Abstract
To understand the effect of hydration on protein dynamics, inelastic neutron-scattering experiments were performed on staphylococcal nuclease samples at differing hydration levels: dehydrated, partially hydrated, and hydrated. At cryogenic temperatures, hydration affected the collective motions with energies lower than 5 meV, whereas the high-energy localized motions were independent of hydration. The prominent change was a shift of boson peak toward higher energy by hydration, suggesting a hardening of harmonic potential at local minima on the energy landscape. The 240 K transition was observed only for the hydrated protein. Significant quasielastic scattering at 300 K was observed only for the hydrated sample, indicating that the origin of the transition is the motion activated by hydration water. The neutron-scattering profile of the partially hydrated sample was quite similar to that of the hydrated sample at 100 K and 200 K, whereas it was close to the dehydrated sample at 300 K, indicating that partial hydration is sufficient to affect the harmonic nature of protein dynamics, and that there is a threshold hydration level to activate anharmonic motions. Thus, hydration water controls both harmonic and anharmonic protein dynamics by differing means.
INTRODUCTION
Proteins, molecular machines that contribute to almost all living phenomena, primarily function in an aqueous milieu at ambient temperatures. It has been reported that proteins cannot function at cryogenic temperatures or at low hydration levels (1,2), which strongly implies that both thermal fluctuations and hydration affect protein function. To understand the molecular mechanisms that underlie cellular biology, it is essential to examine protein dynamics in aqueous environments. Low-temperature protein crystallography was used to show that hydration water molecules encircle a protein to form a hydrogen-bond network (3). Although many experimental and theoretical studies were performed to reveal the relationship between hydration water and protein dynamics (2,4–12), a complete understanding of how hydration water affects protein dynamics has yet to be achieved.
Inelastic neutron scattering (INS) allows protein dynamics to be examined at picosecond to nanosecond timescales (13). This technique was successfully used to examine characteristic protein dynamic properties (2,5,6,8,13–15), including the protein boson peak and dynamical transitions. The boson peak is characterized by excess densities of states G(ω) from the Debye model, in which G(ω) is proportional to ω2. The protein boson peak is a broad peak observed in a low-frequency region of INS and Raman spectra (16,17). This peak is also commonly observed in synthetic polymers and glassy materials. The origin of the boson peak is a subject of debate in the field of condensed-matter physics. A molecular dynamics (MD) simulation showed that the protein backbone, the nonpolar side chains in the interior of the protein, and the polar side chains exposed to a solvent equally contribute to the boson peak (18). Experimentally, the frequency of a boson peak depends on the protein molecular weight (16). These studies suggest that the origin of the boson peak is closely correlated with the motions that extend over the whole protein molecule. The boson peak is suggested to reflect water-coupled librations of polar side chains that are depressed in the hydrated protein by strong intermolecular hydrogen bonding (5). A more recent MD simulation demonstrated that the protein boson peak originates in the energy landscape of a protein with rugged multiple minima, and that the ruggedness is affected by hydration (7). That study implied that the fine structure of the energy landscape should be investigated to characterize the nature of the boson peak.
Another important property of protein dynamics is the dynamical transition, which is also dependent on hydration. The averaged mean-square displacement (MSD) of the motion was measured by experimental techniques such as Mossbauer spectroscopy (10), x-ray crystallography (1), and neutron scattering at various temperatures (6,8). As the temperature increases, deviation from linearity of the temperature dependence of the MSD is observed, suggesting the onset of anharmonic motions. A major dynamical transition at 180–240 K was observed for hydrated proteins (6,8,9). The existence of another transition at ∼150 K was also identified (2,8). The anharmonic motion responsible for the 150 K transition was independent of hydration, and the origin of the anharmonic motion was assigned to the methyl rotation (2). These dynamical transitions should be accompanied by the appearance of quasielastic scattering attributable to the relaxation process in forms such as rotational jumps between defined sites, or to diffusion in a confined volume (6).
In this study, we investigated the effects of hydration on the dynamics of a globular protein, staphylococcal nuclease (SNase). Solvent and temperature are crucial variables for determining the protein dynamics. We found that hydration affects both harmonic and anharmonic properties of the protein dynamics. At cryogenic temperatures, a protein is trapped in a local energy minimum, and behaves as a harmonic oscillator. We observed that the boson peak position shifts toward higher energy via hydration. The slope of temperature dependence of the MSD in the low-temperature region is decreased by hydration. These results are well-explained by the increase in the force constant, indicating that hydration water causes a protein to harden at cryogenic temperatures. Hydration water also affects the major dynamical transition. The major transition was evident for the hydrated sample only, indicating that the anharmonic motions responsible for the transition are activated by hydration water. Hydration water causes a protein to soften at room temperature. Thus, hydration water controls both the harmonic and anharmonic protein dynamics.
MATERIALS AND METHODS
Sample preparation
The SNase was expressed in Escherichia coli and purified as described previously (15). The purified protein was dialyzed against D2O and lyophilized. The hydration level of the dehydrated SNase was 0.12 g D2O/g protein, as estimated by thermogravimetry. Two higher-hydration samples, partially hydrated and hydrated SNase, were prepared by equilibration of the dehydrated sample with a saturated NaBr and KCl solution of D2O. The hydration levels of the partially hydrated and hydrated SNase were 0.29 and 0.44 g D2O/g protein, respectively. The hydration level of the dehydrated SNase roughly corresponds to the level estimated by considering the number of crystal water molecules, and that of the hydrated SNase is consistent with the estimated total number of water molecules per protein in the crystal.
Neutron-scattering experiment
Neutron-scattering measurements were performed with LAM-40 and LAM-D inverted geometry time-of-flight (TOF) spectrometers installed at the pulsed spallation neutron source at the High Energy Accelerator Research Organization (Tsukuba, Japan). The energy resolution and energy window for LAM-40 are ∼200 μeV and <15 meV, respectively. The scattering angles were at 16° intervals between 16.3° and 112.3°. The energy resolution of LAM-D was ∼350 μeV, and its energy window was <300 meV. The neutron-scattering measurements were performed between 10 K and 300 K on LAM-40, and at 20 K on LAM-D. The observed TOF spectrum was converted to the dynamical structure factor 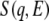 after correction for the background contribution, the counterefficiency, and the incident neutron spectrum. Here, q and E correspond to the momentum and energy transfers, respectively, between incident neutron and sample. The data analysis was performed without correcting for multiple scattering because the sample transmission value was relatively large, at ∼90%. Because both the hydrated and dehydrated samples contained the same protein mass,
after correction for the background contribution, the counterefficiency, and the incident neutron spectrum. Here, q and E correspond to the momentum and energy transfers, respectively, between incident neutron and sample. The data analysis was performed without correcting for multiple scattering because the sample transmission value was relatively large, at ∼90%. Because both the hydrated and dehydrated samples contained the same protein mass, 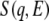 values could be compared without scaling. Incoherent neutron scattering by nonexchangeable hydrogen atoms of SNase provides information on the global dynamics of the protein (13).
values could be compared without scaling. Incoherent neutron scattering by nonexchangeable hydrogen atoms of SNase provides information on the global dynamics of the protein (13).
Analysis of quasielastic scattering
As demonstrated previously (19), quasielastic scattering was estimated as:
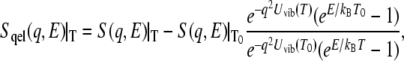 |
(1) |
where 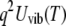 is the vibrational Debye-Waller factor, and T0 is a reference temperature at which quasielastic scattering does not appear. In this study, T0 = 100 K. The estimated quasielastic scattering spectra were directly fitted with a single Lorentzian model function convoluted with the resolution function of the spectrometer, which was obtained from vanadium measurements.
is the vibrational Debye-Waller factor, and T0 is a reference temperature at which quasielastic scattering does not appear. In this study, T0 = 100 K. The estimated quasielastic scattering spectra were directly fitted with a single Lorentzian model function convoluted with the resolution function of the spectrometer, which was obtained from vanadium measurements.
RESULTS AND DISCUSSION
Effect of hydration on harmonic dynamics
We used the lyophilized powder of purified SNase as the dehydrated sample, with a hydration level of 0.12 g D2O/g protein as confirmed by thermogravimetry. The hydrated sample was prepared by equilibration with saturated KCl-D2O solution. After incubation, the weight of the powder increased by 0.44 g D2O/g protein. Fig. 1 a shows the neutron spectra of the dehydrated and hydrated SNase at 20 K in a wide energy region observed with the LAM-D spectrometer. The spectrum shown in Fig. 1 is essentially identical to the spectrum at 25 K observed with TOSCA at ISIS (14). The peaks measured with TOSCA were assigned using normal mode analysis (14). The peaks at ∼29.0 meV and 160.5 meV were assigned to CH3 torsion and CH2 twisting, respectively (14). Furthermore, vibrational modes higher than 90 meV were determined to be contributed by alkyl protons. The modes between 50 meV and 90 meV were mainly assigned to skeletal angle bending and dihedral displacement. The ratio of the dehydrated to the hydrated protein spectrum is given in the inset of Fig. 1 a. The high-energy vibrational modes that are attributable to covalent bonds were not markedly affected by hydration, suggesting that these modes are rather localized and not damped significantly by hydration friction. We observed small differences between 40 meV and 65 meV, which are likely due to the bending motion of hydrated heavy water on the protein (14). The inset in Fig. 1 a shows that hydration strongly affects the modes below 5 meV. The harmonic modes of the protein in the low-energy range were highly affected by hydration. According to normal mode analysis, low-energy modes at a few meV are collective motions (20). The hydration-dependent INS spectrum demonstrated that the collective motions of protein are highly coupled with hydration.
FIGURE 1.
INS spectra of dehydrated and hydrated SNase obtained by (a) LAM-D at 20 K, and those obtained by (b) LAM-40 at 100 K. (a) Data representing the hydrated sample are shifted along the ordinate by 0.01 for the sake of clarity. (a, inset) Ratio of dehydrated protein spectrum to hydrated spectrum.
Fig. 1 b shows the INS spectra in the lower energy range observed with the LAM-40 spectrometer at 100 K. A boson peak was observed for both the hydrated and dehydrated protein samples. For the hydrated sample, the scattering intensity below ∼5 meV is depressed, and then the boson peak position shifts from 3 to 4 meV by hydration. A hydration-induced shift of boson peak was observed for other proteins (5,9,21–24). The q dependence of the integrated intensity of the boson peak shows a q2 trend (25), indicating that the protein dynamics in the corresponding energy range are harmonic. This shows that protein motion is trapped in a local minimum on the energy landscape at this low temperature. Therefore, the boson peak position reflects the property of harmonic potential. If the boson peak comes from harmonic vibration, the peak shift toward higher energy by hydration suggests the increase of the average force constant of harmonic motions. The frequency of the boson peak is very sensitive to environmental conditions (22). The frequency-upshift of the boson peak with hydration at low temperature suggests an increase in the number and strength of protein-water hydrogen bonds (5). A computational study showed that solvent water affects the shape of the potential energy surface of the collective modes (4). The hydrogen-bond interaction should deform the harmonic potential at the local minima, causing the characteristic frequency to shift higher.
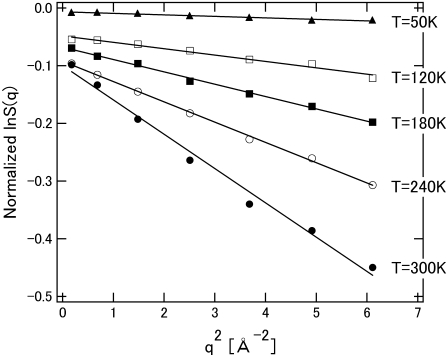
Fig. 2 shows the elastic intensity of the hydrated protein at various temperatures. The elastic intensity is summed up with the scattering within the energy resolution of the spectrometer, ∼200 μeV, corresponding to observable motions faster than ∼20 ps. The analysis of elastic incoherent neutron scattering gives the MSD of individual atoms in the timescale by Gaussian approximation as:
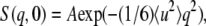 |
(2) |
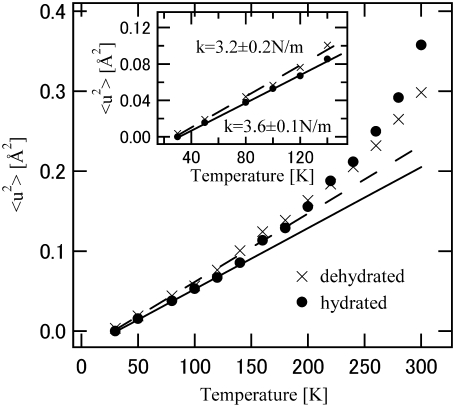
where A and 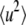 are a constant and the MSD averaged over hydrogen atoms, respectively. Fig. 3 shows the MSD of the hydrated and dehydrated samples at various temperatures. At temperatures lower than 140 K, the MSD of the hydrated protein is systematically smaller than that of the dehydrated protein. Up to ∼140 K, the MSDs of both samples increased linearly as a function of temperature (Fig. 3, inset), indicating that the protein motion is harmonic in this temperature range, regardless of hydration. The slope of the regression line of the temperature-dependent MSD gives the force constant of the harmonic potential 〈k〉 (6):
are a constant and the MSD averaged over hydrogen atoms, respectively. Fig. 3 shows the MSD of the hydrated and dehydrated samples at various temperatures. At temperatures lower than 140 K, the MSD of the hydrated protein is systematically smaller than that of the dehydrated protein. Up to ∼140 K, the MSDs of both samples increased linearly as a function of temperature (Fig. 3, inset), indicating that the protein motion is harmonic in this temperature range, regardless of hydration. The slope of the regression line of the temperature-dependent MSD gives the force constant of the harmonic potential 〈k〉 (6):
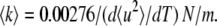 |
(3) |
FIGURE 2.
Logarithm of integrated elastic intensity versus q2 for hydrated SNase. Data, obtained using LAM-40, were normalized by using the data at 10 K. Temperatures (T) of measurements are as indicated. Only five sets of data are shown. Errors are indicated by the size of data points. Solid curves represent Gaussian approximation fittings of data points. Their slopes give the corresponding MSD.
FIGURE 3.
Temperature dependence of MSDs of dehydrated (×) and hydrated (•) SNase. Errors are indicated by the size of data points. (Inset) Low-temperature region where force constants were estimated from regression lines. Dashed and solid lines represent regression lines for dehydrated and hydrated SNase, respectively. The fitting regions are below 140 K.
The amplitude of fluctuation is averaged over all motions, but theoretically a small number of low-frequency modes dominantly contribute to the average amplitude (20). Therefore, the force constant should describe the nature of the low-frequency modes. The force constants of the hydrated and dehydrated proteins are estimated to be 3.6 ± 0.1 N/m and 3.2 ± 0.2 N/m, respectively. This is consistent with the boson peak shift. The increase of the force constant by hydration indicates that the energy potential of the hydrated protein is more resilient. The larger force constant in the hydrated state indicates that the parabolic energy surface at the local minima is sharpened by hydration in the low-frequency modes. In the hydrated state, hydration water encircles the protein, and hydrogen bonds are formed at the protein-water interface (3). The interaction restricts protein fluctuation, indicating the hardening of the potential surface. The MD simulation demonstrated that hydration makes the energy landscape rugged, that protein fluctuation is restricted in the small space of a local minimum by hydration, and that the boson peak originates in the rugged energy landscape (7). Glycerol and trehalose also cause protein to harden at a low temperature (22). This suggests that the hydrogen-bond interaction of glassy solvent with the embedded protein affects the flexibility of the protein structure. In this sense, the origin of the protein boson peak is the hydrogen bond-coupled low-energy mode. A computational study showed that the MD simulation for a single protein molecule in vacuo did not result in a boson peak (7). The MD simulation in crystal, however, quantitatively reproduced the boson peak, because the crystal situation mimics the experimental powder sample condition (18,26–28). This indicates that the boson peak is also sensitive to the protein-protein interaction. Thus, protein environments such as the neighbor proteins, as well as hydration, affect the low-frequency collective modes. Such interactions deform the energy landscape to become more rugged, and such effects essentially produce the boson peak observed.
Effect of hydration on anharmonic dynamics
Fig. 3 shows the temperature dependence of MSDs in the hydrated and dehydrated SNase samples. Regardless of hydration, the deviation from the harmonic line becomes remarkable at ∼140 K, indicating the onset of certain anharmonic motions. This anharmonicity was attributed to the β-process, implying that hydration-independent anharmonic dynamics are attributable to local motions of atoms (8). One study suggested that the rotation of methyl groups contributes to this transition, and the averaged activation energy of these rotations is ∼16.6 kJ/mol (2). The estimated timescale of the rotational transition is ∼280 ns at 140 K. This motion at this timescale is too slow to be resolved using the LAM-40 spectrometer, which resolves motions faster than 20 ps. Timescales of motions depend on the locations of the protons (2). The methyl rotations, with lower energy barriers, may contribute to the transition, because they have faster rotational relaxation. Other possible origins may include CH2-twisting and CH-bending motions. Therefore, the faster local dynamics in the polypeptide chain should contribute to the observed transition. It is reasonable that these motions are unaffected by hydration water.
Another dynamical transition was observed at ∼240 K for the hydrated samples. The significant increase in MSD for the hydrated sample indicates the activation of anharmonic dynamics by hydration. Hydration water changes the energy landscapes of the low-frequency modes because of the hydrogen-bond network interaction between protein and water (4). This suggests that the hydration-dependent transition should be reasonably explained by hydration-coupled low-frequency dynamics. Based on the experiments, it was deduced that librational modes of side chains lead to the hydration-related neutron spectral features (5).
The activation of anharmonicity should be reflected in the appearance of quasielastic scattering. To examine temperature-dependent dynamics further, inelastic scattering spectra in the low-energy region were examined in hydrated and dehydrated samples at 100 K, 200 K, and 300 K. Fig. 4 shows the dynamical structure factor 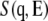 at 100 K, 200 K, and 300 K. The spectrum at 100 K is equivalent to the curve shown in Fig. 1 b. Each spectrum is scaled by the Bose factor at 300 K. Therefore, the excess intensity over the spectrum at 100 K represents the contribution from quasielastic scattering (Eq. 1). A slight increase in quasielastic scattering was evident at 200 K for both the hydrated and dehydrated samples. At 300 K, however, a prominent increase in quasielastic scattering was observed only for the hydrated protein. This corresponds to the onset of anharmonicity responsible for the transition at ∼240 K. Fig. 5 shows the temperature dependence of quasielastic scattering intensity, defined as the area of excess intensity in the spectrum between 1 meV and 4 meV. The quasielastic intensities for both states increased when the temperature was increased. The quasielastic intensities for the hydrated and dehydrated protein were almost identical up to ∼200 K. This suggests the presence of anharmonic motions that were not markedly affected by hydration below 200 K (2). The anharmonicity is responsible for the transition at ∼140 K commonly observed for hydrated and dehydrated proteins. Fig. 5 shows that the hydrated sample produced additional, strong quasielastic scattering above 200 K. This increase in quasielastic scattering indicates the appearance of hydration-dependent anharmonic motions (8), which should be closely coupled to the hydration-dependent dynamical transition. The hydration-dependent dynamical transition indicates that hydration water activates the anharmonic motions, suggesting that this anharmonicity is determined by solvent water rather than by a dynamical property of a polypeptide chain.
at 100 K, 200 K, and 300 K. The spectrum at 100 K is equivalent to the curve shown in Fig. 1 b. Each spectrum is scaled by the Bose factor at 300 K. Therefore, the excess intensity over the spectrum at 100 K represents the contribution from quasielastic scattering (Eq. 1). A slight increase in quasielastic scattering was evident at 200 K for both the hydrated and dehydrated samples. At 300 K, however, a prominent increase in quasielastic scattering was observed only for the hydrated protein. This corresponds to the onset of anharmonicity responsible for the transition at ∼240 K. Fig. 5 shows the temperature dependence of quasielastic scattering intensity, defined as the area of excess intensity in the spectrum between 1 meV and 4 meV. The quasielastic intensities for both states increased when the temperature was increased. The quasielastic intensities for the hydrated and dehydrated protein were almost identical up to ∼200 K. This suggests the presence of anharmonic motions that were not markedly affected by hydration below 200 K (2). The anharmonicity is responsible for the transition at ∼140 K commonly observed for hydrated and dehydrated proteins. Fig. 5 shows that the hydrated sample produced additional, strong quasielastic scattering above 200 K. This increase in quasielastic scattering indicates the appearance of hydration-dependent anharmonic motions (8), which should be closely coupled to the hydration-dependent dynamical transition. The hydration-dependent dynamical transition indicates that hydration water activates the anharmonic motions, suggesting that this anharmonicity is determined by solvent water rather than by a dynamical property of a polypeptide chain.
FIGURE 4.
INS spectra of (a) dehydrated and (b) hydrated SNase at 100 K, 200 K, and 300 K observed with LAM-40. The sum of measured spectra over each scattering vector q was used in the analysis because the position and shape of the boson peak do not depend on q.
FIGURE 5.
Temperature dependence of integrated quasielastic intensity between 1 meV and 4 meV of dehydrated and hydrated SNase.
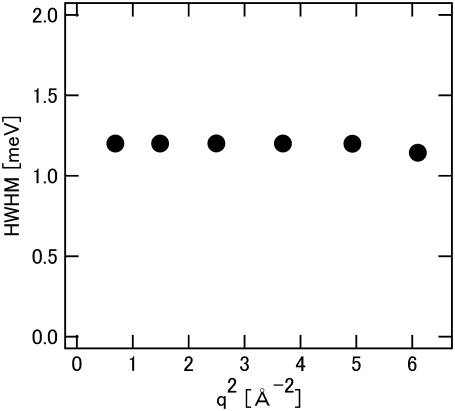
To characterize the hydration-induced dynamical transition, the quasielastic scattering of the hydrated sample estimated by Eq. 1 was analyzed. Fig. 6 shows that the width of quasielastic scattering is independent of q, indicating that this process is localized within the 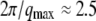 Å displacement. The q-independence suggests that the average amplitude of the motions of side chains is expected to be restricted because of the unique tertiary structure of the protein, and that the hydration-coupled collective motions of the protein contribute to the dynamical transition.
Å displacement. The q-independence suggests that the average amplitude of the motions of side chains is expected to be restricted because of the unique tertiary structure of the protein, and that the hydration-coupled collective motions of the protein contribute to the dynamical transition.
FIGURE 6.
q dependence of half-width at half-maximum (HWHM) of the Lorentzian function (quasielastic scattering) with hydrated SNase at 300 K.
Effect of hydration level
Hydration water affects both harmonic and anharmonic protein dynamics. To examine the hydration effect in detail, the inelastic spectrum of a partially hydrated sample was obtained and compared with those of hydrated and dehydrated samples. Fig. 7 shows the dynamical structure factors 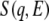 of the hydrated, partially hydrated, and dehydrated samples at 100 K, 200 K, and 300 K. The spectra for the hydrated and dehydrated samples are the same as in Figs. 1 and 4. At 100 K, the spectrum for the partially hydrated sample is quite similar to that of the hydrated sample. The boson peak shift was evident in the partially hydrated sample. The boson peak position was reported to be dependent on the hydration level of the protein (23,24), suggesting that the effect of hydration on harmonic motion is additive. The peak position for the partially hydrated sample seems to be located at a lower energy than for the hydrated samples, as also observed for the spectrum at 200 K. Within the experimental error, however, we cannot insist that the boson peak position is hydration level-dependent. It should be noted that partial hydration is sufficient to affect protein dynamics.
of the hydrated, partially hydrated, and dehydrated samples at 100 K, 200 K, and 300 K. The spectra for the hydrated and dehydrated samples are the same as in Figs. 1 and 4. At 100 K, the spectrum for the partially hydrated sample is quite similar to that of the hydrated sample. The boson peak shift was evident in the partially hydrated sample. The boson peak position was reported to be dependent on the hydration level of the protein (23,24), suggesting that the effect of hydration on harmonic motion is additive. The peak position for the partially hydrated sample seems to be located at a lower energy than for the hydrated samples, as also observed for the spectrum at 200 K. Within the experimental error, however, we cannot insist that the boson peak position is hydration level-dependent. It should be noted that partial hydration is sufficient to affect protein dynamics.
FIGURE 7.
INS spectra of dehydrated, partially hydrated, and hydrated SNase at (a) 100 K, (b) 200 K, and (c) 300 K observed with LAM-40. The sum of measured spectra over each scattering vector q was used in the analysis because the shapes of inelastic and quasielastic scattering spectra do not depend on q in this study.
At 200 K, the spectrum of the partially hydrated sample is again similar to that of the hydrated sample. However, at 300 K, the spectrum of the partially hydrated sample is essentially the same as that of the dehydrated sample. This suggests that the dynamical transition at ∼240 K is observed only for the hydrated protein. Therefore, a hydration level higher than 0.29 g D2O/g protein is required for the onset of anharmonic motions responsible for the 240 K transition, in contrast to the effect on harmonic motions at low temperatures. Hydration water affects both harmonic and anharmonic dynamics of proteins, but the origins of the effects are completely different.
The hydration environment of a protein significantly affects its dynamics. However, the hydration effects are strongly dependent on both temperature and hydration. At cryogenic temperatures, hydration stiffens protein structure because of the hydrogen-bond interaction, whereas at physiological temperatures, hydration softens the structure through the activation of anharmonic motion. At low temperatures, hydration waters are strongly bound to the hydration sites on the protein (3), and therefore protein structure becomes less flexible by hydration. As temperature increases, the hydrogen bond-breaking dynamics become the main contributors to the internal molecular flexibility. It was shown that fluctuations in the hydration shell control fast fluctuations in the protein (10). Translational hydration water dynamics drive the protein transition (11). These results indicate that, to understand protein dynamics, the hydration water dynamics should be revealed. Another interesting result (as well as an unsolved problem in this study) is that a boson peak upshift was observed even in the partially hydrated samples, but the dynamical transition was not evident. The appearance of a dynamical transition requires a hydration level of >0.29 but <0.44 g D2O/g protein hydration. This result indicates that hydration effects at cryogenic temperatures may be additive, but the dynamical transition has a threshold hydration level. The difference in hydration effects at cryogenic and physiological temperatures should be attributable to the dynamical properties of the protein hydration water, which are hydration level-dependent and temperature-dependent. The hydration water dynamics and their dynamical coupling with the protein are probably essential for protein dynamics and biological function.
CONCLUSIONS
We studied the effects of hydration on the picosecond to nanosecond dynamics of SNase in the wide temperature range from cryogenic to room temperature. At cryogenic temperature, hydration water causes a shift of boson peak position toward higher energy, which is consistent with the hardening of protein dynamics because of hydrogen-bond formations between residues on the protein surface and hydration water molecules. The hydration-dependent boson peak shift was observed for myoglobin (5) and lysozyme (5,29). On the other hand, hydration water also activated anharmonic motion to cause a dynamical transition at ∼200 K, indicating that hydration water causes a softening of protein dynamics above 200 K. This hydration-activated dynamical transition was also observed for myoglobin (8), lysozyme (2), bacteriorhodopsin (6), and α-amylase (21). Therefore, the hydration effects are not unique properties of SNase, but are common for proteins in general. We revealed a threshold hydration level for the dynamical transition, as also reported for lysozyme [2]. The threshold level is >0.29 g D2O/g protein. Hydration-level dependence on the boson peak shift was different from that on the dynamical transition. Partial hydration below the threshold level for a dynamical transition was sufficient for the boson peak. Therefore, the origins of both effects are completely different. To discriminate between hydration effects on harmonic and anharmonic motions, the hydration-water dynamics should be examined. Hydration-water motion is expected to be slowed down and spatially restricted by the interactions of the protein surface. Thus, the neutron instrument accessible to the higher energy, ∼μeV, and wider q-range is required for studying hydration-related protein dynamics. Because currently accessible (q,ω) ranges are limited and insufficient, the next-generation neutron sources such as the Japan Proton Accelerator Research Complex and Spallation Neutron Source in the United States are expected to cover hydration-related slow dynamics with ∼μeV resolution and wide q-range, which will open the new era for protein dynamics study.
Acknowledgments
We thank Kaoru Shibata and Atsushi Tokuhisa for their help with the experiments, and Nobuhiro Go for many stimulating discussions.
This work was partly supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports, Culture, and Technology of Japan to H.N. (18031042), Y.J. (18031006), A.K. (15300103, 16041208, and 16087202), and M.K. (15076208). The INS experiments were performed under the auspices of the B1 Project of KENS.
Editor: Arthur G. Palmer 3rd.
References
- 1.Rasmussen, B. F., A. M. Stock, D. Ringe, and G. A. Petsko. 1992. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature. 357:423–424. [DOI] [PubMed] [Google Scholar]
- 2.Roh, J. H., V. N. Novikov, R. B. Gregory, J. E. Curtis, Z. Chowdhuri, and A. P. Sokolov. 2005. Onsets of anharmonicity in protein dynamics. Phys. Rev. Lett. 95:038101. [DOI] [PubMed] [Google Scholar]
- 3.Nakasako, M. 1999. Large-scale networks of hydration water molecules around bovine β-trypsin revealed by cryogenic x-ray crystal strucuture analysis. J. Mol. Biol. 289:547–564. [DOI] [PubMed] [Google Scholar]
- 4.Kitao, A., F. Hirata, and N. Go. 1993. Effects of solvent on the conformation and the collective motions of a protein. 3. Free energy analysis by the extended RISM theory. J. Phys. Chem. 97:10231–10235. [Google Scholar]
- 5.Diehl, M., W. Doster, W. Petry, and H. Schober. 1997. Water-coupled low-frequency modes of myoglobin and lysozyme observed by inelastic neutron scattering. Biophys. J. 73:2726–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaccai, G. 2000. Biochemistry—how soft is a protein? A protein dynamics force constant measured by neutron scattering. Science. 288:1604–1607. [DOI] [PubMed] [Google Scholar]
- 7.Joti, Y., A. Kitao, and N. Go. 2005. Protein boson peak originated from hydration-related multiple minima energy landscape. J. Am. Chem. Soc. 127:8705–8709. [DOI] [PubMed] [Google Scholar]
- 8.Doster, W., S. Cusack, and W. Petry. 1989. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 337:754–756. [DOI] [PubMed] [Google Scholar]
- 9.Steinback, P. J., R. J. Loncharich, and B. R. Brooks. 1991. The effects of environment and hydration on protein dynamics: a simulation study of myoglobin. Chem. Phys. 158:383–394. [Google Scholar]
- 10.Fenimore, P. W., H. Frauenfelder, B. H. McMahon, and R. D. Young. 2004. Bulk-solvent and hydration-shell fluctuations, similar to α- and β-fluctuations in glass, control protein motions and functions. Proc. Natl. Acad. Sci. USA. 101:14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tournier, A. L., J. C. Xu, and J. C. Smith. 2003. Translational hydration water dynamics drives the protein glass transition. Biophys. J. 85:1871–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paciaroni, A., A. R. Bizzarri, and S. Cannistraro. 1999. Neutron scattering evidence of a boson peak in protein hydration water. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 60:R2476–R2479. [DOI] [PubMed] [Google Scholar]
- 13.Smith, J. C. 1991. Protein dyanmics: comparison of simulations with inelastic neutron scattering experiments. Q. Rev. Biophys. 24:227–291. [DOI] [PubMed] [Google Scholar]
- 14.Goupil-Lamy, A. V., J. C. Smith, J. Yunoki, S. F. Parker, and M. Kataoka. 1997. High-resolution vibrational inelastic neutron scattering: a new spectroscopic tool for globular proteins. J. Am. Chem. Soc. 119:9268–9273. [Google Scholar]
- 15.Nakagawa, H., H. Kamikubo, I. Tsukushi, T. Kanaya, and M. Kataoka. 2004. Protein dynamical heterogeneity derived from neutron incoherent elastic scattering. J. Phys. Soc. Jpn. 73:491–495. [Google Scholar]
- 16.Kataoka, M., H. Kamikubo, J. Yunoki, F. Tokunaga, T. Kanaya, Y. Izumi, and K. Shibata. 1999. Low energy dynamics of globular proteins studied by inelastic neutron scattering. J. Phys. Chem. Solids. 60:1285–1289. [Google Scholar]
- 17.Leyser, H., W. Doster, and M. Diehl. 1999. Far-Infrared emission by boson peak vibrations in a globular protein. Phys. Rev. Lett. 82:2987–2990. [Google Scholar]
- 18.Tarek, M., and D. J. Tobias. 2001. Effects of solvent damping on side chain and backbone contributions to the protein boson peak. J. Chem. Phys. 115:1607–1612. [Google Scholar]
- 19.Cusack, S., and W. Doster. 1990. Temperature dependence of the low frequency dynamics of myoglibin: measurement of the vibrational frequency distribution by inelastic neutron scattering. Biophys. J. 58:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go, N., T. Noguti, and T. Nishikawa. 1983. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc. Natl. Acad. Sci. USA. 80:3696–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitter, J. 1999. The temperature dependence of internal molecular motions in hydrated and dry α-amylase: the role of hydration water in the dynamical transition of proteins. Biophys. J. 76:1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caliskan, G., A. Kisliuk, A. M. Tsai, C. L. Soles, and A. P. Sokolov. 2003. Protein dynamics in viscous solvents. J. Chem. Phys. 118:4230–4236. [Google Scholar]
- 23.Kurkal, V., R. M. Daniel, J. L. Finney, M. Tehei, R. V. Dunn, and J. C. Smith. 2005. Enzyme activity and flexibility at very low hydration. Biophys. J. 89:1282–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurkal, V., R. M. Daniel, J. L. Finney, M. Tehei, R. V. Dunn, and J. C. Smith. 2005. Low frequency enzyme dynamics as a function of temperature and hydration: a neutron scattering study. Chem. Phys. 317:267–273. [Google Scholar]
- 25.Nakagawa, H., M. Kataoka, Y. Joti, A. Kitao, K. Shibata, A. Tokuhisa, I. Tsukushi, and N. Go. 2006. Hydration-coupled protein boson peak measured by incoherent neutron scattering. Physica B (Amsterdam). 385–86:871–873. [Google Scholar]
- 26.Tarek, M., G. J. Martyna, and D. J. Tobias. 2000. Amplitudes and frequencies of protein dynamics: analysis of discrepancies between neutron scattering and molecular dynamics simulations. J. Am. Chem. Soc. 122:10450–10451. [Google Scholar]
- 27.Kurkal-Siebert, V., and J. C. Smith. 2006. Low-temperature protein dynamics: a simulation analysis of interprotein vibrations and the boson peak at 150 K. J. Am. Chem. Soc. 128:2356–2364. [DOI] [PubMed] [Google Scholar]
- 28.Joti, Y., H. Nakagawa, M. Kataoka, and A. Kitao. 2008. Hydration effects on low-frequency protein dynamics observed in simulated neutron scattering spectra. Biophys. J. 94:4435–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paciaroni, A., A. Orecchini, S. Cinelli, G. Onori, R. E. Lechner, and J. Pieper. 2003. Protein dynamics on the picosecond timescale as affected by the environment: a quasielastic neutron scattering study. Chem. Phys. 292:397–404. [Google Scholar]