C-GFP in a cycle 9 embryo from prophase (0 s) to telophase (240 s). Bar, 10 μm. (B) Plot profiles show absence of γTub37C at the metaphase plate and distribution along spindle fibers at anaphase. (C) Antibody-stained Ore R embryo spindle. Merge, γTub37C (green; yellow, colocalization with α-tubulin), α-tubulin (red), DNA (blue). Bar, 5 μm. (D) Mitotic spindles in γTub37C-gfp embryos showing γTub37C-labeled spindle fibers; labeling by a centromere-specific histone, Cid-GFP, at metaphase; NCD-RFP (red) and γTub37C-GFP (green; yellow, colocalization with NCD); and NCD. Bar, 5 μm.
γ-Tubulin levels in the spindle and centrosome were estimated by modeling the spindle as two half-ellipsoids and the centrosomes as spheres, and measuring the fluorescence of an optical section through the spindle. Mean fluorescence was higher in centrosomes (175 ± 5, n = 19) than spindles (66 ± 1, n = 19) by ∼2.7-fold, but total fluorescence was higher in each spindle half (11,975 ± 1,622, n = 19) than centrosome (996 ± 105, n = 19) by ∼12-fold because of the larger volume of the spindle. γ-Tubulin mean fluorescence was higher in the spindle than the cytoplasm (56 ± 2, n = 19) by only ∼1.2-fold.
Flies expressing γTub37C mutated in a conserved residue (E116R) of helix H3, which is thought to form lateral interactions between γ-tubulin molecules in γTuRC (31), were analyzed to determine γTub37C E116R interactions with the spindle. γTub37C+ embryos expressing the mutant protein fused to GFP showed centrosome and cytoplasm fluorescence, but spindle fluorescence was not above cytoplasm levels, although outlines of spindles due to exclusion of fluorescence by the chromosomes and spindle were apparent (Fig. S1 A, Data S1). Mean fluorescence in centrosomes was low (75 ± 4, n = 20) and was the same in spindles (41 ± 1, n = 20) as the cytoplasm (43 ± 1, n = 20) (Fig. S1 B, Data S1), indicating that the mutant γTub37C protein is not being trapped in the spindle. Failure to accumulate in the spindle was also observed for another protein, αTubΔC, which is present in the cytoplasm and enters the nucleus after nuclear envelope breakdown; αTubΔC showed the same level of fluorescence in mitotic spindles as the cytoplasm (Fig. S2, Data S1). The failure of both the mutant γTub37C E116R and αTubΔC to accumulate in the spindle indicates that the spindle viscosity and its fibrous structure do not differ sufficiently from the cytoplasm to trap the proteins. This implies that wild-type γTub37C in the spindle must be binding weakly, rather than being trapped.
FRAP bleach profiles and fluorescence recovery
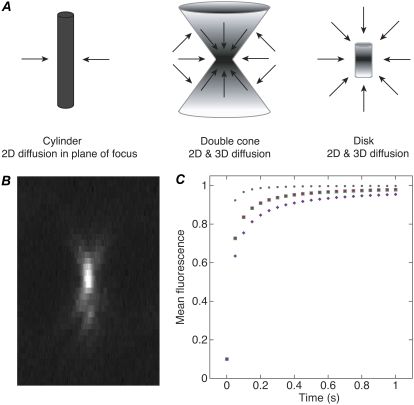
Before analyzing our FRAP data, we compared fluorescence recovery in a bleach profile made with a high NA objective and small ROI like those used in our assays to bleach profiles assumed by available models (Fig. 2 A). The profiles included a double cone that resembles the profile of a small bead made by our high NA objective (Fig. 2 B), a cylinder like the ones assumed by several models (14,15), and a disk (16). The mean fluorescence values corresponding to the diffusional phase of FRAP recovery were calculated at time t = 0–1 s. As a control, values were calculated by the Soumpasis equation for fluorescence recovery in a photobleached cylinder (26). The analysis showed that the double cone fluorescence recovery values did not overlap with either the disk or the cylinder values, but they were much closer to the cylinder values (Fig. 2 C). In contrast to the effects of diffusion, recovery that is rate-limited by binding should not be affected by the geometry of the bleach profile. Given that the overall shape of the diffusion curve is similar for the cylinder and double cone profiles, errors in the kinetic parameters due to assumption of a cylinder bleach profile are likely to be small. These considerations suggested that the Sprague et al. models (15), which assume a photobleached cylinder to model diffusion, could be used to reliably infer kinetic constants for γ-tubulin binding to the spindle and centrosomes from our confocal microscope data.
FIGURE 2.
Fluorescence recovery in bleach profiles assumed by different models. (A) Depictions of bleach profiles. Arrows, diffusional recovery. The double cone approximates the bleach profile for a high NA objective and small ROI like those used in this study. Models are based on a photobleached cylinder (14,15) or disk (16). (B) Image profile of a small bead by the high NA objective used in this study. Z-scan of a line through a 200-nm fluorescent bead. (C) Diffusional recovery over time calculated for a cylinder (green squares), double cone (purple diamonds) and disk (gray circles) bleach profile. Values calculated using the Soumpasis equation (26) (pink circles) for a cylinder, for comparison. Mean fluorescence = 0.1 at t = 0 for all the curves.
Fluorescence recovery of γ-tubulin in the spindle and centrosomes
γ-Tubulin binding interactions with the spindle and centrosomes were analyzed by fluorescence recovery kinetics. The concentration of γTub37C-GFP in embryo cytoplasm was estimated to be ∼0.5–1 μM, indicating the presence of a large cytoplasmic pool of protein to replace photobleached protein. FRAP assays were performed by rapidly photobleaching an ROI under high laser power using two different-sized bleach spots, then imaging at low power and high time resolution to monitor fluorescence recovery. The smaller bleach spot is expected to recover more rapidly by diffusion; thus the two ROIs test for recovery due to diffusion. Normalized, corrected mean data from 7 to 14 data sets were plotted versus time and fit to kinetic models based on a cylinder bleach profile for fluorescence recovery by pure diffusion (14,26), binding-dominant interactions (15), or differing contributions of diffusion and binding (15). Where possible, data for the large and small ROIs were fit concurrently to a given model to obtain single values for the kinetic parameters for the two data sets. This method of concurrent fitting provides additional confidence in the estimates because of the constraints of fitting two curves simultaneously. Analysis of the recovery curves yielded kinetic parameters for γ-tubulin binding interactions with the spindle and centrosomes, including association and dissociation rate constants, and a value for the diffusion coefficient, D, in the cytoplasm.
Fluorescence recovery of γTub37C-GFP in the spindle was rapid with a fast early phase (Fig. 3, A and B, Movie S2) that differed for the two bleach spot sizes, indicating that diffusion is a major factor. The curves were not fit well by pure diffusion or single-state binding-dominant (Fig. 3 B, inset) models; the best fits were to a two-term model with a rapid diffusional phase, followed by a weak-binding phase. This model, referred to here as the diffusion-binding model, closely approximates the binding curve when k*onw2/D ≪ 1, where k*on is the pseudo first-order binding rate constant, w is the bleach spot radius, and D is the diffusion coefficient (15), as observed for the fit parameters. The fast early phase had a turnover time of t1/2 ∼ 1 s and comprised ∼85% of the recovery, whereas the weak binding phase had k*on = 0.030 ± 0.004 s−1, a dissociation rate constant of koff = 0.16 ± 0.01 s−1 and t1/2 ∼ 4 s, and represented the remaining ∼15% of the recovery (Table 1). The k*on ≪ koff for the binding phase, together with the small fraction of γ-tubulin bound at equilibrium, Ceq ∼ 0.15, indicate that γ-tubulin binds weakly to the spindle.
FRAP assays were also performed on γTub37C-GFP at centrosomes during interphase of the rapid embryo cleavage divisions. Fluorescence recovery curves for centrosomes differed from spindles (Fig. 3, C and D, Movie S3) in that the data points for the small and large ROIs almost completely overlapped and the fast early phase was detectable only by the failure of a few early points to fit to a single-state binding-dominant curve (Fig. 3 D, inset). These features indicate that diffusion is not significantly limiting during recovery of γ-tubulin at centrosomes; instead, recovery is dominated by binding interactions. The diffusion-binding model gave an on-rate, k*on = 0.030 ± 0.001 s−1, that was the same as for spindles, whereas the dissociation rate constant, koff = 0.0272 ± 0.0008 s−1, was ∼6-fold slower, giving k*on ≈ koff, indicating relatively tight binding of γ-tubulin to the centrosome. Fluorescence recovered to ∼55% with t1/2 ∼ 26 s, differing from spindles and from the finding by others of a very slow (5–6 h) recovering centrosome fraction of ∼50% in vertebrate cells (2), presumably reflecting differences in cell types. The k*on ≈ koff and Ceq ∼ 0.55 indicate that binding interactions, rather than diffusion, dominate during fluorescence recovery of γ-tubulin at centrosomes.
Assays of the kinesin-14 NCD were performed to compare γ-tubulin with a protein in the spindle that binds to and dissociates from microtubules. NCD-GFP in the spindle (Fig. S3, A and B, Data S1) showed a strong dependence on bleach spot size, as for γTub37C-GFP in the spindle, but with more pronounced differences between the small and large ROIs. The data were best fit by the diffusion-binding model with a fast initial diffusional phase slowed by binding interactions, followed by a weak-binding phase, as for γ-tubulin in the spindle; however, the kinetics of the two phases differed from those for γ-tubulin and the values obtained from the curve fits indicated that the diffusion-binding model was not the appropriate model, as discussed in Materials and Methods. For this reason, we do not include the NCD spindle kinetic parameters in Table 1, although it is apparent from the recovery curves that NCD and γ-tubulin binding interactions in the spindle differ significantly from one another.
Assays of GFP-α-tubulin (32) were performed to compare fluorescence recovery of γ-tubulin with a protein that incorporates into spindle microtubules, rather than binding to microtubules. The recovery curves for the small and large ROIs both showed a fast early phase that differed somewhat in timescale, indicating significant contributions by diffusion (Fig. S3, C and D, Data S1). The data were best fit by the diffusion-binding model, as for γ-tubulin in the spindle, with a rapid initial diffusional phase, followed by a weak-binding phase. The curve fits showed that the fast diffusion-dominant phase corresponded to ∼40% of the recovery and k*on < koff for the weak binding phase, which comprised ∼60% of the recovery. The koff for α-tubulin was slower than for γ-tubulin by ∼2-fold (Table 1). Thus, both α-tubulin, which incorporates rapidly into spindle microtubules and releases by depolymerization, and NCD, a motor that binds to and dissociates from microtubules, differ from γ-tubulin in the spindle in their fluorescence recovery kinetics.
γ-Tubulin cytoplasmic diffusion coefficient
We also measured fluorescence recovery of γTub37C-gfp cytoplasm with two different-sized bleach spots to estimate D for γTub37C-GFP in the cytoplasm. The curves showed a rapid early phase that differed for the two ROIs, indicating a strong dependence on diffusion, followed by a slower phase that was similar for the two ROIs, indicating weak binding interactions (Fig. 4, A and B, Movie S4). Although pure diffusion and single-state binding models did not fit well, the diffusion-binding model, representing a transition between pure diffusion and binding-dominant states, gave a good fit with k*on ≪ koff and k*onw2/D ≪ 1, as for γ-tubulin in the spindle, indicating that this model was appropriate to describe the recovery (15). The fits gave a value for γ-tubulin in the cytoplasm of D = 19.1 ± 2.2 μm2/s (Table 1).
This value is close to the range of 6–18 μm2/s estimated for cytoplasmic γTuSC or γtub37C-GFP, based on the measured D for GFP, protein mass M, and D ∝ M−1/3 (15) or calculated from the Stokes' radius for γTuSC without GFP (Rs = 7 nm) (33), corrected by a factor of 2–5 for cytoplasm viscosity (34). NCD-GFP in the cytoplasm gave D = 9.5 ± 0.8 μm2/s (Fig. 4, C and D, Table 1), half the value of γTub37C-GFP. The Stokes' radius of dimeric full-length NCD (Rs = 7.6 nm) (35) is similar to that of γTuSC, predicting a diffusion constant of similar value. The NCD D value is probably an effective diffusion constant slowed by binding interactions, e.g., with the extensive network of astral microtubules in the early embryo (36). γ-Tubulin does not bind to astral microtubules (Fig. 1), but k*on and koff for γ-tubulin in the cytoplasm do not differ from NCD in the cytoplasm; they also do not differ from k*on and koff for γ-tubulin in the spindle. Given this and the poor fit by a pure diffusion model, the D value for γ-tubulin in the cytoplasm is unlikely to be Dfree; instead, γ-tubulin in the cytoplasm probably exists in a weak-binding, diffusional state that is weaker binding than γ-tubulin in the spindle. FRAP assays of cytoplasmic GFP-α-tubulin gave D = 14.6 ± 0.9 μm2/s (Fig. S3, E and F, Data S1; Table 1), which could also represent an effective diffusion constant, due to its existence as α-β-tubulin dimers and incorporation into cytoplasmic microtubules.
The D for γ-tubulin is consistent with a major cytoplasmic form existing as γTuSC or γ-tubulin monomers or dimers, rather than the much larger γTuRC (Rs = 15 nm) (33) with an estimated D = 3–8 μm2/s. The estimates for D obtained using these methods are not exact enough to distinguish between γTuSC or γ-tubulin monomers/dimers in the cytoplasm. The finding that γTub37C E116R does not specifically label spindles indicates that assembly into higher-order complexes may be needed for γ-tubulin spindle binding. Others have reported that γ-tubulin association with the spindle requires the four γTuRC-specific proteins that have been identified so far (6) and depletion of one of them in human cells blocks chromatin-mediated microtubule nucleation (4). This implies that γ-tubulin binds to spindles complexed with γTuRC proteins to nucleate microtubules in the spindle. If so, γ-tubulin in the cytoplasm may assemble rapidly with γTuRC proteins before binding to spindles.
SUMMARY
The distribution of γ-tubulin along the length of bundled microtubules in early embryo mitotic spindles is consistent with its binding to the microtubule lattice. Although this does not exclude incorporation into microtubules by nucleation from sites along the spindle fibers, γ-tubulin interactions with the spindle during fluorescence recovery in FRAP assays are dominated by diffusion, as indicated by the initial rapid rise of the recovery curves and slower recovery of the large bleach spot. Fits to kinetic models indicate weak binding interactions in which the dissociation rate is much higher than the pseudo on-rate. These observations imply that γ-tubulin associates with spindles by diffusing rapidly into the spindle and binding weakly to spindle microtubules. γ-Tubulin is not merely trapped by the spindle, as a mutant that binds weakly to centrosomes does not show spindle binding above cytoplasmic levels and this is also true of a truncated α-tubulin protein. By contrast, FRAP recovery curves for γ-tubulin at centrosomes are dominated by binding interactions, consistent with the idea that γ-tubulin binds to centrosomes and remains bound as it nucleates new microtubules. Evidence for a population of γ-tubulin at centrosomes in vertebrate cells that is tightly bound has been obtained by others using FRAP assays (2).
The transient interactions of γ-tubulin with the spindle thus differ from the centrosome, where γ-tubulin nucleates microtubules. It also differs from NCD, a motor that binds to spindle microtubules and dissociates after ATP hydrolysis. γ-Tubulin further differs from α-tubulin, a protein that is incorporated into microtubules and shows tighter binding with a faster on-rate and slower dissociation rate. γ-Tubulin interactions with the spindle indicate that its role in the early embryo spindle may be associated with transient, weak binding, e.g., bundling and organizing microtubules rather than nucleating microtubules, although its interactions could differ later in development, where the mitotic divisions are not as rapid as in early embryos. Alternatively, γ-tubulin in the spindle could nucleate microtubules by a mechanism that differs from centrosomes, transiently associating with chromosomes or microtubules in the spindle, nucleating, and then rapidly dissociating from newly nucleated microtubules. Our findings indicate that γ-tubulin interactions with the mitotic spindles of early embryos differ from centrosomes, implying a weak binding role or different mechanism of microtubule nucleation in spindles and centrosomes. Further studies will be essential to determine the mechanism by which γ-tubulin functions in the spindle and potentially nucleates microtubules.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank V. Bennett for the use of a Zeiss LSM 510 confocal microscope, Y. Zheng and W. Sullivan for antibodies, R. Tsien for mRFP, E. Shumsky for fluorescent bead imaging, and G. Hammes for discussions.
This work was supported by a National Institutes of Health grant (S.A.E.) and a Howard Hughes Medical Institute Summer Fellowship (J.H.). M.A.H. is a 2008 Goldwater Scholar.
Editor: E. Michael Ostap.
References
- 1.Lajoie-Mazenc, I., Y. Tollon, C. Detraves, M. Julian, A. Moisand, C. Gueth-Hallonet, A. Debec, I. Salles-Passador, A. Puget, H. Mazarguil, and others. 1994. Recruitment of antigenic γ-tubulin during mitosis in animal cells: presence of γ-tubulin in the mitotic spindle. J. Cell Sci. 107:2825–2837. [DOI] [PubMed] [Google Scholar]
- 2.Khodjakov, A., and C. L. Rieder. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haren, L., M.-H. Remy, I. Bazin, I. Callebaut, M. Wright, and A. Merdes. 2006. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication. J. Cell Biol. 172:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lüders, J., U. K. Patel, and T. Stearns. 2006. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147. [DOI] [PubMed] [Google Scholar]
- 5.Mahoney, N. M., G. Goshima, A. D. Douglass, and R. D. Vale. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569. [DOI] [PubMed] [Google Scholar]
- 6.Vérollet, C., N. Colombie, T. Daubon, H.-M. Bourbon, M. Wright, and B. Raynaud-Messina. 2006. Drosophila melanogaster γ-TuRC is dispensable for targeting γ-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172:517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goshima, G., R. Wollman, S. S. Goodwin, N. Zhang, J. M. Scholey, R. D. Vale, and N. Stuurman. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, B., J. Marc, H. C. Joshi, and B. A. Palevitz. 1993. A γ-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104:1217–1228. [DOI] [PubMed] [Google Scholar]
- 9.Murata, T., S. Sonobe, T. I. Baskin, S. Hyodo, S. Hasezawa, T. Nagata, T. Horio, and M. Hasebe. 2005. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol. 7:961–968. [DOI] [PubMed] [Google Scholar]
- 10.Janson, M. E., T. G. Setty, A. Paoletti, and P. T. Tran. 2005. Efficient formation of bipolar microtubule bundles requires microtubule-bound γ-tubulin complexes. J. Cell Biol. 169:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiato, H., C. L. Rieder, and A. Khodjakov. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basto, R., J. Lau, T. Vinogradova, A. Gardiol, C. G. Woods, A. Khodjakov, and J. W. Raff. 2006. Flies without centrioles. Cell. 125:1375–1386. [DOI] [PubMed] [Google Scholar]
- 13.Cole, N. B., C. L. Smith, N. Sciaky, M. Terasaki, M. Edidin, and J. Lippincott-Schwartz. 1996. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 273:797–801. [DOI] [PubMed] [Google Scholar]
- 14.Axelrod, D., D. E. Koppel, J. Schlesinger, E. Elson, and W. W. Webb. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprague, B. L., R. L. Pego, D. A. Stavreva, and J. G. McNally. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braeckmans, K., L. Peeters, N. N. Sanders, S. C. De Smedt, and J. Demeester. 2003. Three-dimensional fluorescence recovery after photobleaching with the confocal scanning laser microscope. Biophys. J. 85:2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heim, R., A. B. Cubitt, and R. Y. Tsien. 1995. Improved green fluorescence. Nature. 373:663–664. [DOI] [PubMed] [Google Scholar]
- 18.Endow, S. A., S. Henikoff, and L. Soler-Niedziela. 1990. Mediation of meiotic and early mitotic chromosome segregation in Drosophila by a protein related to kinesin. Nature. 345:81–83. [DOI] [PubMed] [Google Scholar]
- 19.Endow, S. A., and D. J. Komma. 1996. Centrosome and spindle function of the Drosophila NCD microtubule motor visualized in live embryos using NCD-GFP fusion proteins. J. Cell Sci. 109:2429–2442. [DOI] [PubMed] [Google Scholar]
- 20.Endow, S. A., and D. J. Komma. 1997. Spindle dynamics during meiosis in Drosophila oocytes. J. Cell Biol. 137:1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endow, S. A., R. Chandra, D. J. Komma, A. H. Yamamoto, and E. D. Salmon. 1994. Mutants of the Drosophila NCD microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 107:859–867. [DOI] [PubMed] [Google Scholar]
- 22.Schüpbach, T., and E. Wieshaus. 1989. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 121:101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson, P. 1999. γTub37CDPL10. Personal communication to FlyBase.
- 24.Endow, S. A., and D. J. Komma. 1998. Assembly and dynamics of an anastral:astral spindle: the meiosis II spindle of Drosophila oocytes. J. Cell Sci. 111:2487–2495. [DOI] [PubMed] [Google Scholar]
- 25.Endow, S. A., and D. W. Piston. 1998. Methods and protocols. In Green Fluorescent Proteins: Properties, Applications and Protocols. M. Chalfie and S. Kain, editors. Wiley-Liss, New York. 271–369.
- 26.Soumpasis, D. M. 1983. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41:95–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murthy, M. S. N., M. G. Jones, J. Kulka, J. D. Davies, M. Halliwell, P. C. Jackson, D. R. Bull, and P. N. T. Wells. 1995. Scanning confocal near-infra-red microscopy: a new microscopy technique for three-dimensional histopathology. Eng. Sci. Educ. J. 4:223–230. [Google Scholar]
- 28.Bulinski, J. C., D. J. Odde, B. J. Howell, T. D. Salmon, and C. M. Waterman-Storer. 2001. Rapid dynamics of the microtubule binding of ensconsin in vivo. J. Cell Sci. 114:3885–3897. [DOI] [PubMed] [Google Scholar]
- 29.Shrager, R., A. Jutan, and R. Muzic. 1994. leasqr.m. Version 3.beta.
- 30.Henikoff, S., K. Ahmad, J. S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA. 97:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldaz, H., L. M. Rice, T. Stearns, and D. A. Agard. 2005. Insights into microtubule nucleation from the crystal structure of human γ-tubulin. Nature. 435:523–527. [DOI] [PubMed] [Google Scholar]
- 32.Grieder, N. C., M. de Cuevas, and A. C. Spradling. 2000. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 127:4253–4264. [DOI] [PubMed] [Google Scholar]
- 33.Oegema, K., C. Wiese, O. C. Martin, R. A. Milligan, A. Iwamatsu, T. J. Mitchison, and Y. Zheng. 1999. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144:721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berland, K. M., P. T. So, and E. Gratton. 1995. Two-photon fluorescence correlation spectroscopy: method and application to the intracellular environment. Biophys. J. 68:694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao, L., A. Mogilner, G. Civelekoglu-Scholey, R. Wollman, J. Evans, H. Stahlberg, and J. Scholey. 2006. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes NCD in motility assays. Curr. Biol. 16:2293–2302. [DOI] [PubMed] [Google Scholar]
- 36.Foe, V. E., G. M. Odell, and B. A. Edgar. 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In The Development of Drosophila melanogaster. M. Bate and A. M. Arias, editors. CSHL Press, Cold Spring Harbor, NY. 149–300.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.