Abstract
The two highly conserved RAS genes of the budding yeast Saccharomyces cerevisiae are redundant for viability. Here we show that haploid invasive growth development depends on RAS2 but not RAS1. Ras1p is not sufficiently expressed to induce invasive growth. Ras2p activates invasive growth using either of two downstream signaling pathways, the filamentation MAPK (Cdc42p/Ste20p/MAPK) cascade or the cAMP-dependent protein kinase (Cyr1p/cAMP/PKA) pathway. This signal branch point can be uncoupled in cells expressing Ras2p mutant proteins that carry amino acid substitutions in the adenylyl cyclase interaction domain and therefore activate invasive growth solely dependent on the MAPK cascade. Both Ras2p-controlled signaling pathways stimulate expression of the filamentation response element-driven reporter gene depending on the transcription factors Ste12p and Tec1p, indicating a crosstalk between the MAPK and the cAMP signaling pathways in haploid cells during invasive growth.
INTRODUCTION
When starved for nitrogen, diploid strains of Saccharomyces cerevisiae undergo a developmental transition from growth as single yeast form cells to a multicellular form consisting of filaments of pseudohyphal cells. Filamentous growth, also referred to as pseudohyphal growth, is a composite of several distinct and genetically dissectable processes: cell elongation, filament formation, and agar penetration (Gimeno et al., 1992; Mösch and Fink, 1997). A related phenomenon, invasive growth, occurs in haploid cells (Roberts and Fink, 1994). Haploid invasiveness is distinct from diploid filamentous growth, because haploids are able to invade agar substrate rich in nitrogen, whereas diploids are not. In addition, haploid invasion does not require BUD gene functions, which regulate the budding patterns of haploid and diploid cells.
Haploid invasive growth requires some of the signaling components that also regulate diploid filamentous growth. These include the p65PAK kinase homologue Ste20p, Ste11p (MAPK kinase kinase), and Ste7p (MAPK kinase) protein kinases and the transcription factor Ste12p proteins that constitute part of the MAPK module, which is also required for mating (Liu et al., 1993). In diploids, the transcription factor Tec1p, a regulator originally identified to control expression of yeast transposons (Laloux et al., 1990), is also required for filamentous growth (Gavrias et al., 1996; Mösch and Fink, 1997). Tec1p contains the TEA/ATTS DNA-binding domain, which is sheared by several eukaryotic transcription factors, including the human TEF-1 and Aspergillus abaA (Andrianopoulos and Timberlake, 1991; Burglin, 1991). One of the functions of Tec1p is to target Ste12p to filamentation response elements (FREs) through cooperative binding (Madhani and Fink, 1997). FREs are necessary and sufficient to direct filamentation-specific gene expression and are present in the promoter regions of at least two genes required for pseudohyphal development, TEC1 (Madhani and Fink, 1997) and FLO11 (Lo and Dranginis, 1998). The role of Tec1p in haploid invasive growth has not been studied.
Activated alleles of CDC42 (CDC42Val12 and CDC42Leu61) encoding a small G protein, induce diploid filamentous growth, and also increase transcription of the FRE reporter gene (Mösch et al., 1996). Therefore, they are presumably located upstream of the MAPK module in the filamentous growth signal transduction pathway. The function of Cdc42p in regulation of haploid invasive growth has not yet been studied.
Recently it was found that the dominant active form of RAS2, RAS2Val19, stimulates expression of the transcriptional reporter that contains the FRE filamentous growth response element (Mösch et al., 1996). Full induction of the reporter and of filamentous growth of diploid yeast requires STE20, STE11, STE7 and STE12. Genetic studies suggest that RAS2 is located upstream of CDC42 in the signal transduction pathway. It is unknown whether RAS2 is also involved in haploid invasive growth. The Ras guanine nucleotide–binding proteins play a key role in signal processes that regulate proliferation and differentiation in eukaryotes (McCormick, 1995). The budding yeast S. cerevisiae possesses two Ras protein homologues encoded be the two genes RAS1 and RAS2. The best characterized target of Ras proteins in S. cerevisiae is adenylyl cyclase encoded by the CYR1 gene (for review, see Broach, 1991). Activation of Ras in S. cerevisiae results in elevated intracellular cAMP levels that in turn activate the cAMP-dependent protein kinase (PKA or A kinase). This enzyme is composed of an inhibitory subunit Bcy1p and a catalytic subunit encoded by the three redundant genes, TPK1, TPK2, and TPK3. Activation of the A kinase by expression of the dominant RAS2Val19 allele also confers sensitivity to heat shock and nutrient starvation, loss of carbohydrate reserves, and a block to sporulation. In addition, Ras2 protein seems to be involved in the control of cell division (Morishita et al., 1995) and the yeast lifespan (Sun et al., 1994).
Several recent studies indicate that filamentous growth is also regulated by the cAMP/A kinase pathway (Ward et al., 1995; Kübler et al., 1997; Lorenz and Heitman, 1997). This suggests that Ras2p regulates filamentous growth by two separate pathways, one involving the MAPK cascade and another mediated by cAMP. The function of RAS1, the second RAS gene in S. cerevisiae, in regulating pseudohyphal development or haploid invasive growth is not known.
Elements of the invasive growth signaling network are highly conserved in evolution and may provide an interesting paradigm for the understanding of signal transduction in all eukaryotes. Here we studied the role of Ras proteins in haploid invasive growth of yeast. We present evidence that Ras proteins perform distinct functions in regulating invasive growth development. We show that 1) only RAS2 but not RAS1 is sufficiently expressed to regulate invasive growth development; 2) invasive growth signaling by Ras2p is mediated by both effector pathways, the Cdc42p/Ste20/MAPK cascade and the A kinase pathway; 3) expression of specific RAS2 alleles can uncouple these two effector pathways; and 4) the A kinase stimulates expression of the MAPK cascade–controlled FRE reporter gene, indicating a crosstalk between both signaling pathways.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
All yeast strains used in this study are congenic to the Σ1278b genetic background (Table 1). The ras1Δ::HIS3, ste20Δ::HIS3, ste12Δ::HIS3, tec1Δ::HIS3 deletion mutations were introduced using deletion plasmids pras1Δ::HIS3, pste20Δ::HIS3, pste12Δ::HIS3, and ptec1Δ::HIS3 (Table 2). The ras2Δ::ura3::HIS3 deletion mutation was obtained by 1) using the deletion plasmid pras2Δ::URA3 (Kataoka et al., 1984) and 2) subsequent disruption of the ras2Δ::URA3 locus using the linear ura3::HIS3 URA3 disruption cassette (from Yona Kassir, Technion, Haifa, Israel). The bcy1 mutation was introduced using the disruption plasmid pbcy1::URA3 (Toda et al., 1987a). The FG(Ty1)::lacZ reporter cassette was integrated at the URA3 locus using plasmid YIp356R-FG(Ty1)::lacZ (from Steve Kron, University of Chicago, Chicago IL). The FRE(Ty1)::lacZ reporter was integrated at the LEU2 locus using YIpFRE(Ty1)::lacZ.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YHUM86 | MATα, ras2Δ::ura3::HIS3, FG(Ty1)::lacZ::URA3, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM108 | MATα, ras1Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM114 | MATα, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM120 | MATα, ras2Δ::ura3::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM143 | MATα, ste12Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM145 | MATα, tec1Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM158 | MATα, ras2Δ::ura3::HIS3, ste20Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM167 | MATa, ras2Δ::ura3::HIS3, tec1Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM175 | MATα, ras2Δ::ura3::HIS3, ste12Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
| YHUM184 | MATa, ste20Δ::HIS3, FRE(Ty1)::lacZ::LEU2, ura3-52, leu2::hisG, his3::hisG, trp1::hisG | This study |
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pras1Δ::HIS3 | Cassette for full deletion of RAS1-open reading frame | This study |
| pras2Δ::URA3 | Cassette for full deletion of RAS2-open reading frame | M. Wigler |
| pste20Δ::HIS3 | Cassette for full deletion of STE20-open reading frame | This study |
| pste12Δ::HIS3 | Cassette for full deletion of STE12-open reading frame | This study |
| ptec1Δ::HIS3 | Cassette for full deletion of TEC1-open reading frame | This study |
| pRS316-RAS2 | 3-kb fragment containing RAS2 in pRS316 | This study |
| YCplac22-RAS2 | 3-kb fragment containing RAS2 in YCplac22 | This study |
| YEplac112-RAS2 | 3-kb fragment containing RAS2 in YEplac112 | M. Wigler |
| YCp50-RAS2Val19 | 3-kb fragment containing RAS2Val19 in YCp50 | M. Wigler |
| YCplac22-RAS2Val19 | 3-kb fragment containing RAS2Val19 in YCplac22 | This study |
| YCplac22-RAS2Gly41 | 3-kb fragment containing RAS2Gly41 in YCplac22 | This study |
| YCplac22-RAS2Asn45 | 3-kb fragment containing RAS2Asn45 in YCplac22 | This study |
| YCplac22-RAS2Val19Gly41 | 3-kb fragment containing RAS2Val19Gly41 in YCplac22 | This study |
| YCplac22-RAS2Val19Asn45 | 3-kb fragment containing RAS2Val19Asn45 in YCplac22 | This study |
| pRS314-RAS2ΔC | 3-kb fragment containing RAS2 with C-terminal deletion in pRS314 | This study |
| pRS314-R2N-R1C | 3.5-kb fragment containing RAS2-RAS1 chimeric gene in pRS314 | This study |
| YCplac22-R2p-R1orf | 3.6-kb fragment containing RAS2-promoter fused to RAS1-ORF in YCplac22 | This study |
| YEplac112-RAS1 | 3-kb fragment containing RAS1 in YEplac112 | This study |
| pRS316-GAL-CDC42Val12 | 2.5-kb fragment containing CDC42Val12 fused to GAL1-10-promoter in pRS316 | This study |
| pRS316-GAL-CDC42Leu61 | 2.5-kb fragment containing CDC42Leu61 fused to GAL1-10-promoter in pRS316 | This study |
| pRS426-STE20 | 4.9-kb fragment containing STE20 in pRS426 | Mösch et al., 1996 |
| pYGEX-STE20 | GST-STE20 fusion gene under control of the GAL1-10-promoter | Roberts et al., 1997 |
| B2616 | 5.6-kb fragment containing STE11-4 in YCp50 | G. Sprague |
| pRS426-STE12 | 5-kb fragment containing STE12 in pRS426 | This study |
| B2065 | GAL1-10::STE12 fragment in URA3 marked 2-micron plasmid | B. Errede |
| pRS202-TEC1 | 6.5-kb fragment containing TEC1 in pRS202 | Mösch and Fink, 1997 |
| pGAL-TEC1 | TEC1-cDNA fused to GAL1-10-promoter in pRS316 | This study |
| YEplac195-TPK1 | 2.5-kb fragment containing TPK1 in YEplac195 | This study |
| pRS426-TPK2 | 5.3-kb fragment containing TPK2 in pRS426 | This study |
| pRS426-TPK3 | 3.9-kb fragment containing TPK3 in pRS426 | This study |
| YIpFRE(Ty1)::lacZ | Integrative FRE(Ty1)::lacZ reporter construct | This study |
| YIp356R-FG(Ty1)::lacZ | Integrative FG(Ty1)::lacZ reporter construct | S. Kron |
Standard methods for genetic crosses and transformation were used, and standard yeast culture medium was prepared essentially as described (Guthrie and Fink, 1991). Synthetic complete (SC) medium lacking appropriate supplements was used for scoring invasive growth or β-galactosidase assays with either 2% glucose or a mixture of 3% raffinose and 0.1% galactose as carbon sources. Invasive growth tests were performed as described previously using solid SC medium lacking appropriate supplements (Roberts and Fink, 1994).
Plasmids
Plasmids are listed in Table 2. Deletion alleles for RAS1, STE20, STE12, and TEC1 were created by replacement of coding sequences by either the HIS3- or the TRP1-selectable marker. Plasmids pRS316-RAS2 and YCplac22-RAS2 were constructed by insertion of a 3-kb RAS2 EcoRI–HindIII fragment into vectors pRS316 (Sikorski and Hieter, 1989) and YCplac22 (Gietz and Sugino, 1988), respectively. YCplac22-RAS2Val19 was obtained by subcloning of a 3-kb RAS2Val19 EcoRI–HindIII fragment from YCp50-RAS2Val19 (Michael Wigler, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) into YCplac22. The RAS2Gly41, RAS2Asn45, RAS2Val19Gly41, and RAS2Val19Asn45 alleles were constructed by oligonucleotide-directed mutagenesis using a two-step PCR technique. pRS314-RAS2ΔC and pRS314-R2N-R1C were obtained by subcloning of a 3-kb SalI–NotI fragment from p413-RAS2Δ or a 3.5-kb SalI–NotI fragment from p413-r2r1 (Hurwitz et al., 1995) into pRS314 (Sikorski and Hieter, 1989). Plasmid YCplac22-R2p-R1orf, carrying the RAS1 ORF under the control of the RAS2 promoter, was constructed by 1) separate amplification of both the RAS1 ORF and the RAS2 promoter region using PCR and 2) insertion of both PCR products into YCplac22 (Gietz and Sugino, 1988). YEplac112-RAS1 was obtained by subcloning of a 3-kb BamHI–EcoRI fragment carrying RAS1 from p413-RAS1 (Hurwitz et al., 1995) into YEplac112 (Gietz and Sugino, 1988). pRS316-CDC42Val12 and pRS316-CDC42Leu61 were constructed by subcloning of 2.4-kb fragments containing either CDC42Val12 or CDC42Leu61 under control of the GAL1-10 promoter from plasmids pGAL-CDC42Val12 and pGAL-CDC42Leu61 (Ziman et al., 1991) into pRS316. pRS426-STE12 was obtained by insertion of a 5-kb SalI–HindIII fragment carrying the STE12 locus into pRS426 (Christianson et al., 1992). YEplac195-TPK1 was constructed by insertion of a 2.5-kb SphI–HindIII fragment carrying TPK1 from plasmid YEp-TPK1 (Toda et al., 1987b) into YEplac195 (Gietz and Sugino, 1988). pRS426-TPK2 and pRS426-TPK3 were obtained by subcloning of a 5.3-kb SalI–EcoRI fragment containing TPK2 from pMB05 (Maria Mazón, Universidad Autonoma de Madrid, Madrid, Spain) or a 3.9-kb SalI–SacI fragment carrying TPK3 from plasmid pSA103 (Kelly Tatchell, Louisiana State University, Shreveport, LA) into pRS426. pGAL-TEC1 was isolated from the yeast cDNA library constructed by (Liu et al., 1992) using colony hybridizations with a TEC1-specific 32P-radiolabeled probe. YIpFRE(Ty1)::lacZ was constructed by replacing a 5-kb SalI–HindIII fragment carrying both the yeast 2μ DNA region and URA3 for a 2.5-kb LEU2 fragment in plasmid FRE(Ty1)::lacZ (Madhani and Fink, 1997).
β-Galactosidase Assay
Assays were performed on extracts of cultures grown on solid media for 30 h as described previously (Mösch et al., 1996). Specific β-galactosidase activity was normalized to the total protein in each extract and equals (OD420 × 1.7)/(0.0045 × protein concentration × extract volume × time). Assays were performed on at least three independent transformants, and the mean value is presented. SD did not exceed 15%.
Cell Extracts and Western Blot Analysis
Protein extracts were prepared from 107 cells of cultures grown on solid SC medium lacking the appropriate supplements for 30 h according to a method described earlier (Yaffe and Schatz, 1984). Proteins were then separated using SDS-PAGE and transferred onto nitrocellulose membranes. Ras proteins were detected using ECL technology (Amersham, Buckinghamshire, United Kingdom) after incubation of membranes with the rat monoclonal anti-H-Ras antibody (259) (Santa Cruz Biotechnology, Santa Cruz, CA) and a peroxidase-coupled goat anti-rat IgG secondary antibody (Dianova, Hamburg, Germany).
Northern Blot Analysis
Total RNA was prepared from cultures grown on solid media for 30 h according to the method described by Cross and Tinkelenberg (1991). Total RNA was separated on a 1.4% agarose gel containing 3% formaldehyde and transferred onto nylon membranes as described earlier (Mösch et al., 1992). RAS1, RAS2, and ACT1 transcripts were detected using gene-specific 32P-radiolabeled DNA probes.
RESULTS
RAS2 Is Required for Invasive Growth Development in Haploid Yeast
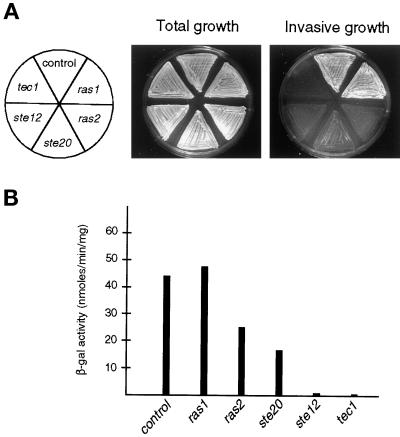
The requirement of both RAS genes, RAS1 and RAS2, for haploid invasive growth was determined, because Ras2p is a potent regulator of pseudohyphal differentiation of diploid cells under nitrogen starvation conditions (Gimeno et al., 1992; Mösch et al., 1996). Haploid strains carrying complete deletions of RAS1 or RAS2 were constructed and tested for invasive growth on rich medium. Yeast strains with full deletions of either STE20, STE12, or TEC1 were used as controls. Figure 1A shows that deletion of RAS2 prevents invasive growth to the same extent as inactivation of STE20, STE12, or TEC1. However, deletion of RAS1 does not affect invasive growth, because a ras1 strain still penetrates agar indistinguishable from a control strain carrying both RAS genes. Thus, haploid yeast strains require RAS2 but not RAS1 for their invasive growth phase.
Figure 1.
RAS2 is required for haploid invasive growth and FRE-dependent transcription. (A) Yeast strains YHUM114 (control), YHUM108 (ras1), YHUM120 (ras2), YHUM184 (ste20), YHUM143 (ste12), and YHUM145 (tec1) were patched on SC-Leu medium. After incubation the plate was photographed before (Total growth) and after (Invasive growth) washing the cells of the plate. (B) FRE(Ty1)::lacZ expression levels. β-Galactosidase activity was measured on the strains shown in A. Units are nanomoles per minute per milligram. Bars depict means of three independent measurements.
Ras2 mutants are phenotypically identical to ste20, ste12, and tec1 strains. Therefore, we tested whether RAS2 is required for FRE-dependent reporter gene expression. A single copy of the FRE(Ty1)::lacZ reporter gene (Madhani and Fink, 1997) was integrated at the LEU2 locus in ras1, ras2, ste20, ste12, and tec1 mutants, and expression was measured on rich medium. FRE-dependent reporter gene expression is reduced twofold in the absence of RAS2, threefold without STE20, and ∼50-fold when either STE12 or TEC1 is deleted (Figure 1B). The same twofold decrease in FRE-dependent reporter gene expression in the absence of RAS2 is found when the FG(Ty1)::lacZ reporter gene (Mösch et al., 1996) is used (Figure 2D). Furthermore, expression of the dominant active RAS2Val19 allele induces transcription of an FRE reporter gene sevenfold when compared with a strain lacking RAS2 (Figure 3B). These data show that RAS2 is essential for invasive growth and suggest a function of Ras2p in the haploid invasive growth signaling pathway described earlier (Roberts and Fink, 1994; Madhani et al., 1997).
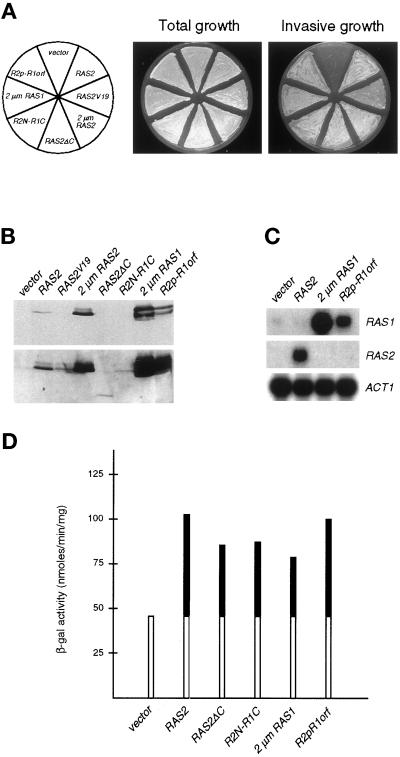
Figure 2.
Expression of different RAS gene constructs in a ras2 mutant strain. (A) Shown is invasive growth of yeast strain YHUM86 (ras2) containing YCplac22 (vector), YCplac22-RAS2 (RAS2), YCplac22-RAS2Val19 (RAS2V19), YEplac112-RAS2 (2 μm RAS2), pRS314-RAS2ΔC (RAS2ΔC), pRS314-R2N-R1C (R2N-R1C), YEplac112-RAS1 (RAS1 2 μm) or YCplac22-R2p-R1orf (R2p-R1orf). Plates were incubated for 4 d and photographed before (Total growth) and after (Invasive growth) washing cells of the agar surface. (B) Expression levels of Ras proteins was determined in strains described in A by Western blot analysis using monoclonal anti-H-Ras antibody (259) that cross-reacts equally well with either Ras1p or Ras2p (upper panel, short exposure; lower panel, long exposure). (C) Autoradiogram showing steady-state levels of RAS1 and RAS2 mRNA in strains described in A. ACT1 gene expression was used as internal standard. (D) FG(Ty1)::lacZ expression levels. β-Galactosidase activity was measured on strains described in A. Units are nanomoles per minute per milligram. Bars depict means of three independent measurements. White bars indicate RAS2-independent expression of FG(Ty1)::lacZ.
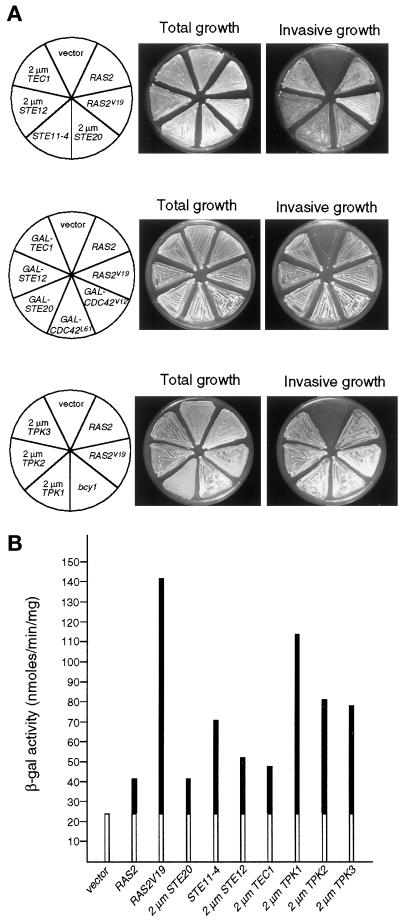
Figure 3.
Activation of either the Cdc42p/Ste20p/MAPK pathway or the PKA is sufficient to induce invasive growth in strains lacking RAS2. (A) Invasive growth of yeast strain YHUM120 (ras2) containing pRS426 (vector), pRS316-RAS2 (RAS2), YCp50-RAS2Val19 (RAS2V19), pRS426-STE20 (2 μm STE20), B2616 (STE11-4), pRS426-STE12 (2 μm STE12), pRS202-TEC1 (2 μm TEC1), pRS316-GAL-CDC42Val12 (GAL-CDC42V12), pRS316-GAL-CDC42Leu61 (GAL-CDC42L61), pYGEX-STE20 (GAL-STE20), B2065 (GAL-STE12), pGAL-TEC1 (GAL-TEC1), YEplac195-TPK1 (2 μm TPK1), pRS426-TPK2 (2 μm TPK2), pRS426-TPK3 (2 μm TPK3), or a bcy1 disruption mutation (bcy1), respectively. Strains were grown on solid SC-Ura medium containing either 2% glucose (upper and lower panels) or a mixture of 3% raffinose and 0.1% galactose (middle panel). After incubation for 4 d, plates were photographed before (Total growth) or after (Invasive growth) washing cells from the agar surface. (B) FRE(Ty1)::lacZ expression levels. β-Galactosidase activity was measured on strains described in A. Units are nanomoles per minute per milligram. Bars depict means of three independent measurements. White bars indicate RAS2-independent expression of FG(Ty1)::lacZ.
Expression of the Conserved N Terminus of Ras2p Is Sufficient to Activate Haploid Invasive Growth
In contrast to RAS2, RAS1 is not required for invasive growth. We determined whether this distinct requirement of both RAS genes in invasive growth regulation is due to structural differences between the two Ras proteins. Although the N-terminal 180 amino acids of Ras1p and Ras2p are 91% homologous, no similarity is found between the hypervariable C-terminal regions. Thus, different RAS gene constructs were transformed into a ras2 mutant carrying a copy of the FG(Ty1)::lacZ reporter gene (Mösch et al., 1996). Resulting strains were assayed for invasive growth and expression of the FRE-driven FG(Ty1)::lacZ reporter gene. Expression levels of the different RAS gene constructs were monitored using Western blot analysis (Figure 2).
We find that expression of RAS2ΔC, a construct lacking the sequences encoding the hypervariable region of Ras2p (amino acids 175–300) (Hurwitz et al., 1995) is sufficient to complement a ras2 strain for both invasive growth and expression of the FG(Ty1)::lacZ reporter gene. This result suggests that the conserved N terminus alone conveys activation of haploid invasive growth and that for this function the hypervariable C terminus of Ras2p is dispensable. We further tested whether the hypervariable C terminus of Ras1p has an inhibiting function on invasive growth. Therefore, a Ras2p–Ras1p chimera consisting of the Ras2p N-terminal 151 residues followed by amino acids 152–309 of Ras1p (Hurwitz et al., 1995) was expressed in a ras2 strain. Invasive growth of the resulting strain is indistinguishable from a strain expressing the full Ras2p or the Ras2ΔCp deletion form. In summary, our findings indicate that the hypervariable C terminus of yeast Ras proteins is not required for regulating invasive growth.
High Expression of RAS1 Restores Invasive Growth in ras2 Mutants
Whereas RAS2 is expressed under invasive growth conditions resulting in clearly detectable amounts of Ras2p, only very low levels of Ras1p are detectable (Figure 2B). In addition, only low levels of RAS1 transcripts are detectable under these conditions (Figure 2C), suggesting that RAS1 is poorly expressed. When assayed in a wild-type strain, expression of RAS2 is ∼10 times higher that expression of RAS1 (our unpublished results). To test whether Ras1p can substitute for Ras2p in invasive growth regulation when expressed at appropriate levels, RAS1 was expressed from a high-copy plasmid or under the control of the RAS2 promoter (Figure 2). We find that strains lacking RAS2 but overexpressing RAS1 were restored for invasive growth as well as FRE-dependent transcription (Figure 2, A and D). This result shows that both Ras proteins Ras1p and Ras2p are functionally redundant for invasive growth signaling, but that only RAS2 is sufficiently expressed under these conditions.
Activation of the Cdc42p/Ste20p/MAPK Cascade Can Suppress Defective Invasive Growth of a ras2 Mutant
In diploids Ras2p induces filamentous growth by activating the Cdc42p/Ste20p/MAPK signaling pathway (Mösch et al., 1996). Because we found that RAS2 is required for regulation of haploid invasive growth and FRE-dependent transcription, we examined whether in haploids Ras2p activates invasive growth development also via the MAPK pathway. Therefore, we tested whether activation of the MAPK pathway is sufficient to suppress the invasive growth defect of a ras2 mutant strain. A strain lacking RAS2 was transformed with an array of different plasmids that either express activated alleles of CDC42 or STE11 or that overexpress STE20, STE12, or TEC1, respectively. Invasive growth of the resulting strains was assayed on rich medium. Expression of the different CDC42 alleles was controlled by growing strains with pGAL::CDC42 constructs on raffinose plus 0.1% galactose to avoid growth inhibition caused by high levels of these mutant proteins. Under all conditions tested, expression of either the dominant active CDC42 alleles CDC42Val12 and CDC42Leu61 or the hyperactive STE11 allele STE11-4 or overexpression of STE20, STE12, or TEC1 was sufficient to suppress defective invasive growth caused by a deletion of RAS2 (Figure 3A). In addition, activation of the MAPK pathway in ras2 mutant strains induces FRE-dependent transcription at least to the levels found in strains harboring a functional RAS2 gene (Figure 3B). These results further corroborate that in haploids Ras2p regulates invasive growth by activating the Cdc42p/Ste20p/MAPK pathway.
Hyperactive PKA Induces Invasive Growth in Strains Lacking RAS2
Recent studies indicate that filamentous growth of diploid yeast is regulated by cAMP (Ward et al., 1995; Kübler et al., 1997; Lorenz and Heitman, 1997). Thus, we determined whether regulation of haploid invasive growth by Ras2p can also be achieved by activation of PKA. A ras2 mutant strain was either disrupted at the BCY1 locus, encoding the inhibitory subunit of PKA, or was transformed with plasmids overexpressing any of the three TPK genes. All constructions result in high A kinase activity in the cell (Toda et al., 1987a,b). Resulting strains were tested for agar penetration and expression of the FRE(Ty1)::lacZ reporter (Figure 3). We find that overexpression of any of the catalytic A kinase subunits encoding TPK1, TPK2, or TPK3 genes induces invasive growth in the absence of Ras2p.
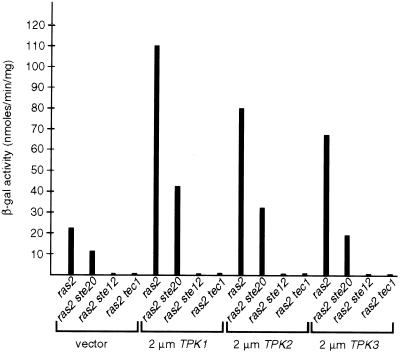
Surprisingly, in haploid cells high A kinase activity not only induces invasive growth in the absence of RAS2 but also stimulates expression of the FRE(Ty1)::lacZ reporter gene to levels comparable to strains with an activated MAPK pathway (Figure 3B). This suggests that the MAPK cascade and the A kinase pathway are interconnected. Therefore, we examined whether the A kinase induces expression of the FRE reporter gene depending on elements of the MAPK cascade. All three TPK genes, TPK1, TPK2, and TPK3, were overexpressed in either ras2, ras2 ste20, ras2 ste12, or ras2 tec1 mutants, and resulting strains were assayed for expression of the FRE(Ty1)::lacZ reporter gene (Figure 4). The ras2 mutation was introduced to prevent crosstalk between the A kinase and the MAPK pathway via Ras2p. Stimulation of FRE-dependent transcription by all three Tpk subunits completely depends on the presence of both Ste12p and Tec1p but is only partially attenuated by deletion of STE20. These findings suggest that regulation of FRE-dependent gene expression by the A kinase and the Cdc42p/Ste20/MAPK cascade might be interconnected at a level downstream of Ste20p.
Figure 4.
Activation of FRE-dependent transcription by the A kinase depends on Ste12p and Tec1p. Yeast strains YHUM120 (ras2), YHUM158 (ras2 ste20), YHUM175 (ras2 ste12), and YHUM167 (ras2 tec1) containing pRS426 (vector), YEplac195-TPK1 (2 μm TPK1), pRS426-TPK2 (2 μm TPK2), or pRS426-TPK3 (2 μm TPK3) were assayed for FRE(Ty1)::lacZ expression levels by measuring β-galactosidase activity. Units are nanomoles per minute per milligram. Bars depict means of three independent measurements.
Alleles of Activated RAS2Val19 Carrying Second Site Mutations in the Adenylyl Cyclase-interacting Domain Require STE20, STE12, or TEC1 for Activation of Haploid Invasive Growth
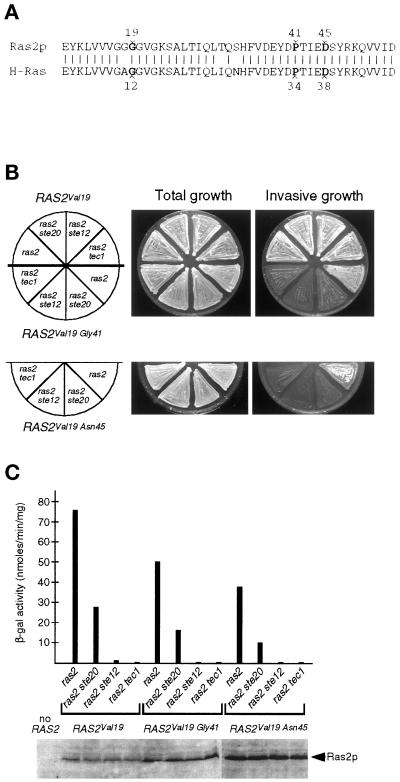
Ras2p regulates invasive growth by activating both the Cdc42p/Ste20p/MAPK and the A kinase pathways. Therefore, we determined whether these two functions of Ras2p are separable. We created RAS2 mutant alleles RAS2Gly41, RAS2Asn45, RAS2Val19Gly41, and RAS2Val19Asn45, encoding single amino acid substitutions at positions 41 and 45 either alone or in combination with the activating mutation of codons for valine instead of glycine at position 19 (Figure 5A). Analogous mutant proteins of the highly conserved human Ha-Ras, Ha-RasVal12Gly34 and Ha-RasVal12Asn38, are specifically blocked for binding and activating yeast adenylyl cyclase Cyr1p (Akasaka et al., 1996). Yeast Ras2p and human Ha-Ras share 90% identity in their N terminus, and both amino acids Pro34 and Asp38 of Ha-Ras required for Cyr1p binding are conserved in yeast Ras2p (Figure 5A). In addition, yeast RAS2 effector mutants RAS2Ser42 and RAS2Ala42, coding for single amino acid substitutions at position 42 in Ras2p, are specifically attenuated for adenylyl cyclase-stimulating activity (Marshall et al., 1988; Sun et al., 1994).
Figure 5.
Properties of RAS2 mutants. (A) Sequence alignment between N-terminal regions of yeast Ras2p (amino acids 10–75) and human H-Ras (amino acids 3–68). Identical residues are indicated by vertical lines. Substitution of Gly19 in yeast Ras2p or Gly12 in H-Ras for Val leads to activated mutant proteins. Amino acids Pro34 and Asp38 of H-Ras and corresponding residues Pro41 and Asp45 of Ras2p are indicated. (B) Invasive growth of yeast strains YHUM120 (ras2), YHUM158 (ras2 ste20), YHUM175 (ras2 ste12), and YHUM167 (ras2 tec1) containing the plasmid YCplac22-RAS2Val19 (RAS2Val19), YCplac22-RAS2Val19Gly41 (RAS2Val19Gly41) or YCplac22-RAS2Val19Asn45 (RAS2Val19Asn45) on solid SC-Trp medium. After incubation, plates were photographed before (Total growth) and after (Invasive growth) washing under a stream of water. (C) Expression levels of FRE(Ty1)::lacZ and Ras2 proteins. β-Galactosidase activity was measured on strains described in B, and expression of Ras2 mutant proteins was analyzed by Western blot analysis.
The two single mutant alleles RAS2Gly41 and RAS2Asn45 were expressed in a ras2 mutant strain and tested for induction of invasive growth. We find that expression of either RAS2Gly41 or RAS2Asn45 leads to strains exhibiting reduced invasive growth, although the amount of invasively growing cells is clearly above levels of strains lacking RAS2 (our unpublished results). Thus, amino acid residues Pro41 and Asp45 of Ras2p are required for full induction of invasive growth. We concluded that part of the invasive growth–stimulating activity of Ras2p is achieved by interaction with Cyr1p, because these residues have been shown to be required for interaction of H-Ras with adenylyl cyclase. Therefore, the remaining invasive growth activation potential of Ras2p mutant proteins carrying the P41G or D45N substitutions should depend on elements of the Ste20p/MAPK pathway. However, testing this prediction by expression of the RAS2Gly41 and RAS2Asn45 single mutant alleles in strains with a defective MAPK pathway is not conclusive, because, e.g., ste20 and tec1 mutants are defective for invasive growth, although they carry an intact RAS2. Expression of the dominant RAS2Val19 allele, however, activates invasive growth in strains that lack STE20, STE12, or TEC1 (Figure 5B). Therefore, we asked whether an activated RasVal19 protein with an additional defect in the adenylyl cyclase interaction domain is still able to overcome the defects in invasive growth caused by inactivation of the MAPK pathway. For this purpose, we expressed the double mutant alleles RAS2Val19Gly41 and RAS2Val19Asn45 in ras2, ras2 ste20, ras2 ste12, and ras2 tec1 mutant strains. Invasive growth behavior, FRE-dependent reporter gene expression, and intracellular Ras2 mutant protein levels were measured in the resulting strains (Figure 5, B and C). We find that expression of either RAS2Val19Gly41 or RAS2Val19Asn45 in the ras2 single mutant leads to invasive growth induction comparable to that achieved by wild-type RAS2 but below that induced by RAS2Val19. The same result is found for stimulation of the FRE-dependent reporter gene expression (Figure 5C). This suggests that the Ras2Val19Gly41 and Ras2Val19Asn45 effector mutant proteins activate invasive growth independent of Cyr1p interaction by stimulating other downstream effector pathways, e.g., the MAPK pathway. Indeed, expression of the Ras2Val19Gly41 and Ras2Val19Asn45 effector mutants is not sufficient to restore the invasive growth defect of the ras2 ste20, ras2 ste12, and ras2 tec1 double mutant strains with a defective MAPK pathway, whereas it is restored by expression of Ras2Val19 (Figure 5B). The failure of Ras2Val19Gly41 or Ras2Val19Asn45 to induce invasive growth in MAPK mutant strains cannot be due to reduced expression of the different Ras2 proteins, because expression of Ras2p is independent of STE20, STE12, or TEC1 (Figure 5C). Thus, both Ras2Val19Gly41 and Ras2Val19Asn45 depend on an intact Ste20p/MAPK cascade to induce invasive growth, whereas Ras2Val19 does not. This indicates that amino acid substitutions P41G or D45N disrupt the ability of Ras2p to activate the cAMP/PKA pathway by interaction with Cyr1p, but not the effector domain of Ras2p responsible for activation of the MAPK cascade.
In summary, results in this section indicate that the two functions by which Ras2p activates invasive growth can be separated. Amino acid substitutions within Ras2p uncouple activation of the Cdc42p/Ste20p/MAPK cascade from the PKA pathway.
DISCUSSION
Differential Transcriptional Expression of RAS Genes in Yeast Growth and Development
We have investigated the role of Ras proteins in cellular growth and development using the developmental switch of S. cerevisiae from growth as single haploid yeast cells to invasive filaments as a paradigm. Our studies show that the two RAS homologues of S. cerevisiae, RAS1 and RAS2, fulfill divergent roles in regulating yeast growth and development. Both ras1 and ras2 single mutant strains exhibit normal growth, whereas a ras1 ras2 double mutant is not viable. Therefore, at least one of the RAS genes must be expressed to ensure normal growth. Already very low levels of Ras1p in a ras2 mutant are sufficient to allow growth but not invasive growth development. High levels of either Ras1p or Ras2p are required to induce invasive growth development. In vivo, yeast cells are only able to make use of RAS2 but not RAS1 to regulate invasive growth. The two Ras proteins are differentially expressed at the level of transcription resulting in RAS1 mRNA levels that are too low during invasive growth. When the RAS1 ORF is under the control of the RAS2 promoter, both RAS1 mRNA and Ras1 protein levels are sufficient to induce invasive growth. Thus, expressional divergence of the two Ras proteins is achieved by transcriptional regulators that differentially control the RAS1 and RAS2 promoters.
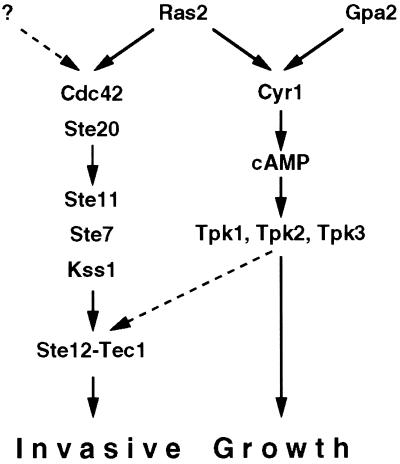
Ras2p Stimulates Distinct Pathways to Induce Haploid Invasive Growth
In diploid yeast, Ras2p signals via the filamentous growth MAPK cascade to induce pseudohyphal development under starvation conditions (Mösch et al., 1996). Here we extend this finding by demonstrating that in haploid yeast Ras2p is required for invasive growth differentiation under nonstarvation conditions. Loss of Ras2p function can be overcome by activation of either of two signaling units, the Cdc42p/Ste20p/MAPK cascade or the PKA pathway. Therefore, Ras2p constitutes a junction that affects both of these signaling units as effector pathways (Figure 6). This is supported by our finding that expression of activated RAS2 alleles, RAS2Val19Gly41 and RAS2Val19Asn45 carrying effector mutations in the adenylyl cyclase–stimulating domain activate invasive growth solely depending on elements of the MAPK cascade. Thus, similar to higher eukaryotes, yeast Ras proteins control cell growth and development by inducing distinct effector pathways. In mammalian cells, ectopic expression of activated H-RAS, H-RASVal12, induces at least two distinct RAS effector pathways, a MAPK cascade and a signal pathway inducing membrane ruffling. Stimulation can be separated by expression of specific effector mutants of H-RAS, H-RASVal12Ser35 and H-RASVal12Cys40 (Joneson et al., 1996). H-RASVal12Cys40 is defective for MAPK activation but has retained the ability to induce membrane ruffling, whereas H-RASVal12Ser35 still activates the MAPK but is defective for stimulation of membrane ruffling (Joneson et al., 1996). The effector protein for stimulation of the MAPK cascade, which directly binds to activated H-RAS, is the RAF protein kinase (McCormick, 1994). Stimulation of membrane ruffling by H-RAS involves the RAC pathway, but the effector protein directly binding H-RAS remains to be determined. In yeast, only the adenylyl cyclase protein Cyr1p has been identified as an effector protein directly binding to activated forms of Ras2p. No direct effector protein of Ras2p has been identified for stimulation of filamentous invasive growth via the Cdc42p/Ste20p/MAPK pathway. Our results indicate that Ras2p must target effectors distinct from adenylyl cyclase to activate the MAPK cascade.
Figure 6.
Model for the regulatory network controlling invasive growth in S. cerevisiae (see DISCUSSION for details).
Crosstalk between the cAMP-dependent Kinase and the Invasive MAPK Pathway
We have demonstrated that in haploid yeast the A kinase can stimulate expression of the FRE-dependent reporter gene depending on the transcription factors Ste12p and Tec1p. This appears to be in contrast to earlier findings showing that hyperactive A kinase only slightly induces expression of an FRE-dependent reporter gene (Mösch et al., 1996) or that very high A kinase activity inhibits FRE-dependent transcription (Lorenz and Heitman, 1997). However, these earlier measurements were all performed in diploid cells under conditions of nitrogen starvation and not in haploid cells under conditions in which supply with ammonia is sufficient. In addition, one major difference between haploid and diploid cells is that expression of both the FRE-dependent reporter gene and the FRE-controlled FLO11 gene is ∼10 times higher in haploids than in diploids (Mösch et al., 1996; Lo and Dranginis, 1998). Thus, although the signaling pathways for diploid filamentous growth and haploid invasive growth share identical components, signaling by these pathways in response to the different nutritional stimuli must not be the same. The mating type or starvation for nitrogen could control the activity of the A kinase toward the MAPK pathway, switching from activating to repressing by a specificity factor.
The finding that in haploid cells the A kinase stimulates expression of an FRE-dependent reporter gene implies the existence of a crosstalk between PKA and the invasive MAPK cascade. How does the A kinase act on FRE-controlled transcription? Activated A kinase could target any of the protein kinases of the invasive MAPK cascade, thereby stimulating their activity toward the transcriptional activators Ste12p and Tec1p. However, deletion of the Ste20p protein kinase only partially attenuates transcriptional induction of the FRE-dependent reporter gene by PKA, suggesting a link at a level downstream of Ste20p. At the moment, we favor a model in which Ste12–Tec1p forms a junction connecting the A kinase pathway to the invasive MAPK cascade (Figure 6). In this model PKA stimulates FRE-dependent transcription by acting on the Ste12p–Tec1p transcription factor, thereby inducing target genes of the invasive growth MAPK cascade. The exact molecular mechanism by which PKA acts on the MAPK pathway to induce FRE-dependent gene expression remains to be elucidated.
Induction of FRE-dependent transcription does not appear be the only way by which PKA induces invasive growth, because expression of RAS2Val19 induces invasion in strains that lack STE12 (Figure 4). This suggests that activation of the cAMP pathway by Ras2p stimulates invasive growth by targets distinct from Ste12p. The transcription factor Sfl1p is a good candidate for such a target, because genetic evidence indicates that Tpk2p acts upstream of Sfl1p in the regulation of Flo11p (Robertson and Fink, 1998). However, the exact interplay between PKA and the different transcriptional factors that form putative downstream targets regulating invasive growth is not clear.
A Highly Regulated Signaling Network for Invasive Growth Development
Our study shows that invasive growth development is under the control of a highly regulated signaling network (Figure 6). Two central signal transduction pathways, the Cdc42p/Ste20p/MAPK cascade and the Cyr1p/cAMP/PKA pathway, control invasive growth by targeting a number of downstream transcription factors. Thus, any regulator affecting the activity of these two pathways is able to control invasive growth. This study defines Ras2p as a central regulator for stimulating both the Cdc42p/Ste20p/MAPK cascade and the Cyr1p/cAMP/PKA pathway. However, a number of recent studies show that apart from Ras2p additional proteins are able to control both growth and filamentous development via the cAMP/PKA pathway. These proteins include Gpa2p, a G-protein alpha subunit (Kübler et al., 1997; Lorenz and Heitman, 1997), Gpr1p, a putative G protein–coupled receptor that associates with Gpa2p (Xue et al., 1998), and Mep2p, a high-affinity ammonium permease with a putative ammonium sensor function (Lorenz and Heitman, 1998). How these genes regulate invasive growth of haploids under conditions of high ammonia, however, has not been carefully studied.
What are the factors acting upstream of Ras2p to regulate invasive growth development? A number of proteins are known that regulate Ras2p activity, including Cdc25p, which functions as guanine nucleotide exchange factor for Ras2p (Engelberg et al., 1990; Jones et al., 1991), and Ira1p and Ira2p, which are thought to act as GTPase-activating proteins (Tanaka et al., 1990). Thus, changes in Cdc25p or Ira1p/Ira2p intracellular levels or activity may be mechanisms to regulate invasive growth. Interestingly, the cellular content of Cdc25p is regulated by destabilization through a cyclin destruction box (Kaplon and Jacquet, 1995), opening the possibility that modulation of Cdc25p stability is an important mechanism to control Ras2p-mediated yeast development. Alternatively, invasive growth may be regulated by modulation of intracellular Ras2p content rather than variation in activity or localization control. Indeed, we have found that expression of both Ras proteins is strictly regulated during invasive growth. Therefore, factors that control development-specific expression of both Ras1p and Ras2p might be important regulators of yeast development.
In summary, our study shows that the control of yeast growth and development by Ras is mediated by a highly regulated signaling network. Because elements of this network are conserved in evolution, the molecular mechanisms underlying their function provide a paradigm for signal transduction in all eukaryotes.
ACKNOWLEDGMENTS
We thank Maria Meyer for brilliant technical assistance during the course of this work. We thank M. Wigler, S. Kron, I. Marbach, A. Levitzki, M. Mazón, K. Tatchell, H. Madhani, and D. Johnson for generously providing plasmids. We also thank S. Irniger for helpful comments on the manuscript and S. Rupp for fruitful discussions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Volkswagen-Stiftung.
REFERENCES
- Akasaka K, Tamada M, Wang F, Kariya K, Shima F, Kikuchi A, Yamamoto M, Shirouzu M, Yokoyama S, Kataoka T. Differential structural requirements for interaction of Ras protein with its distinct downstream effectors. J Biol Chem. 1996;271:5353–5360. doi: 10.1074/jbc.271.10.5353. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A, Timberlake WE. ATTS, a new and conserved DNA binding domain. Plant Cell. 1991;3:747–748. doi: 10.1105/tpc.3.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991;7:28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- Burglin TR. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991;66:11–12. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Engelberg D, Simchen G, Levitzki A. In vitro reconstitution of cdc25 regulated S. cerevisiae adenylyl cyclase and its kinetic properties. EMBO J. 1990;9:641–651. doi: 10.1002/j.1460-2075.1990.tb08156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrias V, Andrianopoulos A, Gimeno CJ, Timberlake WE. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-bp restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and. RAS Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Hurwitz N, Segal M, Marbach I, Levitzki A. Differential activation of yeast adenylyl cyclase by Ras1 and Ras2 depends on the conserved N terminus. Proc Natl Acad Sci USA. 1995;92:11009–11013. doi: 10.1073/pnas.92.24.11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Vignais ML, Broach JR. The CDC25 protein of Saccharomyces cerevisiae promotes exchange of guanine nucleotides bound to ras. Mol Cell Biol. 1991;11:2641–2646. doi: 10.1128/mcb.11.5.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joneson T, White MA, Wigler MH, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- Kaplon T, Jacquet M. The cellular content of Cdc25p, the Ras exchange factor in Saccharomyces cerevisiae, is regulated by destabilization through a cyclin destruction box. J Biol Chem. 1995;270:20742–20747. doi: 10.1074/jbc.270.35.20742. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kübler E, Mösch HU, Rupp S, Lisanti MP. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- Laloux I, Dubois E, Dewerchin M, Jacobs E. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol Cell Biol. 1990;10:3541–3550. doi: 10.1128/mcb.10.7.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Marshall MS, Gibbs JB, Scolnick EM, Sigal IS. An adenylate cyclase from Saccharomyces cerevisiae that is stimulated by RAS proteins with effector mutations. Mol Cell Biol. 1988;8:52–61. doi: 10.1128/mcb.8.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. Raf: the holy grail of Ras biology? Trends Cell Biol. 1994;4:347–350. doi: 10.1016/0962-8924(94)90075-2. [DOI] [PubMed] [Google Scholar]
- McCormick F. Ras-related proteins in signal transduction and growth control. Mol Reprod Dev. 1995;42:500–506. doi: 10.1002/mrd.1080420419. [DOI] [PubMed] [Google Scholar]
- Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 1995;270:1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- Mösch HU, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch HU, Graf R, Braus GH. Sequence-specific initiator elements focus initiation of transcription to distinct sites in the yeast TRP4 promoter. EMBO J. 1992;11:4583–90. doi: 10.1002/j.1460-2075.1992.tb05560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H-U, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes & Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kale SP, Childress AM, Pinswasdi C, Jazwinski SM. Divergent roles of RAS1 and RAS2 in yeast longevity. J Biol Chem. 1994;269:18638–18645. [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, Matsumoto K, Kaziro Y, Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Scott JD, McMullen B, Hurwitz M, Krebs EG, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987a;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987b;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- Ward MP, Gimeno CJ, Fink GR, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, O’Brien JM, Ouellette LA, Church WR, Johnson DI. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]