Abstract
Impaired transport of mitochondria, in dendrites and axons of neurons, and bioenergetic deficit are increasingly recognized to be of pathological importance in neurodegenerative diseases. To study the relationship between transport and bioenergetics, we have developed what to our knowledge is a novel technique to quantify organelle velocity in cultured cells. The aim was to combine measurement of motion and bioenergetic parameters while minimizing photodynamic oxidative artifacts evoked by fluorescence excitation. Velocity determination from sequential fluorescence images is not trivial, and here we describe an application of “optical flow”, the flow of gray values in grayscale images, to this problem. Based on the principles of photon shot noise occurring in low light level fluorescence microscopy, we describe and validate here an optical flow-based, robust method to measure velocity vectors for organelles expressing fluorescent proteins. This method features instantaneous velocity determination from a pair of images by detecting motion of edges, with no assumptions about the separation or shapes of the objects in the image. Optical flow was used in combination with single mitochondrion assay of mitochondrial thiol redox status by mitochondrially targeted redox-sensitive green fluorescent protein and measurement of mitochondrial membrane potential by tetramethylrhodamine methyl ester. Mitochondrial populations of resting cultured hippocampal neurons were analyzed. It was found that mitochondria with more oxidized thiol redox status have lower membrane potentials and are smaller in size. These mitochondria are more motile than the average; however, mitochondrial motility is only slightly dependent on the observed bioenergetic parameters and is correlated the best to the size of the mitochondria.
INTRODUCTION
The mechanism by which neurons distribute their mitochondria to axonal and dendritic sites of ATP demand is incompletely understood (see (1), or (2,3,4)). Mitochondrial location and therefore motion is also critical for Ca2+ buffering (5,6), the distribution of newly synthesized mitochondrial DNA and proteins by fusion and fission (7,8), and in the elimination of old or damaged mitochondria by autophagy (3,9,10). The pathological relevance of disturbed mitochondrial transport is illustrated by axonal clogging in Alzheimer's disease and Huntington's disease (11–13) and by the general failure of mitochondrial transport during pathological glutamate exposure of cortical neurons (14,15).
There is an increasing body of evidence that individual mitochondria within the hundreds or thousands populating a single cell show heterogeneous responses to pathologic stimuli (16–19). The aim of this study is to provide a technique for the instantaneous determination of velocities of single mitochondria in combination with single mitochondrial measurements of key physiological/bioenergetical parameters such as membrane potential and redox status.
We introduce here a technique for the unsupervised and robust determination of the velocities and directions of organelle transport, providing a snapshot of motion at the time point of the measurement. The technique is designed to calculate velocity vectors from as little as two image acquisitions, minimizing phototoxicity. Since these acquisitions are closely spaced in time, they can be correlated with functional assays, such as determination of mitochondrial membrane potential (ΔΨm) and thiol redox potential. By utilizing stage motors and multiple imaging positions, the technique allows velocities to be determined for multiple view fields in parallel.
The approach involves the implementation and tuning of an image processing technique called optical flow (20), which to our knowledge has not previously been used in fluorescence microscopic data analysis. Optical flow (OptFlow) is defined as the “flow” of gray values in the image plane, has the dimension of velocity (see page 406 in Jahne (20)) and reflects the real physical motion of the imaged objects parallel with the image plane when these objects do not change their brightness and have spatial intensity gradients, edges, or corners. These criteria are fulfilled when imaging cells expressing mitochondrially targeted fluorescent proteins, which do not change their fluorescent properties within the time frame of the image acquisition and where cells or their processes are relatively thin. Importantly, we have refined the technique to handle low light level, noisy fluorescence images, establishing robustness over a wide range of signal/noise ratios. The algorithm calculates a velocity vector field from a grayscale image sequence. In practice, the simplest output of the OptFlow calculation is an image representing the absolute velocities of the objects by grayscale values, enabling simple region of interest averaging to determine the mean mitochondrial velocity in a given cell or subcellular region.
We show here that the OptFlow technique accurately measures velocities with subpixel resolution. It can resolve anterograde from retrograde motion and distinguish between directional transport and mitochondrial “wiggling”. The velocity measurements are combined with single mitochondrial determination of ΔΨm, thiol redox potential, and mitochondrial size. We analyze populations of mitochondria in resting cultured hippocampal neurons, showing that mitochondria with more oxidized thiol redox status have lower membrane potentials and are smaller in size, and remarkably, these mitochondria are more motile than the average. However, mitochondrial motility is only slightly dependent on the observed bioenergetic parameters, and correlates the best to the size of mitochondria. Directionality of mitochondrial motion does not correlate with the assayed parameters in dendrites of hippocampal neurons.
METHODS AND MODELING
Optical flow to measure motion of fluorescence microscopic objects
Velocities and directions of movements of mitochondria were determined by computation of optical flow from image sequences. Optical flow is defined as the “flow” of gray values in the image plane and has the dimension of velocity (see page 406 in Jahne (20)). The OptFlow reflects the real physical motion of the imaged objects between frames of an image sequence when the following criteria are met: the imaged objects move in the plane of the image, do not change their intensity, and have spatial gray value gradients, i.e., edges or corners.
The OptFlow is defined by the brightness change constraint equation (Eq. 1 and Fig. 1), which assumes that the total intensity of the image does not change during the image sequence, i.e., gray values (g) in the image are conserved. Equation 1 relates the slope of intensity versus distance for a given edge (∂g/∂x) moving with a velocity f (≡OptFlow) to the rate at which the intensity changes with time (−∂g/∂t) as the edge passes over a fixed point in the image (Fig. 1). Equation 1 is shown in its one-dimensional form for simplicity:
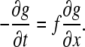 |
(1) |
FIGURE 1.
Illustration of the brightness change constraint equation (Eq. 1). The light gray contour is the fluorescence intensity profile (g(x, t)) of the edge of an object moving in the x direction. The dark gray contour is the same object at a time Δt later, when the object has moved to the right by a distance Δx. The motion is observed at a given pixel (dashed line) where the intensity changes (decreases) by Δg as the object moves (to the right). The brightness change constraint equation can be derived from the definitions of the slope and the velocity (f).
The discrete least-squares method of OptFlow determination for two-dimensional motion
For the general case where the object is moving simultaneously in x and y dimensions, f cannot be unambiguously determined from a single equation. To solve this in two dimensions, the assumption is made that the OptFlow is constant in a small square window around the pixel of interest. This “neighborhood” consisting of N pixels is called the aperture. Equation 1 is written for each pixel of the aperture, yielding a linear equation system. The solution of this equation system using the method of least squares provides f (Eqs. 2 and 3; for details see pages 414–415 in Jahne (20)):
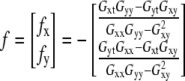 |
(2) |
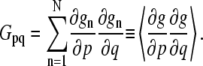 |
(3) |
In Eq. 3, gn is the n th pixel of the aperture and p,q are x, y, or t. The operator 〈〉 denotes the sum within the aperture neighborhood.
Two important constraints are required for the validity of Eq. 2 (see page 415 in Jahne (20)); First, (∂g/∂x) and (∂g/∂y) cannot be both zero in all points of the aperture neighborhood, i.e., there has to be an object with edges in the aperture (constraint No. 1). Second, the gradients must not point into the same direction (constraint No. 2). These constraints will be considered in the section dealing with the handling of images with noise.
Image processing steps to calculate OptFlow are detailed in Appendix A. Briefly, separate copies of the image sequence were differentiated in x, y, and t directions (∂g/∂x, ∂g/∂y, and ∂g/∂t, respectively) using kernel convolution filtering. Gpq images (Eq. 3) were then calculated as the product of the differentiated images pixel by pixel, followed by the application of the operator 〈〉, which was also implemented as a kernel convolution (“the aperture kernel”). Finally, f was calculated by Eq. 2 performing the calculation with the Gpq images, pixel by pixel. Separate fx and fy images were generated, where the gray values reflected the motion in the respective dimensions. This is the “raw OptFlow” in the scheme shown in Fig. 7. Absolute OptFlow velocity images could then be calculated as 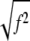 for each pixel.
for each pixel.
FIGURE 7.
Scheme of the OptFlow calculation. Each box corresponds to an image where pixels can take values of real numbers. The input is an image sequence g(x,y,t) (top middle).  ∂g/∂x, and ∂g/∂y images are calculated from the image in the middle of the image sequence (shown for wt = 3). ∂g/∂t and the minimum intensity projected (MinIP) mask are calculated from the whole image sequence. Brackets indicate inputs and outputs of each computation step (arrows) given above by equations or explained in text. Images denoted by boxes here are processed pixel by pixel in algebraic formulas (Eqs. 2, 3, and 6). In inequalities (Eqs. 5 and 7–9), masks are generated that are 0 (rejecting) where the inequality is true and 1 (transmitting) where the inequality is false. The output of the calculation is a pair of images fx and fy corresponding to the velocities over each pixel in x and in y directions, respectively. Absolute and radial velocities (scalar values) are calculated from the fx and fy image pair. When wt = 2avg is used, the whole scheme is computed twice by taking the first or the second image as middle image and finally averaging the results.
∂g/∂x, and ∂g/∂y images are calculated from the image in the middle of the image sequence (shown for wt = 3). ∂g/∂t and the minimum intensity projected (MinIP) mask are calculated from the whole image sequence. Brackets indicate inputs and outputs of each computation step (arrows) given above by equations or explained in text. Images denoted by boxes here are processed pixel by pixel in algebraic formulas (Eqs. 2, 3, and 6). In inequalities (Eqs. 5 and 7–9), masks are generated that are 0 (rejecting) where the inequality is true and 1 (transmitting) where the inequality is false. The output of the calculation is a pair of images fx and fy corresponding to the velocities over each pixel in x and in y directions, respectively. Absolute and radial velocities (scalar values) are calculated from the fx and fy image pair. When wt = 2avg is used, the whole scheme is computed twice by taking the first or the second image as middle image and finally averaging the results.
Validation of the discrete least-squares method of OptFlow determination using model “mitochondria”
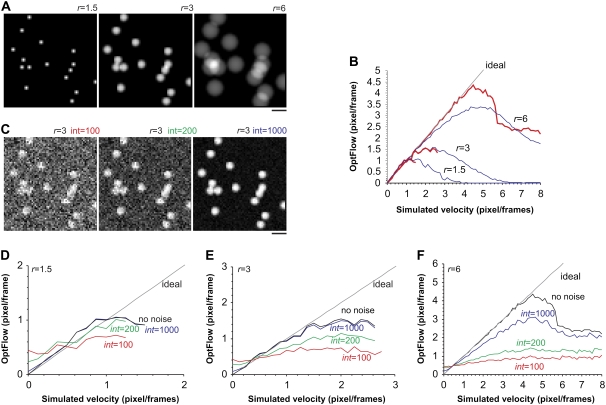
Computer-generated image sequences of moving model “mitochondria” were drawn as projections of fluorescent spheres and capsules (see Appendix B). Fig. 2 A shows spheres with radii (r) of 1.5, 3, and 6 pixels. These values correspond to observed mitochondrial radii visualized below in Fig. 8 A. Image sequences were generated in which the spheres were moving with a constant speed varying between 0 and 8 pixels/frame. One image sequence was generated for each velocity at 1:8 pixel/frame increments. Subpixel positioning of the models in the image was enabled by using bilinear interpolation when drawing shapes (see also Fig. 6 E). The OptFlow calculation was performed on each of these image sequences, using symmetric differentiating kernels with a width of 3 (see Appendix A).
FIGURE 2.
Effect of photon shot noise on the OptFlow. To test the OptFlow algorithm, image sequences of moving model mitochondria were computer generated and the measured velocities were compared to the simulated ones. To simulate low light level imaging, noisy models were used. (A) Model mitochondria were images of solid fluorescent spheres with radii (r) from 1.5 to 6 pixels over a zero background. Scale bar, 10 pixels. (B) OptFlow as a function of simulated velocity: effects of object radius and masking. Blue traces: the middle frame of the image sequence was used as a mask; red traces: the minimum intensity projection of the image sequence was used as a mask. OptFlow was determined by using simple (width = 3) kernels. Note that the minimum intensity projection masking (red trace) avoids underestimation of higher velocities. (C) Mitochondria of r = 3 are modeled with simulated photon shot noise at intensities (int) of 100, 200, and 1000 over a background of 1000 photo electron/pixel. Scale bar, 10 pixels. (D–F) OptFlow as a function of modeled velocity for different sized mitochondria (r = 1.5–6) as a function of noise (int = 100–1000). OptFlow was determined by using simple kernels (width = 3; see Appendix A) in model images containing photon shot noise. Note that the nonoptimized raw OptFlow algorithm is not robust, overestimates low velocities, and underestimates high velocities in noisy images.
FIGURE 8.
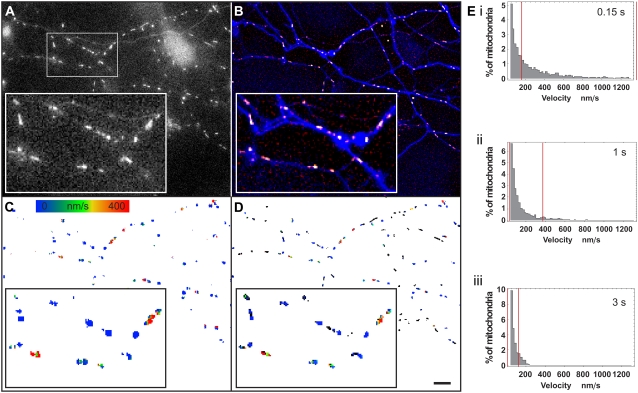
Determination of optimal frame rate to follow the motion of mitochondria in dendrites of hippocampal neurons. To match the dynamic range of the OptFlow calculation and the velocities of mitochondria in dendrites of hippocampal neurons, OptFlow calculation was performed at various image acquisition intervals. (A) Typical mitochondria at 0.32 μm/pixel resolution had gradients with 1.5–3.5 pixels radii. (B) Raw fluorescent image of a hippocampal neuron expressing mito-roGFP1. (C) Minimum intensity projected, high pass filtered, and segmented image of the same neuron. Each segment is shown in a distinct color. (D) Absolute OptFlow image calculated using the parameters described in Fig. 6, shown with no intensity image masking. White pixels were masked by the constraint criteria. The time lapse interval was 1 s/frame. (E) Masked absolute OptFlow image overlaid on the shapes of the segments in black. Black pixels show those parts of mitochondria that are masked by the constraint criteria, whereas colored pixels show valid OptFlow determinations (see scale in D). (F) Histograms of mean velocities of mitochondria. The histograms are truncated at velocities <50 nm/s for improved visualization of higher velocities, whereas percents refer to the total mitochondrial population. The acquisition interval (0.15 s–3 s) used for recording images for OptFlow calculation is indicated in the diagrams. Note that there are no distinguished mitochondrial velocities, i.e., the velocity distribution is continuous. Vertical red lines indicate the estimated boundaries of the sensitivity. These data are also shown in Table 1. Note that large parts of the histograms are outside of the red lines in i and iii, whereas the 1 s/frame acquisition interval ii brackets most of the velocities. Data are from ∼17,500 mitochondria pooled from 12 neurons and 20 repeats per cell. Scale bar, 10 μm.
FIGURE 6.
Test of the optimized OptFlow algorithm. Traces of OptFlow were plotted as functions of simulated velocity of noisy model mitochondria, at various radii and noise levels. The following parameters were used for the OptFlow algorithm: wt = 2avg, wxy = 3, wsm =7, wap = 5, ∂t cutoff correction (Eq. 5) with k = 1.5 (k stands for the constant in Eq. 9), noise bias correction (Eq. 6) at k = 1.5, constraint No.1 (Eq. 7) at k = 1.5 and constraint No. 2 (Eq. 8) at k = 0.045 and using minimal intensity projection masking. (A–C) Effects of varying sphere radii (r) or intensity (int) on the corrected OptFlow values. The int = 100 trace (red) is missing due to excessive masking by constraints 1 and 2 in A and B. Note that the accuracy of velocity measurement is practically independent of the noise level. (D) Statistical analysis of the smallest detectable velocity in r = 3 int = 1000 model mitochondria. Five image sequence sets were generated for velocities below 1/8 pixels/frames. The asterisk indicates the first data point, which was p < 0.05 significant with ANOVA and Tukey post hoc testing. (E) Illustration of 1/8 pixels shift to the right of a group of model mitochondria of r = 3. Green arrows indicate temporal changes (∂g/∂t) of the gray values, which results in measurable OptFlow at subpixel dislocations.
To measure the velocity of the moving objects without the diluting effect of the stationary background, images were masked and only the OptFlow over the bright objects was averaged. This was necessary, because the constraints of Eq. 2 were not handled at this point. Absolute values of velocity vectors were calculated and spatially averaged in the OptFlow images. The blue traces in Fig. 2 B plot the mean absolute OptFlow rates against the simulated velocity.
Because the OptFlow technique measures the translocation of spatial grayscale gradients in time, and these gradients are determined from images captured at discrete time points (frames), this implies that the gradients must retain significant overlap over a given point of the image during the full image sequence (as illustrated in Fig. 1, observed pixel). Therefore the OptFlow images were masked by the binarized minimum intensity t-projection image (analogous to z-projection of image stacks) of the image sequences ensuring that the OptFlow function is only determined at those pixels where the model “mitochondria” are present throughout the entire image sequence (see Appendix C).
The red traces in Fig. 2 B show that the OptFlow determination provides an accurate measure of velocity for subpixel per frame velocities, when minimum intensity projected mask is used. This way of masking prevents the underestimation of higher velocities for the model particles of r = 1.5 and r = 3 pixels, and partially at r = 6. Consequently, the OptFlow signal is lost at higher velocities due to the lack of overlap of the objects between adjacent frames.
The effect of noise on the OptFlow
When images contain negligible noise, OptFlow is not dependent on the fluorescence intensity. However, typical low light level fluorescence signals are affected by photon shot noise, and the signal/noise ratio decreases when the fluorescence intensity decreases. Since it is essential to minimize phototoxicity in live-cell imaging, it is important to establish the extent to which shot noise affects the OptFlow calculation. Photon shot noise follows a Poisson distribution with the hallmark that the variance of the noise (σ2) is proportional to the intensity gray values (g) with a proportionality constant equal to 1/AD, where the AD (analog to digital conversion factor) is the number of photo electrons per gray value unit of the camera (Eq. 4). Because AD varies by camera type, gain, and binning settings, the intensities (int) in the simulations below are given in photo electrons. This equals gray value units if AD = 1:
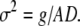 |
(4) |
Photon shot noise was modeled by simulating the Poisson distribution of photo electrons in the charge-coupled device camera. Sets of noisy model image sequences for each “mitochondrial” radius and velocity were generated at photon counts (int) of 100, 200, and 1000 photo electron/pixel over a background of 1000 photo electrons (Fig. 2 C; see also Appendix B.) In our CoolSNAP HQ camera (Photometrics, Tucson, AZ; at 2 × 2 binning and 2× gain), the AD was measured to be 2.7 (Fig. 3 in the Supplementary Material, Data S1) so these values translate to 37, 74, and 370 gray value units. For comparison, typical intensities in our experimental conditions are given in Fig. 10, D and E.
FIGURE 3.
Optimization of the OptFlow technique for robustness. Traces of OptFlow were plotted as functions of simulated velocity of noisy model mitochondria. The aim of the optimization was to decrease the low velocity overestimation and high velocity underestimation, i.e., to get the OptFlow traces close to the gray (ideal) line corresponding to the accurate velocity determination. (A and B) Effects of increased width of spatial differentiation (wxy) or smoothing (wsm) kernels for large (r = 6 pixels) or small (r = 1.5) objects. OptFlow was determined by using the indicated kernels and simple symmetric (wt = 3) temporal differentiation kernel in model mitochondria containing photon shot noise at intensity of 200 above a background of 1000 photo electrons. (C–F) OptFlow was determined by using the indicated kernels in r = 6 model mitochondria containing photon shot noise at int = 200. (C) Effects of increased temporal differentiation kernel width (wt) or frame rate (in frames per seconds; fps). The default frame rate was 1 fps. For the 7/3 fps trace, a separate set of image sequences was generated to model the higher frame rate. (D) Temporal differentiation with wt = 2 kernel (i.e., two using frames rather than three) and width of 2 with averaging (wt = 2avg; see text). (E) Effect of increased aperture kernel size (wap). For this and the following plots, wt = 2avg was used. (F) Effect of cutting off temporal derivatives that are smaller than the effect of the noise (∂t cut off; Eq. 5). (G) Effect of correction for the systematic underestimation bias caused by the noise (Eq. 6). The corrected noisy (int = 200) trace (red) overlaps with the uncorrected high intensity, therefore less noisy (int = 20,000; blue) trace.
FIGURE 10.
Dynamics of OptFlow during artificial cessation of motion. (A) Scheme of OptFlow time lapse calculation, corresponding to Fig. 7. A time lapse of absolute OptFlow images was calculated from short image sequences (two frames, 1 s/frame) recorded in 2 min intervals as summarized in Fig. 7 with the parameters described in Fig. 6. (B) Each trace indicates the mean velocity of mitochondrial movement in the proximal dendrites of a single hippocampal neuron. Cell cultures were fixed by 4% paraformaldehyde at the indicated time point. Data were pooled from three experiments. (C) 0.2 μm diameter solidly embedded fluorescent beads (TetraSpeck slide, Molecular Probes, Invitrogen) were time-lapse imaged in similar multidimensional image acquisition setting to the live cell imaging. Each trace indicates the mean velocity of the beads in a view field from three experiments and three view fields per experiment. Note that there is a residual OptFlow over stationary objects likely originating from incomplete cutoff of photon shot noise-related ∂g/∂t signal. The fluorescently labeled mitochondria (D) and fluorescent beads (E) had similar intensity profiles, so the noise level and the gradients were similar. The indicated photo electron counts were calculated based on Fig. 3 in the Supplementary Material, Data S1. F and G show the images corresponding to the intensity profile measurements (D and E, respectively). Scale bar, 5 μm.
Fig. 2, D–F, indicates that noise can introduce two errors: a nonzero estimate of OptFlow for stationary objects and an underestimation of velocities of moving objects. To minimize these errors, we used a variety of temporal and spatial differentiation, smoothing (see below), and aperture kernels of increasing width (wt, wxy, wsm, and wap, respectively). The effects of these kernels on the estimated OptFlow of the “noisy” model mitochondrion image sequences were explored. The aim was to achieve a minimal absolute OptFlow for stationary objects and a more accurate estimate of high velocities (Fig. 3). A severe test was imposed in that the low intensity (int = 200) noisy models and a very high background of 1000 photo electrons were used.
Since differentiation is noise sensitive, wider spatial differentiation kernels (Savitzky-Golay kernels for each wxy width are given in Appendix A) support better noise tolerance due to their additional smoothing effect. However, although this improved the OptFlow estimation at higher velocities (Fig. 3 A), it led to an increased overestimation at lower velocities and also resulted in a general OptFlow overestimation for small objects (Fig. 3 B).
Spatial smoothing kernel filtering (see Appendix A) perpendicular to the spatial differentiation kernel (e.g., differentiating in the x direction with a row kernel while smoothing in the y direction with a column kernel) decreased the underestimation of the velocity of the large (r = 6) objects as effectively as a wider spatial differentiation kernel (Fig. 3 A), but avoided the overestimation of the velocities of small (r = 1.5) objects (Fig. 3 B). A width of 7 pixels for both spatial (wxy = 7) and smoothing (wsm = 7) kernels was used in the following tests and measurements. These tests gave similar results for different radii, therefore only r = 6 is shown (Fig. 3, C–G).
A temporal differentiation kernel wider than wt = 3, which was used above, (Fig. 3 C, black trace) can be used when the number of acquired frames is increased. At the same frame rate, this requires a longer image sequence, or an increased frame rate at the same acquisition length. The former resulted in a more accurate OptFlow at small velocities, but an earlier saturation or loss of signal due to the zero area of the minimum intensity projected mask (Fig. 3 C, red trace). Increasing the frame rate had little effect on the low velocity overestimation (Fig. 3 C, green trace).
When imaging live cells, it is essential to minimize phototoxicity. Reducing the number of frames required for the temporal differentiation from 3 to 2 will decrease exposure significantly. However, when wt = 2 is applied to the model under the conditions of Fig. 3 C, an overestimation of OptFlow is found for all velocities (Fig. 3 D). When the motion of the model mitochondria is visualized by a pseudo color scale (Fig. 4 F), it is seen that the OptFlow gives different values close to the leading and trailing edges of the models. This is because the spatial derivatives obtained from either the first or second frame are different, thus results are inherently asymmetric. In contrast, at wt = 3, the middle image is used to calculate spatial derivatives (Appendix A). To avoid asymmetry, OptFlow was calculated as the average of the OptFlow obtained using the first and the second frame for spatial differentiation (wt = 2avg; Fig. 4 E). This reestablishes the symmetry of leading and trailing edges and reduces the overestimate at higher velocities while not affecting the zero velocity artifact in the OptFlow (Fig. 3 D).
FIGURE 4.
Absolute velocity images of noisy model mitochondria. The OptFlow algorithm optimized in Fig. 3 G was used to visualize motion by coloring each pixel according to the velocity of the underlying object. (A) Spherical model mitochondria of r = 6 and int = 1000 with simulated photon shot noise are shown in the 64 × 64 pixel image. (B–E) Pseudo color-coded absolute OptFlow images calculated from image sequences showing model mitochondria (A) moving with the indicated velocities to the right. Parameters of OptFlow calculation correspond to Fig. 3 G: wt = 2avg, wxy = 3, wsm = 7, wap = 5, corrections for ∂t cutoff (Eq. 5), and the bias caused by noise (Eq. 6) were applied. (F) is same as E but wt = 2 was used instead of wt = 2avg. Note that as a result of the simulated photon shot noise, velocities are not uniform over the pixels. However, when mean velocities are measured (e.g., Fig. 3 G) this causes little error. Scale bar, 10 pixels.
Since minimizing phototoxicity was considered to be of paramount importance, it was decided to utilize the wt = 2avg averaged differentiation kernel for subsequent refinements. First the effect of increasing the size of the aperture kernel (wap) from 3 × 3 to a 5 × 5 or 7 × 7 matrix of ones was considered (Fig. 3 E). This significantly reduced the overestimation of low velocities, although stationary objects still maintained a nonzero OptFlow. An aperture kernel of 5 × 5 was chosen to be used in the following iterations.
Estimation of photon shot noise and its effects on temporal differentiation
As shown in (Fig. 3, D–F), photon shot noise at the camera generates an artifactual OptFlow over stationary objects. This is because noise-related fluctuation of intensity over the edge of a stationary object results in OptFlow and mimics motion. To eliminate OptFlow over stationary objects, first  was calculated for each aperture neighborhood based on the proportionality given by Eq. 4 (see Eq. 9 in Appendix D). Then, given that the mean-square of differentiated noise equals the (σ2) of the noise (at unit Δt), we set up the following constraint (∂t cutoff). A zero ∂g/∂t was assigned to those pixels where Eq. 5 was satisfied; thus the mean-square of temporal derivatives (Gtt) could not be distinguished from values expected to occur due to noise (a k = 1.5 was used in Eq. 9 to obtain
was calculated for each aperture neighborhood based on the proportionality given by Eq. 4 (see Eq. 9 in Appendix D). Then, given that the mean-square of differentiated noise equals the (σ2) of the noise (at unit Δt), we set up the following constraint (∂t cutoff). A zero ∂g/∂t was assigned to those pixels where Eq. 5 was satisfied; thus the mean-square of temporal derivatives (Gtt) could not be distinguished from values expected to occur due to noise (a k = 1.5 was used in Eq. 9 to obtain  ). This procedure is indicated as “Mask ∂t cut off” in Fig. 7. The effect of “∂t cut off” in abolishing the zero velocity artifact is shown in Fig. 3 F:
). This procedure is indicated as “Mask ∂t cut off” in Fig. 7. The effect of “∂t cut off” in abolishing the zero velocity artifact is shown in Fig. 3 F:
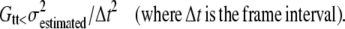 |
(5) |
Correction for the systematic underestimation of the OptFlow by noise
The use of a simple spatial differentiation kernel resulted in more underestimation of velocity for large objects (Fig. 3 A versus 3 B) than for small objects of the same intensity, because the gray value gradients are smaller for the larger objects. This is because noise also introduces a systematic error into f calculated by Eq. 2. such that f is biased toward lower values by noise. Correction for this bias was done by multiplying f by the following factor (see page 443 in Jahne (20)):
 |
(6) |
Fig. 3 G shows that this noise bias correction (with k = 1.5 in Eq. 9) makes the simple spatial differentiation kernel suitable for OptFlow determination of larger faint or noisy objects. Below (see Fig. 6) we will show that this does not affect the OptFlow determination on small objects. Fig. 4, A–E, shows raw and absolute OptFlow images for model mitochondria corresponding to the optimized parameters used in Fig. 3 G (see also bias corrected OptFlow in Fig. 7).
Violation of constraints of Eq. 2 by noise
The two constraints to the validity of the two-dimensional solution of the brightness change constraint equation (Eq. 2) could be violated by excessive photon shot noise. First, the neighborhood aperture has to have gray value gradients that are distinguishable from ones expected to rise from noise. This follows from constraint No. 1 that ∂g/∂x and ∂g/∂y must be nonzero in the neighborhood. Secondly, violation of constraint No. 2 is considered at the problem of elongated shapes below.
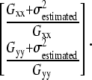
The most noise-sensitive terms in Eq. 2 are the squares of derivatives Gxx and Gyy. Products of different derivatives Gxy, Gxt, and Gyt are close to zero, because these values can be considered as cross correlations of independent noise, which is zero. Therefore, aperture neighborhoods where Gxx and Gyy values are less than  (with k = 4 in Eq. 9 see Appendix D) were discarded by masking (Eq. 7). Of note, masking with the minimum intensity t-projected image removes most of the pixels satisfying Eq. 7 in the case of the model mitochondrion images; therefore, it has no effect in the above modeling paradigms (not shown, but see below in Fig. 5, D and E). However, application of this constraint is necessary in the case of the less clean real microscopic images (e.g., due to out-of-focus blur):
(with k = 4 in Eq. 9 see Appendix D) were discarded by masking (Eq. 7). Of note, masking with the minimum intensity t-projected image removes most of the pixels satisfying Eq. 7 in the case of the model mitochondrion images; therefore, it has no effect in the above modeling paradigms (not shown, but see below in Fig. 5, D and E). However, application of this constraint is necessary in the case of the less clean real microscopic images (e.g., due to out-of-focus blur):
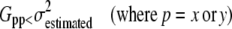 |
(7) |
FIGURE 5.
Elongated mitochondria and the constraints of the least squares approach. To accurately measure velocities of elongated objects, pixels corresponding to areas that do not fulfill the mathematical criteria of valid OptFlow calculation (constraints of Eq. 2), like the middle of rod shapes, were rejected by masking. (A) Model mitochondria of r = 3 and length = 15 pixels moving at 1.4 pixel/frame according to the arrow were generated with simulating photon shot noise at int = 1000 over background of 1000 photo electron/pixel. (B) Absolute OptFlow image masked with the binarized minimum intensity projection of the raw image sequence. Note that velocities measured over the middle of rod shapes (blue color) are inaccurately low. (C) absolute OptFlow image after enforcement of constraint No. 2 by masking pixels satisfying Eq. 8 and also using the minimum intensity projection mask as in C. Note that the invalid (blue) pixels of B were masked. (D) absolute OptFlow image after enforcement of both constraints (Eqs. 7 and 8), but not the minimum intensity projection mask. Scale bar, 10 pixels. (E) OptFlow as a function of modeled velocity: effects of the masking according to the constraints of the least squares approach. Red trace: identical results were obtained with or without application of constraint No. 1. Note that constraint No. 2 reduces underestimation.
The problem of elongated mitochondria
Mitochondria are typically elongated rather than spherical structures, and move parallel to their longest axis on trajectories provided by the microtubule cytoskeletal system. It is therefore necessary to consider the accuracy of the OptFlow calculation for such elongated objects. Constraint No. 2 of Eq. 2 is that the gradients must not point into the same direction within a neighborhood. This means in the mitochondrial context that the OptFlow cannot be determined at the middle of a rod-like mitochondrion, where the intensity will not change at a given pixel between adjacent frames as the mitochondrion moves along, but is valid at the tips. Therefore these parts of the OptFlow image have to be masked to maintain accuracy.
This second constraint requires the evaluation of gradient vectors ∇g (see definition in Eq. 10) in each neighborhood. To this end, we consider that if all gradient vectors of the neighborhood are parallel, then their vector products are zero in the absence of noise. This can be simplified as a calculation of the vector products between a mean (or sum) vector and each of the vectors of the aperture neighborhood (Eq. 8, left side numerator; see details in Appendix E). For a valid OptFlow determination in the presence of noise, this vector product sum has to be smaller than a threshold, defined above as  To be independent of gray value intensities and of the number of pixels in the aperture kernel (N), the summed vector product was normalized by
To be independent of gray value intensities and of the number of pixels in the aperture kernel (N), the summed vector product was normalized by 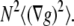 The left side of Eq. 8 was raised to a 0.75th power that was determined empirically to further lessen intensity dependence (data not shown). Finally, constraint No. 2 was enforced by masking OptFlow images where Eq. 8 was satisfied:
The left side of Eq. 8 was raised to a 0.75th power that was determined empirically to further lessen intensity dependence (data not shown). Finally, constraint No. 2 was enforced by masking OptFlow images where Eq. 8 was satisfied:
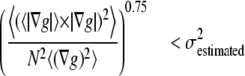 |
(8) |
Rod-like model mitochondria were generated (Fig. 5 A), and the effect of implementing Eq. 8 (Appendix E) was tested. Fig. 5, B and E, show that with no constraints, velocities are underestimated because of the contribution of inaccurately low OptFlow in the middle of the rod shape. Application of constraint No. 2 (with k = 0.045 in Eq. 9) allows the underestimated middle section of the model mitochondrion to be masked (Fig. 5 C), resulting in a more accurate mean OptFlow at velocities up to 2 pixel/frame (Fig. 5 E, red trace). Equation 8 proved to be a valid correction, since OptFlow was independent of kernel selections (not shown), and to have little dependence on intensity and noise (see below). Fig. 5 D illustrates the effect of applying constraint No. 1 of Eq. 7 in addition to constraint No. 2, but using no masking with the binarized minimum intensity projected image. However, this had little effect on the OptFlow (Fig. 5 E, blue trace).
The dynamic range of velocity determination
The above data indicate that short spatial differentiation kernels can be used on large objects with minimal error when the bias correction is applied (Eq. 6 and Fig. 3 G). Fig. 6, A–C, demonstrates that the OptFlow algorithm tuned in the above sections is accurate for the three sizes of model mitochondria at various “fluorescence” intensities or noise levels using a single set of parameters. Fig. 6, A–C, also indicate that in the case of the modeled spheres it was possible to accurately measure velocities (in pixel/frame) up to the approximate square root of the radius.
The smallest velocities that can be distinguished from zero vary with the noise level (Figs. 6, A–C). The least noisy models (intensities 1000 and 20000) seem to yield linear simulated versus measured velocity relationship at the bottom of the simulated velocity range. To establish the smallest velocity that can be distinguished from zero, OptFlow was calculated for simulated velocities increasing at 1/64 pixel/frame increments for the r = 3, int = 1000 model mitochondria. The smallest velocity that was significantly different from zero was 5/64 pixel/frame for wt = 2avg (Fig. 6 D) and 3/64 pixel/frame for the wt = 3 (not shown). Fig. 6 E illustrates this by showing that a 1/8 pixel shift of a group of model mitochondria yields a sufficient ∂g/∂t (green arrows) to result in a significant OptFlow. Thus the OptFlow technique is exquisitely sensitive for the detection of slight displacements. The OptFlow algorithm with all corrections and constrains introduced above is summarized in Fig. 7.
Targeted fluorescent proteins
The plasmid vector encoding mito-roGFP1 was a gift of James Remington (21) and the mito-mPlum was from Ella Bossy-Wetzel (Burnham Institute, La Jolla, CA). These constructs selectively mark mitochondria by using the N-terminal targeting sequence of the cytochrome oxidase subunit VIII. These fluorophores localize to the mitochondrial matrix. The myristoyl palmitoyl mCFP was a gift of Roger Tsien (22). This latter construct, due to its acylation, enriches in membranes, as in the plasma membrane.
Cell culture and transfection
Primary hippocampal neurons were prepared from E18 rat hippocampi (BrainBits LLC, Springfield, IL). Hippocampi were dissociated with 2 mg/ml papain (Worthington, Lakewood, NJ) in Hibernate (BrainBits) for 30 min at 37°C, and were dispersed manually by 5–10 strokes with a 1 ml pipette. Cells were plated onto poly-D-lysine-coated Lab-Tek I 8-well chambered coverglasses (Nunc, Rochester, NY) at a density of 105 cells/well in neurobasal medium (Invitrogen, Carlsbad, CA) containing 2% B27 supplement, 0.5 mM GlutaMAX, 25 μM glutamate, 1% dialyzed fetal bovine serum, and 1% penicillin/streptomycin; 3 h later, plating medium was replaced by a serum-free medium. Neurons were maintained at 37°C in 92% N2/5% CO2/3% O2 atmosphere. Cultures were fed after 4 days by replacing half of the medium with fresh medium lacking serum and glutamate. Cultures were transfected at days 11 and 12 using Lipofectamine 2000 (Invitrogen) in Neurobasal medium at a 3:2 ratio of Lipofectamine (μl) to plasmid DNA (μg); 0.2 μg of DNA was transfected per well in 20 μl Neurobasal medium over the top of 100 μl culture medium. Saved culture medium diluted in 1:1 with fresh medium was replaced over the cells after 3–4 h. Experiments were performed 2–4 days posttransfection at 37°C in a medium containing (in mM) 120 NaCl, 3.5 KCl, 1.3 CaCl2, 1 MgCl2, 0.4 KH2PO4, 5 NaHCO3, 1.2 Na2SO4, 20 TES, 15 glucose and 0.4% fatty acid free bovine serum albumin at pH 7.4.
Image acquisition
Time-lapse fluorescence microscopy was performed on an Olympus IX-81 inverted microscope equipped with an UAPO 40× oil 1.3 NA lens, a Lambda LS Xe-arc light source (175 W, using an ND1.5 filter to attenuate intensity), Lambda 10-2 excitation and emission filter wheels (Sutter Instruments, Novato, CA), CoolSNAP HQ cooled digital camera (−30°C, 10 Mhz readout, 2× gain, 12 bit depth) and a Proscan linear encoded xy-stage (Prior, Rockland, MA). At 2 × 2 binning of the camera, 512 × 512 pixel images were recorded at ∼0.32 μm/pixel resolution. To image mito-roGFP1, a 438/24 nm (Semrock, Rochester, NY) or a 480/20 nm (Chroma, Rockingham, VT) excitation filter, a 505LP dichroic mirror (Chroma 74100) and a 528/38 nm (Chroma) emitter were used. Although mito-roGFP1 exhibits redox potential-dependent fluorescence (21), intensity changes at the 438 nm excitation are small, because this wavelength is close to the redox-insensitive isosbestic point. Therefore, only the 438 nm excitation wavelength of the mito-roGFP1 was used for OptFlow calculation. In addition, physiological redox-sensitive changes in roGFP1 fluorescence are too slow to affect the OptFlow calculation (data not shown). For mito-mPlum, a 582/15 nm exciter (Semrock), a 500/580 nm dichroic mirror (Chroma 51019), and a 610LP emitter (Omega) were used. For myrpalm-CFP, the 438/24 nm exciter, a 458LP dichroic mirror, and a 483/32 nm emitter (Semrock) were used. For tetramethylrhodamine methyl ester (TMRM), a 555 nm (Chroma) excitation filter, a 475/560 nm dichroic mirror (Chroma 73000), and a 588/21 nm (Semrock) emission filter were used.
The whole microscope enclosure was maintained at 37°C in air. Image acquisition was controlled by the Multi Dimensional Acquisition application and journal programming in Metamorph 6.3 (Molecular Devices, Downingtown, PA). In each acquisition cycle, the sample was autofocused on green fluorescent beads placed in an empty well of the eight-well chamber. Then 4–16 cells at different stored locations were visited and imaged at 1–3 wavelengths. Image acquisition for OptFlow calculation was performed by recording short sequences of images (two frames at one frame/s; except for Figs. 8 and 9) at each location. Image sequences were repeated at intervals of ∼2 min. Because there was no change in stage position or optics of the microscope for the duration of the short image sequence, mechanical motion artifacts related to the multidimensional image acquisition were avoided (i.e., imprecision of xy stage and focus motors during revisiting stage positions and emission filter/dichroic mirror changers when acquiring multiple wavelengths). Mechanical motion artifacts originating from stage or focus drift, vibrations, and heat dilation/contraction of the microscope frame could lead to the appearance of uniform motion vectors over all stationary objects of the view field, but this was not found. Image data were stored on the hard drive and analyzed off-line.
FIGURE 9.
Determination of optimal frame rate to follow the motion of mitochondria in axons of hippocampal neurons. To match the dynamic range of the OptFlow calculation and the velocities of mitochondria in axons of hippocampal neurons, OptFlow calculation was performed at various image acquisition intervals. (A) Raw fluorescent image of axons of hippocampal neurons expressing mito-mPlum. (B) High pass-filtered image of mito-mPlum (red) and myrpalm-CFP (blue). For the latter, a lower cut on high pass filter was used. (C) Absolute OptFlow image. The time lapse interval was 0.15 s/frame. (D) Masked absolute OptFlow image overlaid on the shapes of the segments in black. Black pixels show those parts of mitochondria that are masked by the constraint criteria, whereas colored ones show valid OptFlow determinations. (E) Histograms of mean velocities of mitochondria. The histograms are truncated at velocities <50 nm/s for improved visualization of higher velocities, whereas percents refer to the total mitochondrial population. The acquisition interval (0.15 s–3 s) used for recording images for OptFlow calculation is indicated in the diagrams. Vertical red lines indicate the estimated boundaries of the sensitivity. These data are also shown in Table 2. Data are from ∼6300 mitochondria from six view fields. Scale bar, 10 μm.
Measurements of mean OptFlow over the mitochondrial population (Fig. 10) were done in the same way as detailed above for model mitochondria, processing raw fluorescence image sequences of mito-roGFP1 438 nm excitation. Measurements of single mitochondrion fluorescence intensities and OptFlow (Figs. 8, 9, 11–13) were based on binarization and segmentation of both mito-roGFP1 images (see Appendix C). Although the OptFlow was calculated in raw image sequences, mean fluorescence intensities were determined for each mitochondrion in local background subtracted, aligned images. Dehazing and local background removal were done by high-pass filtering (23). The high-pass filter had a characteristic of a gradual cut on between ω = 0.05–0.5 cycles/μm (24).
FIGURE 11.
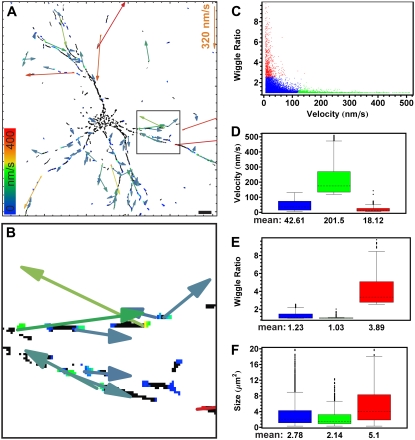
OptFlow as vector field: mitochondrial transport and wiggle. Hippocampal neurons expressing mito-roGFP1 were imaged and OptFlow was calculated as detailed in Figs. 6 and 7. Velocity vectors allow the distinction of directional transport from local, “wiggling” motion. (A and B) Velocity vectors shown over the absolute OptFlow image corresponding to Fig. 8 E. Arrows (pseudo color coded and sized according to the velocity) indicate mean velocities in each patch of valid, in range, OptFlow determination. (A) Scale bar, 10 μm. (B) magnification corresponding to the rectangle in A. Note that velocity vectors are close to parallel with the dendrites. (C) Scatter diagram of “wiggle ratio” (calculated as absolute value of mean vectors divided by mean of absolute vectors) as a function of velocity (calculated as absolute value of mean vectors). Each point indicates a single mitochondrion. Different colors indicate the result of cluster analysis, and correspond to the diagrams D–F. A total of 14,510 mitochondria were analyzed (mean velocities smaller than bit noise were excluded). (D–F) Box-and-whisker plots indicate the median of the data sets by dashed lines, the ±33 percentile around the median by the boxes, and the entire data set excluding outlying points by the whiskers. Outlying points were defined as points farther from the edge of the box than 150% of the range that the box spans. Below the diagrams, the corresponding means are shown. Note that higher velocities are associated with directional transport (small “wiggle ratio”; green in C–F), whereas high “wiggle ratio” (red) with larger size. Data were pooled from 27 neurons and 20 repeats in three experiments.
FIGURE 13.
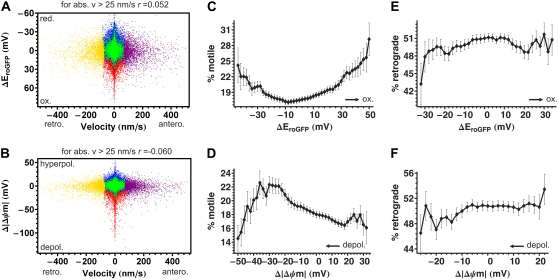
Relationships among mitochondrial membrane potential, thiol redox potential, and motility. Data correspond to the cluster analysis introduced in Fig. 12. (A and B) Scatter plots of ΔEroGFP or Δ| ΔΨm| against velocity. Each data point represents a single mitochondrion and color coding corresponds to the clusters defined in Fig. 12 B. The Spearman rank correlation coefficient (r) is indicated above the diagrams. Note that the correlation was calculated for absolute velocities. Oxidation or depolarization is less correlated to velocity than the size of mitochondria in Fig. 12, D and E. (C and D) Percentage of motile mitochondria (defined as faster than the bottom detection limit; 25 nm/s, and not wiggling; i.e., “wiggle ratio” <1.2) as a function of ΔEroGFP or Δ| ΔΨm|, respectively. Note the increasing motility with depolarization in D, and the high error of the less motile, very depolarized mitochondria that are small in numbers. (E and F) Percentage of mitochondria undergoing retrograde rather than anterograde transport as a function of ΔEroGFP or Δ| ΔΨm|, respectively. For each point, motility percentage was averaged ±10 mV around the indicated ΔEroGFP or Δ| ΔΨm| value, and then the mean ± SE of individual neurons was calculated. Note that very close to half of mitochondria moves in anterograde or retrograde directions, and this does not depend on ΔEroGFP or Δ| ΔΨm|. Data of 83,848 mitochondria were pooled from 102 neurons and 15 repeats per cell in four different cell culture preparations.
Calculation of relative thiol redox potential and ΔΨm
The relative redox potential of the mito-roGFP1 (ΔEroGFP) as compared to the cellular mean was calculated from the 438:480 nm fluorescence excitation ratio (R) as 60 log ((R-Rmin)/(Rmax-R)). These potentials were normalized by the subtraction of the mean redox potential calculated separately for each neuron. Rmin (0.6) and Rmax (1.4) minimal and maximal fluorescence ratios corresponding to fully reduced and fully oxidized states, respectively, were determined in separate experiments.
To measure ΔΨm, TMRM (7.5 nM) was preincubated for 1 h and was continuously present in the experimental medium together with tetraphenylboron (1 μM) facilitating its equilibration through cellular membranes (25). This concentration of TMRM was below the threshold for fluorescence quenching of the dye in the mitochondria (25,26); thus differences of ΔΨm (Δ| ΔΨm|) can be directly calculated in millivolts from the relative fluorescence intensities. The difference of ΔΨm in millivolts from the mean of the total mitochondrial population of each neuron was calculated for individual mitochondria as follows. Because of the high-pass filtering, the measured signal reflected the mitochondrially originated TMRM fluorescence (23). Individual mitochondria can be smaller or thinner than the resolution of the microscope, so their fluorescence is diluted by the background (27). To normalize TMRM fluorescence intensity for each mitochondrion, the intensity measured in the high-pass filtered image was divided by the calculated fluorescence intensity corresponding to the isosbestic wavelength of roGFP1 (see Appendix F). The log of this ratio was then multiplied by 60 to give a value corresponding to Δψm + K for that mitochondrion in millivolts, where K is a constant for a given cell reflecting the level of roGFP1 expression. Finally, the extent to which each individual mitochondrion deviated from the mean was calculated by subtraction of the mean value of Δψm + K calculated for each neuron (Δ| ΔΨm|). Therefore, the Δ| ΔΨm| is independent both of the dimensions of mitochondria and the level of roGFP1 expression.
Radial projection of velocity vectors
To assess and distinguish between anterograde and retrograde motion of mitochondria, the radial projection of the velocity vectors from the soma of the neuron was calculated. For this, the scalar product of the velocity vector and a unit vector pointing from a manually selected point in the soma to the given pixel was calculated for each pixel of the OptFlow vector field. Anterograde motion is therefore defined as a positive radial velocity.
Cluster analysis
To find subpopulations of mitochondria differing in any of the above quantified parameters, multidimensional cluster analysis was performed by the FindClusters function of Mathematica 5.2, using it with its default options. For clustering, all data were normalized to span a unit length between the 10th and 90th percentile for each parameter, and individual mitochondria were optimized into the given number of clusters based on the two- or four-dimensional squared Euclidian distance between the data points in an unsupervised manner.
Software environment
Image processing was carried out using custom developed image analysis software written in Pascal language (Delphi 6.0; Borland, Austin, TX) and Mathematica 5.2 (Wolfram Research, Champaign, IL). All further data analysis was done in Mathematica 5.2.
MATERIALS
Reagents were obtained from Sigma (St. Louis, MO) or EMD Chemicals (Gibbstown, NJ) unless otherwise stated.
EXPERIMENTAL RESULTS
Measurement of velocity of individual mitochondria in dendrites of hippocampal neurons
Cultured rat hippocampal neurons expressing mitochondrially targeted redox sensitive GFP (mito-roGFP1 (21)), were imaged to record mitochondrial motion in the proximal ∼100 μm of dendrites (Fig. 8 B). Mitochondria in these dendrites were sufficiently plentiful for their fluorescence to reveal the shape of the cell, and no additional fluorescence marker was required. Mitochondria appeared as rods and spheres with apparent radii of 1.5–3.5 pixels (Fig. 8 A).
First, the optimal image acquisition interval was assessed for the measurement of mitochondrial velocities. Short image sequences for the OptFlow calculation consisting of seven frames with 0.15 s, 0.5 s, or 2 s intervals were recorded every 2 min. OptFlow was calculated by selecting two frames from the image sequence separated by 0.15–4 s. Temporal differentiation was done as described for wt = 2avg. The minimum intensity-projected image sequence was segmented to identify individual mitochondria (shown in false color in Fig. 8 C) before determination of their velocity. Segmentation sometimes detected conglomerates of mitochondria as single entities; however, these were very few compared to the large number of smaller mitochondria and were therefore not expected to bias the calculation. Mean velocity of each segment in Fig. 8 C was measured in the absolute OptFlow images (Fig. 8 D, where the velocity of the individual mitochondria is represented by the color coding). In Fig. 8 E, the absolute OptFlow image was masked by the segmented image and overlaid on the segments shown in black. The black pixels correspond to the parts of each mitochondrion that were masked by the constraint criteria in Fig. 8 D, whereas colored pixels show valid OptFlow determinations that counted into the mean velocity of each given segment.
Since individual mitochondria frequently change velocity and direction, stop and restart, 20 measurements were done for each neuron at 2 min intervals and the data were pooled. Each mitochondrion in the repeated measurements was treated as a separate, individual data point. Velocity distributions were plotted in histograms (Fig. 8 F and summarized in Table 1). Remarkably, these histograms confirm that mitochondria do not have preferred velocities, but rather exhibit a continuous distribution (28,29). The power of the technique beyond conventional manual rate determination is exemplified by the fact that the histograms represent velocity determinations from ∼17,500 mitochondria pooled from 12 neurons and 20 repeats per cell.
TABLE 1.
Velocities of dendritic mitochondria and the dynamic range of the OptFlow measurement as a function of image acquisition interval
| Acquisition interval (s) | Percent lost by minimum intensity projection | Percent lost by constraints | Percent stationary | Velocity of moving mitochondria (nm/s)
|
Detection range (nm/s)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | 95% | 99% | Bottom | Top | ||||
| 0.15 | 1.8 ± 0.4 | 4.8 ± 2.5 | 56.0 ± 2.0 | 103 ± 4.6 | 42.3 ± 2.1 | 412 ± 20.3 | 800 ± 79.1 | 167 | 2560 |
| 0.3 | 2.1 ± 0.3 | 5.1 ± 2.7 | 54.6 ± 2.4 | 69.8 ± 3.4 | 32.1 ± 1.7 | 263 ± 16.0 | 489 ± 36.5 | 83 | 1280 |
| 0.5 | 2.2 ± 0.3 | 4.8 ± 2.5 | 51.3 ± 2.6 | 57.8 ± 3.7 | 27.4 ± 1.8 | 206 ± 12.8 | 442 ± 54.2 | 50 | 768 |
| 1 | 2.6 ± 0.3 | 5.1 ± 2.6 | 46.2 ± 2.9 | 44.5 ± 3.5 | 24.8 ± 2.1 | 154 ± 12.0 | 286 ± 26.4 | 25 | 384 |
| 1.5 | 2.5 ± 0.3 | 5.0 ± 2.5 | 42.9 ± 3.1 | 38.2 ± 3.0 | 22.7 ± 2.2 | 127 ± 9.6 | 222 ± 14.4 | 17 | 256 |
| 2 | 2.9 ± 0.4 | 5.0 ± 2.4 | 39.6 ± 3.1 | 33.3 ± 2.6 | 21.2 ± 2.0 | 107 ± 8.1 | 179 ± 12.3 | 13 | 192 |
| 2.5 | 3.1 ± 0.3 | 5.1 ± 2.5 | 37.5 ± 3.2 | 30.5 ± 2.4 | 20.9 ± 1.9 | 92.5 ± 6.9 | 159 ± 14.0 | 10 | 154 |
| 3 | 3.2 ± 0.3 | 5.1 ± 2.5 | 34.1 ± 3.3 | 28.2 ± 1.8 | 20.1 ± 1.5 | 84.1 ± 5.1 | 134 ± 7.9 | 8 | 128 |
| 4 | 3.3 ± 0.4 | 5.4 ± 2.7 | 31.0 ± 3.1 | 24.2 ± 1.8 | 17.6 ± 1.5 | 69.7 ± 5.2 | 111 ± 6.6 | 6 | 96 |
Velocities in bold indicate values within the detection range. Data are shown as mean ± SE of neurons (n = 12). Corresponds to Fig. 8 F.
To determine the optimal acquisition interval, we considered that the OptFlow measurement is accurate between 5/64 and 1.2 pixels/frames based on Fig. 6. The actual calibrated dynamic range is marked by the red lines on the histograms in Fig. 8 F. As would be expected, increasing the acquisition interval increased the sensitivity for the detection of small velocities. Thus the percentage of stationary mitochondria (defined here as <1.3 nm/s corresponding to the least significant bit of the integer data storage) decreased with increasing acquisition interval (Table 1; histograms in Fig. 8 F were truncated to hide the large number of stationary mitochondria). As Table 1 indicates, shorter acquisition intervals (0.15–2 s) are suitable for detection of fast movements in dendrites since the 99th percentile of measured velocities is substantially below the top detection limit. However, at acquisition intervals <1 s, the mean and median velocities are below the bottom detection limit, and these velocities are considered to be inaccurate. Therefore, we conclude that acquisition intervals of 1–2 s are optimal for overall assessment of mitochondrial motion in dendrites of hippocampal neurons.
We have also determined the proportion of mitochondria excluded by the OptFlow algorithm. Loss of data corresponding to certain mitochondria occurs because of elimination of objects, which move faster than the algorithm can measure, by the minimum intensity projection masking. The number of such mitochondria was determined by binarization and segmentation of both maximum and minimum intensity projection images, followed by counting those mitochondria that show up only in the maximum but not in the minimum intensity projected images. Table 1, column 2, shows that although the numbers of excluded mitochondria are small, they increase with increasing acquisition interval. A second group of mitochondria is excluded due to the constraints of the optical flow algorithm, independently of the acquisition interval (Table 1, column 3).
Measurement of velocity of individual mitochondria in axons of hippocampal neurons
Mitochondria in axons are shorter and more sparsely distributed than in dendrites (Fig. 9 A). Therefore, the plasma membrane marker myristoyl palmitoyl mCFP (myrplam-CFP) was used to define the axon, together with the mitochondrially targeted red fluorescent protein mito-mPlum to study mitochondrial movement. Axons were defined by their thin, spineless appearance shown by the myrpalm-CFP (Fig. 9 B, blue). OptFlow was calculated from image sequences of mito-mPlum (Fig. 9, A–D). Histograms of axonal mitochondrial velocities (Fig. 9 E) are distinguished from the dendritic ones (Fig. 8 E) by the longer tail of the velocity distribution toward higher values. Table 2 summarizes the statistical parameters of velocity distributions obtained by different image acquisition intervals. For accurate measurement of high velocities of axonal mitochondrial transport, only the shortest acquisition intervals (0.15–0.3 s) were suitable as the 99th percentile of velocities was below the top detection limit.
TABLE 2.
Velocities of axonal mitochondria and the dynamic range of the OptFlow measurement as a function of image acquisition interval
| Acquisition interval (s) | Percent lost by minimum intensity projection | Percent lost by constraints | Percent stationary | Velocity of moving mitochondria (nm/s)
|
Detection range (nm/s)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | 95% | 99% | Bottom | Top | ||||
| 0.15 | 6.5 ± 1.4 | 2.7 ± 1.1 | 54.8 ± 1.2 | 185 ± 19.2 | 67.3 ± 8.2 | 751 ± 101 | 1299 ± 104 | 167 | 2560 |
| 0.3 | 6.4 ± 0.9 | 3.2 ± 1.2 | 53.8 ± 1.0 | 129 ± 19.2 | 48.1 ± 6.1 | 544 ± 131 | 1030 ± 121 | 83 | 1280 |
| 0.5 | 6.3 ± 1.2 | 2.7 ± 1.1 | 54.0 ± 1.3 | 105 ± 16.1 | 38.4 ± 5.9 | 439 ± 74.5 | 867 ± 91.4 | 50 | 768 |
| 1 | 7.3 ± 1.8 | 3.0 ± 1.2 | 54.5 ± 1.7 | 70.3 ± 6.6 | 28.8 ± 2.6 | 293 ± 28.6 | 510 ± 34.4 | 25 | 384 |
| 1.5 | 7.8 ± 1.3 | 3.0 ± 1.2 | 52.0 ± 1.7 | 54.0 ± 4.9 | 25.6 ± 2.7 | 206 ± 14.2 | 344 ± 12.0 | 17 | 256 |
| 2 | 8.5 ± 1.2 | 2.9 ± 1.2 | 52.2 ± 1.9 | 46.3 ± 4.9 | 22.9 ± 3.0 | 173 ± 13.7 | 272 ± 9.1 | 13 | 192 |
| 2.5 | 9.0 ± 1.5 | 3.2 ± 1.3 | 52.0 ± 2.0 | 40.1 ± 4.5 | 21.2 ± 3.0 | 144 ± 13.9 | 236 ± 16.1 | 10 | 154 |
| 3 | 9.6 ± 1.4 | 3.3 ± 1.4 | 48.6 ± 2.0 | 35.0 ± 3.3 | 18.6 ± 1.7 | 124 ± 7.9 | 188 ± 10.6 | 8 | 128 |
| 4 | 9.7 ± 1.0 | 3.3 ± 1.3 | 44.3 ± 2.4 | 28.5 ± 2.5 | 17.4 ± 1.9 | 92.0 ± 8.0 | 142 ± 11.5 | 6 | 96 |
Velocities in bold indicate values within the detection range. Data are shown as mean ± SE of view fields (n = 6). Corresponds to Fig. 9 E.
Dynamics of OptFlow after artifactual cessation of motion
To test the dynamics of the OptFlow, we recorded time lapses of short image series (Fig. 10 A) during the addition of paraformaldehyde (4% in phosphate-buffered saline) to fix the specimen while recording (Fig. 10 B). Paraformaldehyde caused a large drop in OptFlow, but did not decrease it completely to zero. To observe completely stationary objects, solidly embedded 0.2 μm fluorescent beads were imaged in similar conditions (Fig. 10 C). The remaining OptFlow was similar to the one measured in fixed cells. The bead specimen had similar size and intensity to the mito-roGFP expressing mitochondria (Fig. 10, D–G). Observation of this remaining OptFlow as velocity vectors indicated random directions within a view field, thus this OptFlow arises from noise rather than form microscope stage or focus drift or fluctuations of illumination intensity (not shown). Moreover, it was possible to eliminate the OptFlow over stationary objects by increasing the k-factor of Eq. 9 for ∂t cut off correction. Nevertheless the remaining OptFlow was below the safe lower detection limit (25 nm/s for the 1 s acquisition interval; see Table 1), therefore, we did not change the parameters of the OptFlow calculation tuned with model mitochondria. These results support the satisfactory application of the OptFlow technique to microfluorimetric recordings.
Mitochondrial transport vs. ‘wiggle’
The primary output of the OptFlow calculation is a velocity vector field, assigning a velocity vector to each (nonmasked) pixel. The mean OptFlow over a single mitochondrion was calculated above as the mean of absolute velocities (Figs. 8–10). Alternatively, velocities can be averaged as vectors. In Fig. 11, A and B, mean velocity vectors are visualized at each region of valid OptFlow determination by arrows, typically at the tips of mitochondria as a consequence of constraint No. 2.
If the whole mitochondrion moves in the same direction (i.e., transport movement with no shape change), then the absolute value of the mean velocity vector and mean of absolute velocities for the whole mitochondrion are the same. However, if the mitochondrion is undergoing shape change or makes an uneven, rotating, or wiggling motion, then the vectors may partially cancel each other; therefore, the absolute value of the mean of vectors will be smaller than the mean of absolute values. For easy reference, we denote the ratio of the mean of absolute vectors over the absolute value of the mean vector as the “wiggle ratio”, which has a value of ∼1 for linear transport movement and is larger for wiggling movement. Fig. 11 C shows the relationship of velocity (as mean of vectors) and the wiggle ratio. To analyze distinct subpopulations of mitochondria, three groups were selected by two-dimensional cluster analysis. The cluster analysis groups (clusters) data points (individual mitochondria) based on the similarity of their parameters. In our case, the similarity of parameters is defined by distance of the data points in the two-dimensional space of velocity and wiggle ratio. 13.5% of mitochondria from a total of 14,510 had faster velocities (>120 nm/s; green in Fig. 11 C) and were typically associated with small wiggle ratios (<1.2). Slower or stationary (<50 nm/s; red) mitochondria (5%) typically showed a wiggle ratio >2.5. However, most of the mitochondria (81.5%) were slow moving or stationary and not wiggling (blue, Fig. 11, D–F). The high wiggling ratio is associated with larger mitochondria (Fig. 11 F).
Parallel measurement of mitochondrial motion, membrane potential, and thiol redox potential in hippocampal neurons
ΔΨm was measured by TMRM in nonquench mode (26) in mito-roGFP1 expressing cultured hippocampal neurons (Figs. 12 and 13). Imaging was performed by sequentially recording one TMRM image, one of mito-roGFP 480 nm excitation, and a sequence of 2 images at 1 s interval for the 438 nm excitation for the near isosbestic point image of mito-roGFP1 (Fig. 12 A). Velocities of mitochondria were determined from this last sequence. Cultured hippocampal neurons have mostly radial neurites (Fig. 11 A). Therefore, the direction of the velocity vectors, whether pointing toward the soma or away, were calculated by radial projection from the soma. This calculation results in scalar velocities; positive for anterograde and negative for retrograde motion.
FIGURE 12.
Cluster analysis of mitochondrial velocity, membrane potential, thiol redox potential, and size in hippocampal neurons. (A) Scheme of single mitochondrion quantification. Thick black arrows denote image processing maneuvers, gray arrows stand for reading out scalar values corresponding to each segment, and thin black arrows indicate calculation done with the obtained scalar values. (B) The pooled mitochondrial population was classified into five groups by four-dimensional clustering in the space of Δ| ΔΨm|, ΔEroGFP, size and velocity. Each cluster represents a set of individual mitochondria with similar properties, and indicated by a different color. Solid and open bars show numbers and the sizes as total area of mitochondria in the fluorescence images, respectively, for each cluster. (C–E) Scatter plots of absolute velocity, Δ| ΔΨm| and ΔEroGFP, respectively, against the size of mitochondria. (F) Scatter plot of Δ| ΔΨm| against ΔEroGFP. (C–F) Each point represents a single mitochondrion and color coding corresponds to B. The Spearman rank correlation coefficient (r) is indicated in the diagrams. Note that the signs of correlation coefficients suggests that (C) larger mitochondria are slower, (D) are more depolarized, (E) oxidized mitochondria are smaller, and (F) oxidation correlates with depolarization. (G) Membrane potential as a function of redox potential. For each point, Δ| ΔΨm| was averaged ±10 mV around the indicated ΔEroGFP value, and then the mean ± SE of individual neurons was calculated. (H) Redox potential as a function of membrane potential. Note that above a certain oxidation level (∼ΔEroGFP > +10mV), mitochondria increasingly loose their ΔΨm. Data of 83,848 mitochondria were pooled from 102 neurons and 15 repeats per cell in four different cell culture preparations.
The mito-roGFP1 images were segmented and the radial velocity and the fluorescence intensities corresponding to single mitochondria were logged (Fig. 12 A). In addition, the segmentation data also resulted the size of the mitochondria (apparent area in images in μm2). The mito-roGFP1 fluorescence intensity ratios for individual mitochondria were converted into thiol redox potentials (ΔEroGFP; in mV) expressed relative to the mean cell redox potential. Similarly, TMRM fluorescence expressed as the ΔΨm for each mitochondrion was calculated relative to the cell mean (i.e., as Δ| ΔΨm| in millivolts). To detect overlapping or erroneously segmented mitochondria, the variance over the mean of the TMRM fluorescence was measured, with the assumption that an individual mitochondrion has a uniform potential (30,31). The shot noise-related standard deviation as determined by error propagation from the measured intensities and the noise characteristics of the camera in 99% of mitochondria was for ΔEroGFP and Δ|ΔΨm| <1.5 mV and 3.5 mV, respectively (Appendix G). Table 3 summarizes how valid data points were filtered.
TABLE 3.
Parameters used for selection of subpopulations of mitochondria
| Parameter | General filtering | Motile | Direction measurement |
|---|---|---|---|
| Absolute velocity (nm/s) | <400 | >25 | >25 |
| Wiggle ratio | <1.2 | <1.2 | |
| TMRM σ2/mean | <90 percentile | ||
| Radial/absolute velocity | >0.75 | ||
| ΔEroGFP (mV) | SD <6 mV | ||
| ΔΨm (mV) | SD <10 mV |
Corresponds to graphs shown in Figs. 12 and 13. SD is the standard deviation propagated from the photon shot noise converted to millivolts (see Appendix G).
To assess resting neurons, imaging was done in the presence of the NMDA receptor inhibitor MK-801 (10 μM), the AMPA receptor inhibitor NBQX (10 μM), and the voltage gated Ca2+ channel inhibitor nifedipine (1 μM) to suppress spontaneous activity; 102 neurons were imaged 15 times at 3 min intervals. Importantly, only those mediocre mito-roGFP1 expressor neurons were selected for image acquisition, which had similar TMRM fluorescence intensity to their nontransfected neighbors, indicating healthy, well-polarized mitochondria. Notably, the neurons with the brightest mito-roGFP1 fluorescence, especially early after transfection, showed diminished ΔΨm.
To find subpopulations of mitochondria differing in any of the four measured parameters (velocity, Δ|ΔΨm|, ΔEroGFP, and size), four-dimensional cluster analysis was performed on a pooled data set consisting of 83,848 mitochondria (Figs. 12 and 13). Because the pooled mitochondrial population did not have significantly distinct subpopulations in the above four-dimensional space, the margins between them are considered arbitrary. The number 5 for the clusters was chosen because this resulted in biologically meaningful categorization for visualization of the data set. The cluster analysis indicated the following:
The majority of mitochondria (in number and in total size; Fig. 12 B) are stationary and of small to medium size (coded blue in Fig. 12 B; see also Fig. 1 in the Supplementary Material, Data S1).
The second largest group for total size consists of large mitochondria and these are also stationary (Fig. 12 C, green) and have a small variability in Δ| ΔΨm| and ΔEroGFP (Fig. 12, D and E).
The most distinct group consists of more oxidized, less polarized, small mitochondria (Fig. 12, D and E, red).
and e. Mitochondria performing retrograde (yellow) or anterograde (purple) motion, had indistinguishable properties (Fig. 13, A and B). Of note, similar clustering results were achieved by processing data from individual neurons, so the above classification reflects real heterogeneity of mitochondria within single hippocampal neurons.
To refine the analysis, parameters were compared pairwise. To find linear correlations or nonlinear trends between pairs of mitochondrial parameters, Spearman rank correlation was used. Correlation coefficients are indicated in the scatter diagrams in Figs. 12 and 13. This comprehensive multiparameter analysis of individual mitochondria allows a range of relevant hypotheses to be tested.
Is there a correlation between the size and the velocity of mitochondria?
Although the correlation coefficient between size and velocity is small (r = −0.12; excluding stationary mitochondria), the size is the strongest correlating parameter to the velocity as compared to Δ|ΔΨm| or ΔEroGFP (see below). The negative sign of the correlation coefficient indicates that larger mitochondria are slower (Fig. 12 C).
Is there a correlation between membrane potential and thiol redox potential?
Fig. 12 F shows marked correlation at r = −0.33 between the Δ|ΔΨm| and the ΔEroGFP. To further investigate this, the mean Δ|ΔΨm| was plotted as a function of ΔEroGFP (Fig. 12 G). To this end, for each given ΔEroGFP ± 10 mV range, the mean Δ|ΔΨm| was calculated and the mean ± SE is shown. This was done only in those neurons where at least 5 mitochondria with the given ΔEroGFP range were present during the entire experiment, and only those data points are shown where at least a third of the total 102 neurons was such. Fig. 12 H shows the converse relationship. Note that there is an approximately linear relationship between ΔEroGFP and mean Δ| ΔΨm| in the middle of the range. Interestingly, Δ|ΔΨm| was only slightly decreased by increasing ΔEroGFP until ∼10 mV oxidation, but at more oxidized redox potentials Δ|ΔΨm| steeply decreased. Of note, even though the mito-roGFP1 fluorescence was used for normalization of the TMRM fluorescence, the above correlation cannot be a normalization artifact, because it was also found between ΔEroGFP and unnormalized TMRM fluorescence intensity data (not shown).
Does the matrix thiol redox potential or the membrane potential predict the direction and motility of individual mitochondria?
ΔEroGFP and Δ|ΔΨm| correlated very poorly with the absolute value of velocities of motile mitochondria (r = 0.052 and r = −0.060, respectively; Fig. 13, A and B). However, the indicated correlation coefficients were markedly larger than the coefficients calculated from scrambled data, where |r| was always <0.005. Therefore, these results suggest that more oxidized and less polarized mitochondria have a tendency to move faster.
To facilitate the detection of relationships between Δ| ΔΨm| or ΔEroGFP and velocity, mitochondria were classified as motile or nonmotile (see criteria in Table 3). Fig. 13, C and D, show the mean percentage of motile mitochondria per neuron as functions of ΔEroGFP and Δ|ΔΨm|, respectively, calculated in the same way as detailed for Fig. 12 G. Subpopulations of oxidized or reduced mitochondria were marginally more motile than ones with average redox potential. Motility was increased by depolarization until ∼−40 mV but mitochondria more depolarized than this threshold had lower motility (Fig. 13 D).
No correlation could be detected between the direction of movement and either ΔEroGFP or Δ| ΔΨm| (Fig. 13, E and F), i.e., oxidized mitochondria were not preferentially returned to the soma from the dendrites. It was, however, observed that the almost all of depolarized (Δ| ΔΨm| < −40 mV), oxidized (ΔEroGFP > 40 mV) small (size <5 μm2) mitochondria (representing 0.1% of the population) failed to line up with other mitochondria in the neurites of the observed neuron, suggesting that they are likely to belong to other neurons that could be injured or dead (Fig. 2 in the Supplementary Material, Data S1). In contrast, the motile, oxidized, but only partially depolarized mitochondria appeared within dendrites. Therefore, in resting, healthy cultured hippocampal neurons, the number of damaged, discharged, and oxidized mitochondria is virtually zero, suggesting an efficient quality control of mitochondria (see (32)).
DISCUSSION
The transport of mitochondria within neuronal processes and their targeting to specific sites such as synapses are clearly critical to maintaining neuronal integrity. Thus defective mitochondrial transport resulting from mutations in the mitochondrial fusion gene Mitofusin-2 leads to the peripheral neuropathy Charcot-Marie-Tooth Type 2A in which long motor neurons become dysfunctional, whereas a similar fusion deficit due to OPA1 mutation causes dominant optic atrophy (see Chen and Chan (33)). Genetic screens in Drosophila for visual dysfunction have led to the identification of proteins such as milton (34) and miro (35) that are required for mitochondrial attachment to kinesin motors required for anterograde transport. Despite these advances, it is still unclear how mitochondria are delivered to the energy-demanding sites where they are required, and whether a mechanism exists for the return of damaged or senescent mitochondria to the cell body for destruction. Such studies would be facilitated by a convenient and robust technique for quantifying mitochondrial motion in neuronal processes that minimizes selection bias and phototoxicity, particularly since the latter may induce artifactual mitochondrial behavior.
Although several techniques exist, each has some limitation. Early quantitative techniques included phase contrast or fluorescence video microscopy and manual or computer assisted tracking of individual mitochondria (28,36,37). These studies shed light on the role of microtubule and actin cytoskeletal elements in the transport of mitochondria in neuronal axons and dendrites. Single particle tracking, either manual or semiautomatic, is still the most commonly used technique applied to wide-field or confocal microscope-based digital image acquisition techniques (2,29,38–41). A limitation, however, is that particle tracking relies on the clear recognition of individual mitochondria, which is often compromised by their overlap in mitochondria-rich regions of the cell, or by the dynamic morphological changes of mitochondria such as fusion and fission. Although sophisticated algorithms exist for handling such problems (42), there are also more robust approaches, which are less affected by these morphological factors. Kymograms (time-space or line-scan diagrams) and manual line fitting to the characteristic diagonal trails of objects in motion have proved to be useful to measure velocities of mitochondria with different fluorescence intensities in crowded areas (3,43). However, by using only a single spatial dimension, kymograms work only in long cellular processes, such as axons.
With these limitations, single particle tracking and kymograms can give accurate data on velocity, direction, run length, and saltatory motion for individual mitochondria. However, both require the acquisition of a large number of images to track objects in time. This could result in phototoxicity, especially during high resolution fluorescence imaging of multiple fluorophores, or in the presence of photosensitizing mitochondrial membrane potential dyes such as TMRM. Kymograms and single particle tracking rely on short mitochondria typically present in axons (44) or in injured cells (45). Healthy, elongated, eventually branched mitochondria, which are present in nonneuronal cells or in proximal dendrites and in the soma of neurons, are unsuitable for such analysis.
Simple techniques for the evaluation of general motion include counting the variation of binarized pixels in consecutive images of fluorescently labeled mitochondria (4), or using the difference of consecutive images and counting pixels changing more than a threshold (6,14,46). However these techniques do not give information about single mitochondria and their velocities and directions of movements and cannot distinguish transport from local, “wiggling” motion or shape changes.
OptFlow has been used in cell biology to detect subpixel dislocations of the organ of Corti of the inner ear (47) and for muscle contraction of the leech (48), but we are not aware of any prior organelle transport or fluorescence microscopic applications.
We have illustrated two basic applications of the OptFlow technique: measurement of mean velocity of the entire mitochondrial population of a neuron in the field of view (Fig. 10) and single mitochondrion assays (Figs. 8, 9, and 11–13). Two major advantages of the OptFlow technique can be emphasized. First, the two-dimensional velocity vectors can be determined from a pair of low light level, noisy images minimizing photodynamic oxidative artifacts evoked by fluorescence excitation. The instantaneous velocity determination is also exploited in the calculation of the correlations of data from other fluorescence assays to the motion, and in the possibility of cyclic/revisiting imaging of different view fields using stage motors. Second, OptFlow measures motion irrespective of shapes and overlaps between mitochondria. This is because gray value gradients corresponding to the individual tips of mitochondria are visible for the algorithm even when mitochondria form conglomerates. This allows the measurement of mean absolute mitochondrial velocities in typical cell lines (A. A. Gerencser, unpublished results), where unlike neuronal dendrites, mitochondria form overlapping networks (31) and individual organelles cannot be distinguished; therefore, these specimens are unsuitable for single particle tracking. The most critical property of the technique is the relatively narrow dynamic range as compared to the wide distribution of mitochondrial velocities (Figs. 8 and 9). The OptFlow measures dislocations with subpixel precision and resolution, but the maximum velocities are limited by the sizes of the gradients. It is concluded that by careful selection of the resolution of microscopy and the acquisition interval, the biologically relevant range of velocities can be matched by the dynamic range of the OptFlow technique. In addition, the dynamic range can be extended by calculating a combination of OptFlow from multiple pairs of images recorded at different intervals (not shown). In the case of single mitochondrion measurements, an unavoidable step is the binarization and segmentation of the images. This imposes similar problems that exist for particle tracking: binarization may oversee dim details or fail to distinguish individual mitochondria within conglomerates. When OptFlow is combined with segmentation, the average velocities are measured within the conglomerates. Conglomerates were excluded from statistical analysis in Figs. 11–13, based on size and heterogeneity of TMRM fluorescence.
The principle of the OptFlow is that the brightness of the objects does not change in time (brightness change constraint equation, Eq. 1). This excludes the use of those fluorophores, which may change their fluorescence intensity in the time frame of the acquisition of the short image sequence for the OptFlow calculation, like mitochondrial membrane potential dyes or calcium probes. Trivially, the illumination intensity and the exposure timing of the camera also have to be precise, but this is not an obstacle in research grade instrumentation. Our implementation of the OptFlow is strictly two-dimensional; therefore, in thicker samples, motion in an angle to the image plane could lead to erroneous velocity measurements.
The OptFlow technique is typically based on wide-field fluorescence microscopy. The scanning mechanism of laser scanning confocal microscopes can have insufficient precision at the subpixel level preventing accurate gradient calculations. Importantly, the maximal resolution of fluorescence microscopy does not limit the precision or sensitivity of velocity measurements. Considering one fluorescent spot (any sized sub- or super resolution object), the point spread function of the microscope only determines the sharpness or steepness of the edges of the objects, but not that where the gradients are. Out of focus blur and slightly unfocused objects cause no problem for the OptFlow determination in wide-field microscopy; whereas out of focus blur is typically rejected by masking according to constraint No. 1, slightly unfocused objects are normally detected as long as their edge gradients are significant compared to the photon shot noise. Image acquisition for OptFlow calculation has to be designed as the camera resolution (in Figs. 10–13, 0.32 μm/pixel) divided by the frame interval (1 s/frame) and multiplied by the detection range of ∼0.08–1.2 pixel/s = 0.025–0.38 μm/s brackets the biologically expected values. The differentiation and smooth kernels used for OptFlow calculation were optimized to work on a wider range of object sizes (1.5–6 pixels radii), which we expect to occur in typical fluorescence micrographs of labeled mitochondria. In a general case, when the moving objects are bigger than this and small objects are absent, selection of wider spatial differential and smoothing kernels can be beneficial in terms of noise insensitivity and dynamic range.
Mean velocities for anterograde and retrograde transport of mitochondria of 0.44 μm/s and 0.51 μm/s, respectively, have been reported in hippocampal neurons (29). In that study, a range from 0.2 to 2.2 μm/s was measured (counting only moving mitochondria) and about half moved with velocities between 0.2 and 0.4 μm/s. Similar values were determined in immature hippocampal neuronal processes (39). Our theoretical upper detection limit at a 0.15 s frame interval was 2.6 μm/s. Due to the long tail of the distribution of mitochondrial velocities toward the high values, it is difficult to accurately represent the entire population. Estimating from the short frame interval acquisitions (Tables 1 and 2), only 5% of the moving mitochondria (with in-range velocities) was faster than 0.4 and 0.75 μm/s in dendrites and axons, respectively. Conversely, half of the moving mitochondria were slower than ∼0.02 μm/s both in dendrites and axons based on the longest acquisition intervals in Tables 1 and 2. It is possible that the above cited articles overestimate mean mitochondrial velocities by selecting clearly visible, moving mitochondria for the particle tracking method. Conversely, the single mitochondrion application of the OptFlow technique may underestimate motion when averaging velocities over aggregated mitochondria and vector means or radial velocity are calculated.
Small, depolarized, and oxidatively damaged mitochondria have been thought to play a role in mitochondrial turnover ((32), see Kim et al. (49)). Autophagocytotic removal of mitochondria belonging to such a subpopulation has been recently showed by tracking mitochondria with photoactivated green fluorescent protein (50). These authors found in a pancreatic β-cell line that mitochondrial fission is often asymmetric (in ∼19%), resulting in a sustained partial depolarization in one of the daughter mitochondria, and some of these small, depolarized mitochondria undergo autophagy hours later. The high rate of asymmetric fission and the long latency to removal suggests the existence of a larger population of small, depolarized mitochondria. This depolarization is slight (−6.5 mV) (50), so this mitochondrial population may be identical to the one identified by cluster analysis in our study in hippocampal neurons as small, less polarized mitochondria (Fig. 12 Bc; red). However, due to the arbitrary boundaries between the clusters, the size of this cluster is a rough estimate. Importantly, we showed here that although these mitochondria are slightly more motile than the average (Fig. 13 D), they do not exhibit distinct transport patterns from the rest of the population (Fig. 1 in the Supplementary Material, Data S1).
Although the focus of this article is on mitochondrial movement, it is suggested that the technique could be used for any other organellar movement, e.g., vesicle transport, or because it does not rely on shape recognition, e.g., on pleiomorph structures like the endoplasmic reticule.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
This work was supported in the Buck Institute by the National Institutes of Health (PL1 AG032118).
APPENDIX A: IMAGE PROCESSING IMPLEMENTATION OF THE LEAST SQUARES METHOD OF OPTFLOW
In the image processing implementation, g(x,y,t) and Gpq(x,y) refer to a gray value image sequence and images, respectively, with real numbers as pixel values. The OptFlow is calculated from the g(x,y,t), as illustrated in Fig. 7. First, separate copies of the image sequence are differentiated in x, y, and t directions (∂g(x,y,0)/∂x, (∂g(x,y,0)/∂y and (∂g(x,y,0)/∂t|t=0, respectively; considering a three-dimensional x, y, t data set). The differentiation was performed by kernel convolution, using simple ([1,−1]) or second polynomial, first derivative order Savitzky-Golay kernels (51). These kernels, due to their additional smoothing effects, perform well in noisy data sets. The width of these kernels was w = 3, 5, or 7 for both spatial and temporal differentiation ([0.5, 0, −0.5], [0.2, 0.1, 0, −0.1, −0.2] or [0.107, 0.071, 0.036, 0, −0.036, −0.071, −0.107]). Spatially differentiated images were also smoothed in the direction perpendicular to the differentiation using a wsm = 5 or 7 width Savitzky-Golay nondifferentiating smoothing kernel ([−0.0857, 0.343, 0.486, 0.343, −0.0857] or [−0.0952, 0.143, 0.286, 0.333, 0.283, 0.143, −0.0952]). In practice a rectangular, combined differentiation-smooth kernel was used, which was obtained by the outer product of the differentiation and smooth kernels. Whereas ∂g/∂x|t=0 was a result of calculation from 2 to 7 image planes (equal to wt), ∂g/∂x and ∂g/∂y were calculated from the image corresponding to the middle (0) of the symmetric (3, 5, and 7 width) temporal differentiation kernels. The asymmetric, simple wt = 2 ([1,−1]) temporal differentiation kernel resulted in biased OptFlow. Therefore, when using an asymmetric kernel, the OptFlow is calculated for spatial derivatives of both frames and then averaged (wt = 2avg), whereas constraint masks (see below) are merged with logical AND.
Gpq images were calculated according to Eq. 3 by multiplication of differentiated images pixel by pixel. The operator 〈〉 was implemented as kernel convolution, using a kernel of 3 × 3, 5 × 5, or a 7 × 7 matrix of ones, denoted as the aperture kernel (wap = 3, 5, or 7 and N = 9, 25, or 49, respectively). This was followed by calculation of f by Eq. 2 performing image arithmetics with the Gpq images, pixel by pixel.
APPENDIX B: MODEL MITOCHONDRIA FOR THE DEMONSTRATION OF THE OPTFLOW TECHNIQUE
Images modeling fluorescent mitochondria as fluorescent spheres and capsule bodies were generated by Mathematica 5.2 as previously described (24). Briefly, zero length (l) mitochondria were drawn as spheres of given radius (r), or elongated mitochondria as l-long r-radius cylinders capped with r-radius half spheres on both ends; 64 × 64 pixels images were generated by considering that the spheres and capsule bodies are solid and fluorescent, and the projected fluorescence intensity on the image was calculated using bilinear interpolation to simulate diameters and positions with subpixel accuracy. OptFlow was calculated from the model images with the same algorithm used for processing fluorescence microscopy data. Image noise corresponding to photon shot noise typical at low intensity fluorescence microscopic image acquisition was simulated in the model images. Model images were generated with intensity values corresponding to photo electron counts. Then, each pixel was replaced with a random number, which followed Poisson distribution with an expected value of the photo electron count. The dark current and readout noise of the camera was not included into this model.
APPENDIX C: MASKING AND IMAGE SEGMENTATION FOR SINGLE MITOCHONDRION MEASUREMENTS
Image acquisition of TMRM and mito-roGFP1 required different dichroic mirrors and emission filters leading to a shift of images. Therefore, TMRM images were first corrected by subpixel alignment using affin transformation with bilinear interpolation. Local background-subtracted (23) images were scaled between 50 and 99.2 percentiles for dendrites or between 85 and 99.99 percentiles for axonal mitochondria at a γvalue of 1.2 to enhance dim details. Only the region of interest of the measurement (the dendritic structure of the neuron, avoiding the soma or other nearby cells) was processed. The scaled and γcorrected image (and not the fluorescence intensity data) was binarized at the optimal threshold calculated by Otsu's method (52) frame by frame. To remove speckle-like noise from the binarized images, only those pixels which had at least 7 “1”-s among the surrounding 48 pixels were kept as “1”. In practice this was done by kernel filtering a copy of the binarized image (considering zeros and ones as grayscale values) with a 7 × 7 kernel of ones, and thresholding the filtered image at 8. This latter image was multiplied pixel by pixel with the original binarized image. This resulted in the elimination of objects <8 pixels. Segmentation was done by separating spatially contiguous objects >7 and <500 pixels.
To avoid errors in fluorescence intensity readout due to dislocation of faster moving mitochondria between the acquisition of different fluorescence channels (TMRM, roGFP 480 nm, and roGFP 438 nm), both roGFP 480 nm and 438 nm images were binarized and segmented (see also Fig. 12 A). The roGFP 438 nm intensity was measured in the first frame of the short image sequence recorded for OptFlow calculation. To read out TMRM fluorescence, the segmented roGFP 480 nm channel was used, because the recording of this channel followed the TMRM channel closest in time. The roGFP fluorescence intensities were determined using their own segmented images. Next, a new set of images was generated by substituting the measured mean fluorescence intensity value into each pixel of the given segment. Finally, to log fluorescence intensities and velocities for each individual mitochondrion, the same minimum intensity projected, binarized, and segmented image was used as the one used for masking the OptFlow. Therefore, when a segment corresponding to a moving mitochondrion maintains spatial overlap between the binarized, segmented images of the different acquisition channels, the final intensity readout reflects true mean intensity regardless of the extent of the dislocation and the overlap. Cases where multiple segments of the TMRM, roGFP 480 nm, or roGFP 438 nm were overlapping with the minimum intensity projected readout segment were rejected based on the increased variance.
APPENDIX D: ESTIMATION OF PHOTON SHOT NOISE
The noise characteristics of the camera were assessed by acquiring images of evenly illuminated background at different intensities (Fig. 3 in the Supplementary Material, Data S1). First, the mean intensity measured at zero illumination (camera offset) was subtracted from all images (68 in our case). The  corresponds to the σ2 of pixel values at zero illumination (3.3 in our case).
corresponds to the σ2 of pixel values at zero illumination (3.3 in our case).  also contains noise arising from the dark current of the camera; however, it was independent of exposure times used in this study (100–200 ms), suggesting negligible contribution. 1/AD was determined as the slope of intensity versus variance diagram (0.37 in our case).
also contains noise arising from the dark current of the camera; however, it was independent of exposure times used in this study (100–200 ms), suggesting negligible contribution. 1/AD was determined as the slope of intensity versus variance diagram (0.37 in our case).
The mean-square of differentiated noise equals the σ2 of the noise (for pixels and frames as space and time units, respectively). When using kernel convolution for differentiation, the σ2 of the differentiated noise is multiplied by the square sum of the convolution kernel (∑ ker2). The σ2 of differentiated photon shot noise is estimated by Eq. 9 based on the proportionality defined in Eq. 4. Equation 9 gives  for each neighborhood from the offset subtracted raw fluorescence intensities (g(x,y)) and corrects for the effect of kernel filtering and summing within the aperture:
for each neighborhood from the offset subtracted raw fluorescence intensities (g(x,y)) and corrects for the effect of kernel filtering and summing within the aperture:
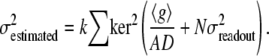 |
(9) |
 reflects the summed σ2 within the aperture to accommodate to the 〈〉 operator of Eq. 3. In Eq. 9, the resultant
reflects the summed σ2 within the aperture to accommodate to the 〈〉 operator of Eq. 3. In Eq. 9, the resultant  (x,y) is an image and N is the number of pixels in the aperture. To tune the noise elimination, an empirical factor (k) was used. Different k factors and
(x,y) is an image and N is the number of pixels in the aperture. To tune the noise elimination, an empirical factor (k) was used. Different k factors and  values were used to calculate
values were used to calculate  in the different criteria (Eqs. 5, 7, and 8).
in the different criteria (Eqs. 5, 7, and 8).
APPENDIX E: IMAGE PROCESSING IMPLEMENTATION OF THE CONSTRAINT NO. 2 OF EQ. 2 FOR NOISY IMAGES
It is considered that if all gradient vectors (∇g) of the neighborhood are parallel, then the sum of the vector products of the gradient vectors multiplied with their sum vector is zero in the absence of noise (Eq. 8). To avoid zero vector sum over point symmetric objects, the absolute value of the vector components was taken before computation. The numerator of Eq. 8 left side (Eq. 10) cannot be generalized for image processing, because 〈|∇g|〉 should to be multiplied by each |∇g| of the same neighborhood, whereas, if 〈〉 is implemented as kernel convolution, a distinct local 〈|∇g|〉 is multiplied by each |∇g|. In other words, 〈〉s must not be embedded into each other. Therefore, we have expanded the left side of Eq. 10 and distributed the 〈〉. The right side of Eq. 11 contains only arithmetics between 〈〉 of gradient images; therefore, the sum vector and the products is calculated from the same neighborhood. This latter formula can be used for image processing as a combination of kernel convolutions and image arithmetics:
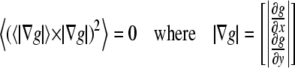 |
(10) |
 |
(11) |
APPENDIX F: CALCULATION OF FLUORESCENCE INTENSITY AT THE ISOSBESTIC WAVELENGTH OF MITO-ROGFP1
The fluorescence corresponding to the isosbestic wavelength (FroGFPiso) was calculated by the linear combination of fluorescence intensities F438 and F480 acquired by the 438 nm and 480 nm excitations of mito-roGFP1, respectively (Eq. 12):
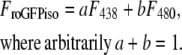 |
(12) |
Here we introduce what we consider a novel method for calculation of coefficients a and b from a short time lapse experiment, where the fluorescence ratio R = F438/F480 is changed by oxidation or reduction of the roGFP, and FroGFPiso does not have to be necessarily constant. We assume that the time course of R and FroGFPiso are not correlated in time, thus their cross correlation at 0 time lag or their Pearson's correlation coefficient is zero (Eq. 13). Substituting Eq. 12 into Eq. 13, we obtain the solution Eq. 14 for a:
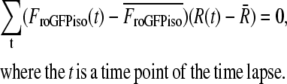 |
(13) |
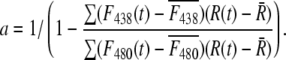 |
(14) |
In our microscopic settings. a and b were 1.37 and −0.37, respectively.
APPENDIX G: CALCULATION OF PROPAGATED ERROR FROM PHOTON SHOT NOISE
In low light level fluorescence imaging, the determination of mean fluorescence intensities in small regions consisting of only a few tens of pixels is burdened by the photon shot noise. To assess the effect of photon shot noise on the calibrated (millivolt) values of Δ| ΔΨm| and ΔEroGFP, error propagation (24) of variances was performed (Eq. 15). To this end 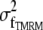 and
and 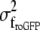 were obtained by substitution of Eqs. 16 and 18 or Eqs. 17 and 18 into Eq. 15:
were obtained by substitution of Eqs. 16 and 18 or Eqs. 17 and 18 into Eq. 15:
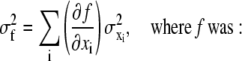 |
(15) |
 |
(16) |
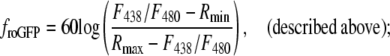 |
(17) |
and xi denotes FTMRM, F480 or F438 fluorescence intensities
 |
(18) |
where xi_original is the intensity without background removal and n is the number of pixels in the segment; values correspond to Fig. 3 in the Supplementary Material, Data S1.
Editor: David W. Piston.
References
- 1.Hollenbeck, P. J. 2005. Mitochondria and neurotransmission: evacuating the synapse. Neuron. 47:331–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mironov, S. L. 2007. ADP regulates movements of mitochondria in neurons. Biophys. J. 92:2944–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller, K. E., and M. P. Sheetz. 2004. Axonal mitochondrial transport and potential are correlated. J. Cell Sci. 117:2791–2804. [DOI] [PubMed] [Google Scholar]
- 4.Muller, M., S. L. Mironov, M. V. Ivannikov, J. Schmidt, and D. W. Richter. 2005. Mitochondrial organization and motility probed by two-photon microscopy in cultured mouse brainstem neurons. Exp. Cell Res. 303:114–127. [DOI] [PubMed] [Google Scholar]
- 5.Wang, G. J., J. G. Jackson, and S. A. Thayer. 2003. Altered distribution of mitochondria impairs calcium homeostasis in rat hippocampal neurons in culture. J. Neurochem. 87:85–94. [DOI] [PubMed] [Google Scholar]
- 6.Yi, M., D. Weaver, and G. Hajnoczky. 2004. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J. Cell Biol. 167:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono, T., K. Isobe, K. Nakada, and J. I. Hayashi. 2001. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 28:272–275. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., J. M. McCaffery, and D. C. Chan. 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 130:548–562. [DOI] [PubMed] [Google Scholar]
- 9.Scherz-Shouval, R., and Z. Elazar. 2007. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 17:422–427. [DOI] [PubMed] [Google Scholar]
- 10.Kundu, M., and C. B. Thompson. 2005. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell Death Differ. 12(Suppl. 2):1484–1489. [DOI] [PubMed] [Google Scholar]
- 11.Gunawardena, S., L. S. Her, R. G. Brusch, R. A. Laymon, I. R. Niesman, B. Gordesky-Gold, L. Sintasath, N. M. Bonini, and L. S. Goldstein. 2003. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 40:25–40. [DOI] [PubMed] [Google Scholar]
- 12.Trushina, E., R. B. Dyer, J. D. Badger, D. Ure, L. Eide, D. D. Tran, B. T. Vrieze, V. Legendre-Guillemin, P. S. McPherson, B. S. Mandavilli, H. B. Van, S. Zeitlin, M. McNiven, R. Aebersold, M. Hayden, J. E. Parisi, E. Seeberg, I. Dragatsis, K. Doyle, A. Bender, C. Chacko, and C. T. McMurray. 2004. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell. Biol. 24:8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, D. T., G. L. Rintoul, S. Pandipati, and I. J. Reynolds. 2006. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis. 22:388–400. [DOI] [PubMed] [Google Scholar]
- 14.Rintoul, G. L., A. J. Filiano, J. B. Brocard, G. J. Kress, and I. J. Reynolds. 2003. Glutamate decreases mitochondrial size and movement in primary forebrain neurons. J. Neurosci. 23:7881–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds, I. J., L. M. Malaiyandi, M. Coash, and G. L. Rintoul. 2004. Mitochondrial trafficking in neurons: A key variable in neurodegeneration? J. Bioenerg. Biomembr. 36:283–286. [DOI] [PubMed] [Google Scholar]
- 16.Pivovarova, N. B., H. V. Nguyen, C. A. Winters, C. A. Brantner, C. L. Smith, and S. B. Andrews. 2004. Excitotoxic calcium overload in a subpopulation of mitochondria triggers delayed death in hippocampal neurons. J. Neurosci. 24:5611–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, H., A. Chomyn, and D. C. Chan. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280:26185–26192. [DOI] [PubMed] [Google Scholar]
- 18.Kuznetsov, A. V., J. Troppmair, R. Sucher, M. Hermann, V. Saks, and R. Margreiter. 2006. Mitochondrial subpopulations and heterogeneity revealed by confocal imaging: possible physiological role? Biochim. Biophys. Acta. 1757:686–691. [DOI] [PubMed] [Google Scholar]
- 19.Wikstrom, J. D., S. M. Katzman, H. Mohamed, G. Twig, S. A. Graf, E. Heart, A. J. Molina, B. E. Corkey, L. M. de Vargas, N. N. Danial, S. Collins, and O. S. Shirihai. 2007. β-Cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes. 56:2569–2578. [DOI] [PubMed] [Google Scholar]
- 20.Jahne, B. 1997. Digital Image Processing. Springer-Verlag, Berlin.
- 21.Dooley, C. M., T. M. Dore, G. T. Hanson, W. C. Jackson, S. J. Remington, and R. Y. Tsien. 2004. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 279:22284–22293. [DOI] [PubMed] [Google Scholar]
- 22.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916. [DOI] [PubMed] [Google Scholar]
- 23.Gerencser, A. A., and V. Adam-Vizi. 2001. Selective, high-resolution fluorescence imaging of mitochondrial Ca2+ concentration. Cell Calcium. 30:311–321. [DOI] [PubMed] [Google Scholar]
- 24.Gerencser, A. A., J. Doczi, B. Töröcsik, E. Bossy-Wetzel, and V. Adam-Vizi. 2008. Mitochondrial swelling measurement in situ by optimized spatial filtering: astrocyte-neuron differences. Biophys. J. 95:2583–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholls, D. G. 2006. Simultaneous monitoring of ionophore- and inhibitor-mediated plasma and mitochondrial membrane potential changes in cultured neurons. J. Biol. Chem. 281:14864–14874. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls, D. G., and M. W. Ward. 2000. Mitochondrial membrane potential and cell death: mortality and millivolts. Trends Neurosci. 23:166–174. [DOI] [PubMed] [Google Scholar]
- 27.Loew, L. M., R. A. Tuft, W. Carrington, and F. S. Fay. 1993. Imaging in five dimensions: time-dependent membrane potentials in individual mitochondria. Biophys. J. 65:2396–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris, R. L., and P. J. Hollenbeck. 1995. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J. Cell Biol. 131:1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ligon, L. A., and O. Steward. 2000. Role of microtubules and actin filaments in the movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J. Comp. Neurol. 427:351–361. [DOI] [PubMed] [Google Scholar]
- 30.Diaz, G., A. M. Falchi, F. Gremo, R. Isola, and A. Diana. 2000. Homogeneous longitudinal profiles and synchronous fluctuations of mitochondrial transmembrane potential. FEBS Lett. 475:218–224. [DOI] [PubMed] [Google Scholar]
- 31.Gerencser, A. A., and V. Adam-Vizi. 2005. Mitochondrial Ca2+ dynamics reveals limited intramitochondrial Ca2+ diffusion. Biophys. J. 88:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatsuta, T., and T. Langer. 2008. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 27:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, H., and D. C. Chan. 2006. Critical dependence of neurons on mitochondrial dynamics. Curr. Opin. Cell Biol. 18:453–459. [DOI] [PubMed] [Google Scholar]
- 34.DiMauro, S. 2004. Mitochondrial diseases. Biochim. Biophys. Acta Bio-Energetics. 1658:80–88. [DOI] [PubMed] [Google Scholar]
- 35.Guo, X., G. T. Macleod, A. Wellington, F. Hu, S. Panchumarthi, M. Schoenfield, L. Marin, M. P. Charlton, H. L. Atwood, and K. E. Zinsmaier. 2005. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 47:379–393. [DOI] [PubMed] [Google Scholar]
- 36.Morris, R. L., and P. J. Hollenbeck. 1993. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J. Cell Sci. 104:917–927. [DOI] [PubMed] [Google Scholar]
- 37.Overly, C. C., H. I. Rieff, and P. J. Hollenbeck. 1996. Organelle motility and metabolism in axons vs. dendrites of cultured hippocampal neurons. J. Cell Sci. 109:971–980. [DOI] [PubMed] [Google Scholar]
- 38.Pilling, A. D., D. Horiuchi, C. M. Lively, and W. M. Saxton. 2006. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 17:2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez-Mateos, E. M., C. Gonzalez-Billault, H. N. Dawson, M. P. Vitek, and J. Avila. 2006. Role of MAP1B in axonal retrograde transport of mitochondria. Biochem. J. 397:53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belzacq, A. S., H. L. Vieira, F. Verrier, G. Vandecasteele, I. Cohen, M. C. Prevost, E. Larquet, F. Pariselli, P. X. Petit, A. Kahn, R. Rizzuto, C. Brenner, and G. Kroemer. 2003. Bcl-2 and Bax modulate adenine nucleotide translocase activity. Cancer Res. 63:541–546. [PubMed] [Google Scholar]
- 41.Chada, S. R., and P. J. Hollenbeck. 2004. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr. Biol. 14:1272–1276. [DOI] [PubMed] [Google Scholar]
- 42.Racine, V., M. Sachse, J. Salamero, V. Fraisier, A. Trubuil, and J. B. Sibarita. 2007. Visualization and quantification of vesicle trafficking on a three-dimensional cytoskeleton network in living cells. J. Microsc. 225:214–228. [DOI] [PubMed] [Google Scholar]
- 43.Miller, K. E., and M. P. Sheetz. 2006. Direct evidence for coherent low velocity axonal transport of mitochondria. J. Cell Biol. 173:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popov, V., N. I. Medvedev, H. A. Davies, and M. G. Stewart. 2005. Mitochondria form a filamentous reticular network in hippocampal dendrites but are present as discrete bodies in axons: a three-dimensional ultrastructural study. J. Comp. Neurol. 492:50–65. [DOI] [PubMed] [Google Scholar]
- 45.Barsoum, M. J., H. Yuan, A. A. Gerencser, G. Liot, Y. Kushnareva, S. Graber, I. Kovacs, W. D. Lee, J. Waggoner, J. Cui, A. D. White, B. Bossy, J. C. Martinou, R. J. Youle, S. A. Lipton, M. H. Ellisman, G. A. Perkins, and E. Bossy-Wetzel. 2006. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 25:3900–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang, D. T., A. S. Honick, and I. J. Reynolds. 2006. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J. Neurosci. 26:7035–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fridberger, A., J. Widengren, and J. Boutet de Monvel. 2004. Measuring hearing organ vibration patterns with confocal microscopy and optical flow. Biophys. J. 86:535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoccolan, D., A. Giachetti, and V. Torre. 2001. The use of optical flow to characterize muscle contraction. J. Neurosci. Methods. 110:65–80. [DOI] [PubMed] [Google Scholar]
- 49.Kim, I., S. Rodriguez-Enriquez, and J. J. Lemasters. 2007. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462:245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twig, G., A. Elorza, A. J. Molina, H. Mohamed, J. D. Wikstrom, G. Walzer, L. Stiles, S. E. Haigh, S. Katz, G. Las, J. Alroy, M. Wu, B. F. Py, J. Yuan, J. T. Deeney, B. E. Corkey, and O. S. Shirihai. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Press, W. H., and S. Teukolsky. 1990. Savitzky-Golay smoothing filters. Comput. Phys. 4:669. [Google Scholar]
- 52.Otsu, N. 1979. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9:66–66. [Google Scholar]