Abstract
Amyloids are associated with a number of protein misfolding disorders, including prion diseases. In this study, we used single-molecule force spectroscopy to characterize the nanomechanical properties and molecular structure of amyloid fibrils formed by human prion protein PrP90-231. Force-extension curves obtained by specific attachment of a gold-covered atomic force microscope tip to engineered Cys residues could be described by the worm-like chain model for entropic elasticity of a polymer chain, with the size of the N-terminal segment that could be stretched entropically depending on the tip attachment site. The data presented here provide direct information about the forces required to extract an individual monomer from the core of the PrP90-231 amyloid, and indicate that the β-sheet core of this amyloid starts at residue ∼164–169. The latter finding has important implications for the ongoing debate regarding the structure of PrP amyloid.
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs), more commonly referred to as prion diseases, are a group of fatal neurodegenerative disorders that affect several mammalian species and include bovine spongiform encephalopathy in cattle, scrapie in sheep, chronic wasting disease in cervids, and Creutzfeldt-Jakob's disease and kuru in humans (1–6). These devastating brain-wasting diseases are known to arise through sporadic, inherited, or infectious mechanisms, and in all cases result in a neuropathology that shows characteristic spongiform degeneration and astrogliosis (2,4,5). Like a number of other neurodegenerative disorders, such as Alzheimer's, Parkinson's, and Huntington's diseases, prion diseases are associated with cerebral accumulation of abnormal protein deposits (1–8). In many (though not all) cases, these deposits are largely composed of amyloid fibrils—highly ordered protein aggregates that show a characteristic “cross-β” x-ray diffraction pattern (7). In the case of TSEs, these deposits result from conformational conversion of normal cellular prion protein, PrPC, to an abnormal isoform, PrPSc. According to the protein-only hypothesis, the transmission of TSEs does not require nucleic acids, and PrPSc itself represents the infectious pathogen that self-replicates by binding to PrPC and templating its conversion to the PrPSc state (1–6). This model is gaining growing acceptance, particularly in the light of recent successes in generating infectious PrPSc in vitro (9–11).
Cellular human PrPC is a 209 residue membrane-associated protein, with a C-terminally attached glycosylphosphatidylinositol anchor and two Asn-linked glycosylation sites (2–5). High-resolution NMR structures of recombinant PrP, a nonglycosylated model of PrPC, reveal a flexible N-terminal region and a C-terminal globular domain containing three α-helices and two short β-strands, with a disulfide bond bridging helices 2 and 3 (12–14). Upon conversion of monomeric PrPC to PrPSc aggregate, the protein becomes insoluble in nonionic detergents and acquires remarkable resistance to proteinase K digestion, with the PK-resistant core typically starting at residue ∼90 (1–4). Furthermore, low-resolution optical spectroscopic data indicate that, in contrast to PrPC, PrPSc is rich in β-sheet structure (15,16).
Given that higher-resolution structural studies with brain-derived protein deposits present enormous experimental challenges and the molecular structure of PrPSc associated with TSE infectivity is still poorly defined, a number of laboratories have focused on PrPSc-mimicking amyloid fibrils formed by the recombinant prion protein. In particular, recent structural studies using H/D exchange (17) and site-directed spin labeling (18) indicate that the stable H-bonded “β-core” of the recombinant PrP fibril encompasses residues ∼160–220, and these residues form single-molecule layers that stack on top of one another with parallel, in-register alignment of β-strands. Furthermore, the entire region N-terminal to this amyloid core does not appear to possess a stable secondary structure (17,18).
Other important questions relate to the energetics of amyloid fibril assembly, the stability of these structures, and their nanomechanical properties. Recently, single-molecule force spectroscopy using an atomic force microscope (AFM) has emerged as a powerful tool for studying the nanomechanical and thermodynamic properties of proteins and other biomolecules (19). In this study we adapted this approach to probe amyloid fibrils formed by the recombinant polypeptide corresponding to human prion protein 90-231 (huPrP90-231). This C-terminal region of PrP appears to play a critical role in TSE pathogenesis. It encompasses the entire sequence of the protease-resistant fragment of aggregate PrPSc found in diseased brain, contains all point mutations associated with hereditary prion disorders, and is sufficient for propagation of the disease (2,4).
MATERIALS AND METHODS
Generation of the Cys variants of huPrP90-231 and protein purification
Cysteine was incorporated at positions 103 and 90 of huPrP90-231 by site-directed mutagenesis, and proteins were expressed and subjected to our purification and refolding protocol as previously described (20). The correctness of folding of the Cys variants was verified by far-UV circular dichroism spectroscopy.
Preparation of amyloid fibrils
Amyloid fibrils were obtained by incubation of the prion protein at 37°C with gentle rotation essentially as described previously (20). To prevent intermolecular disulfide formation between introduced Cys residues, the disulfide-specific reducing agent Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was added to the conversion buffer at a concentration of 1 mM.
AFM imaging and force measurements
For AFM experiments, 5 μl of fibrilized protein were deposited on freshly cleaved mica and left to adsorb for 1 min. Afterward, the sample was rinsed three times with 50 μl of 50 mM phosphate buffer, pH 6.4, to remove traces of TCEP. Imaging and force measurements were performed on a Nanoscope IV AFM (Veeco, Santa Barbara, CA) using an E-scanner (calibrated on a standard grid) and gold-covered tips mounted on a rectangular cantilever with a nominal spring constant of 0.027 N/m (Olympus Bio-Lever BL-RC 150VB). These tips were calibrated using the thermal tune procedure on a PicoForce MultiMode AFM (Veeco). Some experiments (as indicated in the text) were performed using oxide-sharpened Si3N4 tips attached to a triangular cantilever with a spring constant of 0.06 N/m (NanoProbe, Santa Barbara, CA). A fluid cell without O-ring was filled with 50 mM phosphate buffer, pH 6.4, and the sample was scanned in tapping mode. In this mode, individual contact events between the tip and sample are very short, on the order of tenths of milliseconds at typical oscillation frequencies (∼8.5–9.0 kHz). We further decreased the contact time and interaction forces by adjusting the “amplitude setpoint” to the highest possible value at which the image was still stable. Next, we zoomed in on the spot of interest (increasing the magnification in two to three consecutive steps to reduce mechanical drift and position the tip precisely over a fibril), switched to force mode, and collected force-distance curves on the selected spot. The contact force used during these measurements was limited to 0.3–0.4 nN to minimize any nonspecific adsorption of protein to the tip.
Force-curves analysis
The force curves were analyzed according to the worm-like chain (WLC) model for elasticity of a polymer chain, using the equation: F(x) = (kT/p)[0.25(1 − x/Lc)−2 −0.25 + x/Lc] (21), where F(x) is the force at position x, p is the persistent length, kT is the thermal energy, and Lc is the contour length. The loading rate dependency was fit to the equation: F = (kT/Δx)ln [r/((kT/Δx)koff)] (19), where F is the measured force, r is the loading rate, Δx is the distance between the ground and transition state, koff is the natural off-rate of the system, and kT is the thermal energy.
RESULTS
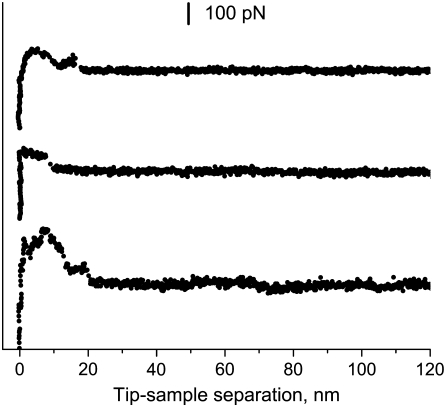
Our initial attempts to perform single-molecule measurements with huPrP90-231 amyloid deposited on a mica surface relied on nonspecific adsorption of protein to the AFM tip. However, the resulting force spectra showed great variability between individual measurements (see Fig. 1). This variability, which most likely results from random and/or multiple attachment sites of the protein to the tip, precluded any meaningful interpretation of the force-distance curves. To overcome this problem, we functionalized the protein by replacing Ser at position 103 with Cys, and performed force measurements using a gold-coated AFM tip. The thiol group of Cys is known to form a covalent bond with gold, facilitating single-molecule AFM experiments with systems requiring a well-defined attachment site (22,23). Placement of the engineered Cys residue near the N-terminus of huPrP90-231 was dictated by recent H/D exchange and site-directed spin labeling data indicating that the entire region of the protein N-terminal to residue ∼160 is outside the amyloid core (17,18). Thus, residue 103 should be easily accessible to the Au-covered AFM tip.
FIGURE 1.
Examples of force curves obtained in experiments relying on nonspecific adsorption of wild-type huPrP90-231 (without engineered Cys at position 103) to the AFM tip.
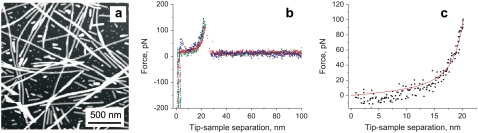
For each sample preparation, we first imaged the surface to visualize fibrils (Fig. 2 a). To minimize potential collection of protein molecules on the tip surface, we performed AFM imaging in a tapping mode using the parameters that minimize the contact time and interaction force. Next, we zoomed in on the selected spot and pushed the tip onto the fibril with a contact force of 0.3–0.4 nN for ∼0.5 s, and recorded force spectra during retraction of the tip as described in Materials and Methods. A large proportion (∼35%) of force-distance curves recorded for the Cys-mutant probed with the gold-coated tip had characteristics typical of elastic stretching (Fig. 2 b). This is in sharp contrast to the irregular and poorly reproducible curves observed for the wild-type protein or the S103C variant probed with Si3N4 tip, where the elastic stretching events were very rare, corresponding to <5% of the measurements. The elastic stretching-type curves for the Cys-mutant probed with a gold-coated tip are reminiscent of force extension spectra observed for unfolding of individual units of modular proteins such as titin (24). These curves could be well described by the WLC model for entropic elasticity of a polymer chain. A representative fit of experimental data according to this model is shown in Fig. 2 c.
FIGURE 2.
AFM image and force-extension curves for S103C huPrP90-231 amyloid fibrils. (a) AFM height image obtained in tapping mode. Z-scale is 10 nm. (b) Superposition of three representative force-extension curves showing elastic stretching behavior (at a pulling rate of 9.4 nN/s). (c) A fit of one of these curves according to the WLC model with a persistence length of 0.4 nm. The contour length derived from this particular fit is 24.2 nm.
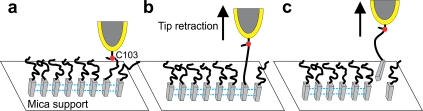
What is the nature of molecular interactions that give rise to the force curves observed during pulling of PrP90-231 from amyloid fibrils? The elastic character of these curves implies that the segment of the polypeptide chain between the covalent attachment site to the tip (residue 103) and the β-sheet core of the amyloid must be able to undergo purely entropic stretching, strongly suggesting that this region of the protein is largely disordered (see Discussion). Further retraction of the tip resulted in a “rupture” of the system that occurred as an “all-or none” event (Fig. 2 b). Since the force of this rupture is one order of magnitude lower than the strength of a covalent Au-Cys bond [∼1.4 nN (25)], and given the control experiments described below, we conclude that this latter event most likely represents mechanical extraction of the prion protein from the core of the amyloid fibril (Fig. 3). We also considered the possibility that, rather than extracting individual monomers from the fibril, our force measurements correspond to lifting of the whole fibril from the mica surface. However, simple calculations of attractive van der Waals forces can be made using the formula for the force of interaction between a cylinder and flat surface (26). Using the Hamaker constant that is typical for protein-surface interactions in electrolyte solution (4 × 10−21 J (27)), and a separation distance of 0.4 nm (28), we estimate that fibrils attach to mica with a van der Waals force of ∼7 pN per nanometer (assuming a fibril radius of 5 nm). Therefore, even for fibrils as short as 500 nm, the attractive force is estimated to be ∼3.5 nN (i.e., much higher than those registered in our experiments), making the possibility that our measurements correspond to lifting of the whole fibril highly unlikely. Consistent with this estimate, strong nonspecific attachment of amyloid fibrils to a mica surface was found in previous studies with Aβ peptide, as indicated by a similar force response in experiments using fibrils attached nonspecifically or chemically cross-linked to the surface (29).
FIGURE 3.
Schematic representation of the experiment, with the core of amyloid fibril depicted in gray, and blue lines representing intermolecular hydrogen bonds between the monomers. (a) The gold-covered AFM tip attaches covalently to C-103 on one of the monomers of the mica-adsorbed amyloid. (b) Retraction of the tip results in elastic stretching of the monomer between residue 103 and the beginning of the amyloid core region (black line). (c) When the pulling force exceeds a critical value, the monomer is extracted from the amyloid as an “all-or-none” event. This cartoon is for illustrative purposes only; it does not imply any specific structure of the PrP amyloid core, or any knowledge of pulling geometry in these experiments. The relative dimensions of the tip and protein molecules are not to scale.
One source of potential artifacts in interpretation of our experimental data is the relatively large radius of curvature inherent to the gold-coated tip, which raises the possibility that multiple PrP monomers covalently attach during individual pulling events. To address this issue, force measurements were repeated using fibrils in which the Cys-containing mutant was comixed (at a 1:3 ratio) with the wild-type protein. Such a dilution of the Cys-variant would be expected to greatly reduce the probability for the attachment of multiple molecules. For these comixed fibrils, the frequency of specific attachment was reduced to ∼7–10%; however, the force histogram was nearly identical to that in Fig. 4 a (data not shown for brevity), strongly indicating that the elastic stretching curves observed represent a single-molecule event. Indeed, the frequency of specific events in force measurements on the order of ∼10% is usually considered as indicative of a single-molecule level manipulation (30).
FIGURE 4.
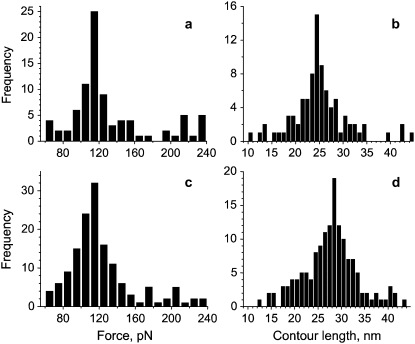
Histograms of forces and contour lengths (at a tip retraction rate of 9.4 nN/s) for C-103 huPrP90-231 (a and b) and C-90 huPrP90-231 (c and d). (a and c) Force-frequency histograms. (b and d) Contour length-frequency histograms. Only experiments that show an elastic stretching behavior are included in this analysis.
A potential complication of the measurements presented here is that PrP monomers extracted in earlier cycles remained attached to the gold-coated tip throughout the experiment. In principle, this could influence force measurements since many pulling events are monitored with a single tip. However, our analysis revealed that the registered forces do not show variation with time (i.e., there is no difference between elastic curves observed in the first stretch cycle (with a new tip) and subsequent ones). To further probe this issue, we examined the interaction between Au-covered tips and mica covered with S103C PrP monomers, as well as the interaction between the free mica surface and the tip preincubated in the protein monomer solution. In both cases, we found that only a miniscule fraction (<2%) of the force curves showed any elastic behavior. Altogether, these control experiments strongly indicate that the force-distance curves shown in Fig. 2 indeed represent single-molecule events in which elastic stretching precedes removal of PrP monomers from the amyloid.
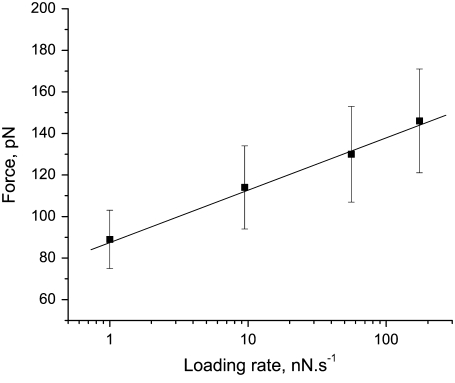
The histogram of forces recorded in over 100 measurements resulting in elastic curves, such as those exemplified in Fig. 2, shows a well-defined peak at 115 ± 5 pN (Fig. 4 a). As discussed above, we attribute this force to mechanical extraction of an individual prion protein monomer from the amyloid fibril. The forces required to trigger mechanical unfolding of individual domains of modular monomeric proteins are known to depend on the rate at which the force is applied (31–33). A similar dependency on the loading rate was found in this study with the PrP90-231 amyloid (Fig. 5). Analysis of the force versus loading rate plot, according to the formalism of Evans and Ritchie (34) allowed us to estimate the rate of the reaction corresponding to PrP monomer dissociation from the amyloid at zero force (koff) as 2.4 × 10−2 s−1, and the position of the transition state (distance in configurational space from the native geometry to the transition state of the monomer extraction event) as Δx = 0.38 nm. The latter value is remarkably similar to those reported for the transition state in mechanical unfolding of modular β-sheet proteins (31–33,35).
FIGURE 5.
Pulling-speed dependence of forces measured for extraction of individual monomers from huPrP90-231 amyloid fibrils.
The contour-length histogram derived from data analysis for C-103 huPrP90-231 according to the WLC model shows a well-defined maximum around 24–25 nm (Fig. 4 b), implying that ∼63–66 residues can be stretched entropically before the protein is extracted from the amyloid core of PrP90-231 fibrils (assuming 0.38 nm per residue in a fully extended conformation). The ability to stretch entropically strongly suggests that the region encompassing these residues lacks any stable, systematically H-bonded structure. Since the protein is covalently attached to the tip at residue 103, this would place the beginning of the H-bonded core of the amyloid at residue ∼166–169. To further test this model, we repeated the force measurements using a huPrP90-231 variant in which the Cys residue for specific attachment to the gold-coated tip was moved N-terminally to position 90. The histogram of forces recorded for C-90 huPrP90-231 amyloid fibrils has a maximum at precisely the same value as observed for protein with Cys at position 103 (Fig. 4, a and c). However, the contour length is significantly longer, with a maximum in the histogram around 28–29 nm (Fig. 4 d). This contour length (as compared to 24–25 nm in the case of C-103 huPrP90-231) indicates that, when the protein is attached to the tip at position 90, ∼74–76 residues can be stretched entropically, placing the beginning of the H-bonded core region of the amyloid at residue ∼164–166. A very good correlation between the number of residues that can be stretched entropically and the tip attachment site (and thus a very consistent estimate for the beginning of the amyloid core) provides an important internal control, validating our interpretation of force spectroscopy data.
DISCUSSION
In this study, we used AFM to probe the nanomechanical properties of amyloid fibrils formed by recombinant human prion protein 90-231. Numerous control experiments strongly indicate that the force-distance curves obtained in our experiments represent single-molecule events in which elastic stretching precedes removal of the PrP monomer from the amyloid (Fig. 3). This force of mechanical extraction of the monomer from the amyloid fibril (115 ± 5 pN at a pulling rate of 9.4 nN/s) is within the range of values reported for mechanical unfolding (at comparable pulling rates) of individual units of modular β-sheet proteins, such as different domains of titin (80-300 pN) (31–33,35) or tenascin (∼137 pN) (36), and is significantly higher than those typically observed for the unfolding of α-helical proteins (33,37,38) (e.g., 15–25 pN for spectrin (37) or 64 pN for lysozyme (38)). Recent studies indicate that the mechanical resistance of β-sheet proteins is strongly dependent on pulling geometry (23,39–42) (the direction of the applied force), with longitudinal “shearing” of β-strands generally requiring much larger forces than orthogonal “peeling” (39,40). For instance, in the case of an all-β protein, E2lip3, the force needed for mechanical unfolding was as high as ∼180 pN when the protein was subjected to shearlike extension, dropping below the detection limit of AFM (∼15 pN) when peeling force was applied (39). A similar dependence on extension directionality was reported for proteins such as ubiquitin (41) and GFP (42). The experimental design of this study did not allow us to control the pulling geometry. However, in the context of data reported for mechanical unfolding of modular β-sheet proteins, the magnitude of forces measured here is surprisingly low, especially given the size of the H-bonded core of PrP amyloid (17,18). This suggests that the extraction of the monomer from amyloid fibrils in our experiments likely occurs by a mechanism other than a simple shearlike mechanism.
Perhaps the most important finding of this study is that the N-terminal part of huPrP90-231 in amyloid fibrils up to residue ∼164–169 is able to undergo purely entropic stretching, indicating a largely disordered structure in this region of the protein. This has important implications for the ongoing debate regarding the structure of PrP amyloid. The models frequently discussed in the literature postulate that the β-core region of the amyloid maps to the N-terminal part of PrP90-231 (residues ∼90–175 and 116–164 for the β-helix (43) and spiral model (44), respectively), with at least two C-terminal α-helices remaining largely preserved. The experimental data presented here are clearly at odds with both of these largely in silico models, since such an N-terminal location of the core region would not allow for an entropic stretching as observed in the force-extension curves. On the other hand, single-molecule AFM measurements correlate remarkably well with recent structural studies that used hydrogen/deuterium exchange and site-directed spin labeling (17,18). Both of these studies indicate that amyloid fibril formation involves major refolding of the entire α-helical region of PrP90-231, mapping the β-sheet core of the amyloid to the C-terminal part of the protein starting at residue ∼161–169.
Although recombinant PrP amyloid fibrils similar to those used in this study have been reported to induce a neurological disease in transgenic mice overexpressing N-terminally truncated PrP (9), the infectivity titer was exceedingly low, suggesting that perhaps only a tiny proportion of these synthetic aggregates represent the infectious entity. Furthermore, TSE prions are known to form different strains, and this strain variability is believed to be encoded by distinct conformations of PrPSc (2–6). Clearly, the recombinant fibrils used in this study represent only a small part of the spectrum of the misfolded PrP conformers—one that does not fully recapitulate structural properties of the highly infectious TSE agent. Nevertheless, studies with these synthetic fibrils are fundamentally important for guiding the design and interpretation of future, experimentally far more challenging studies with brain-derived PrPSc.
Previous attempts to use force spectroscopy in amyloid research were limited to probing the global properties of these biologically important but poorly understood structures (29,45,46). In particular, studies with Aβ amyloid fibrils (29,45) reported complex force curves featuring a nonlinear elastic response (attributed by the authors to “elasticity of the β-sheet that has been liberated from its lateral confinement within the fibril”) and long force plateaus (attributed to the “unzipping of β-sheets from the fibril”). In this study, with PrP amyloid, we did not observe any such complex force curves. The different response types observed in force measurements with Aβ fibrils is not surprising, given that the disordered part of the Aβ peptide outside the H-bonded amyloid core (i.e., available for entropic stretching) is very short (47). Furthermore, apart from using an entirely different protein, the other major difference between the previous studies with Aβ peptide (29,45) and the present study with PrP is methodological. Whereas the experiments with Aβ fibrils reported on more-global events that occur upon nonspecific attachment of the protein to the tip (presumably at multiple sites), the work presented here exploited specific attachment to an engineered recognition site, providing direct information about the energetics and nanomechanical properties of amyloid fibrils at a single-molecule level. The approach employed herein should be applicable for probing a wide range of fundamental questions in amyloid research, including those related to the mechanism of fibril assembly and stability, as well as the structural and/or thermodynamic basis of prion and amyloid strain variability (2,48).
Acknowledgments
This work was supported by National Institutes of Health grants NS 44158 and NS 38604 (to W.K.S.).
Editor: Jane Clarke.
References
- 1.Prusiner, S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science. 1982:136–144. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA. 95:13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughey, B., and G. S. Baron. 2006. Prions and their partners in crime. Nature. 443:803–810. [DOI] [PubMed] [Google Scholar]
- 4.Collinge, J. 2001. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24:519–550. [DOI] [PubMed] [Google Scholar]
- 5.Agguzi, A., and M. Polymenidou. 2004. Mammalian prion biology: one century of evolving concepts. Cell. 116:313–327. [DOI] [PubMed] [Google Scholar]
- 6.Weissmann, C. 2004. The state of the prion. Nat. Rev. Microbiol. 2:861–871. [DOI] [PubMed] [Google Scholar]
- 7.Dobson, C. M. 2003. Protein folding and misfolding. Nature. 426:884–889. [DOI] [PubMed] [Google Scholar]
- 8.Caughey, B., and P. T. Lansbury. 2003. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26:267–298. [DOI] [PubMed] [Google Scholar]
- 9.Legname, G., I. V. Baskakov, H. O. Nguyen, D. Riesner, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2004. Synthetic mammalian prions. Science. 305:673–676. [DOI] [PubMed] [Google Scholar]
- 10.Castilla, J., P. Saá, C. Hetz, and C. Soto. 2005. In vitro generation of infectious scrapie prions. Cell. 121:195–206. [DOI] [PubMed] [Google Scholar]
- 11.Deleault, N. R., B. T. Harris, J. R. Rees, and S. Supattapone. 2007. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. USA. 104:9741–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riek, R., S. Hornemann, G. Wider, M. Billeter, R. Glockshuber, and K. Wüthrich. 1996. NMR structure of the mouse prion protein domain PrP(121–321). Nature. 382:180–182. [DOI] [PubMed] [Google Scholar]
- 13.Zahn, R., A. Liu, T. Lührs, R. Riek, C. von Schroetter, F. López García, M. Billeter, L. Calzolai, G. Wider, and K. Wüthrich. 2000. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. USA. 97:145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donne, D. G., J. H. Viles, D. Groth, I. Mehlhorn, T. L. James, F. E. Cohen, S. B. Prusiner, P. E. Wright, and H. J. Dyson. 1997. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl. Acad. Sci. USA. 94:13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caughey, B. W., A. Dong, K. S. Bhat, D. Ernst, S. F. Hayes, and W. S. Caughey. 1991. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 30:7672–7680. [DOI] [PubMed] [Google Scholar]
- 16.Pan, K. M., M. Baldwin, J. Nguyen, M. Gasset, A. Serban, D. Groth, I. Mehlhorn, Z. Huang, R. J. Fletterick, F. E. Cohen, et al. 1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA. 90:10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, X., P. L. Wintrode, and W. K. Surewicz. 2007. β-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc. Natl. Acad. Sci. USA. 104:1510–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobb, N. J., F. D. Sönnichsen, H. McHaourab, and W. K. Surewicz. 2007. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc. Natl. Acad. Sci. USA. 104:18946–18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinterdorfer, P., and Y. Dufrêne. 2006. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 3:347–355. [DOI] [PubMed] [Google Scholar]
- 20.Apetri, A. C., D. L. Vanik, and W. K. Surewicz. 2005. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90–231. Biochemistry. 44:15880–15888. [DOI] [PubMed] [Google Scholar]
- 21.Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 22.Oesterhelt, F., D. Oesterhelt, M. Pfeiffer, A. Engel, H. E. Gaub, and D. J. Müller. 2000. Unfolding pathways of individual bacteriorhodopsins. Science. 288:143–146. [DOI] [PubMed] [Google Scholar]
- 23.Dietz, H., and M. Rief. 2006. Protein structure by mechanical triangulation. Proc. Natl. Acad. Sci. USA. 103:1244–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 25.Grandbois, M., M. Beyer, M. Rief, H. Clausen-Schaumann, and H. E. Gaub. 1999. How strong is a covalent bond? Science. 283:1727–1730. [DOI] [PubMed] [Google Scholar]
- 26.Bhushan, B. 1999. Handbook of Micro/Nanotribology. CRC Press, Boca Raton, FL.
- 27.Roth, C. M., B. L. Neal, and A. M. Lenhoff. 1996. Van der Waals interactions involving proteins. Biophys. J. 70:977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freitas, R. A. 1999. Nanomedicine, Vol. 1. Basic Capabilities. Landes Bioscience, Austin, TX.
- 29.Kellermayer, M. S. Z., L. Grama, A. Karsai, A. Nagy, A. Kahn, Z. L. Datki, and B. Penke. 2005. Reversible mechanical unzipping of amyloid β-fibrils. J. Biol. Chem. 280:8464–8470. [DOI] [PubMed] [Google Scholar]
- 30.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53. [DOI] [PubMed] [Google Scholar]
- 31.Li, H., M. Carrion-Vazquez, A. F. Oberhauser, P. E. Marszalek, and J. M. Fernandez. 2000. Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 7:1117–1120. [DOI] [PubMed] [Google Scholar]
- 32.Oberhauser, A. F., C. Badilla-Fernandez, M. Carrion-Vazquez, and J. M. Fernandez. 2002. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319:433–447. [DOI] [PubMed] [Google Scholar]
- 33.Brockwell, D. J., G. S. Beddard, E. Paci, D. K. West, P. D. Olmsted, D. A. Smith, and S. E. Radford. 2005. Mechanically unfolding the small, topologically simple protein L. Biophys. J. 89:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rief, M., M. Gautel, A. Schemmel, and H. E. Gaub. 1998. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys. J. 75:3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 393:181–185. [DOI] [PubMed] [Google Scholar]
- 37.Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. 1999. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561. [DOI] [PubMed] [Google Scholar]
- 38.Yang, G., C. Cecconi, W. A. Baase, I. R. Vetter, W. A. Breyer, J. A. Haack, B. W. Matthews, F. W. Dahlquist, and C. Bustamante. 2000. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc. Natl. Acad. Sci. USA. 97:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brockwell, D. J., E. Paci, R. C. Zinober, G. S. Beddard, P. D. Olmsted, D. A. Smith, R. N. Perham, and S. E. Radford. 2003. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat. Struct. Biol. 10:731–737. [DOI] [PubMed] [Google Scholar]
- 40.Rohs, R., C. Etchebest, and R. Lavery. 1999. Unraveling proteins: a molecular mechanics study. Biophys. J. 76:2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrion-Vazquez, M., H. Li, H. Lu, P. E. Marszalek, A. F. Oberhauser, and J. M. Fernandez. 2003. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10:738–743. [DOI] [PubMed] [Google Scholar]
- 42.Dietz, H., F. Berkemeier, M. Bertz, and M. Rief. 2006. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 103:12724–12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Govaerts, C., H. Wille, S. B. Prusiner, and F. E. Cohen. 2004. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci. USA. 101:8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeMarco, M. L., and V. Daggett. 2004. From conversion to aggregation: protofibril formation of the prion protein. Proc. Natl. Acad. Sci. USA. 101:2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karsai, A., Z. Martonfalvi, A. Nagy, L. Grama, B. Penke, and M. S. Z. Kellermayer. 2006. Mechanical manipulation of Alzheimer's amyloid β1–42 fibrils. J. Struct. Biol. 155:316–326. [DOI] [PubMed] [Google Scholar]
- 46.Smith, J. F., T. P. Knowles, C. M. Dobson, C. E. Macphee, and M. E. Welland. 2006. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA. 103:15806–15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petkova, A. T., Y. Ishii, J. J. Balbach, O. N. Antzutkin, R. D. Leapman, F. Delaglio, and R. Tycko. 2002. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 99:16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones, E. M., and W. K. Surewicz. 2005. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 121:63–72. [DOI] [PubMed] [Google Scholar]